Abstract
The conserved RNA binding protein La recognizes UUU-3′OH on its small nuclear RNA ligands and stabilizes them against 3′-end-mediated decay. We report that newly described La-related protein 4 (LARP4) is a factor that can bind poly(A) RNA and interact with poly(A) binding protein (PABP). Yeast two-hybrid analysis and reciprocal immunoprecipitations (IPs) from HeLa cells revealed that LARP4 interacts with RACK1, a 40S ribosome- and mRNA-associated protein. LARP4 cosediments with 40S ribosome subunits and polyribosomes, and its knockdown decreases translation. Mutagenesis of the RNA binding or PABP interaction motifs decrease LARP4 association with polysomes. Several translation and mRNA metabolism-related proteins use a PAM2 sequence containing a critical invariant phenylalanine to make direct contact with the MLLE domain of PABP, and their competition for the MLLE is thought to regulate mRNA homeostasis. Unlike all ∼150 previously analyzed PAM2 sequences, LARP4 contains a variant PAM2 (PAM2w) with tryptophan in place of the phenylalanine. Binding and nuclear magnetic resonance (NMR) studies have shown that a peptide representing LARP4 PAM2w interacts with the MLLE of PABP within the affinity range measured for other PAM2 motif peptides. A cocrystal of PABC bound to LARP4 PAM2w shows tryptophan in the pocket in PABC-MLLE otherwise occupied by phenylalanine. We present evidence that LARP4 expression stimulates luciferase reporter activity by promoting mRNA stability, as shown by mRNA decay analysis of luciferase and cellular mRNAs. We propose that LARP4 activity is integrated with other PAM2 protein activities by PABP as part of mRNA homeostasis.
The RNA binding domain of the conserved La protein consists of a La motif (LaM) and an RNA recognition motif (RRM) that work together to recognize UUU-3′OH on small nascent transcripts and to protect them from 3′ exonucleases (7, 45). In addition to this, La proteins can modulate mRNA translation (30, 63-65). The LaM-RRM arrangement has been found in La-related proteins 1 (LARP1), 1b, 4, 4b, 6, and 7, which have been separately conserved during evolution (8, 10) (LARP4b is also referred to as LARP5 in multiple databases and here will be designated LARP5/4b). LARP7 is specific for 7SK snRNA, which it recognizes in part via UUU-3′OH (29, 46). LARP6 binds to a stem-loop in the 5′ untranslated regions (UTRs) of collagen mRNAs in a uracil-dependent manner (15), and LARP1 was shown to bind poly(U) and to a lesser extent poly(G), but not poly(A) or poly(C) (51). Consistent with these specificities, LARP1, -6, and -7 have conserved all of the amino acids involved in UUU-3′OH recognition in La-RNA crystals (37, 66), while LARP4 and -5/4b have diverged, suggesting alternative RNA binding (8). Moreover, an invariant divergence in all of the LARP4 and -5/4b sequences available occurs in a most critical residue involved in base-specific recognition seen in La-RNA crystals, corresponding to human La Q20, suggesting a conserved difference in RNA recognition (8). Although the LaM-RRM in La protein recognizes RNA in a unique way (8, 45), whether LARPs share this or have adopted alternative modes of RNA recognition is unknown.
Of the LARP families studied for function, LARP1, -5/4b, and -6 appear to be involved in mRNA metabolism and/or translation (9, 14, 15, 51, 57). Of these, LARP1 and -5/4b interact with poly(A) binding protein (PABP), although the precise mechanisms were not reported (9, 14, 57), whereas LARP6 appears to block assembly of its associated mRNAs with initiating ribosomes (15).
Translation is facilitated by interactions of the 5′ cap and 3′ poly(A) of the mRNA by eukaryotic initiation factor 4E (eIF4E) and PABP (47). The translation initiation activity of PABP can be regulated by PABP-interacting protein 1 (Paip1), which stabilizes initiation complexes via interactions with the 40S ribosome (18, 47). Multiple molecules of PABP can bind poly(A) tails (5, 44), and some data suggest that at least two molecules of poly(A)-associated PABP are required for efficient translation initiation (2). PABP can engage a variety of protein partners via their common PABP interaction motif 2 (PAM2) sequences, including Paip1, Paip2, eRF3, GW182 (TNRC6C), ataxin 2, Tob2, and poly(A) nuclease, representing different mechanisms of control involving translation initiation and termination, as well as mRNA stability (19, 33, 34, 41, 44, 54, 60, 69). Since the PAM2 motifs make direct contacts with the MLLE domain of PABP (39, 40), proper signal integration presumably involves their competition for PABP (25). A model in which the PAM2 motifs of eRF3 and poly(A) nucleases compete for PABP reflects a balance of translation termination and mRNA deadenylation activities (25, 39, 41, 55).
In rat neurogenic cells, LARP5/4b (KIAA0217) was a component of an mRNA-protein (mRNP) complex associated with PABP that could bind poly(A) in a Northwestern blotting assay, although no other RNAs were tested (3). A recent report demonstrated that two broad regions of LARP5/4b interact with PABP to stimulate translation, although RNA binding was not examined (57). While human LARP4 and -5/4b are most homologous in their LaM-RRMs, they share patchy homology in the ∼500 amino acids outside this region (8). Here, we identify variant PAM2 motifs in LARP4 and -5/4b that contain Trp in place of the otherwise critical invariant Phe (8) found in all of the other ∼150 PAM2 sequences examined (1), which we refer to as PAM2w hereafter.
We show that the LaM-RRM of human LARP4 preferentially binds poly(A) and exhibits other characteristics that suggest a recognition mode different from that of La proteins. Screening of a human cDNA library for yeast 2-hybrid interactions with LARP4 yielded RACK1, a 40S ribosome- and mRNA-associated kinase (17, 50), which was confirmed by reciprocal immunoprecipitations (IPs) from HeLa cells. LARP4 is cytoplasmic and interacts with PABP via two regions, PAM2w and a region following the RRM that includes ∼70 residues with significant homology to LARP5/4b. A peptide representing PAM2w of human LARP4 and the MLLE domain of PABP was examined by binding, nuclear magnetic resonance (NMR), and crystallography, which showed direct interactions similar to those of other PAM2-MLLE complexes. Further consistent with a translation-related function, LARP4 cosediments as two peaks on polysome profiles, with 40S ribosomes and with PABP on polysomes. After LARP4 knockdown, polysome profiles indicate deficiency in translation initiation, with [35S]methionine incorporation into newly synthesized protein diminished by ∼20%. LARP4 appears to promote translation, in part, by stabilizing mRNA, as suggested by our decay analyses.
MATERIALS AND METHODS
Cloning.
HeLa poly(A)+ RNA (Ambion) was used to make LARP4 cDNA, which was cloned into the HindIII-BamHI sites of pFlag-CMV2 (Sigma-Aldrich). Deletions and substitutions were done by PCR and with QuikChange XL (Stratagene). The F-LARP4-M3 mutations were K124D, K125A, Q126D, D139K, and Y141D in the La motif and I202D and R204D in the RRM1 β-1. All constructs were verified by sequencing.
Immunofluorescence used anti-Flag, followed by anti-mouse IgG-Cy3 or anti-LARP4, followed by anti-rabbit IgG-Cy3. Hoechst (blue) was used to stain nuclei.
Immunoblotting.
The secondary antibody (Ab) used for detection was anti-rabbit IgG (GE, United Kingdom) or anti-mouse IgG (Cell Signaling), and processing for chemiluminescence was done using a SuperSignal West Dura kit (Thermo Scientific). In most cases, a single membrane was probed, exposed, and stripped (Restore Western Blot Stripping buffer; Thermo Scientific) before probing with a different Ab was done.
Immunoprecipitation was by anti-FLAG M2 affinity gel (Sigma-Aldrich) using 20 μl of gel bead volume and 1 mg total cellular protein. One milliliter of cell lysate in polysome lysis buffer (PLB) containing 25 mM EDTA, 2 mM dithiothreitol (DTT), and proteinase and RNase inhibitors was incubated with beads prewashed with NT2 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl, 0.05% NP-40) for 4 h at 4°C, followed by 5 washes with NT2. After elution with hot SDS buffer, fresh beta-mercaptoethanol was added and samples were loaded.
Antibodies.
Anti-rpS6 and -PABP were from Cell Signaling, anti-Flag and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) was from Sigma Aldrich, anti-rpL28 was from Santa Cruz, anti-RACK1 was from TRansduction Laboratories, and anti-Paip1 was a gift from N. Sonenberg. Rabbit anti-LARP4 was obtained using the LARP4 C-terminal sequence CGVTRRNGKEQYVPPRSPK as the antigen and affinity purified using the same peptide (Open Biosystems).
Polysome profiles were prepared from fresh extract made from HEK-293 cells by standard methods (6) using a Programmable Density Gradient Fractionation System Spectrophotometer (Foxy Jr. model; Teledyne Isco, Lincoln, NE). Gradients were made using a Gradient Master (Biocomp). After 15 min of exposure to cycloheximide (100 μg/ml), cells were lysed with PLB (10 mM HEPES, pH 7.0, 100 mM KCl, 5 mM MgCl, 5% NP-40). Cell lysate (150 μl; 2 mg total protein) was layered on 5 to 45% sucrose gradients and centrifuged at 39,000 × g for 2 h.
RNA binding with recombinant La-NTD(1-235) and LARP4-NTD(1-286) purified from Escherichia coli was described using an electrophoretic mobility shift assay (EMSA) (31). The sequence of the 36-mer RNA was 5′-GAACACUUUGGCGCUCAAUGCGCCCUUCGUUAAAAA-3′. Each 32P-labeled RNA was purified by excision of a tight band from a denaturing 20% polyacrylamide gel.
