Abstract
Objective
Autoimmune diseases often have susceptibility genes in common, indicating similar molecular mechanisms. Increasing evidence suggests that rs6822844 at the IL2–IL21 region is strongly associated with multiple autoimmune diseases in individuals of European descent. This study was undertaken to attempt to replicate the association between rs6822844 and 6 different immune-mediated diseases in non-European populations, and to perform disease-specific and overall meta-analyses using data from previously published studies.
Methods
We evaluated case–control associations between rs6822844 and celiac disease (CD) in subjects from Argentina; rheumatoid arthritis (RA), type 1 diabetes mellitus (DM), primary Sjögren's syndrome (SS), and systemic lupus erythematosus (SLE) in subjects from Colombia; and Behçet's disease (BD) in subjects from Turkey. Allele and gene distributions were compared between cases and controls. Meta-analyses were performed using data from the present study and previous studies.
Results
We detected significant associations of rs6822844 with SLE (P = 0.008), type 1 DM (P = 0.014), RA (P = 0.019), and primary SS (P = 0.033) but not with BD (P = 0.34) or CD (P = 0.98). We identified little evidence of population differentiation (FST = 0.01) within cases and controls from Argentina and Colombia, suggesting that association was not influenced by population substructure. Disease-specific meta-analysis indicated significant association for RA (Pmeta = 3.61 × 10–6), inflammatory bowel disease (IBD; Crohn's disease and ulcerative colitis) (Pmeta = 3.48 × 10–12), type 1 DM (Pmeta = 5.33 × 10–5), and CD (Pmeta = 5.30 × 10–3). Overall meta-analysis across all autoimmune diseases reinforced association with rs6822844 (23 data sets; Pmeta = 2.61 × 10–25, odds ratio 0.73 [95% confidence interval 0.69–0.78]).
Conclusion
Our results indicate that there is an association between rs6822844 and multiple auto-immune diseases in non-European populations. Meta-analysis results strongly reinforce this robust association across multiple autoimmune diseases in both European-derived and non-European populations.
Genetic susceptibility plays an important role in autoimmune diseases. Many susceptibility genes/loci contributing to disease development have been identified. Several genetic studies have also shown that susceptibility genes/loci are commonly shared by many autoimmune diseases. For example, there is evidence of the involvement of interleukin genes in several autoimmune diseases, such as IL12B, which is associated with systemic lupus erythematosus (SLE) (1) and inflammatory bowel disease (IBD) (2); and IL18RAP, which is associated with both celiac disease (CD) (3) and IBD (4).
The IL2–IL21 region at 4q27 has shown increasing evidence of association with multiple autoimmune diseases. This region is strongly associated with CD (5,6), type 1 diabetes mellitus (DM) (7,8), rheumatoid arthritis (RA) (9,10), ulcerative colitis (11), juvenile idiopathic arthritis (JIA) (12), psoriatic arthritis (13), psoriasis (13), and SLE (14). Although several genes in this region (IL2, IL21, TENR, and KIAA1109) are associated with these diseases (9), identifying causative genes and the respective predisposing single-nucleotide polymorphisms (SNPs) is challenging. SNPs in this susceptibility region may be independently associated (11) and are likely to play both causative and protective roles in these diseases. Several SNPs, rs13151961, rs13119723, rs6840978, and rs6822844 (in the intergenic region between IL21 and IL2), have been shown to be significantly associated with multiple autoimmune diseases (5,11). Of these SNPs, rs6822844 has been shown to be the most significantly associated with multiple autoimmune diseases. We checked the linkage disequilibrium pattern for the IL2–IL21 region in CEU (Caucasian) and MEX (Mexican) HapMap III data, and we found extensive linkage disequilibrium (r2) between SNPs (r2 = 0.52–0.85 for CEU and r2 = 0.31–0.99 for MEX). However, the primary involvement and functional significance of SNP rs6822844 in gene expression is unknown (15).
Genes located in this region may have related immune functions. IL21 plays a global regulatory role in T cell homeostasis (16) and rapidly up-regulates messenger RNA (mRNA) synthesis for interferon-γ, T-bet, interleukin-2 receptor α (IL-2Rα), IL-12Rβ2, IL-18R, and myeloid differentiation factor 88 (17). These genes are important in activation of innate immunity and Th1 response in natural killer and T cells. However, intestinal epithelial cells are a target of IL-21, and the involvement of IL-21 in the crosstalk between epithelial and immune cells in the gut (18) could indicate the molecular basis for Crohn's disease and IBD. IL-21 also induces insulin growth factor 1 expression (19) and may have consequences in the development of type 1 DM, since it is shown that IL-21 signaling is critical in the development of type 1 DM in NOD mice (20).
