Abstract
Australia has efficient and visible plant quarantine measures, which through various border controls and survey activities attempt to prevent the entry of unwanted pests and diseases. The ability to successfully perform this task relies heavily on determining what pathogens are present and established in Australia as well as those pathogens that are exotic and threatening. There are detailed checklists and databases of fungal plant pathogens in Australia, compiled, in part, from surveys over many years sponsored by Federal and State programmes. These checklists and databases are mostly specimen-based, which enables validation of records with reference herbarium specimens and sometimes associated cultures. Most of the identifications have been based on morphological examination. The use of molecular methods, particularly the analysis of DNA sequence data, has recently shown that several well-known and important plant pathogenic species are actually complexes of cryptic species. We provide examples of this in the important plant pathogenic genera Botryosphaeria and its anamorphs, Colletotrichum, Fusarium, Phomopsis / Diaporthe and Mycosphaerella and its anamorphs. The discovery of these cryptic species indicates that many of the fungal names in checklists need scrutiny. It is difficult, and often impossible, to extract DNA for sequence analysis from herbarium specimens in order to validate identifications that may now be considered suspect. This validation can only be done if specimens are recollected, re-isolated and subjected to DNA analysis. Where possible, herbarium specimens as well as living cultures are needed to support records. Accurate knowledge of the plant pathogens within Australia’s borders is an essential prerequisite for the effective discharge of plant quarantine activities that will prevent or delay the arrival of unwanted plant pathogens.
Keywords: disease associated fungi, disease threats, molecular phylogeny, quarantine, taxonomy
INTRODUCTION
Most records of plant pathogenic fungi in Australia are derived from, and substantiated by, dried herbarium specimens and relatively few are based on living cultures. There are several reasons for this including, i) many groups of pathogens are obligate and cannot be cultured; ii) many pathogens can be identified with confidence in situ based on morphology and thus cultures are not necessary for diagnosis; iii) living cultures are often difficult to preserve, especially over time and may loose their ability to sporulate or retain pathogenicity or other physiological properties; iv) maintaining living cultures is relatively costly; and v) isolation of a presumptive causal organism does not demonstrate pathogenicity unless Koch’s postulates are fulfilled. As an example, consider that most plant disease surveys in northern Australia (Hyde & Alcorn 1993, Shivas 1995, Shivas & Alcorn 1996), and neighbouring countries of Papua New Guinea (Hyde & Philemon 1994) and Irian Jaya (Shivas et al. 1996), often as part of the Northern Australian Quarantine Strategy (NAQS), were based almost entirely on herbarium specimens. The NAQS surveys often focused on remote parts of these regions, looking for early signs of pest and disease incursions. The remoteness of many locations meant that facilities were not available to obtain and look after cultures. Furthermore, the scientists themselves were often not allowed to move specimens that might harbour living and exotic pathogens, particularly if the specimens were collected offshore.
The importance of maintaining accurate records of pathogens in Australia is their value in determining those pathogens that are not present (exotic) and potentially a threat if introduced, through a process known as pest risk assessment (Plant Health Australia 2010). The aim of quarantine in Australia is to prevent entry of exotic pests, diseases and weeds that could have serious environmental and economic consequences if introduced. For example, the cost of introducing some exotic plant diseases, e.g. Karnal bunt of wheat, into Australia could cost billions of dollars (http://www.dfat.gov.au/facts/quarantine.html). Several island countries, including Australia, New Zealand, and the Philippines, through their isolation have avoided the introduction of many important exotic threats.
Specimen-based records of most of the plant pathogens that occur in Australia can be accessed through the Australian Plant Disease Database and the Australian Plant Pest Database (Shivas et al. 2006). Both of these important databases are username and password protected which limits their availability. These databases, together with the three large Australian herbaria of plant pathogens that underpin them, are important resources for resolving quarantine issues and facilitating the pest risk assessment process.
Prior to the availability of DNA sequence data, the identification of plant pathogenic fungi was primarily based on morphology, with herbarium specimens serving as proof of identity for future reference. The relatively recent application of molecular phylogenetic analysis to species identification has revealed that many traditionally accepted species actually represent complexes of species. This is especially true for many important plant pathogenic genera such as anamorphic Mycosphaerellaceae (including Cladosporium), Botryosphaeria and its anamorphs, Colletotrichum, Fusarium, Guignardia with its Phyllosticta anamorphs, and Diaporthe with its Phomopsis anamorphs (Crous et al. 2006, 2009a–c, Damm et al. 2007, Alves et al. 2008, Cai et al. 2009, Hyde et al. 2009a, b, Kvas et al. 2009, Phillips et al. 2008, Shivas & Tan 2009, Wulandari et al. 2009, Zhang et al. 2009, Schoch et al. 2009, Phoulivong et al. 2010, Summerell et al. 2010, Walsh et al. 2010). As a result of these recent advances many of the taxa listed in the aforementioned Australian checklists and databases of fungi associated with plant diseases are now outdated. There is a pressing need to address this problem. The purpose of this paper is to provide selected examples where there is a need to carry out a re-inventory of the fungal pathogens of plants in Australia so that the checklist and databases are both accurate and up to date.
MATERIALS AND METHODS
Selected sequences from five plant pathogenic fungal genera were downloaded from GenBank and aligned using either Clustal X or the online MAFFT server (http://mafft.cbrc.jp/alignment/server/index.html). The alignments were optimised manually to allow maximum alignment and maximum sequence similarity. Gaps were treated as missing data or fifth character states (Mycosphaerella and Phomopsis alignments). Phylogenetic analyses were carried out based on the aligned dataset using PAUP v4.0b10 (Swofford 2003). Ambiguously aligned regions were excluded from all analyses, where present. Trees were inferred using the heuristic search option with TBR branch swapping and 100–1 000 random sequence additions. Maxtrees were unlimited, branches of zero length were collapsed, and all multiple parsimonious trees were saved. Trees were drawn in TreeView (Page 1996).
RESULTS AND DISCUSSION
Botryosphaeriaceae and its anamorphs
Several Botryosphaeria species and their anamorphs, such as Lasiodiplodia theobromae, were identified during NAQS quarantine surveys of plant disease associated fungi of northern Australia (Hyde & Alcorn 1993, Shivas 1995, Shivas & Alcorn 1996). However, several recent studies have shown that species of Botryosphaeria such as B. dothidea and anamorph taxa such as Lasiodiplodia theobromae are species complexes (Alves et al. 2008, Phillips et al. 2008, Abdollahzadeh et al. 2010). Botryosphaeria dothidea was epitypified by Slippers et al. (2004) with a specimen from Prunus sp. collected on the Italy-Switzerland border. The taxon has proved to be a complex comprising several species (Smith et al. 2001, Denman et al. 2003, Slippers et al. 2004). Several other Botryosphaeria species and related anamorphs have also been epitypified (see Fig. 1 in black bold) and this has led to advances in understanding the genus (Crous et al. 2006, Alves et al. 2008, Phillips et al. 2008). By epitypifying these taxa with living cultures it is now possible to compare recent collections with that of the type to establish whether they are the same species. This is necessary for all diseases for which Botryosphaeria and its anamorphs are linked to establish accurate disease records.