Yeast 2-hybrid assays used full-length LARP4 and were performed by Hybrigenics S.A. Services (Paris, France) using a human liver cDNA library.
Small interfering RNAs (siRNAs) targeting LARP4 (no. 1 and 2) and PABP were from Invitrogen, and LARP4 no. 3 was from Dharmacon. The siRNA sequences were as follows: LARP4 no. 1, AACAGAGGAAUCUUCUAUUAGAUCC; LARP4 no. 2, AAGAACUGAAGAUGGCUCUUUAGGG; LARP4 no. 3, GAAUGUUGCUGGAACGUAAUU; PABP, AGGUGGUUUGUGAUGAAAAUU. The control siRNA was Stealth RNAi Negative Universal Control Hi GC from Invitrogen.
[35S]methionine pulse-labeling.
Forty-eight hours after siRNA transfection, at ∼80% confluence, the cells were incubated in methionine-free medium for 15 min, followed by medium containing [35S]methionine at 100 μCi/ml. Extracts were prepared, and the total protein concentration was determined (Bio-Rad assay). Equal amounts were loaded for SDS-PAGE. After being Coomassie stained, the gel was photographed and then dried, processed using a Fuji phosphorimager, and quantified using Fuji software.
siRNA-luciferase.
HEK-293 cells growing in 6-well plates were transfected with siRNA (10 nM final concentration), and 24 h later, 100 ng of the luciferase reporter plasmid (pGL3/RLuc/HCVIRES/FLuc) (21, 58) was transfected. The luciferase open reading frame (ORF) and other components of the plasmid, including the simian virus 40 (SV40) promoter and the SV40 late poly(A) addition signal following the firefly luciferase ORF as part of the transcription unit, were confirmed by sequencing (not shown). Forty hours later, cells were harvested and protein was extracted and quantitated for the luciferase assay. Equal amounts of protein were assayed using the Luciferase Reporter Assay System (Promega).
F-LARP4 expression-luciferase.
For each 6-well plate of cells, 1 μg of F-LARP4 or F-vector plasmid, together with 100 ng luciferase plasmid, each quantitated by optical density at 260 nm (OD260) and confirmed by gel electrophoresis, was cotransfected using Fugene 6 (Roche). Forty hours later, total protein was extracted and processed for the luciferase assay.
mRNA decay.
Twenty-four hours after being plated, a mixture of F-LARP4 plasmid and luciferase plasmid was cotransfected. After 40 h (∼80% confluence), cells were treated with actinomycin D (Sigma; used at 10 μg/ml), and RNA was isolated at intervals thereafter.
Proteins for isothermal titration calorimetry (ITC) and structural studies.
La(1-194) was expressed and purified as reported previously (32). LARP4(111-303) was subcloned with N-terminal hexahistidine into a pET-Duet-1 expression vector and expressed in E. coli Rosetta II cells. The cell pellets were lysed by sonication in 20 mM Tris, 300 mM NaCl, 10 mM imidazole, pH 8.0, and centrifuged to separate the soluble and insoluble fractions of the cells. LARP4(111-303) was purified by affinity chromatography on a 5-ml HisTrap column (GE Healthcare) with a gradient of 10 to 300 mM imidazole. After cleavage of the His tag with tobacco etch virus (TEV) protease, the protein was purified from the cleaved tag, the His-tagged TEV, and any undigested product by a second Ni2+ affinity step. The samples were then applied onto a 5-ml Hi-Trap heparin column to remove nucleic acids contaminants. Protein concentrations were calculated based upon the near-UV absorption (ɛ280) using theoretical extinction coefficients derived from ExPASy (26).
The MLLE domain (residues 544 to 626) of human PABPC1 was expressed and purified as described previously (39).
Preparation of RNA for ITC.
A(15), A(10), and U(15) were purchased from IBA GmbH (Göttingen, Germany) and dissolved in 20 mM Tris, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT at pH 7.25. The concentration was determined by UV, using a molar extinction coefficient at 260 nm calculated by the nearest-neighbor model (56).
Peptides.
PAM2w (TGLNPNAKVWQEIA) corresponding to LARP4(13-26) was synthesized by 9-fluorenylmethoxy carbonyl (Fmoc) solid-phase peptide synthesis. PAM2w-long (GPLGSQVASKGTGLNPNAKVWQEIAPGNTDATPVTHGTESSWHEIAAT) corresponding to LARP(7-49) was cloned into pGEX-6p1 and expressed from E. coli BL-21 as a fusion protein that was cleaved with PreScission Protease to release free peptide with a 5-amino-acid extension at the N terminus. Both peptides were purified by reverse-phase chromatography on a C18 column (Vydac, Hesperia, CA) and verified by mass spectroscopy.
ITC experiments.
RNA-protein titrations were carried out in 20 mM Tris, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT, pH 7.25, at room temperature (298 K) using a high-sensitivity iTC-200 microcalorimeter from Microcal (GE Healthcare) following the protocol previously reported (27). In each experiment, 2-μl volumes of an RNA solution were titrated into a protein solution in the same buffer, using a computer-controlled 40-μl microsyringe, with a spacing of 180 s between injections. Each titration was corrected for the heat of dilution by subtracting the measured enthalpies of the injections following saturation. Because La-RNA interactions exhibited large enthalpy of binding, whereas for LARP4-RNA the enthalpy of binding was substantially smaller, higher concentrations were used in the latter experiments to improve the signal-to-noise ratio and to ensure that we were well above the limit of sensitivity of the iTC200 instrument. In particular, for the experiments with La, a protein concentration of 15 μM and an RNA concentration of 120 μM were used; for the experiments with LARP4, we used a protein concentration of 40 μM and an RNA concentration of 380 μM.
ITC of PAM2w-MLLE interactions was carried out in 50 mM NaH2PO4, 100 mM NaCl, pH 7.0, with a protein concentration of 170 μM for 4.5 mM LARP4(13-26) peptide and a protein concentration 85 μM for 1.6 mM LARP4(7-49).
Integrated heat data obtained for the ITCs were fitted using a nonlinear least-squares minimization algorithm to a theoretical titration curve, using the MicroCal-Origin 7.0 software package, from which the binding parameters ΔH° (the reaction enthalpy change in kcal·mol−1), Kb (the binding constant in M−1), and n (the molar ratio between the two proteins in the complex) were derived. The reaction entropy was calculated using the following relationships: ΔG° = −RT·lnKb (R, 1.987 cal·mol−1·K−1; T, 298 K) and ΔG° = ΔH° − TΔS°, where ΔS° is entropy, R is the gas constant, and T is temperature.
Protein crystallization.
The conditions for the MLLE domain in complex with the PAM2w peptide were identified utilizing hanging-drop vapor diffusion with the AmSO4 crystallization suite (Qiagen). The best crystals were obtained at 22°C by seeding microcrystals into a 1-μl drop of PABPC1(544-626)/LARP4(13-26) (10 mg/ml) in 1:2 molar ratio mixed with 1 μl of reservoir solution containing 0.25 M potassium iodide and 1.9 M ammonium sulfate. The solution for cryoprotection contained the reservoir solution with the addition of 15% (vol/vol) glycerol. The crystals contained one MLLE and one PAM2w molecule in the asymmetric unit, corresponding to a Vmax of 1.76 Å3 Da−1 and a solvent content of 30.2%.
Structure solution and refinement.
Diffraction data from a single crystal of the MLLE/PAM2w complex were collected at the McGill Macromolecular X-ray Diffraction Facility and Cornell High-Energy Synchrotron Source (CHESS). Data processing and scaling were performed with HKL2000 (52). The structures were determined by molecular replacement with Phaser (53), using the coordinates of the MLLE/Paip2 complex (Protein Data Bank [PDB] entry 3KUS). The initial MLLE/PAM2w model was completed and adjusted with the program Coot (22) and improved by several cycles of refinement, using the program REFMAC 5.2 (49) and model refitting. At the latest stage of refinement, we also applied the translation-libration-screw (TLS) option (70) with final density for PABPC1 residues 544 to 626 and LARP4 residues 15 to 25. The PBD ID code is 3PKN.
RESULTS
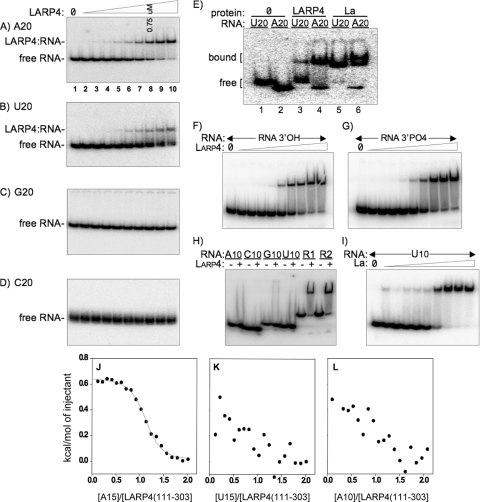
We compared the N-terminal regions containing the LaM-RRM of human La(1-235) and LARP4(1-286), referred to as La-NTD and LARP4-NTD, for RNA binding using EMSA. Of the homopolymers tested, A(20) exhibited the best affinity for LARP4-NTD, as reflected by the ratio of free to bound RNA, whereas U(20) showed less binding and C(20) and G(20) showed no binding (Fig. 1A to D). In this assay, the concentration at which 50% of the probe was shifted into a stable complex reflects the Kd (dissociation constant) (7). Accordingly, the results in Fig. 1A suggest that the Kd of LARP4-NTD for A(20) is ≤750 nM. Less than 50% of U(20) was shifted at 3.0 μM LARP4-NTD, the highest concentration tested (Fig. 1B). Moreover, the smear observed between the bound and free U(20) (Fig. 1B) suggests less stable binding. La-NTD bound more U(20) than A(20), as expected (Fig. 1E).
