Abstract
Spt6 is a highly conserved factor required for normal transcription and chromatin structure. To gain new insights into the roles of Spt6, we measured nucleosome occupancy along Saccharomyces cerevisiae chromosome III in an spt6 mutant. We found that the level of nucleosomes is greatly reduced across some, but not all, coding regions in an spt6 mutant, with nucleosome loss preferentially occurring over highly transcribed genes. This result provides strong support for recent studies that have suggested that transcription at low levels does not displace nucleosomes, while transcription at high levels does, and adds the idea that Spt6 is required for restoration of nucleosomes at the highly transcribed genes. Unexpectedly, our studies have also suggested that the spt6 effects on nucleosome levels across coding regions do not cause the spt6 effects on mRNA levels, suggesting that the role of Spt6 across coding regions is separate from its role in transcriptional regulation. In the case of the CHA1 gene, regulation by Spt6 likely occurs by controlling the position of the +1 nucleosome. These results, along with previous studies, suggest that Spt6 regulates transcription by controlling chromatin structure over regulatory regions, and its effects on nucleosome levels over coding regions likely serve an independent function.
The regulation of transcription by chromatin dynamics is dependent upon distinct classes of factors that function by interacting with nucleosomes. One set of factors directly modifies histones, the protein components of nucleosomes, by either adding or removing covalent modifications, such as methyl or acetyl groups; these modifications can regulate transcription in a positive or negative fashion (12, 61). A second set of factors is comprised of chromatin-remodeling complexes, which control transcription by regulating the occupancy or positions of nucleosomes in an ATP-dependent fashion (16). A third set of factors are believed to act as histone chaperones. These factors function both in transcription initiation, serving to facilitate the removal or deposition of nucleosomes over regulatory regions, and in transcription elongation, to remove nucleosome barriers before transcribing RNA polymerase (RNAP) II or to redeposit nucleosomes in the wake of transcription (9, 20). The integration of the activities of these different classes of factors is critical in transcriptional regulation. Spt6, a putative histone chaperone and a member of the third class, is the subject of this paper.
Spt6 is a highly conserved protein that has been previously shown to control chromatin structure and transcription. Originally identified as an essential factor required for normal transcription in Saccharomyces cerevisiae (17, 54, 55), Spt6 has also been shown to play critical or essential roles in mammalian cells (73, 74), zebrafish (36, 39, 68), Drosophila (5, 23), nematodes (56), Schizosaccharomyces pombe (77), and Candida albicans (2). In addition to chromatin structure and transcription, Spt6 functions in recombination (51), mRNA surveillance and export (4, 22), and histone modifications (13, 15, 75).
Spt6 interacts with several proteins in order to carry out its roles in controlling chromatin structure and transcription. Studies in both S. cerevisiae and mammalian cells have shown that Spt6 interacts directly with histone H3 (10, 71) and that Spt6 can assemble nucleosomes in vitro (10), suggesting that direct Spt6-histone interactions control chromatin structure. In agreement with these studies, spt6 mutations alter chromatin structure in vivo and suppress the loss of the Swi/Snf chromatin-remodeling complex (10, 54, 55). Other studies have shown that Spt6 also controls chromatin structure at the level of histone modifications, as it is required for normal levels of H3 K36 di- and trimethylation (13, 15, 75). In addition, Spt6 has been shown to interact with RNAP II, and this interaction seems to occur predominantly with the elongating form of the polymerase (21, 73), suggesting a direct role in transcription elongation. Spt6 also forms a complex with another essential protein, Spn1/Iws1 (41, 47). This interaction is required for several steps in transcription, from initiation (78) to histone modifications, RNA processing, and mRNA export (73, 74). A role in mRNA processing is also consistent with the fact that Spt6 has been shown to interact with the nuclear exosome in Drosophila (4). Structural studies strongly support the idea that Spt6 contains multiple domains responsible for interactions with several classes of molecules, including RNA, DNA, nucleosomes, and RNAP II (18, 32). The multiple interactions of Spt6 with other proteins suggest that it may play several roles in controlling chromatin structure and transcription; however, the precise nature of these activities remains to be determined.