Fig. 1.
Phylogramme generated from maximum parsimony analysis based on ITS sequences, showing the phylogenetic relationships of B. dothidea with other species of Botryosphaeria, those in yellow highlight are wrongly applied names. Values above the branches are parsimony bootstrap (> 50 %). Ex-type strains are shown in bold.
In Fig. 1 we present a phylogramme comprising 59 ITS sequences downloaded from GenBank including 17 named as Botryosphaeria dothidea with its epitype sequence highlighted in red. Sequences from ex-type strains of Neofusicoccum australe (= B. australis; Crous et al. 2006), N. luteum (= B. lutea), Diplodia seriata (= B. obtusa; Phillips et al. 2007 ), N. parvum (= B. parva), N. ribis (= B. ribis), D. mutila (= B. stevensii), ‘Botryosphaeria’ tsugae, Lasiodiplodia crassispora, L. gonubensis and Phaeobotryosphaeria porosa (= Diplodia porosa; Phillips et al. 2008) are also included. Although the majority of Botryosphaeria dothidea strains cluster around the type sequence in the upper part of the tree, there are five disparate strains scatted in the lower part of the tree. This suggests that there are likely to be many sequences for B. dothidea in GenBank with wrongly applied names. We do not show the data here but there are similar situations for Botryosphaeria rhodina and Lasiodiplodia theobromae. These taxa have not been epitypified and at present we do not know which strain in GenBank (if any) represents B. rhodina or L. theobromae, although this situation is likely to be solved in a future publication (A.J.L. Phillips pers. comm.). Besides the problem with wrongly applied names in GenBank, very few of the GenBank sequences are based on Australian specimens and therefore they need to be recollected, sequenced and checked against verified or typified names. Specific examples of erroneous names concern the species of Botryosphaeria that cause disease of palms. Recent studies in Thailand have shown some species belong to Neodeightonia, a genus that is quite similar to Botryosphaeria (Phillips et al. 2008, Liu et al. in press). Therefore it is essential that every Botryosphaeria-like species associated with disease of palms should be re-assessed and this should also be the case with other hosts.
Colletotrichum
The anamorphic ascomycete genus Colletotrichum contains many well-known plant pathogens that cause a range of diseases worldwide (Crouch & Beirn 2009, Crouch et al. 2009, Damm et al. 2009, Hyde et al. 2009a, b). MycoBank currently contains 676 records of Colletotrichum (www.mycobank.org), while only 66 species names are in current use (Hyde et al. 2009a). Cai et al. (2009) outlined a polyphasic approach for studying Colletotrichum and provided a backbone tree comprising 42 ex-type ITS sequences. Many ubiquitous Colletotrichum species have now been shown to be species complexes containing numerous cryptic species. For example, Colletotrichum acutatum s.lat. has been shown to contain discrete morphological and molecular groups that represent discrete species as found in some Australian isolates (Shivas & Tan 2009).
Perhaps the most commonly known species in the genus is Colletotrichum gloeosporioides (teleomorph Glomerella cingulata), which is represented in Australian collections by more than 5 000 specimens from several hundred host plant species in about 100 different plant families. Very little is known about whether these fungal records represent saprobes, weak or opportunistic pathogens, or genuine pathogens. It is possible that many of these records are misidentified as the morphological characteristics that define C. gloeosporioides are unreliable or even misleading, i.e. having cylindrical conidia with rounded ends, and less than 4.5 μm wide according to the widely used key by Sutton (1980). Cannon et al. (2008) epitypified C. gloeosporioides; conidia of the ex-epitype strain are on average wider, measuring 14.4 × 5.6 μm. The species had been synonymised by von Arx (1957) with about 600 names, some of which might represent discrete species.
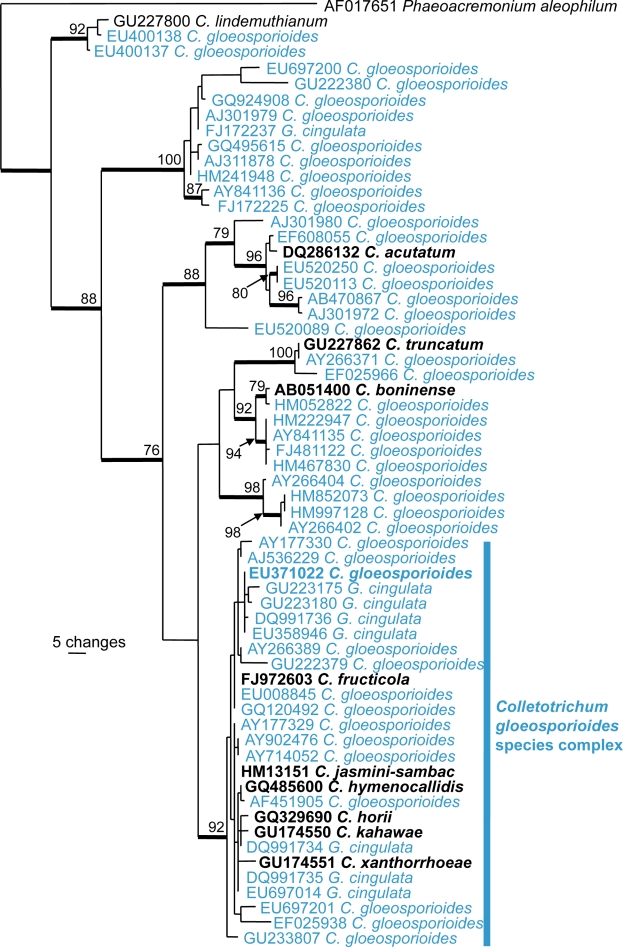
Because of the morphological similarities between C. gloeosporioides and other Colletotrichum species, the close relationship between species within the C. gloeosporioides species complex and the only recent epitypification, most of the sequences lodged in GenBank as C. gloeosporioides are doubtful and belong to many different species. As demonstrated in Fig. 2, ITS sequences lodged in GenBank as C. gloeosporioides and G. cingulata are found throughout the C. gloeosporioides species complex and outside the C. gloeosporioides complex, the latter applies to more than 100 of the about 750 ITS sequences of C. gloeosporioides in GenBank. Furthermore, Fig. 2 shows the difficulty of species recognition within the C. gloeosporioides species complex using ITS sequences only. Only a few of these GenBank sequences were derived from specimens from Australia.