FIG. 1.
Human LARP4(1-286) preferentially binds poly(A) with a length requirement longer than 10 nucleotides. (A to D) EMSA of homopolymeric 20-mer RNAs for binding to the RNA binding domain of LARP4 protein, LARP(1-286), also referred to as LARP4-NTD. Twofold serial dilutions of LARP4(1-286) were used, with a 750-μM final concentration in lane 8, as indicated. (E) EMSA of A(20) and U(20) for La(1-235) and LARP4(1-286). (F to G) Comparison of LARP4(1-286) binding to otherwise identical 36-mer RNAs (see sequences in Materials and Methods) that end with a 3′ OH (F) or a 3′ phosphate (G) (see Materials and Methods). (H) LARP4(1-286) does not bind 10-mer RNAs. R1 and R2 are the same 36-nt RNAs used in panels F and G. (I) La(1-235) binds to U10. (J to L) Isothermal titration calorimetric analysis of LARP4(111-303) interactions with A(15) (J), U(15) (K), and A(10) (L). The Kd and other thermodynamic parameters derived from this analysis are reported in Table 1.
We examined LARP4-NTD for sensitivity to 3′ phosphate on ligand RNA, to which the genuine La proteins from multiple species are sensitive (20, 43, 62, 67, 71). Using otherwise identical 36-nucleotide (nt) RNAs that ended with AAAAA-3′OH or AAAAA-3′PO4, we could observe no difference in binding to the LARP4 protein over a range of concentrations (Fig. 1F and G), whereas the AAAAA-3′PO4 RNA exhibited ∼6-fold less binding to La than the AAAAA-3′OH RNA (not shown). Although we tried repeatedly to find an expected difference in 3′OH and 3′PO4 binding, we could not.
A notable feature of RNA recognition by La is its ready ability to bind short oligo-RNAs as evidenced by the ability of 4- to 6-nt RNAs to compete effectively with longer RNAs for in vitro binding (62), a cocrystal of La with a 5-nt RNA (37), and binding to 9- or 10-nt RNAs as monitored by EMSA (37, 66). LARP4 does not bind A(10) or the other 10-mers (Fig. 1H), even at concentrations well above that required to bind A(20), U(20), and the 36-mer RNAs shown in Fig. 1F and G, designated R1 and R2 in Fig. 1H. Binding to La-NTD shows that U(10) and A(10) are active as a ligand (Fig. 1I and data not shown).
To further examine the RNA binding properties of the LaM-RRM of LARP4, we employed ITC. By measuring the heat generated or absorbed during binding, ITC provides the affinity, stoichiometry, and enthalpy change (ΔH°) of the interaction. Figure 1J to L shows the interactions of a LARP4(111-303) fragment with three RNAs. In the titration of LARP4(111-303) with A(15), binding occurs as one event centered on a molar ratio of 1 (Fig. 1J). Further analysis revealed that LARP4(111-303) interacts with A(15) with a Kd of 714 nM, with entropically driven binding (Table 1; see Fig. S1 and S2 in the supplemental material). In contrast, LARP4(111-303) displayed very weak association with U(15) and A(10) (Fig. 1K and L), with a Kd beyond the threshold that could be rigorously measured by ITC, i.e., ≥0.1 mM. Therefore, LARP4 binds to A(15) at least 200-fold more tightly than U(15) and A(10).
TABLE 1.
Thermodynamic parameters of the interactions of La(1-194) and LARP4(111-303) with single-stranded RNA sequencesa
| Protein | n | Kd (1/Kb) (nM) | ΔH° (kcal mol−1) | −TΔS° (kcal mol−1) | ΔG°298K (kcal mol−1) |
|---|---|---|---|---|---|
| La(1-194)/U(15) | 0.8 | 752 | −39.0 ± 0.2 | 30.7 ± 0.2 | −8.3 ± 0.4 |
| La(1-194)/A(10) | |||||
| La(1-194)/A(15) | |||||
| LARP4(111-303)/U(15) | |||||
| LARP4(111-303)/A(10) | |||||
| LARP4(111-303) A(15) | 1.0 | 714 | 0.7 ± 0.1 | −9.1 ± 0.3 | −8.4 ± 0.4 |
The values represent the averages and the standard deviations over three independent measurements.
LARP4 cosediments with 40S ribosome subunits and polyribosomes.
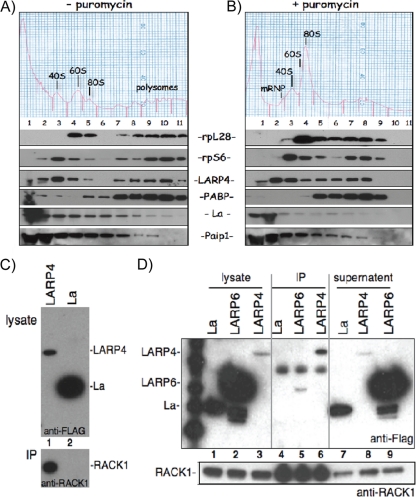
We examined LARP4 distribution in polysome profiles prepared in parallel in the presence and absence of puromycin (Fig. 2A and B). Control proteins rpS6 and rpL28 served as markers of 40S and 60S subunits, respectively, that also cosedimented with polyribosomes (Fig. 2A). La was another marker, most abundant in fractions (fxns) 1 and 2, as expected (12, 16). Endogenous LARP4 was in two peaks, 40S (fxn 3) and polyribosomes (fxn 10), similar to rpS6 (Fig. 2A). In comparison, PABP was most abundant on polysomes and Paip1 was most abundant in fxn 1, decreasing thereafter (Fig. 2A). Puromycin shifted a substantial amount of LARP4 to fxn 2 (Fig. 2B), in a pre-40S fraction that typically contains mRNPs, shifted by one fraction from the bulk of the 40S subunits, as represented by the chromatograph and the rpS6 panel, although a significant amount also remained with the 40S peak in fxn 3 (Fig. 2B).
FIG. 2.
Human LARP4 cosediments with 40S subunits and polyribosomes and associates with RACK1. (A and B) Polysome profiles in the presence (A) and absence (B) of puromycin. Numbered fractions were collected, fractionated by SDS-PAGE, and immunoblotted onto a membrane that was probed, exposed, stripped, and reprobed to detect the proteins indicated between the panels. (C) Co-IP of RACK1 with FLAG-LARP4 (F-LARP4) but not the control protein, F-La, using anti-FLAG Ab. Cells were transfected with F-LARP4 or F-La and processed for IP. The upper gel shows input extracts (lysate) prior to IP using anti-FLAG for immunoblot detection. The lower gel shows the IP material using anti-RACK1 for immunoblot detection. (D) Co-IP of F-LARP4, but not the control protein F-LARP6 or F-La, with RACK1 using anti-RACK1 Ab for the IP. Cells were transfected with F-LARP4, F-LARP6, or F-La and processed for IP. The upper gel shows input extracts (lysate; lanes 1 to 3), the IPed material (lanes 4 to 6), and the supernatant (lanes 7 to 9), using anti-FLAG for immunoblot detection. The lower gel shows a region of the same blot in the upper gel after stripping and reprobing using anti-RACK1 for immunoblot detection.
LARP4 interacts with RACK1 in yeast 2-hybrid assays and in HeLa cells. Sedimentation with 40S suggested that LARP4 may associate with other translation components. This was supported by yeast 2-hybrid screening of a human liver cDNA library for LARP4 association. The library was screened for 5-fold coverage for a total of 81 million interactions. Table 2 lists four independent clones that interacted with LARP4 and that encode RACK1, two of which were isolated multiple times (not shown). The interaction quality was the highest attainable (Table 2, A in the PBS column). The 4 clones collectively encode only the C-terminal portion of RACK1, with the minimal interaction region comprising amino acids 200 to 317, indicating that it is sufficient for LARP4 interaction.
TABLE 2.
Yeast 2-hybrid results from a human cDNA library using LARP4 as baita
| Clone name | Type sequence | Gene name | Start/stop sites | Frame | % identity to: |
PBS | |
|---|---|---|---|---|---|---|---|
| 5p | 3p | ||||||
| pB27_A-123 | 5p/3p | GNB2L1 | 414/988 | IF | 100.0 | 99.5 | A |
| pB27_A-67 | 5p/3p | GNB2L1 | 468/998 | IF | 99.1 | 99.6 | A |
| pB27_A-127 | 5p/3p | GNB2L1 | 510/1036 | IF | 98.4 | 56.8 | A |
| pB27_A-17 | 5p/3p | GNB2L1 | 597/998 | IF | 99.7 | 99.2 | A |
GNB2L1, guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 (50); IF, in frame.
Co-IP from HeLa cells validated the LARP4-RACK1 interaction. The upper gel of Fig. 2C shows the input, and the lower gel shows the IP. RACK1 was co-IPed with F-LARP4, but not F-La, even though the latter was expressed at a higher level (Fig. 2C). Reciprocal IP using anti-RACK1 Ab is shown in Fig. 2D. F-LARP4 was enriched in the anti-RACK1 IP relative to the input lysate and supernatant, whereas F-La and F-LARP6 were not, even though they were expressed at much higher levels than LARP4 (Fig. 2D). Since RACK1 is a 40S-associated protein, its ability to interact with LARP4 may explain, at least in part, the apparent association of LARP4 with 40S subunits described above.
LARP4 knockdown decreases cellular protein synthesis.