Genetic and biochemical studies have suggested that Spt6 controls both transcription initiation and elongation, possibly by a role as a histone chaperone, facilitating nucleosome assembly. For example, spt6 mutants have been shown to be defective for nucleosome reassembly over promoter regions during transcriptional repression (1, 30). In addition, spt6 mutants have been shown to have reduced histone levels over the coding regions of some genes and to have chromatin that is hypersensitive to digestion by micrococcal nuclease (34). These chromatin defects in coding regions have been suggested to cause the widespread cryptic transcription initiation from within coding regions that has been demonstrated in spt6 mutants (14, 34).
Other results are also consistent with a broad role for Spt6 in transcription elongation. Studies in both Drosophila melanogaster and S. cerevisiae have shown that Spt6 is localized to chromosomal regions actively transcribed by RNAP II (3, 33, 35, 37). Furthermore, Spt6 is required for the recruitment of other proteins to transcribed chromatin, including Spt2 (57) and Elf1 (60). In addition, studies have shown that Spt6 facilitates transcription elongation by RNAP II (5, 21). The finding that Spt6 facilitates elongation on a chromatin-free template in vitro (21) indicates that it has a role in transcription elongation beyond effects on nucleosome levels or positions. Taken together, these studies imply varied roles for Spt6 in transcriptional regulation.
Given the broad and critical roles of Spt6 in controlling chromatin structure and transcription, we set out to address the role of Spt6 in controlling nucleosome levels and transcription. First, we tested whether Spt6 is required for normal nucleosome levels by using high-resolution mapping of nucleosome occupancy across S. cerevisiae chromosome III, an expanse of 316.6 kb with over 180 open reading frames (ORFs). Our results show that the requirement for Spt6 is not ubiquitous; rather, in spt6 mutants, there are varying degrees of nucleosome loss across transcribed regions. Second, to determine whether the degree of nucleosome loss correlates with transcription levels, we performed genome-wide chromatin immunoprecipitation (ChIP)-chip analysis for both RNAP II and Spt6; these results show that Spt6 levels correlate with RNAP II levels genome-wide and that, in an spt6 mutant, nucleosomes are preferentially lost from highly transcribed coding regions. This result supports recent studies that suggest that a high level of transcription displaces nucleosomes, whereas a low level of transcription does not (19, 29, 31, 38, 40, 42, 44, 63, 66, 70; for reviews, see references 6, 43, and 69). Our results suggest that these different classes of events are revealed in an spt6 mutant, where restoration of nucleosomes is impaired. Surprisingly, loss of nucleosomes does not correlate with changes in steady-state transcript levels seen in spt6 mutants, nor does it appear to correlate with genes that have cryptic transcription. Finally, to understand the different roles of Spt6 with respect to controlling chromatin structure and mRNA levels, we focused on one gene, CHA1, that is repressed by Spt6. We show that Spt6 repression of CHA1 occurs at the level of initiation, as it is required for the transcriptional repression of CHA1 by remodeling the +1 nucleosome. Taken together, our data suggest that the effects of Spt6 on regulating chromatin structure over transcribed regions are independent of its effects on transcription, which likely involve Spt6 function at promoters.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All FY S. cerevisiae strains used (Table 1) are isogenic with a GAL2+ derivative of S288C (72). The strains were constructed using standard genetics methods (7). All oligonucleotides used for PCR are listed in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm. The Flag-spt6-1004 allele has been described previously (33). The Flag-SPT6 allele was constructed by integrating the EcoNI-digested plasmid pCK40 (a derivative of pRS306 containing amino-terminally tagged SPT6) at the SPT6 genomic locus, resulting in the integration of the auxotrophic marker URA3 flanked by two copies of SPT6. To select for homologous recombinants that have looped out the URA3 marker and contain the Flag tag, Ura− transformants were selected by resistance to 5-fluoroorotic acid (5-FOA). The transformants were further verified by Western blotting for the Flag epitope. The SPT6-HA allele was created by inserting three copies of the hemagglutinin (HA) epitope tag (amplified from vector pFA6 and containing a KanMX6 selection marker) at the C-terminal end of the SPT6 genomic locus (50). SPT6-HA was still fully functional based on testing of Spt phenotypes. The cha4Δ::KanMX4 deletion mutation was constructed by integrative transformation using the amplified CHA4 locus replaced by a KanMX4 marker from the yeast deletion set (25).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| FY85 | MATα his3Δ200 ura3-52 leu2Δ1 lys2-128δ |