Ethnicity-specific genetic associations with autoimmune diseases have been documented. For example, rs1143679 in ITGAM is associated with SLE in European American, Hispanic, and African American populations but is monomorphic in many Asian populations (21,22). Additionally, genetic association varies with geographic distribution. For example, PTPN22 has been associated with multiple autoimmune diseases. However, the strongest association has been found in northern European populations (23). Although genetic association of rs6822844 in the IL2–IL21 intergenic region has been tested in many autoimmune diseases in European-derived populations, to our knowledge its association has not been evaluated in non-European populations. Therefore, in the present study, we examined the genetic associations of rs6822844 with type 1 DM, SLE, RA, and primary Sjögren's syndrome (SS) in samples from Colombia, CD in samples from Argentina, and Behçet's disease (BD) in samples from Turkey. Secondarily, we performed disease-specific and overall meta-analyses using data from previously published studies.
PATIENTS AND METHODS
Sample collection
Written informed consent was obtained from all recruited subjects, and protocols were approved by the appropriate institutional review boards. Population and disease samples used in this study are shown in Table 1.
Table 1.
Allelic association between rs6822844 and multiple inflammatory phenotypes evaluated in the present study*
| No. of cases/No. of controls | MAF in cases | MAF in controls | Genotypic trend |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Population/disease | χ 2 | Allelic P | OR (95% CI) | χ 2 | P | Power† | |||
| Colombian/RA | 354/368 | 0.062 | 0.099 | 5.53 | 0.019 | 0.61 (0.40–0.92) | 6.21 | 0.0127 | 0.70 |
| Colombian/type 1 DM | 131/368 | 0.045 | 0.099 | 6.06 | 0.014 | 0.43 (0.22–0.86) | 7.68 | 0.0056 | 0.42 |
| Colombian/SLE | 200/368 | 0.052 | 0.099 | 7.01 | 0.008 | 0.50 (0.30–0.84) | 7.46 | 0.0063 | 0.55 |
| Colombian/primary SS | 94/368 | 0.048 | 0.099 | 4.54 | 0.033 | 0.46 (0.23–0.95) | 4.85 | 0.0277 | 0.34 |
| Argentinean/CD | 189/222 | 0.079 | 0.078 | 0.001 | 0.978 | 1.01 (0.58–1.74) | 0.02 | 0.8778 | 0.47 |
| Turkish/BD | 193/304 | 0.104 | 0.128 | 0.89 | 0.344 | 0.79 (0.48–1.29) | 0.96 | 0.3262 | 0.53 |
95% CI = 95% confidence interval; RA = rheumatoid arthritis; DM = diabetes mellitus; SLE = systemic lupus erythematosus; SS = Sjögren's syndrome; CD = celiac disease; BD = Behçet's disease.
Power analyses were calculated using P ≤ 0.05 with a minor allele frequency (MAF) of 10%, a disease prevalence of 1%, and an odds ratio (OR) of 0.67.
We recruited 189 patients with CD and 222 controls from Argentina through the Hospital de Clínicas José de San Martín. Diagnosis of CD was based on the presence of clinical features, including characteristic CD enteropathy (24), the presence of a positive CD-related serology (disease-specific antibodies), and response to a strictly monitored gluten-free diet. The controls were generally healthy volunteers and unrelated family friends.
Patients with BD were recruited from the Marmara University Behçet's Clinic in Istanbul, where a large cohort of patients with BD that reflects the ethnic background of the Turkish population is routinely followed up. All patients fulfilled the 1990 International Study Group Criteria for BD (25). Ethnically matched controls were selected randomly from Istanbul, Turkey.
The Colombian sample comprised a total of 1,147 individuals, of whom 689 were patients enrolled at the Corporación para Investigaciones Biológicas in Medellin, 90 were patients enrolled at the Fundación Clínica Valle de Lili in Cali, and 368 were controls. All patients with autoimmune diseases met the international classification criteria for their respective disease (26–29).