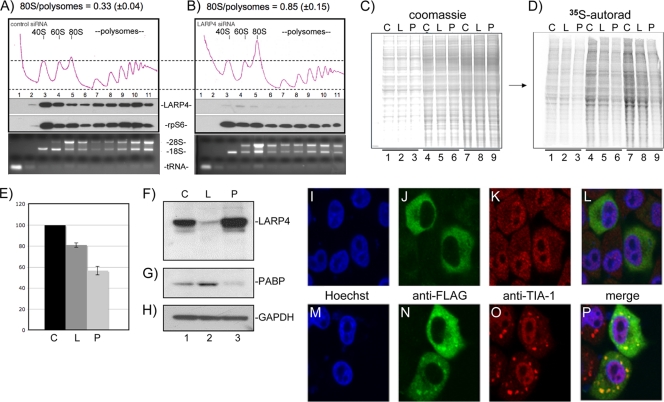
Changes in the relative areas of polysome versus 80S peaks in polysome profiles can reflect alterations in translation (24). We examined polysome profiles after treatment of HeLa cells with control siRNA and siRNA against LARP4 (Fig. 3A and B). LARP4 knockdown led to a decrease in the polysome area with a concomitant increase in the 80S peak (Fig. 3A and B). This change reflects conversion of polyribosomes to monoribosomes upon LARP4 knockdown, suggesting decreased translation initiation (24, 28) with a 10 to 15% decrease in translation.
FIG. 3.
LARP4 knockdown decreases cellular protein synthesis. (A and B) Polysome profiles of extracts made from cells treated with control siRNA (A) or siRNA directed to LARP4 (B). Quantitative areas under the 80S and polysome OD254 tracings are shown as numerical fractions above the panels. Immunoblots of the collected fractions for LARP4 and rpS6 are shown below the OD254 tracings. Ethidium-stained RNA gels showing 28S and 18S rRNAs and tRNA are shown below. (C) Coomassie blue-stained gel containing extracts from cells treated with control siRNA or siRNA directed to LARP4 or PABP (lanes C, L, and P, respectively) and pulsed for 30 min with [35S]methionine. Fifty, 100, and 150 μg of each extract was loaded in lanes 1 to 3, 4 to 6, and 7 to 9, respectively. (D) The gel in panel C was dried and exposed for autoradiography on a Fuji phosphorimager. (E) Quantitation of the total 35S was performed and expressed as [35S]Met incorporation per unit of protein on the y axis for each siRNA-treated cell extract on the x axis; the error bars reflect triplicate data. (F to H) Immunoblot showing relative levels of LARP4, PABP, and GAPDH from the three extracts used for panels C and D; a single membrane was incubated sequentially with the three Abs for panels H to J. LARP localizes to stress granules after exposure to arsenite. (I to P) After transfection with F-LARP4, cells were mock treated (I to L) or treated with arsenite (M to P), which is widely used to induce stress granules, and examined with anti-FLAG (green) and anti-TIA-1 (red) antibodies. LARP4 distribution was homogeneously cytoplasmic in the mock-treated cells (J), whereas a significant fraction became localized in punctate foci after arsenite treatment (N). The stress granule marker TIA-1 also organized into distinct foci after arsenite treatment (K versus O). Image merging revealed superimposed LARP4 foci and TIA-1 foci (P). Endogenous LARP4 also localized to stress granules (see Fig. S7Q to X in the supplemental material).
We also examined incorporation of [35S]methionine into newly synthesized protein. After treatment with control siRNA or siRNA against LARP4 or PABP, cells were pulsed for 30 min with aliquots of a medium containing [35S]Met, extracts were prepared and fractionated by SDS-PAGE, stained with Coomassie blue, and photographed (Fig. 3C). The gel was dried and autoradiographed (Fig. 3D), and 35S was quantified (Fig. 3E). Cells with LARP4 siRNA showed a 15 to 20% reduction in [35S]Met incorporation, and PABP siRNA showed an ∼50% reduction relative to control siRNA (Fig. 3E). Reduction of LARP4 and PABP in these cells relative to the control protein GAPDH was demonstrated by sequential probing of the same immunoblot, which revealed that LARP4 levels appeared to increase upon PABP knockdown (Fig. 3F to H), possibly reflecting some kind of system of homeostatic regulation.
Conditions that inhibit translation lead to accumulation of mRNAs and associated initiation factors, such as PARP, 40S subunits, and RACK1, as well as TIA-1 and certain other RNA binding proteins in stress granules, in which they are transiently inactive but can be reactivated for translation (13, 36). Since LARP4 was found associated with 40S subunits, RACK1, and PABP and was shifted in polysome profiles to a pre-40S mRNP fraction with puromycin, a translation inhibitor known to induce stress granules (36), we examined stress granules for F-LARP4 and detected colocalization with TIA-1, a key component of stress granules (Fig 3I to P). Endogenous LARP4 also colocalized to stress granules with FMRP (57) after treatment with arsenite (see Fig. S7Q to X in the supplemental material), consistent with the idea that LARP4 is associated with mRNPs engaged for translation initiation that assemble into stress granules after exposure to arsenite.
LARP4 interacts with PABP in part through a variant PAM2 sequence.
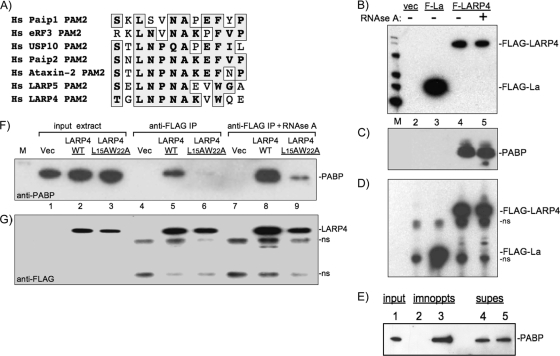
Studies of PAM2 sequences have led to a consensus, SXLNXNAXXF, in which interactions via L and F at positions 3 and 10 mediate much binding to PABC (38). Of all PAM2 sequences catalogued to date, the F at consensus position 10 is the only invariant residue (1, 39, 40). We found sequence near the N terminus of LARP4 that is homologous to PAM2 (Fig. 4A). LARP4 contains a conserved W (LARP4 position 22) in place of F10 and was not previously identified as a PAM2 candidate (1). Potential PAM2 motifs in LARP4 and -5/4b were first noted in a review (8). Here, we show the homologous sequences of LARP4 and -5/4b (Fig. 4A). The LARP4 PAM2 sequence is highly conserved (see Fig. S3 in the supplemental material).
FIG. 4.
LARP4 contains a putative variant PAM2 (PAM2w) motif and interacts with PABP. (A) Sequence alignment of the PAM2 motifs of six Homo sapiens (Hs) proteins, eRF3 (elongation release factor 3), Paip1, Paip2, ataxin 2, and USP10, with the homologous sequences from LARP5, and LARP4. (B) Immunoblot of input extracts from cells transfected with vector only (vec), F-La, and F-LARP4 that were mock treated (−) or treated with RNase A (+) as visualized with anti-FLAG Ab. (C) The extracts shown in panel B were subjected to IP, and the products were examined by immunoblotting using anti-PABP Ab. (D) The blot in panel C was stripped and then probed using anti-FLAG Ab. ns, nonspecific. (E) Immunoblot, using anti-PABP Ab, of input extract (lane 1) and after IP with control IgG (lane 2) and anti-LARP4 Ab raised against a C-terminal peptide of LARP4 (lane 3). PABP remaining in the supernatants is shown in lanes 4 and 5. (F) Immunoblot of input extracts from cells after transfection with empty vector, F-LARP4, and F-LARP4-L15A-W22A (lanes 1 to 3) and after IP (lanes 4 to 6 and 7 to 9) visualized using anti-PABP Ab. (G) The membrane was stripped and reprobed using anti-Flag Ab.
IP using anti-FLAG Ab was done on extracts of cells expressing F-La or F-LARP4 (Fig. 4B). PABP was co-IPed with F-LARP4, but not with F-La, and the interaction was resistant to RNase A (Fig. 4C). In contrast, co-IP of the translation-associated factor eIF4G was sensitive to RNase A, and rpS6 was not in the IP (not shown). Affinity-purified Ab against native LARP4 also co-IPed PABP (Fig. 4E). By quantitative immunodepletion, ∼10% of the total PABP remained associated with LARP4 after IP (not shown). Replacement of LARP4 PAM2w residues L15 and W22 with alanines decreased the amount of PABP that co-IPed with F-LARP4 (Fig. 4F and G).
IP of F-LARP4 followed by microarray analysis identified ∼2,000 LARP4-associated mRNAs, many times more than parallel IPs of other RNA binding proteins, although with no apparent enrichment of any gene ontology (GO) group (not shown). However, despite various strategies and due to technical limitations, we have been unable to decipher which of these are bound directly to LARP4 independently of their binding to PABP. Nonetheless, many mRNAs, including GAPDH mRNA, which we decided to use as a control for some experiments, were not found associated in the anti-LARP4 IP (not shown). The poly(A) specificity of LARP4 and interaction with PABP are consistent with a general association with a large number of mRNAs.
A peptide of LARP4 PAM2w residues 13 to 26 binds the PABP MLLE domain.