| FY2425 | MATα his3Δ200 ura3-52 leu2Δ1 lys2-128δ Flag-spt6-1004 |
| FY1321 | MATα ura3Δ0 |
| FY2788 | MATα ura3Δ0 SPT6-HA3::KanMX6 |
| FY2789 | MATα ura3Δ0 Flag-SPT6 |
| FY2790 | MATα ura3Δ0 cha4Δ::KanMX4 |
| FY2791 | MATα ura3Δ0 cha4Δ::KanMX4 Flag-SPT6 |
| FY2792 | MATα his3Δ200 ura3-52 leu2Δ1 lys2-128δ cha4Δ::KanMX4 |
| FY2793 | MATα his3Δ200 ura3-52 leu2Δ1 lys2-128δ Flag-spt6-1004 cha4Δ::KanMX4 |
For liquid cultures, strains were grown in either YPD rich medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic dropout medium lacking serine (SC-serine) (62). Where indicated, the SC-serine medium was supplemented with either 0.1 mg/ml or 1 mg/ml serine. Unless otherwise indicated, strains were grown to approximately 1 × 107 to 2 × 107 cells/ml at 30°C and then shifted to 37°C for 80 min to inactivate Spt6 function in the spt6-1004 temperature-sensitive strain. For the time course analysis of CHA1 repression, cells were grown in SC-serine supplemented with serine (amounts as indicated) and shifted to 37°C as described above. Following the temperature shift, cells were collected, the medium was discarded, and the cells were resuspended in fresh SC-serine at room temperature for the indicated time.
Northern analysis.
Northern analysis was performed as previously described (7). Strains were grown in YPD or SC-serine medium and supplemented with serine where indicated. Northern hybridization analysis was performed with probes to the coding regions of CHA1, GLK1, PGK1, BUD3, and SNR190 amplified by PCR for random 32P labeling. The oligonucleotides used to synthesize probes are listed in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm.
Chromatin immunoprecipitation.
ChIPs were performed as described previously with minor modifications (52). Briefly, cells were cross-linked in 1% formaldehyde for 30 min and broken by bead beating, and the chromatin fraction was sheared to 200- to 500-bp fragments using a Bioruptor sonicator (Diagenode). Immunoprecipitations were done using 1 μl of anti-H3 antibody (ab1791; Abcam), 5 μl of anti-H2A antibody (generously provided by Jessica Downs, University of Sussex, England), 20 μl of anti-H4 antibody (2960; Cell Signaling Technology), 15 μl of anti-RNAP II antibody (8WG16; Covance), 7 μl of anti-HA antibody (12CA5; generously provided by Brad Cairns, University of Utah), or 1 μl anti-Flag antibody (M2; Sigma Aldrich). The antibodies were incubated with chromatin extracts overnight at 4°C and then coupled to 50 μl of protein G-Sepharose beads (GE Healthcare Life Sciences). For the ChIP-chip analysis of Spt6 and RNAP II localization, the anti-HA and anti-RNAP II antibodies were first preincubated overnight at 4°C with 50 μl of pan-mouse IgG Dynabeads (Invitrogen) before the addition of the chromatin lysates. ChIP DNA was quantified by real-time PCR using a Stratagene MX3000P. Binding of Spt6 and RNAP II to the indicated regions was calculated as the percent IP (IP/input ratio) relative to the binding to an ORF-free region on chromosome I (65). Since the spt6 mutant affects chromatin structure over transcribed regions, for the histone ChIPs, regions within the CDC39 and SNT1 ORFs were chosen as controls because nucleosome levels over the two ORFs were unchanged in the spt6 mutant, according to our nucleosome arrays (see Table S2 in the supplemental material). The data shown are normalized to CDC39, but the values were similar when normalized to SNT1. To determine the specificity of enrichment of the tagged Spt6 protein, the corresponding untagged control samples were included in each experiment (see Fig. 4A and 7E; also see Fig. S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm). For RNAP II and histone ChIPs, a “no-antibody” control was used instead. Each experiment was performed at least three times. The primer sequences are listed in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm.