The common control group for all analyses of Colombian subjects included 368 individuals who did not have a history of chronic inflammatory autoimmune or infectious diseases and were unrelated to the patients. These individuals were healthy volunteers who were asked to donate blood with written consent. Some of these individuals were enrolled in the clinic for autoimmune or infectious diseases. The same controls were used against each disease. This strategy is valid and has been successfully applied previously (30). Colombian patients used in this study were ethnically matched with controls and were mainly recruited from a place in Medellin where populations are more strictly homogenous (31,32).
Genotyping
All genomic DNA samples from Argentina and Colombia were genotyped for rs6822844 at the Oklahoma Medical Research Foundation using the TaqMan assay (Applied Biosystems, Foster City, CA). In order to assess population structure in the Argentinean and Colombian populations, we genotyped 3 additional unlinked SNPs (rs763361, rs3184504, and rs1143679) using the TaqMan assay.
Statistical analysis
For each disease phenotype we estimated Hardy-Weinberg equilibrium separately for cases and controls. Case–control association studies were analyzed by chi-square test using 2 × 3 and 2 × 2 contingency tables of genotype and allele counts, respectively. Allelic odds ratios (ORs), 95% confidence intervals (95% CIs), and genetic models were calculated using Plink, version 1.05 (33). Since we tested only 1 SNP, no correction for multiple testing was required. P values less than 0.05 were considered significant.
To quantify the effect of population structure on genetic association in samples from Argentina and Colombia, we used the statistic FST, which measures variation in allele frequency between populations. We used the unbiased estimator of FST at a biallelic SNP described by Weir and Cockerham (34). It is possible for this unbiased estimator to result in a value <0; since the FST must be a value between 0 and 1, FST values <0 were set to 0.
Meta-analysis
We performed an exhaustive search and considered all previously published studies that examined the association of SNP rs6822844 with autoimmune diseases. A search of the literature was performed using Medline citations to identify available articles in which rs6822844 was determined in patients with autoimmune diseases and controls. References in the studies were reviewed to identify additional studies not indexed by Medline. We used the following medical subject heading terms and/or text words: IL12, IL21, autoimmune disease, and autoimmunity. No restrictions were placed on language, race, ethnicity, or geographic area. Autoimmune diseases were diagnosed using the appropriate diagnosis criteria. A study was included in this meta-analysis if it was published on or before May 1, 2009, it presented original data (was an independent study), and it provided enough data to calculate ORs. We excluded studies that contained overlapping data; studies in which the number of null and wild genotypes could not be ascertained; and studies such as transmission disequilibrium tests, in which family members had been studied, because their analysis was based on linkage considerations. Extraction from each study was conducted independently by 2 authors (AKM and SKN), and consensus was achieved for all data.
Allele frequencies at rs6822844 from each respective study were determined by the allele-counting method. A chi-square test was used to determine if observed frequencies of genotypes in controls conformed to Hardy-Weinberg equilibrium.
We examined the contrast of the allelic effect of T (the associated allele) versus G (the common allele). The point estimates of the risk, the OR, and its 95% CI were estimated for each study. We assessed the within- and between-study variation or heterogeneity by testing Cochran's Q statistic. This heterogeneity test assessed the null hypothesis that all studies were evaluating the same effect. A significant Q statistic (P < 0.05) indicated heterogeneity across studies. The fixed-effects model was used for meta-analysis since ORs were homogeneous in all calculated meta-analyses. Meta-analysis was performed using CatMap software (35).
Power analysis
Power analyses were performed retrospectively for the available samples (cases and controls), using a fixed minor allele frequency of 10%, a disease prevalence of 1%, a Type I error P of 0.05, and an OR of 0.67. We used CATS software (36) for power calculation.
RESULTS
The SNP rs6822844 was in Hardy-Weinberg equilibrium (P ≥ 0.01) in all cases and controls in all study populations. The frequency of the minor allele (T) varied from population to population, with the highest and lowest minor allele frequencies estimated to be 9.9% and 12.8% in our controls.
A summary of the results of allele association tests is shown in Table 1. The strongest allelic association was observed for SLE in subjects from Colombia (P = 0.0081, OR 0.50 [95% CI 0.30–0.84]). Significant associations were also observed for type 1 DM (P = 0.0138, OR 0.43 [95% CI 0.22–0.86]), RA (P = 0.0187, OR 0.61 [95% CI 0.40–0.92]), and primary SS (P = 0.0331, OR 0.46 [95% CI 0.23–0.95]) in subjects from Colombia. However, we did not observe any allelic association of rs6822844 with CD in patients from Argentina or with BD in patients from Turkey. With regard to the effect of population structure on this association, we observed little difference (overall FST = 0.0102 using 4 unlinked SNPs) among samples from Argentina and Colombia. In fact, pairwise FST values between cases and controls were very low. Genetic models for each disease phenotype were assessed under dominant, recessive, genotypic trend (Cochran-Armitage), and multiplicative models. Only the P values from the genotypic trend model are shown (Table 1).