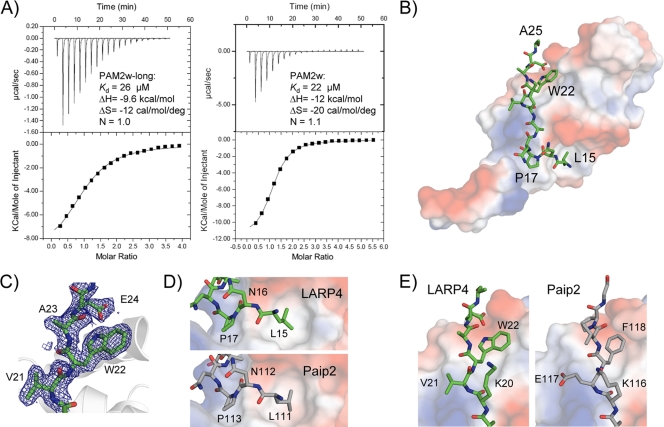
The conserved motif near the N terminus of LARP4 most closely resembles the PAM2 motif found in Paip2 and ataxin 2 (Fig. 4A). We carried out ITC experiments with a fragment of the N terminus of LARP4 (residues 7 to 49) to determine if the putative PAM2 sequence would bind in solution to the MLLE domain of PABPC1 (Fig. 5A). The analysis clearly revealed binding, with a Kd of 26 μM. We then prepared a smaller peptide that contained only the variant PAM2 site (residues 13 to 26). ITC analysis showed even better binding, with a Kd of 22 μM (Fig. 5A). The affinity of the LARP4 PAM2w motif is intermediate between the highest and lowest PAM2 affinities measured for MLLE, which range from 0.12 to 28 μM (39-42). We also carried out NMR analysis (38) to confirm the specificity of the interaction and to map the binding site on the MLLE domain (see Fig. S4 in the supplemental material). Titration of the 15N-MLLE domain with LARP4 PAM2w peptide produced large chemical shift changes that mapped to the canonical PAM2 binding site on the MLLE domain (see Fig. S4 in the supplemental material).
FIG. 5.
Binding of the PAM2w peptide of LARP4 to the MLLE domain of PABP. (A) Isothermal titration calorimetry. Shown are baseline corrected thermograms (top) and integrated areas (bottom) of the heat released for binding of PAM2w-long (left) and PAM2w (right). The dissociation constant (Kd), enthalpy (ΔH), entropy (ΔS), and stoichiometry (N) are indicated. (B) X-ray crystal structure of LARP4 PAM2w bound to the MLLE domain of PABPC1. The PAM2w peptide (green) wraps around the surface of the MLLE domain, colored according to the electrostatic potential (negative in red, positive in blue) (PDB ID, 3PKN). (C) Electron density omit map (2Fo − Fc [where Fo is observed structure factor and Fc is calculated structure factor]; 1-standard deviation contour) of the bound PAM2w peptide showing the tryptophan residue. (D) Side-by-side comparison of PAM2w (green) from LARP4 and PAM2 peptide (gray) (PDB entry 3KUS) from Paip2 bound to MLLE. The peptide N termini show strikingly similar confirmations and interactions with the MLLE domain. (E) Comparison of C termini. In LARP4, tryptophan (W22) replaces the phenylalanine found in other PAM2 peptides (38-41). The larger indole ring is accommodated by a displacement of Gln560 of MLLE (not shown) to generate a shallow hydrophobic pocket.
Structure of the LARP4 PAM2w-MLLE complex.
To obtain structural insight into the LARP4 PAM2w-MLLE interaction, we cocrystallized MLLE (PABP residues 544 to 626) with a peptide representing LARP4 PAM2w (residues 13 to 26) (Table 3 ). The 1.8-Å MLLE/LARP4 diffraction data set was phased by molecular replacement using the crystal structure of MLLE/Paip2. The LARP4 PAM2w peptide wraps around the MLLE domain, similar to the Paip2 and ataxin 2 complexes (Fig. 5B).
TABLE 3.
LARP4 PAM2w-MLLE data collection and refinement statistics
| Parameter | Value in MLLE-LARP4a |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 49.04, 31.14, 52.52 |
| α, β, γ (°) | 90.00, 105.04, 90.00 |
| Resolution (Å) | 50-1.80 (1.83-1.80) |
| Rsym | 0.073 (0.145) |
| I/σI | 19.0 (6.5) |
| Completeness (%) | 98.8 (89.0) |
| Redundancy | 3.8 (2.7) |
| Refinement | |
| Resolution (Å) | 50.8-1.80 |
| No. reflections | 6,871 |
| Rwork/Rfree | 0.226/0.289 |
| No. atoms | |
| MLLE | 603 |
| Peptide | 92 |
| Sulfate | 5 |
| Iodide | 1 |
| Waters | 42 |
| β-Factors | |
| MLLE | 20.7 |
| Peptide | 27.3 |
| Sulfate | 32.5 |
| Iodide | 33.6 |
| Waters | 38.6 |
| RMSb deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.11 |
| Ramachandran statistics (%) | |
| Most favored regions | 98.7 |
| Additional allowed regions | 1.3 |
Highest resolution shell is shown in parentheses.
RMS, root mean square.
Comparison of the LARP4/MLLE and Paip2/MLLE structures revealed a high degree of similarity (Fig. 5B to E). Starting from the N terminus of the LARP4 peptide, Leu15 inserts into the hydrophobic pocket formed by MLLE domain side chains (Fig. 5D). Additional hydrophobic contacts involve Pro17 and Ala19 of LARP4 and the MLLE domain, as observed for the MLLE/Paip2 complex (40). In previous mutagenesis studies, the invariant Phe in PAM2 (Phe118 in Paip2) was shown to be a major binding determinant in the MLLE-PAM2 interaction (38, 39, 41). Since Phe is invariant in all other PAM2 motifs in this position, a major question was how replacement by Trp22 of LARP4 PAM2w would alter the interaction. The cocrystal revealed that the MLLE domain accommodates the larger side chain of Trp22 in almost the same way as Phe118 of Paip2 (Fig. 5D).
Two regions of LARP4 mediate association with PABP and polysomes.
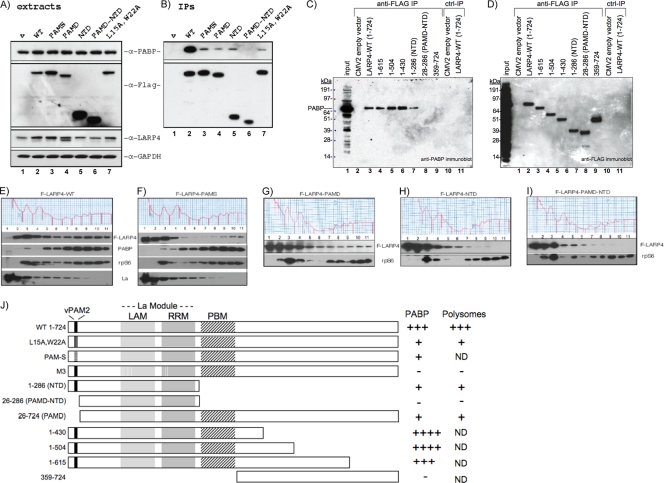
We wanted to know if a region of LARP4 other than PAM2w was required for efficient association with PABP, as is the case for other PAM2-containing proteins. Several F-LARP4 constructs, schematized in Fig. 6 J, were analyzed for co-IP of PABP in the presence of RNase A (Fig. 6A to D) and polysome association (Fig. 6E to I). First, an initial set of proteins was analyzed by probing extracts for PABP, Flag epitope, endogenous LARP4, and GAPDH (Fig. 6A). Deletion of the C-terminal region led to higher levels of F-LARP4-NTD and F-LARP4-PAMD-NTD than the others (Fig. 6A, α-Flag). The other constructs were not grossly overexpressed, as shown in Fig. 6A, lane 4, α-LARP4, in which endogenous LARP4 and F-LARP4-PAMD are distinguished by size and detected by anti-LARP4 Ab, which recognizes the LARP4 C-terminal peptide.
FIG. 6.
Two LARP4 regions are required for association with PABP and polyribosomes. (A) Immunoblot of extracts after transient transfection with a subset of Flag-tagged constructs depicted in panel J. The membrane was sequentially probed with the Abs indicated on the right. (B) Immunoblot of IPed material probed as for panel A. (C) Further mapping of the PABP interaction region of LARP4 C terminal to the RRM; probed with anti-PABP. (D) The blot in panel C was probed with anti-FLAG Ab. (E to I) Five polysome profiles run in parallel after transfection with the indicated constructs. Fractions were analyzed by immunoblotting as indicated. (J) Schematic showing the F-LARP4 constructs used here and in Fig. 7 (N-terminl Flag is not shown) with summarized results on the right (+, positive correlation; −, negative correlation; ND, not determined). The PAM2w, LaM, and RRM motifs are shown. PBM refers to a putative PABP binding motif mapped here. PAM-S contains alanines at LARP4 residues 15 to 22.
These mutated F-LARP4s co-IPed less PABP than wild-type (WT) F-LARP4 (Fig. 6B, top). Only when both PAM2 and the whole C-terminal region downstream of the RRM were deleted, as in PAMD-NTD, was association with PABP completely lost (Fig. 6B).
For more fine mapping, we examined additional constructs. LARP4(1-615), LARP4(1-504), and LARP4(1-430) co-IPed PABP as well as full-length LARP4(1-724) (Fig. 6C, lanes 3 to 6), while LARP4(1-286) again showed less efficient association (lane 7). LARP4(26-286), also known as PAMD-NTD, which lacks the PAM2w and entire C-terminal region, and LARP4(359-724) showed no association with PABP (Fig. 6C, lanes 8 and 9). Figure 6D shows a control with the F-LARP4 proteins pulled down in the IPs. The data indicate that the sequence downstream of the LARP4 RRM required for efficient PABP association resides within or includes LARP4 amino acids 287 to 358. Constructs intermediate between LARP4 1 to 286 and 1 to 430, as well as 287 to 724, designed to map the region further, failed to accumulate in cells and could not be tested (not shown).
Figure 6E to I shows distribution of some of the F-LARP4 constructs in polysome profiles run in parallel. The F-LARP4-WT pattern (Fig. 6E) was similar to that of endogenous LARP4, with a peak at 40S (fxn 3), progressively less in fxns 2 and 1, and a second peak in fxns 8 to 10. F-LARP4-PAMS, in which residues 15 to 22 were replaced by alanines, was more abundant in fxns 1 and 2, with less remaining on polysomes relative to F-LARP4 (Fig. 6F). F-LARP4-PAMD was similar to PAMS, with little remaining on polysome fxns 8 to 11 (Fig. 6G). F-LARP4-NTD and F-LARP4-PAMD-NTD had the most severely diminished polysome association in fxns 8 to 11 (Fig. 6H and I). These results are similar to those for PABP co-IP, namely, that PAM2w, as well as sequences C-terminal to the RRM, are required for optimum association with polysomes. The other constructs used as described above for fine mapping were not examined by polysome profiling. These results, along with the LARP4-M3 mutant in Fig. 7, are summarized in schematic form in Fig. 6J.