Spt6 and RNAP II ChIP-chip.
Strains were grown in YPD to 1 × 107 to 2 × 107 cells/ml at 30°C and were then shifted to 37°C for 80 min. For determining the occupancy of Spt6, DNA was immunoprecipitated using an anti-HA antibody from strains expressing either HA-tagged or untagged Spt6. DNA corresponding to RNAP II binding sites was immunoprecipitated with anti-8WG16 antibody against Rpb1. The specificity of Rpb1 binding was controlled by a “no-antibody” precipitation from the same chromatin extracts. DNA prepared by ChIP as described above was blunt ended, ligated to unidirectional linkers, and amplified by ligation-mediated PCR as described previously (8). 5-(3-Aminoallyl)-2′deoxyuridine-5′-triphosphate was included in the PCR mixture. Labeling was performed post-PCR by coupling to monoreactive Cy5 or Cy3 N-hydroxysuccinimide (NHS) esters. The Cy5 (experiment) and Cy3 (control) samples were mixed and hybridized on DNA microarrays containing melting temperature (Tm)-adjusted oligonucleotides covering the nonrepetitive part of the S. cerevisiae genome. The microarrays (Agilent Technologies) have been described previously (26, 59, 63). They contain 44,290 probes consisting of 60-mer oligonucleotides that cover 85% of the S. cerevisiae genome, including both intergenic and coding regions and excluding highly repetitive regions, with an average probe density of 266 bp.
Micrococcal nuclease (MNase) digestion.
Mononucleosomal DNA was prepared as described previously (76). Briefly, cells were grown to mid-log phase in 440 ml medium (YPD or SC-serine, as indicated), shifted to 37°C for 80 min, and cross-linked with 1% formaldehyde at room temperature for 30 min. The cell pellets were resuspended in 39 ml buffer Z (1 M sorbitol, 50 mM Tris-Cl, pH 7.4), 28 μl of β-mercaptoethanol (β-ME) (14.3 M; final concentration, 10 mM) was added, and the cells were vortexed to resuspend them. One milliliter of zymolyase solution (10 mg/ml in buffer Z; Z1004; U.S. Biological) was added, and the cells were incubated at 30°C for 45 min to make spheroplasts. The spheroplasts were pelleted and resuspended in 3.6 ml of NP buffer (0.5 mM spermidine, 1 mM β-ME, 0.075% NP-40, 50 mM NaCl, 10 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM CaCl2). Aliquots of 600 μl spheroplasts were added to each of six tubes containing micrococcal nuclease (Sigma) in a range of concentrations (from 90 to 525 units of MNase). For the nucleosome-scanning experiment, one sample was kept undigested (stop buffer [5% SDS, 50 mM EDTA] was added immediately) and used as the genomic DNA control. The spheroplasts were incubated at 37°C for 22 min, and the digestion was halted by adding 150 μl of stop buffer. Then, 200 μg of proteinase K was added, and the reaction mixtures were incubated overnight at 65°C to reverse the cross-link and degrade proteins. DNA from fractions containing greater than 90% mononucleosomes was extracted by phenol-chloroform, treated with DNase-free RNase, and extracted three more times to purity.
Linear amplification of DNA and microarray hybridization for nucleosome arrays.
Seventy-five nanograms of DNA corresponding to the mononucleosomal fraction from wild-type or spt6 strains (isolated as detailed above) was amplified using the T7-based linear-amplification method (48). Three milligrams of RNA produced from the linear amplification was used to label probe via the aminoallyl method as described previously (http://www.microarrays.org). Labeled probes were hybridized onto a yeast tiled oligonucleotide microarray (76) at 65°C for 16 h and washed as described previously (http://www.microarrays.org). The arrays were scanned at 5-μm resolution with an Axon Laboratories GenePix 4000B scanner running GenePix 5.1. The values for nucleosome occupancy signals shown in Tables 2 and 3 and in Table S3 at http://genepath.med.harvard.edu/∼winston/supplemental.htm are the averages across each ORF of all the signals for oligonucleotides on the microarray that cover the ORF. In addition, each number is the average of two independent experiments.
TABLE 2.