Fig. 2.
The first of 8 900 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of an ITS alignment for Colletotrichum (PAUP v4.0b10). Bootstrap support values > 69 % are shown at the nodes and strict consensus branches are thickened and type sequences are in bold. GenBank accessions of C. gloeosporioides and Glomerella cingulata are indicated in blue colours. Tree length = 431, CI = 0.719, RI = 0.903 and RC = 0.649.
The C. gloeosporioides aggregate is currently the subject of intensive phylogenetic analysis and Hyde et al. (2009a) noted that it was likely a series of well-supported monophyletic (though not host-specific) clades may be identified. This research has resulted in several publications revealing, describing or typifying species within the C. gloeosporioides species complex (Shivas & Tan 2009, Yang et al. 2009, Phoulivong et al. 2010, Rojas et al. 2010, Weir & Johnston 2010). The taxon was previously considered as a pathogen of many tropical fruits, causing anthracnose (Holliday 1980). Phoulivong et al. (2010) isolated Colletotrichum species from anthracnose symptoms of eight tropical fruits in Laos and Thailand and none of these isolates was C. gloeosporioides. This illustrates the need to re-investigate the Colletotrichum species in Australia using molecular data to establish which species occur in this country.
Diaporthe/Phomopsis
The anamorphic ascomycete genus Phomopsis contains about 1 000 species names (Uecker 1988) with teleomorphic connections in Diaporthe for about 180 species (van der Aa & Vanev 2002). Phomopsis spp. are widespread and occur on a diversity of host plants as pathogens, endophytes and saprobes (Uecker 1988). Plant pathogenic species cause serious diseases of many cultivated plants worldwide, including grapevines (van Niekerk et al. 2005), sunflower (Gulya et al. 1997), strawberry (Maas 1998), and soybean (Li et al. 2010). Interest in Phomopsis has also focused on the secondary metabolites produced by some endophytic and saprobic forms. Two examples are given. Firstly, Diaporthe toxica is known to produce toxic metabolites, phomopsins, on infected Lupinus stubble or seed, which can result in the death of grazing animals (Peterson et al. 1987, Cowley et al. 2010). Secondly, strains of endophytic Phomopsis from healthy plants have been shown to produce taxol, which has strong cytoxicity towards human cancer cells (Kumaran & Hur 2009).
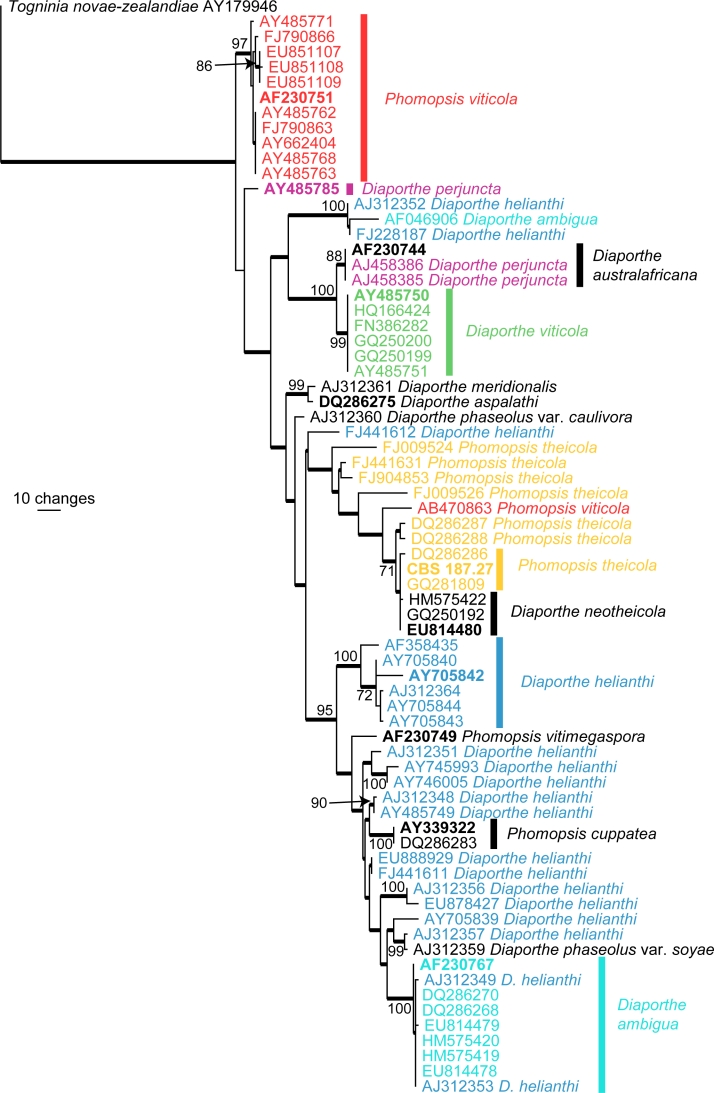
Species delimitation in Phomopsis has been traditionally based on host association as morphological characters are few and not reliable, for example, most species do not produce β-conidia or the teleomorph in culture (Rehner & Uecker 1994). Increasingly molecular phylogenies, especially those derived from the sequences of the internal transcribed spacer (ITS) region of the ribosomal DNA have been used to identify species (Mostert et al. 2001, van Niekerk et al. 2005, van Rensburg et al. 2006, Santos & Phillips 2009, Ash et al. 2010). Fig. 3 shows an ITS phylogeny consisting of 72 sequences (including the outgroup sequence) obtained from NCBIs GenBank nucleotide database. A total of 489 characters were used in the analysis, of which 134 characters were parsimony informative, 267 were constant and 88 variable characters were parsimony uninformative. The tree clearly illustrates the confusion around the application of species names such as Diaporthe ambigua and D. helianthi.
Fig. 3.
The first of 319 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of an ITS alignment for Phomopsis (PAUP v4.0b10). Bootstrap support values > 69 % are shown at the nodes and strict consensus branches are thickened and type sequences are in bold. Names of paraphyletic species are indicated in different colours and clades containing the type sequences of these paraphyletic species are indicated with a coloured bar corresponding to the colour of the species name. Tree length = 568, CI = 0.583, RI = 0.876 and RC = 0.511.
In Australia, specimens of Phomopsis deposited in the major plant pathology herbaria are mostly not identified to species level. The reason for this is that the species concept in Phomopsis needs modernisation, particularly in light of additional biological, biochemical and molecular data (van der Aa & Vanev 2002). Discarding the host-based species concept is the first step in the development of a useful and reliable classification for Phomopsis. This needs to be followed by a major international collaborative effort, as is happening in Botryosphaeria, Colletotrichum and Fusarium, to develop a reliable taxonomy.