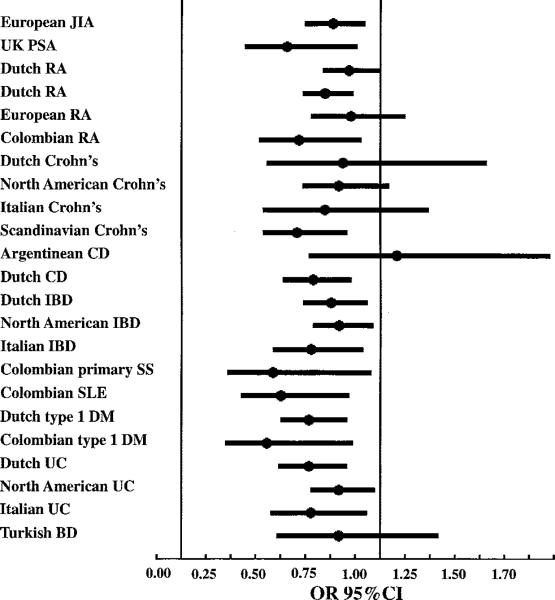
Disease-specific meta-analyses were performed for all diseases for which data were obtained in at least 2 populations (in either the present study or previously published studies) (Figure 1 and Table 2). None of the meta-analyses were significantly heterogeneous for the OR. (Phet varied from 0.10 to 0.79.) The strongest associations were observed in IBD (Pmeta = 3.48 × 10–12; OR 0.74 [95% CI 0.68–0.80]), RA (Pmeta = 3.61 × 10–6; OR 0.77 [95% CI 0.70–0.86]), type 1 DM (Pmeta = 5.33 × 10–5; OR 0.61 [95% CI 0.48–0.78]), and CD (Pmeta = 5.30 × 10–3; OR 0.72 [95% CI 0.58–0.91]). The overall association was significant and consistent with the involvement of the IL2–IL21 region in multiple autoimmune diseases (23 populations; P = 2.61 × 10–25, OR 0.73 [95% CI 0.69–0.78]). The strength of association for this SNP in each of the study populations, using results of the present study as well as those of previous studies, is shown in Table 3.
Figure 1.
Disease- and ethnicity-specific populations used for meta-analysis of rs6822844 across all autoimmune diseases investigated in the present study and in previously published studies. JIA = juvenile idiopathic arthritis; PsA = psoriatic arthritis; RA = rheumatoid arthritis; CD = celiac disease; IBD = inflammatory bowel disease; SS = Sjögren's syndrome; SLE = systemic lupus erythematosus; DM = diabetes mellitus; UC = ulcerative colitis; BD = Behçet's disease. Values are the odds ratio (OR) and 95% confidence interval (95% CI) of the association of rs6822844 with the indicated disease in the indicated population.
Table 2.
Overall and disease-specific meta-analysis for rs6822844*
| Number of populations | Heterogeneity |
|||||
|---|---|---|---|---|---|---|
| OR (95% CI) | χ 2 | P | χ 2 | P | ||
| Overall | 23 | 0.73 (0.69–0.78) | 108.06 | 2.61 × 10–25 | 20.60 | 0.484 |
| RA | 4 | 0.77 (0.70–0.86) | 21.46 | 3.61 × 10–6 | 3.49 | 0.323 |
| Type 1 DM | 2 | 0.61 (0.48–0.78) | 16.33 | 5.33 × 10–5 | 1.12 | 0.291 |
| CD | 2 | 0.72 (0.58–0.91) | 7.77 | 5.30 × 10–3 | 2.71 | 0.100 |
| IBD | 10 | 0.74 (0.68–0.80) | 48.40 | 3.48 × 10–12 | 5.47 | 0.791 |
No meta-analyses were performed for diseases for which data were available from only 1 population, including BD, juvenile RA, psoriatic arthritis, primary SS, and SLE. IBD >= inflammatory bowel disease (see Table 1 for other definitions).