Nucleosome loss is greatest in highly transcribed genesa
| Gene | Nucleosome level in spt6-1004 | Spt6 level | RNAP II level |
|---|---|---|---|
| PGK1 | −1.03 | 2.19 | 4.74 |
| CHA1 | −1.00 | 1.07 | 2.24 |
| PMP1 | −0.89 | 1.67 | 3.57 |
| PDI1 | −0.79 | 1.23 | 2.58 |
| RPS14A | −0.78 | 1.81 | 3.15 |
| GLK1 | −1.36 | 0.76 | 1.96 |
| VAC17 | 0.01 | 0.41 | 0.08 |
| BUD3 | −0.02 | −0.22 | −1.11 |
| DCC1 | −0.14 | −0.34 | −0.33 |
| CDC39 | 0 | 0.32 | −0.37 |
| SNT1 | 0.03 | 0.08 | −0.65 |
The RNAP II occupancy (log2 IP/no Ab) and Spt6 occupancy (log2 HA/no tag) determined by our ChIP-chip assay are compared to the nucleosome occupancy signals (log2 spt6-1004/wild type) across coding regions for representative genes on chromosome III. The genes in boldface in the top half of the table are highly transcribed; those in the bottom half are transcribed at basal levels.
TABLE 3.
Genes on chromosome III with cryptic transcriptsa
| Gene | Nucleosome level in spt6-1004 | Spt6 level | RNAP II level |
|---|---|---|---|
| LDB16 | −0.24 | 0.46 | 1.49 |
| BUD3 | −0.02 | −0.22 | −1.11 |
| DCC1 | −0.14 | −0.34 | −0.33 |
| AGP1 | −0.48 | 0.27 | 0.43 |
| ATG22 | 0.09 | −0.13 | −0.16 |
| YCL042W | −0.71 | 0.69 | 1.92 |
| MRC1 | −0.01 | −0.21 | −1.0 |
| POL4 | 0.02 | −0.05 | 0.23 |
| YCR016W | 0.01 | −0.02 | −0.38 |
| CWH43 | −0.08 | −0.10 | −0.07 |
| MAK32 | −0.05 | −0.17 | −0.68 |
| SYP1 | −0.20 | 0.01 | 0.01 |
| RBK1 | −0.29 | 0.24 | 0.67 |
| RRT12 | −0.29 | 0.30 | 0.09 |
| RAD18 | 0.23 | −0.09 | −0.93 |
| SSK22 | 0.03 | −0.27 | −0.53 |
| SRB8 | 0.11 | −0.36 | −1.0 |
| FIG2 | −0.06 | −0.16 | −0.63 |
| CDC39 | 0.00 | 0.32 | −0.37 |
| RDS1 | 0.14 | −0.42 | −1.4 |
| AAD3 | 0.11 | −0.54 | −0.83 |
The RNAP II occupancy (log2 IP/no Ab) and Spt6 occupancy (log2 HA/no tag) determined by our ChIP-chip assay are compared to the nucleosome occupancy signals (log2 spt6-1004/wild type) across coding regions for genes on chromosome III known to have short transcripts in the spt6-1004 mutant.
Nucleosome scanning.
Mononucleosomal DNA from cells grown in SC-serine medium or the same medium supplemented with 1 mg/ml serine was isolated as described above. Quantitative PCR on MNase-digested DNA was performed using 17 different primer pairs to amplify overlapping 95- to 115-bp fragments spanning 600 bp covering the CHA1 promoter and 5′ coding region. The oligonucleotides used as primers are listed in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm. As controls, we also assayed a well-characterized nucleosome-bound sequence in the GAL1 promoter, as well as an adjacent nucleosome-free region (11, 49). Relative protection of the template for each PCR product in the CHA1 promoter and 5′ ORF was calculated as a ratio to the concentration of the nucleosome-bound template of GAL1.
Microarray data accession number.
ChIP-chip data can be found at the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE21787.
RESULTS
Loss of Spt6 results in nucleosome loss over a subset of coding regions.