Fusarium
Fusarium contains some of the most damaging plant pathogenic fungi as well as species that are important toxin producers and human pathogens (Desjardin 2006, Leslie & Summerell 2006). It is also one of the most actively researched groups of fungi and consequently the taxonomic concepts of many of the key plant pathogens have changed dramatically over the past two decades (Leslie & Summerell 2006). Added to this there have been many new species of Fusarium described from a diversity of environments and host substrates, including a number from Australia, e.g. F. aywerte and F. nurragi (Benyon et al. 2000), F. babinda (Summerell et al. 1995), F. beomiforme (Nelson et al. 1987), F. gaditjirri (Phan et al. 2004), F. lyarnte and F. werrikimbe (Walsh et al. 2010). Like the other genera described above, Fusarium has been shown to be rich in cryptic species, and while some may argue that cryptic speciation is of interest only to taxonomists and evolutionary biologists in Fusarium these species have clearly been demonstrated to have critical importance to plant pathologists, plant breeders and quarantine officials.
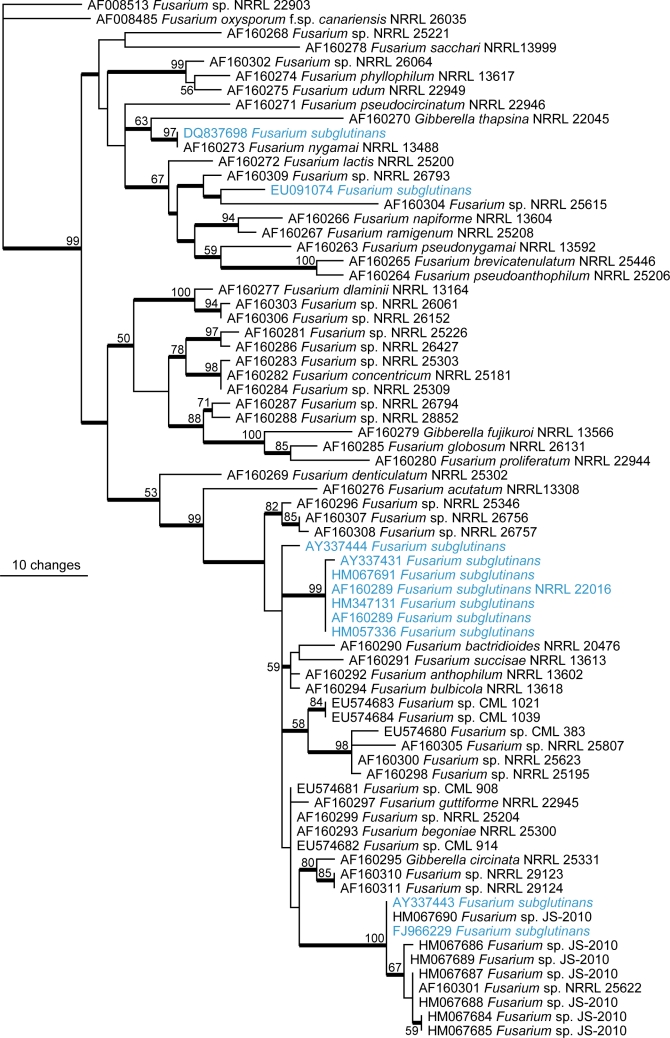
Several species provide good examples of the problems in Fusarium. Stalk rot of maize is caused by F. verticillioides but much of the literature and most of the specimens in Australian collections will use the name F. moniliforme. This latter name we now know refers to at least three species currently described, namely F. verticillioides, F. thapsinum and F. andiyazi (Seifert et al. 2003, Leslie et al. 2005) with a number of undescribed taxa known; all are morphologically identical but differ biologically, ecologically and phylogenetically. We now know that F. verticillioides causes stalk rot of maize and produces the mycotoxin fumonisin, F. thapsinum causes stalk rot of sorghum and does not produce fumonisin, but does produce other less important toxins and F. andiyazi causes some disease in sorghum (Leslie et al. 2005). Clearly in this case accurate identification is critical because of the implications to crop and human health. An even more complex situation occurs within a large group of species previously described as F. subglutinans. Fig. 4 shows a phylogenetic tree comprised of 72 translation elongation factor sequences sourced from GenBank of species either named as F. subglutinans or previously called F. subglutinans and those species closely related in the Gibberella fujikuroi clade. This tree highlights the diversity that exists within this species concept and how names are wrongly applied to data present in GenBank, and also demonstrates the confusion that surrounds a number of the species complexes within Fusarium, especially when using data sourced from databases such as GenBank. Detailed phylogeny-based investigations of the previously broad species concept that applied to F. subglutinans have shown it to include more than 20 species (Steenkamp et al. 2002, Leslie & Summerell 2006) of some extremely important plant pathogens including F. sacchari, F. circinatum and F. mangiferei; the latter two being of high importance to Australia as pathogens of quarantine importance. Several other species such as F. oxysporum (Wang et al. 2004), F. solani (O’Donnell 2000), F. dimerum (Schroers et al. 2009) and F. graminearum (O’Donnell et al. 2004) have been shown to be quite diverse species complexes and the impact of these studies on the identity of cultures in the collections still awaits to be explored.
Fig. 4.
One of 5 000 most parsimonious trees (CI = 0.656, RI = 0.853, RC = 0.559, HI = 0.4463) of the Gibberella fujikuroi species complex inferred from the translation elongation factor-1α gene sequence data. Fusarium sp. NRRL22903 was used as outgroup in the analysis.
Unfortunately, none of the major plant pathogen reference collections or major research collections of Fusarium has completed a full analysis of their holdings and as such urgently need review to determine the status of the isolates held. The preliminary analyses that have been done using DNA based techniques have shown that there is considerable diversity held in these collections that are not reflected in the lists of species present in Australia and as such it is difficult to determine the quarantine status of many species of Fusarium. A checklist including plant pathogenic and quarantine status of Fusarium in Australia has been prepared (Summerell et al. in press) and this will provide a basis on which to analyse the species found in collections in Australia.
Mycosphaerella and it anamorphs
The genus Mycosphaerella s.lat. is commonly accepted as the largest genus of Ascomycetes, containing over 10 000 taxa if anamorph states are included. Recent studies have shown, however, that Mycosphaerella is para- and polyphyletic (Hunter et al. 2006, Schoch et al. 2006, Crous et al. 2007a, b, 2009a, b, c, Arzanlou et al. 2007, Batzer et al. 2008), and in fact contains numerous genera, most of which can only be distinguished based on their unique anamorphs. In most cases anamorph genera are now used as holomorph names for these different clades, and new teleomorph names have not been introduced in an effort to stop the proliferation of dual nomenclature in this complex.