Table 3.
Disease-specific genetic associations with rs6822844 in the present study and previously published studies*
| Population/disease | Author, year (ref.) | OR (95% CI) |
|---|---|---|
| European/JIA | Albers et al, 2009 (12) | 0.76 (0.62–0.92) |
| UK/PsA | Liu et al, 2008 (13) | 0.53 (0.32–0.88) |
| Dutch/RA | Daha et al, 2009 (10) | 0.84 (0.71–1.00) |
| Dutch/RA | Zhernakova et al, 2007 (7) | 0.72 (0.61–0.86) |
| European/RA | Teixeira et al, 2009 (9) | 0.85 (0.65–1.12) |
| Colombian/RA | Present study | 0.61 (0.40–0.92) |
| Dutch/Crohn's disease | Festen et al, 2009 (11) | 0.81 (0.43–1.53) |
| North American/Crohn's disease | Festen et al, 2009 (11) | 0.79 (0.61–1.04) |
| Italian/Crohn's disease | Festen et al, 2009 (11) | 0.72 (0.41–1.24) |
| Scandinavian/Crohn's disease | Amamovic et al, 2008 (6) | 0.58 (0.41–0.83) |
| Argentinean/CD | Present study | 1.01 (0.58–1.74) |
| Dutch/CD | Van Heel et al, 2007 (5) | 0.66 (0.51–0.85) |
| Dutch/IBD | Festen et al, 2009 (11) | 0.75 (0.61–0.93) |
| North American/IBD | Festen et al, 2009 (11) | 0.79 (0.66–0.96) |
| Italian/IBD | Festen et al, 2009 (11) | 0.65 (0.46–0.91) |
| Colombian/primary SS | Present study | 0.46 (0.23–0.95) |
| Colombian/SLE | Present study | 0.50 (0.30–0.84) |
| Dutch/type 1 DM | Zhernakova et al, 2007 (7) | 0.64 (0.50–0.83) |
| Colombian/type 1 DM | Present study | 0.43 (0.22–0.86) |
| Dutch/UC | Festen et al, 2009 (11) | 0.64 (0.49–0.83) |
| North American/UC | Festen et al, 2009 (11) | 0.79 (0.65–0.97) |
| Italian/UC | Festen et al, 2009 (11) | 0.65 (0.45–0.93) |
| Turkish/BD | Present study | 0.79 (0.48–1.29) |
JIA = juvenile idiopathic arthritis; PsA = psoriatic arthritis; IBD = inflammatory bowel disease; UC = ulcerative colitis (see Table 1 for other definitions).
For CD in patients from Argentina, significant associations were not obtained in either allele- or model-based tests, although rs6822844 has primarily been shown to be associated with CD in European populations (5,6). Power analysis demonstrated that these association tests were extremely underpowered (Table 1). The limited sample size used in the present study or a greater stratification of the population may explain why this association was not observed in the subjects from Argentina.
DISCUSSION
Genetic association is a powerful tool used to identify disease susceptibility–associated SNPs. The population-specific allele frequencies and ORs found in the present study provide strong evidence that rs6822844 in the IL2–IL21 region is significantly associated with multiple autoimmune diseases.
Several genetic studies have implicated the IL2–IL21 region as being involved in multiple autoimmune diseases, including type 1 DM, RA, CD, and JIA, in European populations. However, these associations have previously been evaluated only in samples from European-derived populations (11). Often SNPs belong to haplotype blocks in which the SNPs are in high linkage disequilibrium, leading to multiple SNPs being associated with disease. This can make it difficult to tease apart genetic associations, especially within a single ethnic population in which similar genetic backgrounds may be present. There are several strategies to search for causative SNPs (11). First, it may be necessary to fine-map a region in multiple ethnic populations. Transracial mapping (37) can be an important technique to pinpoint the predisposing variant (21). Second, since this region is associated with multiple autoimmune diseases it will also be important to compare haplotype blocks across disease populations to determine if there are multiple causative mutations, or a few common SNPs between autoimmune diseases. Third, once we narrow down this region and identify causative SNP(s), functional studies can be performed to discover how these SNP(s) contribute to the development of autoimmunity.