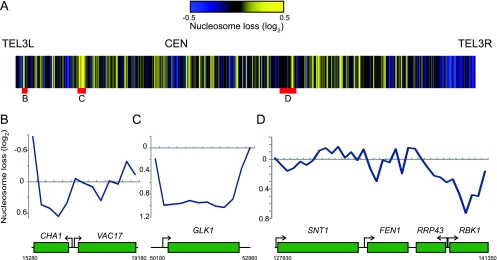
Given the evidence that Spt6 plays a critical role in transcription and in maintaining a proper chromatin environment, we set out to determine the changes that occur in nucleosome occupancy in an S. cerevisiae spt6 mutant, using a previously described procedure (76) (see Materials and Methods). For these studies, we used spt6-1004, a previously characterized temperature-sensitive allele that has been shown to cause widespread changes in transcription and chromatin structure (14, 33, 34). Since spt6-1004 causes reduced levels of Spt6 protein (33), the mutant phenotypes are likely caused, at least in part, by a general loss of Spt6 function. Briefly, mononucleosomal DNA was isolated from both wild-type and spt6-1004 cells, each DNA sample was amplified and fluorescently labeled (one with Cy3 and the other with Cy5), and the two samples were competitively hybridized to a tiled oligonucleotide microarray of S. cerevisiae chromosome III. Our results show that in spt6-1004 mutants there are degrees of change, mostly nucleosome loss, along chromosome III (Fig. 1A; see Table S2 in the supplemental material). Interestingly, the greatest degree of nucleosome loss occurs predominantly over a subset of coding regions, such as CHA1 (Fig. 1B) and GLK1 (Fig. 1C), while nucleosome occupancy in other coding regions, such as VAC17 (Fig. 1B) and SNT1 (Fig. 1D), is unaffected. There are also regions that have an increased nucleosome density, such as near TEL3R. As previous results have shown that the nucleosome density is relatively constant across chromosome III (76), the changes caused by spt6-1004 do not appear to occur in regions of unusual nucleosome distribution.
FIG. 1.
Effects of spt6-1004 on nucleosome occupancy across chromosome III. (A) Heat map of nucleosome occupancy along S. cerevisiae chromosome III, represented as the log2 of the spt6/wild-type signals, where yellow represents relative nucleosome loss in the spt6 mutant compared to the wild type and blue represents nucleosome gain. The data are the average of two independent experiments. (B to D) Higher-resolution views of the nucleosome profiles over the coding regions of CHA1-VAC17 and GLK1 and a cluster of genes containing SNT1 and BUD3. The y axis represents the log2 ratio of the spt6/wild-type signals. TEL3L and TEL3R, left and right telomeres, respectively.
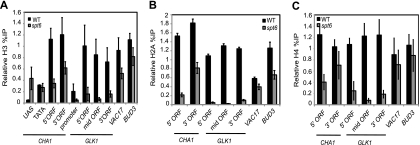
To confirm our microarray results, we determined the levels of histones H3, H4, and H2A physically associated with a set of genes on chromosome III using ChIP. All histone ChIPs were normalized to the histone levels over the middle of the coding regions of CDC39 and SNT1 (not shown), which in our nucleosome microarrays were unchanged in the spt6-1004 mutant compared to the wild type (see Table S2 in the supplemental material). Therefore, a value of 1 suggests that there are equal levels of histones present in the region of interest and the control region, whereas values lower than 1 represent reduced histone levels in the region of interest in the spt6-1004 mutant. The histone ChIP results are consistent with our microarray data; in the spt6-1004 mutant, histone levels were lower over the coding regions of CHA1 and GLK1 and were unaffected over the coding regions of VAC17 and BUD3 (Fig. 2). Histone levels were low in both wild-type and spt6-1004 strains over the promoter regions of CHA1 and GLK1, suggesting that even though there are extensive changes in the chromatin structure over coding regions, the promoters remain nucleosome free (Fig. 2A and data not shown) (45). Taken together, our microarray and histone ChIP results show that in an spt6-1004 mutant, there are varying degrees of nucleosome loss over coding regions, suggesting that Spt6 activity is required for normal chromatin structure in a subset of genes.
FIG. 2.
Histone levels are reduced from specific coding regions in the spt6-1004 mutant. Histone H3 (A), H2A (B), and H4 (C) occupancies were determined by ChIP analysis of chromatin extracts isolated from wild-type (WT) and spt6-1004 mutant strains. The values were normalized to histone levels in a control region (CDC39), which were set to 1. The DNA sequences for each primer pair used in the real-time analysis are listed in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm. Each bar represents the mean percentage immunoprecipitation value ± standard error of the mean (SEM) (n = 3 to 15).