Most species in Mycosphaerella s.lat. (incl. Teratosphaeria) have been described on the assumption that they are host-specific (Chupp 1954, Corlett 1991, Braun 1995, 1998, Crous & Braun 2003, Aptroot 2006). Although this assumption holds true for many species, such as M. fijiensis, M. musicola and M. eumusae on banana (Arzanlou et al. 2008) and M. graminicola on wheat (Stukenbrock et al. 2007), some Mycosphaerella species are able to colonise different and even unrelated hosts (Crous et al. 2004b, Crous & Groenewald 2005). Furthermore, examples are also known of host-specific necrotrophic pathogenic species of Mycosphaerella and Teratosphaeria that appeared to also exhibit a facultative saprobic behaviour (Crous et al. 2009a, b). It is imperative, therefore, that to identify all species occurring in a specific lesion, DNA techniques are also employed.
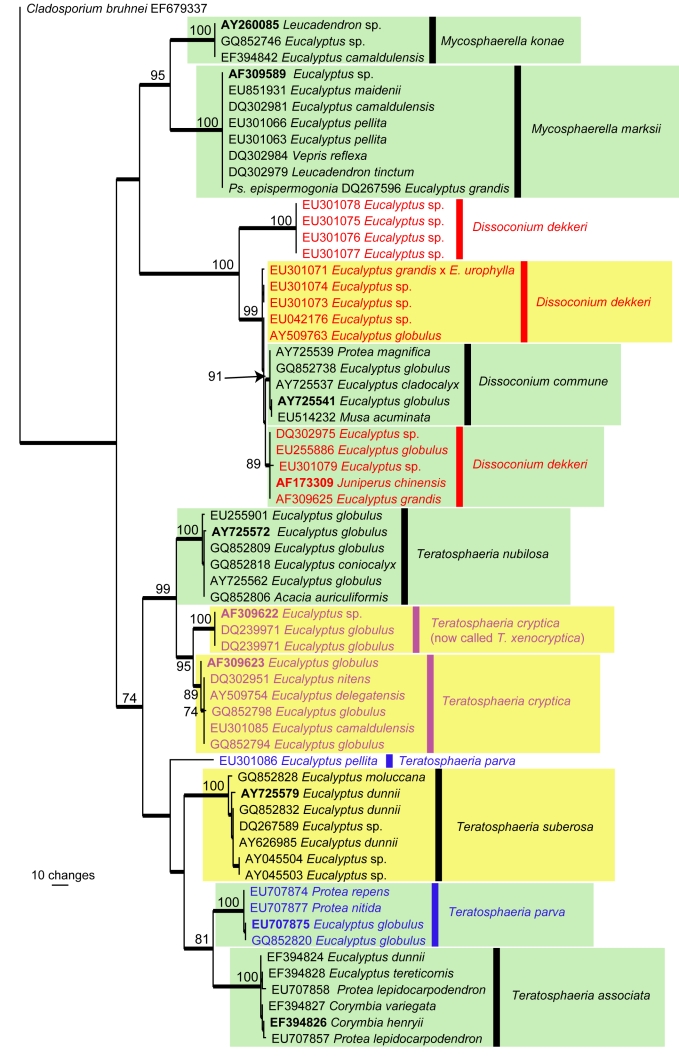
For the purpose of this paper, we chose to focus on the host genus Eucalyptus, which is indigenous to Australia, but also cultivated as exotics in commercial plantations in many countries of the world. Specific examples of the Mycosphaerella complex that occur on eucalypts and have a wider host range include the following: Dissoconium commune (on Eucalyptus in South Africa, Spain, New Zealand, Musa in Trinidad, Protea magnifica in Australia) (Crous et al. 2009c), M. konae (Leucospermum in Hawaii, Eucalyptus in Thailand) (Crous et al. 2007c), M. marksii (Eucalyptus, Australia, Bolivia, China, Ecuador, Ethiopia, Papua New Guinea, New Zealand, South Africa, Spain, Tanzania, Uruguay, Leucadendron on the Madeira Islands, and Musa in Mozambique) (Arzanlou et al. 2008), Teratosphaeria associata (Eucalyptus and Protea in Australia) (Summerell et al. 2006, Crous et al. 2007c), T. parva (Eucalyptus in Australia, Chile, Ethiopia, Portugal, South Africa, Spain, and Protea in South Africa), T. nubilosa (Eucalyptus in Australia, New Zealand, Europe, South America, and Acacia in Thailand (Crous & Groenewald 2005, Hunter et al. 2009), and Mycosphaerella citri (Musa in Florida, Acacia in Thailand, and Eucalyptus in Vietnam, and Aeglopsis, Citrus, Fortunella, Murraya, and Poncirus in North and South America, as well as Asia (Pretorius et al. 2003, Crous et al. 2004a, b, Crous & Groenewald 2005, Burgess et al. 2007) (Fig. 5).
Fig. 5.
The first of four equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions (PAUP v4.0b10). Bootstrap support values > 69 % are shown at the nodes and strict consensus branches are thickened and type sequences are in bold. Names of paraphyletic species are indicated in different colours. Green blocks represent species with a wide host range or host jumping between different host genera whereas the yellow blocks represent those with a wide host range or host jumping across host species (i.e. same genus but different species of the genus). Tree length = 555, CI = 0.674, RI = 0.952 and RC = 0.641.
Furthermore, to illustrate the complexity of the problem, several species occurring on eucalypts were initially described from exotic plantations outside of their native range. These include M. heimii from Madagascar (Crous & Swart 1995), M. fori and M. ellipsoidea from South Africa (Crous & Wingfield 1996, Hunter et al. 2006), Teratosphaeria tasmaniensis from Tasmania (Crous et al. 1998), T. molleriana from Portugal and California (Crous & Wingfield 1997), Dissoconium dekkeri from Europe and Africa (de Hoog et al. 1983, Crous & Wingfield 1996), and T. mexicana from Mexico (Crous 1998), which were only later reported from Australia (Maxwell et al. 2003, Whyte et al. 2005, Jackson et al. 2008). Fig. 5 shows an ITS phylogeny consisting of 119 sequences (including the outgroup sequence) obtained from GenBank. A total of 447 characters were used in the analysis, of which 203 characters were parsimony informative, 230 were constant and 14 variable characters were parsimony uninformative. Although the taxonomy of Mycosphaerella / Teratosphaeria has been subjected to a concerted effort by several research groups to clarify species boundaries, the tree clearly illustrates the confusion around the application of some species names such as Dissoconium dekkeri and Teratosphaeria parva. Furthermore, species with wider host ranges or involved in host jumping are also indicated on the tree.