One challenge for genetic association studies is the presence of population substructure in the samples, which raises the potential for confounding and spurious results, especially in the admixed population. It is known that populations from both Argentina and Colombia have varying degrees of admixture with Native American, European, and African ancestral populations. Therefore, if cases and controls are sampled from several subpopulations with different allele frequencies, the differences in allele frequencies between cases and controls could mimic a statistical association, which might lead to false-positive results. However, the Colombian samples used in this study were mainly collected from Medellin, where populations are more homogenous (Paisa community). Historical documents demonstrate that the Paisa community, to which most of our Colombian subjects belonged, is highly homogenous and endogenous (31,32), and very little stratification is observed. In the present study we addressed this issue of population subdivision by calculating the FST with respect to 4 unlinked SNPs. The homogeneity between our Colombian cases and controls was confirmed with FST analysis. There was little evidence of population substructure, and thus our results on SLE, RA, type 1 DM, and primary SS in Colombian subjects were most likely not affected by population substructure at this locus.
However, we did not find a significant association between rs6822844 and CD, as has previously been identified in European populations. This may be due to small sample size, this SNP may not be associated with CD in the Argentinean population, or ethnicity-specific variations (cultural or other environmental factors) may play a significant role in developing CD in Argentinean populations. Future analysis with a larger, more homogenous sample size could reveal that rs6822844 is associated with CD in both European and Argentinean populations.
We also did not find a genetic association between rs6822844 and BD; however, a larger study is needed to definitively exclude a genetic association between BD and the IL2–IL21 locus. It is also possible that genetic susceptibility to BD does not overlap with that identified in autoimmune diseases. This probably reflects a distinct pathologic mechanism for this inflammatory disease. Indeed, a recent genome-wide association study in this disease revealed genetic associations with 5 novel loci, none of which has previously been shown to be associated with autoimmune diseases (38).
Autoimmune diseases may have related underlying cellular mechanisms involved in immune response cells for humoral and cell-mediated immunity. Interleukins may play a significant role and are often associated with autoimmune phenotypes (5,11). The SNP rs6822844 is in a noncoding region upstream of IL21 and downstream of IL2; the molecular function of this SNP is elusive. It may modulate the gene expression of IL21 or IL2, or it may be linked to causative mutations. Extensive search of this sequence suggests that this SNP-carrying sequence has strong homology with the 13 precursor of mRNA stem loop structure that codes for microRNA. The neighboring sequence shows strong homology with mature microRNA that has been identified in human stem cells (39) and has been shown to be expressed in the human lymphoid BC-1 cell line (40). Moreover, the ancestral allele G is conserved in all microRNA-precursor hairpin structures. This evidence suggests that non-risk G-carrying sequences potentially code for microRNA and modulate cellular function in lymphocytes, whereas the T allele abolishes microRNA-producing ability. It is very likely that this variant affects microRNA generation and modulates expression of genes and, subsequently, their functions. These changes, however, are yet to be characterized. It remains to be explored how this single variation modulates the molecular signaling pathway of proteins and gives rise to different clinical phenotypes that lead to many auto-immune diseases.
ACKNOWLEDGMENTS
We wish to thank the patients and their families for their cooperation and for giving consent to participate in this study.
Supported by the NIH (grants 5R01-AI-063622 and P20-RR-020143), Colciencias (grant 2213-04-16484), the Rosario University School of Medicine, Colombia, and the Colombian Association of Rheumatology.