Spt6 colocalizes with RNA polymerase II at highly transcribed genes.
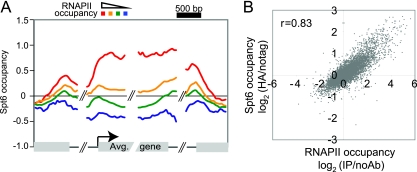
Previous experiments suggested that Spt6 is physically associated with transcribed regions and that its level of association corresponds to the level of transcription (3, 33, 35, 37). To test whether the degree of nucleosome loss in the spt6-1004 mutant corresponds to the level of association of Spt6 with chromatin in wild-type cells, we measured the genome-wide association of Spt6 with chromatin by ChIP-chip. In addition, to test whether Spt6 levels correlate with transcription rates genome-wide, we also measured the genome-wide association of RNAP II. Our results show that Spt6 is primarily localized over coding regions at a level that corresponds with the level of RNAP II (Fig. 3A). As expected, on a genome-wide scale, we found a strong correlation between Spt6 and RNAP II localization (Fig. 3B). These results agree with another recent study of Spt6 (52a).
FIG. 3.
Genome-wide localization of Spt6 and RNAP II. DNA was labeled and hybridized to DNA microarrays as described in Materials and Methods. (A) For Spt6 analysis, genes were binned within four groups based on the level of RNAP II across their open reading frames. The average (Avg.) enrichment signal for Spt6 is shown for each group of genes. The data shown are averages of two independent experiments. (B) Scatter plot of the average enrichment ratio for Spt6 versus that of RNAP II on all ORFs. The average enrichment ratio is represented as the log2 value of the Spt6- or RNAP II-specific IP signal versus background (no tag or no antibody, respectively) signal. The correlation coefficient was 0.83.
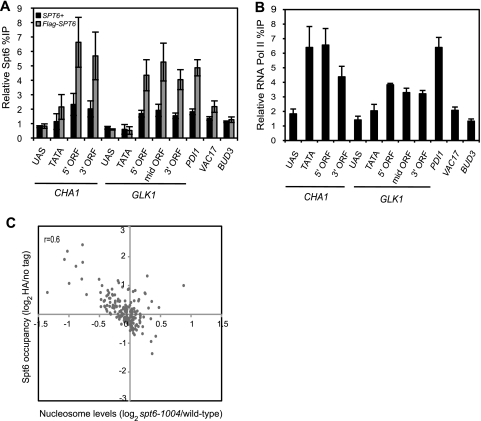
To confirm our ChIP-chip results at specific loci, we measured the levels of Spt6 and RNAP II association at particular genes of interest using gene-specific ChIP. These results showed that Spt6 is localized primarily over the coding regions of highly transcribed genes (Fig. 4A) while RNAP II is found in both promoter and coding regions (Fig. 4B). In contrast, Spt6 fails to localize to the coding regions of genes transcribed at low levels, such as VAC17 and BUD3 (Fig. 4A and B). Taken together, our data suggest that Spt6 colocalizes with RNAP II over the open reading frames of highly transcribed genes.
FIG. 4.
Spt6 colocalizes with RNAP II to coding regions. (A) The occupancy of Spt6 was determined by ChIP analysis of chromatin extracts isolated from strains expressing Flag-tagged Spt6 that were grown to mid-log phase at 30°C and shifted to 37°C for 80 min. The occupancy levels of Spt6 were quantified by real-time PCR of anti-Flag immunoprecipitated DNA as described in Materials and Methods. The black bars represent the background percent immunoprecipitation obtained from ChIP analysis of the corresponding untagged strains. Each bar represents the mean percent immunoprecipitation value ± SEM (n = 3). (B) The relative occupancy of RNAP II was determined as described in Materials and Methods using anti-8WG16 antibody. The bars represent the mean percentage immunoprecipitation value and SEM (n = 4). The DNA sequences for each primer pair used in the real-time analysis are shown in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm. (C) Scatter plot of the average enrichment ratio of Spt6 (represented as the log2 value of Spt6-specific IP signal versus background [no-tag] signal) versus nucleosome levels in spt6-1004 (represented as the log2 value of the ratio of spt6-1004 and wild-type signals) at all ORFs on chromosome III.