CONCLUSIONS
In this paper we have looked at five fungal groups and have shown that in each case the present knowledge of plant disease associated fungi in these genera is often based on names that have now been shown to be species complexes. There are numerous other plant pathogenic genera where recent publications have revealed that what we thought were species now comprise species complexes and the species present in Australia need reassessing. These include Cladosporium (Crous et al. 2007b, Schubert et al. 2007, Bensch et al. 2010), Phoma (Aveskamp et al. 2008, 2010, de Gruyter et al. 2009), Phyllosticta (Wulanderi et al. 2009), and Mycosphaerella and its anamorphs (Crous 2009). We predict the situation to be the same in many other plant pathogenic genera such as Alternaria, Ascochyta, the helminthosporioid genera (including Bipolaris, Drechslera, Exserohilum, Curvularia and their teleomorphs) and Pestalotiopsis.
It is evident based on newly emerging molecular data that checklists of many plant pathogenic genera in Australia are now outdated and in need of revision. There is an urgent need for re-assessment of these plant-associated pathogens in order to preserve the effectiveness of Australia’s biosecurity measures. Unfortunately, it is often difficult or impossible to extract DNA from herbarium specimens in order to validate identifications, and morphology alone cannot always differentiate taxa in species complexes. Mycologists and plant pathologists need to go back to the field and recollect specimens from which fungal pathogens can be isolated and their DNA extracted and sequenced for the purpose of validating identifications. Revised checklists and databases must be supported by herbarium material, living cultures and DNA libraries. In Australia the Biosecurity Bank provides a reference collection of DNA from a range of agriculturally important plant pathogens and pests for molecular analyses, linking DNA specimens to voucher specimens for taxonomic verification (www.biosecuritybank.com). It is only through the combination of molecular and morphological approaches that the plant pathogens within Australia’s borders will be reliably identified. This in turn will preserve the effective role that quarantine plays in keeping unwanted plant pathogens out of Australia.
REFERENCES
- Aa HA van der, Vanev SA.2002. Revision of the species described in Phyllosticta. CBS, Utrecht, the Netherlands: [Google Scholar]
- Abdollahzadeh J, Javadi A, Mohammadi Goltapeh E, Zare R, Phillips AJL.2010. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 25: 1 – 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves A, Crous PW, Correia A, Phillips AJL.2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1 – 13 [Google Scholar]
- Aptroot A.2006. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1 – 231 [Google Scholar]
- Arx JA von.1957. Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift 29: 414 – 468 [Google Scholar]
- Arzanlou M, Groenewald JZ, Fullerton RA, Abeln ECA, Carlier J, et al. 2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia 20: 19 – 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin H-D, Crous PW.2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57 – 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash GJ, Stodart B, Sakuanrungsirikul S, Anschaw E, Crump N.2010. Genetic characterization of a novel Phomopsis sp., a putative biocontrol agent for Carthamus lanatus. Mycologia 102: 54 – 61 [DOI] [PubMed] [Google Scholar]
- Aveskamp MM, Gruyter J de, Crous PW.2008. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Diversity 31: 1 – 18 [Google Scholar]
- Aveskamp MM, Gruyter J de, Woudenberg JHC, Verkley GJM, Crous PW.2010. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology 65: 1 – 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer JC, Diaz Arias MM, Harrington TC, Gleason ML, Groenewald JZ, Crous PW.2008. Four species of Zygophiala (Schizothyriaceae, Capnodiales) are associated with the sooty blotch and flyspeck complex on apple. Mycologia 100: 232 – 244 [DOI] [PubMed] [Google Scholar]
- Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA , et al. 2010. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Studies in Mycology 67: 1 – 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyon F, Burgess LW, Sharp PJ.2000. Molecular genetic investigations and reclassification of Fusarium species in sections Fusarium and Roseum. Mycological Research 104: 1164 – 1174 [Google Scholar]
- Braun U.1995. A monograph of Cercosporella, Ramularia and allied genera (Phytopathogenic Hyphomycetes). Vol. 1. IHW-Verlag, Eching, Germany: [Google Scholar]
- Braun U.1998. A monograph of Cercosporella, Ramularia and allied genera (Phytopathogenic Hyphomycetes). Vol. 2. IHW-Verlag, Eching, Germany: [Google Scholar]
- Burgess TI, Barber PA, Sufaati S, Xu D, Hardy GEStJ, Dell B.2007. Mycosphaerella spp. on Eucalyptus in Asia; new species, new hosts and new records. Fungal Diversity 24: 135 – 157 [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, Weir B, Waller J, Abang MM, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183 – 204 [Google Scholar]
- Cannon PF, Buddie AG, Bridge PD.2008. The typification of Colletotrichum gloeosporioides. Mycotaxon 104: 189 – 204 [Google Scholar]
- Chupp C.1954. A monograph of the fungus genus Cercospora. Ithaca, New York: . Published by the author. [Google Scholar]
- Corlett M.1991. An annotated list of published names in Mycosphaerella and Sphaerella. Mycologia Memoir 18: 1 – 328 [Google Scholar]
- Cowley RB, Ash GJ, Harper JDI, Luckett DJ.2010. Evidence that Diaporthe toxica infection of Lupinus albus is an emerging concern for the Australian lupin industry. Australasian Plant Pathology 39: 146 – 153 [Google Scholar]
- Crouch JA, Beirn LA.2009. Anthracnose of cereals and grasses. Fungal Diversity 39: 19 – 44 [Google Scholar]
- Crouch JA, Clarke BB, White JF, Hillman BI.2009. Systematic analysis of falcate-spored graminicolous Colletotrichum and a description of six new species from warm-season grasses. Mycologia 101: 717 – 732 [DOI] [PubMed] [Google Scholar]
- Crous PW.1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycologia Memoir 21: 1 – 170 [Google Scholar]
- Crous PW.2009. Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Diversity 38: 1 – 24 [Google Scholar]
- Crous PW, Braun U.2003. Mycosphaerella and its anamorphs. 1. Names published in Cercospora and Passalora. CBS Biodiversity Series 1: 1 – 571 [Google Scholar]
- Crous PW, Braun U, Groenewald JZ.2007a. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Schubert K, Groenewald JZ.2007b. Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33 – 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ.2005. Hosts, species and genotypes: opinions versus data. Australasian Plant Pathology 34: 463 – 470 [Google Scholar]