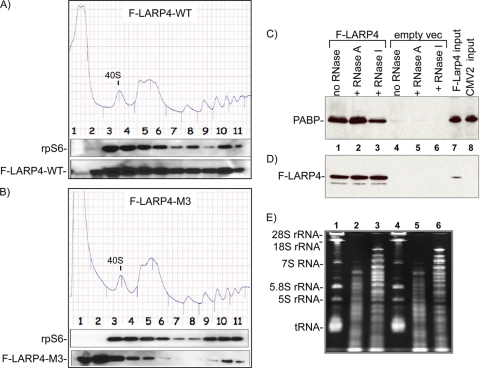
FIG. 7.
The LARP4 RNA binding domain contributes to PABP and polysome association. (A and B) Two polysome profiles run in parallel after transfection with the indicated constructs, F-LARP4-WT and F-LARP4-M3, which contain mutations in the RNA binding domain (see the text); the 40S peaks are indicated. The fractions were analyzed by immunoblotting, as shown under the profiles. (C and D) PABP-LARP4 interaction is resistant to RNase A, which does not cleave poly(A) but is partially sensitive to RNase I. Shown are immunoblots of IPed material after mock treatment (no RNase) or incubation with RNase A or RNase I, as indicated, probed first for PABP (C) and then for F-LARP4 (D). (E) RNAs remaining in supernatants of samples 1 to 6 in panels C and D after IP.
Mutations in LARP4 RNA binding motifs disrupt polysome association.
We examined F-LARP4-M3, which carries point substitutions in key residues of the LaM and RRM designed to impair RNA binding. The identities of the amino acids that replace the wild-type residues were selected to be compatible with the β-strand and α-helix structure of the LARP4 LaM-RRM as predicted by the Protein Structure Prediction Meta Server (35). F-LARP4-WT and F-LARP4-M3 were compared in parallel (Fig. 7A and B). In these gradients, the 40S peaks were split into fxns 3 and 4. F-LARP4-WT was distributed in two peaks, one in fxns 3 and 4 and the other in fxn 10, similar to rpS6 (Fig. 7A). In contrast, F-LARP4-M3 showed a significant decrease in polysome association and a shift to a peak in fxn 2 (Fig. 7A and B), suggesting that LARP4 uses its RNA binding motifs to maintain association with 40S components and polysomes. Consistent with this, F-LARP4-M3 was reproducibly found not to be associated with PABP by co-IP (not shown), further suggesting that the LARP4-PABP interaction is stabilized by RNA binding by LARP4.
The poly(A) binding specificity of LARP4 suggests that it may associate with PABP on the poly(A) tracts of mRNAs. Our IP experiments that used RNase A might not reveal this, since RNase A is pyrimidine specific and has little or no activity for poly(A) (61). To examine this further, we compared the sensitivity of the LARP4-PABP association by co-IP in the presence and absence of the endonucleases RNases A and I, the latter of which exhibits no sequence preference. If bridged by poly(A), the PABP-LARP4 interaction should be relatively insensitive to RNase A but sensitive to RNase I. Wild-type F-LARP4 or empty vector was transfected to HeLa cells, and an extract was made from each. Aliquots were mock treated (no RNase) or treated with RNase A or RNase I during the incubation with anti-Flag Ab. After being washed, the IPed PABP and F-LARP4 proteins were analyzed by immunoblotting (Fig. 7C and D). The amounts of PABP associated with LARP4 in the mock- and RNase A-treated extracts were more than in the RNase I-treated extract (Fig. 7C). This difference was not due to loading or to the amount of extract used, as can be seen by the F-LARP4 in the IP on the same blot (Fig. 7D). The effectiveness of the RNase treatments was evident from the RNA remaining in the extract after IP (Fig. 7E). The cellular RNAs indicated on the left were intact in the mock-treated but degraded in the RNase-treated samples (Fig. 7E). This differential sensitivity to RNases A and I is consistent with the idea that LARP4 interacts with PABP in the presence of poly(A) RNA and that this contributes to the stability of their association.
LARP4 can prolong mRNA half-life.
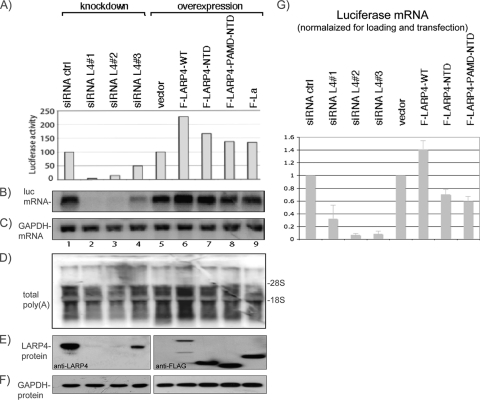
Experiments with a transfected luciferase reporter that contains the SV40 poly(A) addition signal used previously to examine translation (21, 58) suggested that LARP4 stimulated luciferase expression by promoting accumulation of luciferase mRNA (Fig. 8A to C). The results obtained were very reproducible (not shown) but revealed that the extent of the effects of LARP4 knockdown on luciferase activity (Fig. 8A, lanes 1 to 4) were more striking than on endogenous protein synthesis, as monitored by polysome profiles and [35S]Met incorporation (Fig. 3A to E). Although we do not know the basis for this discrepancy, many cellular factors that affect luciferase activity have been noted (59), and it is therefore possible that LARP4 affects a factor(s) that is luciferase specific. The advantage of the luciferase system, however, is that it represents a method by which levels of a specific mRNA can be related to translation, since changes in luciferase mRNA, protein synthesis, and enzyme activity are tightly linked (11). Therefore, we could test if the effects of LARP4 on luciferase are mediated, at least in part, at the level of mRNA accumulation. Our approach was to cover a wide range of LARP4 levels by transfecting cells with any of three siRNAs against LARP4 or with F-LARP4 expression constructs and the appropriate controls.
FIG. 8.
The effects of LARP4 on transfected luciferase reporter activity reflect luciferase mRNA levels. (A to F) Cells transfected with siRNA (lanes 1 to 4) or F-LARP constructs (lanes 5 to 9) and corresponding controls were secondarily transfected with luciferase reporter plasmid, after which extracts were prepared for luciferase activity (A), Northern blotting (B to D), and immunoblotting (E to F). The blots in panels B to D reflect a single membrane sequentially probed for the RNAs indicated on the left. The immunoblots in panel E were processed using anti-LARP (left) or anti-FLAG (right). (G) The experiment represented in panels A to F was repeated in triplicate using a VA1 plasmid together with the luciferase reporter plasmid to normalize for transfection efficiency. A Northern blot was generated from each of the three experiments and probed for luciferase mRNA, GADPH mRNA for normalization for loading, and VA1 RNA for normalization for transfection. The graph shows luciferase mRNA levels after normalization for loading and transfection. The error bars reflect triplicate data.
Luciferase activity and mRNA levels, using GAPDH mRNA on the same blot for normalization (Fig. 8A to C), revealed an apparent good correlation. Three siRNAs each led to decreased luciferase mRNA levels relative to the control siRNA (Fig. 8, lanes 1 to 4). F-LARP4 reproducibly led to the most luciferase mRNA and activity, whereas the NTD and PAMD-NTD mutants and F-La exhibited less (Fig. 8, lanes 5 to 9), even though the last accumulated to higher levels than F-LARP4 (Fig. 8E, lanes 5 to 9).
Figure 8G, shows the results of the above-mentioned experiment repeated in triplicate after equal aliquots of a mixture of luciferase plasmid and VA1 plasmid were used for transfection, the latter of which is transcribed by RNA polymerase III into a small, stable 160-nt VA1 RNA that served as a transfection control. Luciferase mRNA was normalized for loading using endogenous GAPDH mRNA and for transfection using VA1 RNA. The data suggest that LARP4 promotes luciferase mRNA accumulation and, moreover, that F-LARP4 may promote luciferase mRNA stability.
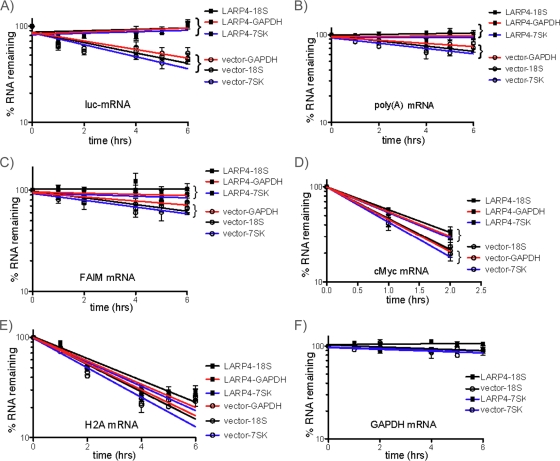
Given the proposed involvement of several PAM2-containing proteins in controlling mRNA turnover via PABP, we examined LARP4 for effects on mRNA stability (25, 39, 41, 55, 68). As noted in the introduction, competition between the PAM2-containing proteins eRF3, PAN2/3, and Tob-Ccr4-Caf1 is thought to impact mRNA homeostasis (25, 41, 55). For this, we chose to express LARP4 ectopically, because this produces robust luciferase mRNA levels that can be followed for decay whereas LARP4 knockdown leads to low luciferase mRNA levels that are technically difficult to follow. We initially examined luciferase mRNA. Actinomycin D was used to inhibit mRNA synthesis, and mRNA decay was followed over time, using the noncoding RNAs, 7SK, and 18S rRNA as controls. GAPDH was chosen as an additional control because it was not among the ∼2,000 mRNAs found associated with F-LARP4 (not shown) and it has a longer half-life than luciferase mRNA. High-quality Northern blots from three independent experiments were generated and sequentially probed (one is shown in Fig. S5 in the supplemental material). Quantitation of each RNA, corrected for loading and using GraphPad Prism to analyze the data (72), is shown in Fig. 9A to F. Each graph contains the results from three Northern blots for one of the mRNAs examined; each line on the graph represents the specific mRNA corrected for by one of the three loading controls on the same blot, 7SK, 18S, and GAPDH mRNAs (the individual decay curves are shown in Fig. S6 in the supplemental material). The error bars reflect triplicate data for the examined RNA corrected for by the loading control RNA. This showed that expression of F-LARP4 stabilized luciferase mRNA compared to the vector control, in which luciferase mRNA decayed with a half-life of ∼4 h (Fig. 9A, vector), in agreement with previous reports (4, 23). We conclude that enhanced expression of LARP4 prolonged luciferase mRNA half-life.