REFERENCES
- 1.Sanchez E, Morales S, Paco L, Lopez-Nevot MA, Hidalgo C, Jimenez-Alonso J, et al. Interleukin 12 (IL12B), interleukin 12 receptor (IL12RB1) and interleukin 23 (IL23A) gene polymorphism in systemic lupus erythematosus. Rheumatology (Oxford) 2005;44:1136–9. doi: 10.1093/rheumatology/keh697. [DOI] [PubMed] [Google Scholar]
- 2.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–2. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–10. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamovic S, Amundsen SS, Lie BA, Gudjonsdottir AH, Ascher H, Ek J, et al. Association study of IL2/IL21 and FcgRIIa: significant association with the IL2/IL21 region in Scandinavian coeliac disease families. Genes Immun. 2008;9:364–7. doi: 10.1038/gene.2008.27. [DOI] [PubMed] [Google Scholar]
- 7.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–8. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teixeira VH, Pierlot C, Migliorini P, Balsa A, Westhovens R, Barrera P, et al. Testing for the association of the KIAA1109/Tenr/IL2/IL21 gene region with rheumatoid arthritis in a European family-based study. Arthritis Res Ther. 2009;11:R45. doi: 10.1186/ar2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daha NA, Kurreeman FA, Marques RB, Stoeken-Rijsbergen G, Verduijn W, Huizinga TW, et al. Confirmation of STAT4, IL2/IL21, and CTLA4 polymorphisms in rheumatoid arthritis. Arthritis Rheum. 2009;60:1255–1260. doi: 10.1002/art.24503. [DOI] [PubMed] [Google Scholar]
- 11.Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58:799–804. doi: 10.1136/gut.2008.166918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albers HM, Kurreeman FA, Stoeken-Rijsbergen G, Brinkman DM, Kamphuis SS, van Rossum MA, et al. Association of the autoimmunity locus 4q27 with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:901–4. doi: 10.1002/art.24296. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, et al. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis. 2008;67:458–61. doi: 10.1136/ard.2007.075424. [DOI] [PubMed] [Google Scholar]
- 15.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 16.Datta S, Sarvetnick NE. IL-21 limits peripheral lymphocyte numbers through T cell homeostatic mechanisms. PLoS One. 2008;3:e3118. doi: 10.1371/journal.pone.0003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strengell M, Sareneva T, Foster D, Julkunen I, Matikainen S. IL-21 up-regulates the expression of genes associated with innate immunity and Th1 response. J Immunol. 2002;169:3600–5. doi: 10.4049/jimmunol.169.7.3600. [DOI] [PubMed] [Google Scholar]
- 18.Caruso R, Fina D, Peluso I, Stolfi C, Fantini MC, Gioia V, et al. A functional role for interleukin-21 in promoting the synthesis of the T-cell chemoattractant, MIP-3α, by gut epithelial cells. Gastroenterology. 2007;132:166–75. doi: 10.1053/j.gastro.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 19.Menoret E, Maiga S, Descamps G, Pellat-Deceunynck C, Fraslon C, Cappellano M, et al. IL-21 stimulates human myeloma cell growth through an autocrine IGF-1 loop. J Immunol. 2008;181:6837–42. doi: 10.4049/jimmunol.181.10.6837. [DOI] [PubMed] [Google Scholar]
- 20.Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc Natl Acad Sci U S A. 2008;105:14028–33. doi: 10.1073/pnas.0804358105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-α(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–4. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 22.Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, et al. Evaluation of imputation-based association in and around the integrin-α-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE). Hum Mol Genet. 2009;18:1171–80. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases–a meta-analysis [review]. Rheumatology (Oxford) 2007;46:49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 24.Marsh MN. Gluten, major histocompatibility complex, and the small intestine: a molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology. 1992;102:330–54. [PubMed] [Google Scholar]
- 25.International Study Group for Behcet's Disease Criteria for diagnosis of Behcet's disease. Lancet. 1990;335:1078–80. [PubMed] [Google Scholar]
- 26.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 28.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjögren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group [review]. Ann Rheum Dis. 2002;61:554–8. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 1997;20:1183–97. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 30.Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–21. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bravo ML, Valenzuela CY, Arcos-Burgos OM. Polymorphisms and phyletic relationships of the Paisa community from Antioquia (Colombia). Gene Geogr. 1996;10:11–7. [PubMed] [Google Scholar]
- 32.Pineda-Trujillo N, Carvajal-Carmona LG, Buritica O, Moreno S, Uribe C, Pineda D, et al. A novel Cys212Tyr founder mutation in parkin and allelic heterogeneity of juvenile Parkinsonism in a population from North West Colombia. Neurosci Lett. 2001;298:87–90. doi: 10.1016/s0304-3940(00)01733-x. [DOI] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 35.Nicodemus KK. Catmap: case-control and TDT meta-analysis package. BMC Bioinformatics. 2008;9:130. doi: 10.1186/1471-2105-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 37.Mijovic CH, Barnett AH, Todd JA. Genetics of diabetes: transracial gene mapping studies. Baillieres Clin Endocrinol Metab. 1991;5:321–40. doi: 10.1016/s0950-351x(05)80130-2. [DOI] [PubMed] [Google Scholar]
- 38.Fei Y, Webb R, Cobb BL, Direskeneli H, Saruhan-Direskeneli G, Sawalha AH. Identification of novel genetic susceptibility loci for Behcet's disease using a genome-wide association study. Arthritis Res Ther. 2009;11:R66. doi: 10.1186/ar2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 40.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]