- Crous PW, Groenewald JZ, Mansilla JP, Hunter GC, Wingfield MJ.2004a. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. Studies in Mycology 50: 195 – 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ.2004b. Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology 50: 457 – 469 [Google Scholar]
- Crous PW, Groenewald JZ, Summerell BA, Wingfield BD, Wingfield MJ.2009a. Co-occurring species of Teratosphaeria on Eucalyptus. Persoonia 22: 38 – 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, et al. 2009b. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17 – 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235 – 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Mohammed C, Himaman W, Groenewald JZ.2007c. Foliicolous Mycosphaerella spp. and their anamorphs on Corymbia and Eucalyptus. Fungal Diversity 26: 143 – 185 [Google Scholar]
- Crous PW, Swart WJ.1995. Foliicolous fungi of Eucalyptus spp. from eastern Madagascar: implications for South Africa. South African Forestry Journal 172: 1 – 5 [Google Scholar]
- Crous PW, Wingfield MJ.1996. Species of Mycosphaerella and their anamorphs associated with leaf blotch disease of Eucalyptus in South Africa. Mycologia 88: 441 – 458 [Google Scholar]
- Crous PW, Wingfield MJ.1997. Colletogloeopsis, a new coelomycete genus to accommodate anamorphs of two species of Mycosphaerella occurring on Eucalyptus. Canadian Journal of Botany 75: 667 – 674 [Google Scholar]
- Crous PW, Wingfield MJ, Groenewald JZ.2009c. Niche sharing reflects a poorly understood biodiversity phenomenon. Persoonia 22: 83 – 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Mohammed C, Yuan ZQ.1998. New foliar pathogens of Eucalyptus from Australia and Indonesia. Mycological Research 102: 527 – 532 [Google Scholar]
- Damm U, Crous PW, Fourie PH.2007. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora spp. nov. Mycologia 99: 664 – 680 [DOI] [PubMed] [Google Scholar]
- Damm U, Woudenberg JHC, Cannon PF, Crous PW.2009. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39: 45 – 87 [Google Scholar]
- Denman S, Crous PW, Groenewald JG, Slippers B, Wingfield BD, Wingfield MJ.2003. Circumscription of Botryosphaeria species associated with Proteaceae based on morphology and DNA sequence data. Mycologia 95: 294 – 307 [PubMed] [Google Scholar]
- Desjardins AE.2006. Fusarium mycotoxins: chemistry, genetics and biology. APS Press, St. Paul, Minnesota, USA: [Google Scholar]
- Gruyter J de, Aveskamp MM, Woudenberg JHC, Verkley GJM, Groenewald JZ, Crous PW.2009. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycological Research: 113: 508 – 519 [DOI] [PubMed] [Google Scholar]
- Gulya T, Rashid KY, Masirevic SM.1997. Sunflower diseases. In: Schneiter AA. (ed), Sunflower technology and production. American Society of Agronomy, Madison, USA: [Google Scholar]
- Holliday P.1980. Fungus diseases of tropical crops. Cambridge University Press, Cambridge: [Google Scholar]
- Hoog GS de, Oorschot CAN van, Hijwegen T.1983. Taxonomy of the Dactylaria complex. II. Proceedings van de Koninklijke Nederlandse Akademie van Wetenschappen, Series C, 86, 2: 197 – 206 [Google Scholar]
- Hunter GC, Crous PW, Carnegie AJ, Wingfield MJ.2009. Teratosphaeria nubilosa, a serious leaf disease pathogen of Eucalyptus spp. in native and introduced areas. Molecular Plant Pathology 10: 1 – 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GC, Wingfield BD, Crous PW, Wingfield MJ.2006. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Studies in Mycology 55: 147 – 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Alcorn JL.1993. Some disease-associated microorganisms on plants of Cape York Peninsula and Torres Strait Islands. Australian Plant Pathology 22: 73 – 83 [Google Scholar]
- Hyde KD, Cai L, Cannon PF, Crouch JA, Crous PW , et al. 2009a. Colletotrichum – names in current use. Fungal Diversity 39: 147 – 182 [Google Scholar]
- Hyde KD, Cai L, McKenzie EHC, Yang YL, Zhang JZ, Prihastuti H.2009b. Colletotrichum: a catalogue of confusion. Fungal Diversity 39: 1 – 17 [Google Scholar]
- Hyde KD, Philemon E.1994. Some disease-associated microorganisms on plants in the Western Province of Papua New Guinea. Australian Plant Pathology 23: 69 – 76 [Google Scholar]
- Jackson SL, Maxwell A, Burgess TI, Hardy GEStJ, Dell B.2008. Incidence and new records of Mycosphaerella species within a Eucalyptus globulus plantation in Western Australia. Forest Ecology and Management 255: 3931 – 3937 [Google Scholar]
- Kumaran RS, Hur BK.2009. Screening of species of the endophytic fungus Phomopsis for the production of the anticancer drug taxol. Biotechnology and Applied Biochemistry 54: 21 – 30 [DOI] [PubMed] [Google Scholar]
- Kvas M, Marasas WFO, Wingfield BD, Wingfield MJ, Steenkamp ET.2009. Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Diversity 34: 1 – 21 [Google Scholar]
- Leslie JF, Summerell BA.2006. The Fusarium laboratory manual. Blackwell Professional, Ames, Iowa, USA: [Google Scholar]
- Leslie JF, Zeller KA, Lamprecht SC, Rheeder JP, Marasas WFO.2005. Toxicity, pathogenicity and genetic differentiation of five species of Fusarium from sorghum and millet. Phytopathology 95: 275 – 283 [DOI] [PubMed] [Google Scholar]
- Li S, Hartman GL, Boykin DL.2010. Aggressiveness of Phomopsis longicolla and other Phomopsis spp. on soybean. Plant Disease 94: 1035 – 1040 [DOI] [PubMed] [Google Scholar]
- Liu JK, Chomnunti P, Cai L, Phookamsak R, Chukeatirote R , et al. In press. Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia 62 [Google Scholar]
- JL Maas. (ed) 1998. Compendium of strawberry diseases, 2nd ed APS Press, St. Paul, Minnesota, USA: [Google Scholar]
- Maxwell A, Dell B, Neumeister-Kemp H, Hardy GEStJ.2003. Mycosphaerella species associated with Eucalyptus in southwestern Australia – new species, new records and a key. Mycological Research 107: 351 – 359 [DOI] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang C-J, Phillips AJL.2001. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 145 – 166 [Google Scholar]
- Nelson PE, Toussoun TA, Burgess LW.1987. Characterization of Fusarium beomiforme sp. nov. Mycologia 79: 884 – 889 [Google Scholar]
- Niekerk JM van, Groenewald JZ, Farr DF, Fourie PH, Halleen F, Crous PW.2005. Reassessment of Phomopsis species on grapevines. Australasian Plant Pathology 34: 27 – 39 [Google Scholar]
- O’Donnell K.2000. Molecular phylogeny of the Nectria haematococca- Fusarium solani species complex. Mycologia 92: 919 – 938 [Google Scholar]
- O’Donnell K, Ward TJ, Geiser DM, Kistler HC.2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology 41: 600 – 623 [DOI] [PubMed] [Google Scholar]