FIG. 9.
LARP4 can promote mRNA stability. Cells were transfected with a mixture of F-LARP4 and luciferase plasmid or empty vector and luciferase plasmid. Forty hours later, the cells were treated with actinomycin D, and RNA was isolated at time zero and intervals thereafter as indicated. Three independent experiments were carried out, and a Northern blot was made from each (one of the blots sequentially probed is shown in Fig. S5 in the supplemental material). The blots were sequentially probed for the RNAs indicated above the x axes. The blots were also probed for 18S rRNA, GAPDH mRNA, and 7SK snRNA, which were used to normalize for loading. Each graph shows the decay curves for LARP4-transfected and empty-vector-transfected cells; each line reflects the time course for the RNA as corrected for by either 18S rRNA, 7SK snRNA, or GAPDH mRNA as indicated; the error bars reflect data from the three data sets.
F-LARP4 prolonged the half-life of total poly(A) RNA, as monitored by oligo(dT) probing, relative to the vector control (Fig. 9B), albeit less so than for luciferase mRNA. F-LARP4 also prolonged the FAIM mRNA half-life (Fig. 9C). c-myc mRNA, which was undetectable after 2 h, was also prolonged by F-LARP4 relative to the vector control (Fig. 9D). After 2 h, c-myc mRNA exhibited distinct decay curves in the LARP4 and control cells. In contrast, the nonpolyadenylated histone H2A mRNA was not significantly different in F-LARP and vector cells at 2 h or at any time (Fig. 9E). GAPDH mRNA was not significantly different in the F-LARP4 and control cells and was highly stable in both (Fig. 9F). These results indicate that overexpression of LARP4 can stabilize some mRNAs.
DISCUSSION
We characterized a new protein, LARP4, that appears to be involved in mRNA metabolism and translation. Multiple data indicate that LARP4 is intimately associated with translating polyadenylated mRNAs. The conserved RNA binding LaM-RRM of LARP4 exhibits a binding preference for poly(A), as well as other unexpected properties of a LaM-RRM-containing protein (Fig. 1 and Table 1). The results expand our appreciation of the potential diversity of the LaM-RRM in RNA recognition.
The LARP4 LaM-RRM is flanked on both sides by PABP-interacting regions. The region N-terminal to the LaM-RRM contains a conserved PAM2w sequence that we showed is important for PABP interaction. PAM2 sequences mediate direct contacts with the MLLE domain of PABP (38-40). Direct binding as monitored by ITC, as well as NMR and crystallography, showed that the LARP4 PAM2w makes contacts with the MLLE domain of PABP.
The region C-terminal to the LaM-RRM that is required for efficient interaction with PABP was mapped to a sequence that resides within or includes LARP4 residues 287 to 358. The RNase I data suggest the possibility that the mechanism of interaction between PABP and this region of LARP4 may involve RNA bridging, although this remains to be determined. We also presented evidence that is consistent with a model in which LARP4 and PABP interact in the presence of RNA and that this contributes to the stability of their interaction. Our characterization represents the first detailed mechanism by which a LARP interacts with PABP.
LARP4 cosediments with 40S ribosome subunits and was found to interact with RACK1, a 40S- and mRNA-associated protein (17, 50). Knockdown of LARP4 led to a decrease in translation, as evidenced by conversion of polyribosomes to monosomes on sedimentation gradients and by a decrease in [35S]Met incorporation into newly synthesized protein, consistent with a 15 to 20% reduction in cellular protein synthesis. The cumulative data indicate that LARP4 is a functional component of a fraction of translating mRNPs.
LARP4 sediments as two peaks on polysome profiles, with 40S ribosome subunits and with polyribosomes. All tested LARP4 mutants that showed decreased association with PABP also showed decreased association with polyribosomes. Substitution of residues in the RNA binding domain of LARP4 predicted to be important for RNA binding also decreased association with polyribosomes.
The results reported here are to be distinguished from the report (57) on a related protein, LARP5/4b. Human LARP4 and -5/4b are distinct proteins encoded on different chromosomes that have maintained distinct characteristics through evolution, including sequence differences in their La motifs in key residues involved in base-specific RNA recognition in human La-RNA crystals, as well as conserved differences in their RRMs. LARP4 and -5/4b are most highly homologous in their LAMs and RRMs but are less homologous in some other regions (see Fig. S3 in the supplemental material). However, even in these most homologous regions, conserved sequence differences occur in key positions of the RRM that are sometimes involved in sequence-specific RNA recognition in other RRM-containing proteins (7), such as the loop connecting the β-2 and β-3 strands of the β-sheet RNA binding surface of the RRM (see Fig. S3 in the supplemental material).
LARP4 and -5/4b share functional relatedness, including interaction with PABP and RACK1 (57). That this was also demonstrated for LARP4 is significant, because although it was reported that the large C-terminal domain of LARP5/4b (∼400 amino acids) interacts with PABP and RACK1, this region shares limited homology with LARP4. The region 287 to 429 of LARP4 that we mapped as important for PABP interaction contains a stretch of about 70 amino acids with significant homology (64% identity and/or similarity) to LARP5/4b (see Fig. S3 in the supplemental material). Fine mapping of the LARP4 region of RACK1 interaction will require further experiments. Our data advanced information regarding the RACK1 interaction, as we found by yeast 2-hybrid analysis that the C-terminal half of RACK1 is sufficient for interaction with LARP4.
Conditions that inhibit translation lead to accumulation of mRNAs and associated initiation factors, such as PABP, 40S subunits, and RACK1, as well as TIA-1 and certain other mRNA binding proteins, in stress granules in which they are transiently inactive but can be reactivated for translation (13, 36). Since LARP4 was found associated with 40S subunits, RACK1 and PABP, and was shifted in polysome profiles to a pre-40S mRNP fraction with puromycin, a translation inhibitor known to induce stress granules (36), we examined stress granules for F-LARP4 and detected colocalization with TIA-1, a key component of stress granules (see Fig. S7 in the supplemental material). Our Ab that detects endogenous LARP4 also demonstrated stress granule localization using anti-FMRP (see Fig. S7 in the supplemental material) (57), consistent with the idea that LARP4 is associated with mRNPs engaged for translation initiation that assemble into stress granules after exposure to arsenite.
While LARP5/4b was shown to bind PABP, its PAM2 motif was not noted or characterized in the previous report (57). Our analysis identified the PAM2w and showed that it indeed interacts directly with the PABP MLLE domain with a Kd within the range found for other PAM2 motifs. This is significant, because it suggests that LARP4 (and probably LARP5/4b) function as parts of a network of proteins that compete for the PABP MLLE domain. Finally, while Schäffler et al. (57) reported that LARP5/4b stimulated protein synthesis, as monitored by luciferase activity, they showed that it did not affect luciferase mRNA levels, whereas our analysis, which included mRNA decay, indicated a role for LARP4 in mRNA stability.
The variant PAM2 sequences found in LARP4 and -5/4b most significantly differ from all other known PAM2 sequences in that they contain a Trp in place of Phe, the latter of which is a most important contributor to MLLE binding (39). While LARP4 PAM2w contains leucine at consensus position 3, which is also very important in MLLE binding (38, 39), we do not yet know if the presence of Trp in place of Phe would increase or decrease binding to MLLE. Occupation of this key position by Trp has been conserved by all LARP4 and -5/4b sequences in the database (not shown), which is especially intriguing in light of hierarchal competition by PAM2 proteins for the PABP MLLE (39).
We note that overexpression of LARP4 may not reflect a physiological role in mRNA stability and that actinomycin D can have untoward effects after 2 h. We therefore examined this after LARP4 knockdown, which appeared to have destabilizing effects on FAIM mRNA (see Fig. S8 in the supplemental material). However, this experiment did not include the luciferase reporter, and other probings were complicated by technical irregularities. Thus, we emphasize that our conclusions regarding mRNA stability are mostly limited to conditions of increased LARP4 expression and that more detailed and mechanistic experiments focusing on this aspect will have to await future endeavors. Nonetheless, given our data showing that modest overexpression of LARP4 can stabilize mRNAs (Fig. 9), it is tempting to speculate that LARP4 may do so by competing with the PAN2/3 and/or the Ccr4-Not-Caf1-Tob2 deadenylation complexes (39). These complexes comprise a biphasic poly(A)-shortening activity of mRNAs in context that appears to be relevant to their antiproliferative properties (reviewed in reference 48).
Supplementary Material
Acknowledgments
We thank M. Fabian, Y. Svitkin, and N. Sonenberg for discussion and for providing the luciferase reporter construct and anti-Paip1 and U. Fischer and K. Schaffler for anti-FMRP. We thank A. Hinnebusch, T. Dever, J. Wilusz, V. Pain, M. Gorospe, T. Dever, and T. Sundaresan for discussion; J. Iben for bioinformatics analysis; S. Tenenbaum and S. Chittur for RIP-chip analysis; G. M. Wilson for discussion and help with GraphPad Prism; and S. Curry for comments. L.M. and M.R.C. thank R. Tata and P. Brown for help with LARP4(111-303) cloning.
This work was supported by Intramural Research Program of the NICHD, NIH, grants to CHESS and Canadian Institutes of Health grant MOP-14219. L.M. is a fellow of the European Molecular Biology Organization (EMBO). M.R.C. acknowledges the Wellcome Trust for the Centre of Biomolecular Spectroscopy.
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albrecht, M., and T. Lengauer. 2004. Survey on the PABC recognition motif PAM2. Biochem. Biophys. Res. Commun. 316:129-138. [DOI] [PubMed] [Google Scholar]
- 2.Amrani, N., S. Ghosh, D. A. Mangus, and A. Jacobson. 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angenstein, F., et al. 2002. A receptor for activated C kinase is part of messenger ribonucleoprotein complexes associated with polyA-mRNAs in neurons. J. Neurosci. 22:8827-8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayala-Breton, C., et al. 2009. Analysis of the kinetics of transcription and replication of the rotavirus genome by RNA interference. J. Virol. 83:8819-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer, B. W., and R. D. Kornberg. 1980. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc. Natl. Acad. Sci. U. S. A. 77:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baroni, T. E., S. V. Chittur, A. D. George, and S. A. Tenenbaum. 2008. Advances in RIP-chip analysis: RNA-binding protein immunoprecipitation-microarray profiling. Methods Mol. Biol. 419:93-108. [DOI] [PubMed] [Google Scholar]
- 7.Bayfield, M. A., and R. J. Maraia. 2009. Precursor-product discrimination by La protein during tRNA metabolism. Nat. Struct. Mol. Biol. 16:430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayfield, M. A., R. Yang, and R. J. Maraia. 2010. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim. Biophys. Acta 1799:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blagden, S. P., et al. 2009. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 334:186-197. [DOI] [PubMed] [Google Scholar]
- 10.Bousquet-Antonelli, C., and J. M. Deragon. 2009. A comprehensive analysis of the La-motif protein superfamily. RNA 15:750-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasier, A. R., and D. Ron. 1992. Luciferase reporter gene assay in mammalian cells. Methods Enzymol. 216:386-397. [DOI] [PubMed] [Google Scholar]
- 12.Brenet, F., N. Socci, N. Sonenberg, and E. Holland. 2009. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene 28:128-139. [DOI] [PubMed] [Google Scholar]
- 13.Buchan, J. R., and R. Parker. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrows, C., et al. 2010. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 38:5542-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai, L., D. Fritz, L. Stefanovic, and B. Stefanovic. 2010. Binding of LARP6 to the conserved 5′ stem-loop regulates translation of mRNAs encoding type I collagen. J. Mol. Biol. 395:309-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardinali, B., C. Carissimi, P. Gravina, and P. Pierandrei-Amaldi. 2003. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J. Biol. Chem. 278:35145-35151. [DOI] [PubMed] [Google Scholar]
- 17.Coyle, S. M., W. V. Gilbert, and J. A. Doudna. 2009. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 29:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig, A. W., A. Haghighat, A. T. Yu, and N. Sonenberg. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392:520-523. [DOI] [PubMed] [Google Scholar]
- 19.Derry, M. C., A. Yanagiya, Y. Martineau, and N. Sonenberg. 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harbor Symp. Quant. Biol. 71:537-543. [DOI] [PubMed] [Google Scholar]
- 20.Dong, G., G. Chakshusmathi, S. L. Wolin, and K. M. Reinisch. 2004. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 23:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowling, R. J., M. Zakikhani, I. G. Fantus, M. Pollak, and N. Sonenberg. 2007. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 67:10804-10812. [DOI] [PubMed] [Google Scholar]
- 22.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 23.Fan, X., et al. 2003. Nitric oxide donors inhibit luciferase expression in a promoter-independent fashion. J. Biol. Chem. 278:10232-10238. [DOI] [PubMed] [Google Scholar]
- 24.Foiani, M., A. M. Cigan, C. J. Paddon, S. Harashima, and A. G. Hinnebusch. 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3203-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funakoshi, Y., et al. 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21:3135-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 27.Hands-Taylor, K. L., et al. 2010. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 38:4052-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartwell, L. H., and C. S. McLaughlin. 1969. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 62:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, N., N. S. Jahchan, E. Hong, Qiang Li, M. A. Bayfield, R. J. Maraia, K. Luo, and Q. Zhou. 2008. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318-327. [DOI] [PubMed] [Google Scholar]
- 31.Huang, Y., M. A. Bayfield, R. V. Intine, and R. J. Maraia. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 13:611-618. [DOI] [PubMed] [Google Scholar]
- 32.Jacks, A., et al. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Camb.) 11:833-843. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson, A. 2005. The end justifies the means. Nat. Struct. Mol. Biol. 12:474-475. [DOI] [PubMed] [Google Scholar]
- 34.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M. N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaján, L., and L. Rychlewski. 2007. Evaluation of 3D-Jury on CASP7 models. Protein Structure Prediction Meta Server, BioInfoBank Institute. http://meta.bioinfo.pl/3djury.pl?meta=v2&id=20292. BMC Bioinform. 8:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kedersha, N., et al. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotik-Kogan, O., E. R. Valentine, D. Sanfelice, M. R. Conte, and S. Curry. 2008. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 16:852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozlov, G., et al. 2004. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 23:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozlov, G., and K. Gehring. 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One 5:e10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozlov, G., M. Menade, A. Rosenauer, L. Nguyen, and K. Gehring. 2010. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J. Mol. Biol. 397:397-407. [DOI] [PubMed] [Google Scholar]
- 41.Kozlov, G., N. Safaee, A. Rosenauer, and K. Gehring. 2010. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J. Biol. Chem. 285:13599-13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim, N. S., et al. 2006. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J. Biol. Chem. 281:14376-14382. [DOI] [PubMed] [Google Scholar]
- 43.Long, K. S., et al. 2001. Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization. RNA 7:1589-1602. [PMC free article] [PubMed] [Google Scholar]
- 44.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maraia, R. J., and M. A. Bayfield. 2006. The La protein-RNA complex surfaces. Mol. Cell 21:149-152. [DOI] [PubMed] [Google Scholar]
- 46.Markert, A., et al. 2008. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 9:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martineau, Y., et al. 2008. The poly(A)-binding protein-interacting protein 1 binds to eIF3 to stimulate translation. Mol. Cell. Biol. 28:6658-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauxion, F., C. Y. Chen, B. Seraphin, and A. B. Shyu. 2009. BTG/TOB factors impact deadenylases. Trends Biochem. Sci. 34:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov, G. N., A. A. Vagin, A. Lebedev, K. S. Wilson, and E. J. Dodson. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55:247-255. [DOI] [PubMed] [Google Scholar]
- 50.Nilsson, J., J. Sengupta, J. Frank, and P. Nissen. 2004. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5:1137-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nykamp, K., M. H. Lee, and J. Kimble. 2008. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 14:1378-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 53.Read, R. J. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373-1382. [DOI] [PubMed] [Google Scholar]
- 54.Roy, G., et al. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 22:3769-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan, L., et al. 2010. Quantitative characterization of Tob interactions provides the thermodynamic basis for translation termination-coupled deadenylase regulation. J. Biol. Chem. 285:27624-27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.SantaLucia, J., Jr., H. T. Allawi, and P. A. Seneviratne. 1996. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35:3555-3562. [DOI] [PubMed] [Google Scholar]
- 57.Schäffler, K., et al. 2010. A stimulatory role for the La-related protein 4B in translation. RNA 16:1488-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shahbazian, D., et al. 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 25:2781-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shifera, A. S., and J. A. Hardin. 2010. Factors modulating expression of Renilla luciferase from control plasmids used in luciferase reporter gene assays. Anal. Biochem. 396:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siddiqui, N., et al. 2007. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 282:25067-25075. [DOI] [PubMed] [Google Scholar]
- 61.Sorrentino, S. 1998. Human extracellular ribonucleases: multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell Mol. Life Sci. 54:785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stefano, J. E. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36:145-154. [DOI] [PubMed] [Google Scholar]
- 63.Svitkin, Y. V., et al. 2009. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 28:58-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svitkin, Y. V., L. P. Ovchinnikov, G. Dreyfuss, and N. Sonenberg. 1996. General RNA binding proteins render translation cap dependent. EMBO J. 15:7147-7155. [PMC free article] [PubMed] [Google Scholar]
- 65.Svitkin, Y. V., A. Pause, and N. Sonenberg. 1994. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 68:7001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teplova, M., et al. 2006. Structural basis for recognition and sequestration of UUU-OH 3′-termini of nascent RNA pol III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 21:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terns, M. P., E. Lund, and J. E. Dahlberg. 1992. 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol. 12:3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tritschler, F., E. Huntzinger, and E. Izaurralde. 2010. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat. Rev. Mol. Cell Biol. 11:379-384. [DOI] [PubMed] [Google Scholar]
- 69.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 70.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300-321. [DOI] [PubMed] [Google Scholar]
- 71.Yoo, C. J., and S. L. Wolin. 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol. 14:5412-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ysla, R. M., G. M. Wilson, and G. Brewer. 2008. Assays of adenylate uridylate-rich element-mediated mRNA decay in cells. Methods Enzymol. 449:47-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.