- Page RDM.1996. TreeView: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357 – 358 [DOI] [PubMed] [Google Scholar]
- Peterson JE, Jago MV, Payne AL, Stewart PL.1987. The toxicity of phomopsin for sheep. Australian Veterinary Journal 64: 293 – 298 [DOI] [PubMed] [Google Scholar]
- Phan HT, Burgess LW, Summerell BA, Bullock S, Liew ECY , et al. 2004. Gibberella gaditjirri (Fusarium gaditjirri) sp. nov., a new species from tropical grasses in Australia. Studies in Mycology 50: 261 – 272 [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, Johnston PR, Ramaley A, et al. 2008. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21: 29 – 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Crous PW, Alves A.2007. Diplodia seriata, the anamorph of “Botryosphaeria” obtusa. Fungal Diversity 25: 141 – 155 [Google Scholar]
- Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, et al. 2010. Colletotrichum gloeosporioides is not a common tropical pathogen. Fungal Diversity 44: doi 10.1007/s13225-010-0046-0. [Google Scholar]
- Plant Health Australia 2010. Pest risk assessment for use in industry biosecurity plans. Plant Health Australia www.planthealthaustralia.com.au [Google Scholar]
- Pretorius MC, Crous PW, Groenewald JZ, Braun U.2003. Phylogeny of some cercosporoid fungi from Citrus. Sydowia 55: 286 – 305 [Google Scholar]
- Rehner SA, Uecker FA.1994. Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666 – 1674 [Google Scholar]
- Rensburg JCV van, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW.2006. Characterisation of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Studies in Mycology 55: 65 – 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas EI, Rehner SA, Samuels GJ, Bael S van, Herre EA, et al. 2010. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panamá: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102: 1318 – 1338 [DOI] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL.2009. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 111 – 125 [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZS, Boehm EWA, Burgess TI, et al. 2009. A class-wide phylogenetic assessment of Dothideomycetes . Studies in Mycology 64: 1 – 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW.2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1043 – 1054 [DOI] [PubMed] [Google Scholar]
- Schroers HJ, O’Donnell K, Lamprecht SC, Kammeyer PL, Johnson S, et al. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101: 44 – 70 [DOI] [PubMed] [Google Scholar]
- Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, et al. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology 58: 105 – 156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Aoki T, Baayen RP, Brayford D, Burgess LW , et al. 2003. The name Fusarium moniliforme should no longer be used. Mycological Research 107: 643 – 644 [Google Scholar]
- Shivas RG.1995. New records of plant pathogens in the Kimberley region of northern Western Australia. Australasian Plant Pathology 24: 188 – 201 [Google Scholar]
- Shivas RG, Alcorn JL.1996. A checklist of plant pathogenic microfungi in the rainforest of the wet tropics of northern Queensland. Australasian Plant Pathology 25: 158 – 173 [Google Scholar]
- Shivas RG, Beasley DR, Pascoe IG, Cunnington JH, Pitkethley R, Priest M.2006. Specimen-based databases of Australian plant pathogens: past, present and future. Australasian Plant Pathology 35: 195 – 198 [Google Scholar]
- Shivas RG, Suyoko S, Raga N, Hyde KD.1996. Some disease-associated microorganisms on plants in Irian Jaya, Indonesia. Australasian Plant Pathology 25: 36 – 49 [Google Scholar]
- Shivas RG, Tan YP.2009. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Diversity 39: 111 – 122 [Google Scholar]
- Slippers B, Crous PW, Denman S, Coutinho TA, Wingfield BD, Wingfield MJ.2004. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96: 83 – 101 [PubMed] [Google Scholar]
- Smith H, Crous PW, Wingfield MJ, Coutinho TA, Wingfield BD.2001. Botryosphaeria eucalyptorum sp. nov., a new species in the B. dothidea-complex on Eucalyptus in South Africa. Mycologia 93: 277 – 285 [Google Scholar]
- Steenkamp ET, Wingfield BD, Desjardins AE, Marasas WFO, Wingfield MJ.2002. Cryptic speciation in Fusarium subglutinans. Mycologia 94: 1032 – 1043 [PubMed] [Google Scholar]
- Stukenbrock EH, Banke S, Javan-Nikkhah M, McDonald BA.2007. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Molecular Biology and Evolution 24: 398 – 411 [DOI] [PubMed] [Google Scholar]
- Summerell BA, Groenewald JZ, Carnegie AJ, Summerbell RC, Crous PW.2006. Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Diversity 23: 323 – 350 [Google Scholar]
- Summerell BA, Laurence MH, Liew ECY, Leslie JF.2010. Biogeography and phylogeography of Fusarium: a review. Fungal Diversity 43: doi 10.1007/s13225-010-0060-2. [Google Scholar]
- Summerell BA, Rugg CA, Burgess LW.1995. Characterization of Fusarium babinda sp. nov. Mycological Research 99: 1345 – 1348 [Google Scholar]
- Sutton BC.1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew, England: [Google Scholar]
- Swofford DL.2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, USA: [Google Scholar]
- Uecker FA.1988. A world list of Phomopsis names with notes on nomenclature, morphology and biology. Contributions from the U.S. National Fungus Collection. Mycologia Memoir 13: 9 – 12 [Google Scholar]
- Walsh JL, Laurence MH, Liew ECY, Sangalang AE, Burgess LW, et al. 2010. Fusarium: two endophytic novel species from tropical grasses of northern Australia. Fungal Diversity 44: doi 10.1007/s13225-010-0035-3 [Google Scholar]
- Wang B, Brubaker CL, Burdon JJ.2004. Fusarium species and Fusarium wilt pathogens associated with native Gossypium populations in Australia. Mycological Research 108: 35 – 44 [DOI] [PubMed] [Google Scholar]
- Weir BS, Johnston PR.2010. Characterisation and neotypification of Gloeosporium kaki Hori as Colletotrichum horii nom. nov. Mycotaxon 111: 209 – 219 [Google Scholar]
- Whyte G, Burgess TI, Barber PA, Hardy GEStJ.2005. First record of Mycosphaerella heimii in Australia. Australasian Plant Pathology 34: 605 – 606 [Google Scholar]
- Wulandari NF, To-anun C, Hyde KD, Duong LM, Gruyter J de, et al. 2009. Phyllosticta citriasiana sp. nov., the cause of Citrus tan spot of Citrus maxima in Asia. Fungal Diversity 34: 23 – 39 [Google Scholar]
- Yang YL, Liu ZY, Cai L, Hyde KD, Yu ZN, McKenzie EHC.2009. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity 39: 123 – 146 [Google Scholar]
- Zhang Y, Schoch CL, Fournier J, Crous PW, Gruyter J de , et al. 2009. Multi-locus phylogeny of the Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85 – 102 [DOI] [PMC free article] [PubMed] [Google Scholar]