Then, to determine whether nucleosome loss in the spt6-1004 mutant occurs primarily over highly transcribed genes, we compared the degree of association of RNAP II (and of Spt6) to the degree of nucleosome loss in the spt6-1004 mutant, examining all open reading frames of chromosome III (Fig. 4C). The data suggest that, in general, genes with high levels of RNAP II and Spt6 in wild-type cells are those with the greatest degree of nucleosome loss in the spt6-1004 mutant. Conversely, those with lower levels of RNAP II and Spt6 show little or no nucleosome loss (examples are shown in Table 2 and in Table S3 at http://genepath.med.harvard.edu/∼winston/supplemental.htm). We note that there is not a strong statistical correlation between RNAP II/Spt6 levels and nucleosome loss (r = 0.6). We speculate that there may not be a strict correlation but rather a threshold level of transcription above which nucleosome loss occurs. This idea is supported by previous studies that suggest that the maintenance of chromatin structure following transcription occurs by mechanisms that differ by the degree of histone loss and exchange and that the level of histone exchange is gene specific rather than transcription rate specific (19, 24) (see Discussion). The simplest conclusion from these data is that, in general, Spt6 is required to maintain a normal level of nucleosomes over genes that are highly transcribed.
Separable requirements for Spt6 for chromatin structure and for transcription.
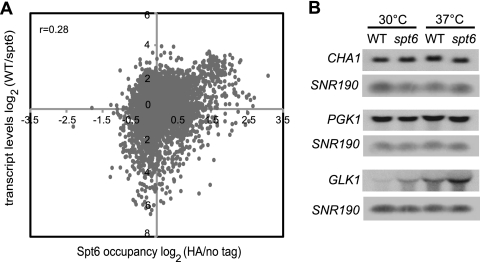
The results presented above show that the levels of association of Spt6 and RNAP II with chromatin strongly correlate genome-wide, suggesting that Spt6 levels are determined by the level of transcription. To test whether this correlation extends to the requirement for Spt6 for normal transcription, we compared our Spt6 ChIP-chip results to previously published microarray results for an spt6-1004 mutant (14). Surprisingly, this comparison showed that there is poor correlation between Spt6 localization and changes in transcript levels in the spt6-1004 mutant (Fig. 5A). Furthermore, this lack of correlation extends to both genes with increased and decreased mRNA levels in the spt6-1004 mutant. These results suggest that changes in nucleosome occupancy over the coding regions in the spt6-1004 mutant are not the cause of the extensive transcriptional alterations that occur.
FIG. 5.
Localization of Spt6 to coding regions does not correlate with altered transcript levels in the spt6 mutant. (A) Scatter plot of transcript levels in the spt6 mutant represented as log2 (wild-type/spt6) versus the average enrichment ratio of Spt6 on all ORFs. Data for transcript levels are from Cheung et al. (14). (B) Northern blot analysis of CHA1, PGK1, and GLK1 mRNA levels. RNA was extracted from wild-type and spt6-1004 strains grown at 30°C or shifted to 37°C for 80 min. SNR190 was used as a loading control. Each Northern blot is representative of three independent experiments.
To examine this issue by a second method, we measured the transcript levels of CHA1, PGK1, and GLK1, three genes that have significant nucleosome loss in an spt6-1004 mutant. Our results (Fig. 5B) show that, under growth conditions identical to those that show nucleosome loss (growth in YPD and a shift to 37°C), the mRNA levels for two of the genes, CHA1 and PGK1, were unchanged in an spt6-1004 mutant compared to the wild type, while GLK1 had elevated mRNA levels (compare Fig. 5B to Table 2). (We show below that CHA1 transcription is regulated by Spt6 under different growth conditions.) Thus, while Spt6 is strongly required for normal chromatin structure over many open reading frames, this activity is apparently independent of the role played by Spt6 in the control of transcription at many genes.
Previous studies have shown that Spt6 also plays a prominent role in the repression of transcription from cryptic promoters within coding regions (14, 34). On chromosome III, which is one of the smallest chromosomes in S. cerevisiae, cryptic transcription was detected at 21 genes in an spt6-1004 mutant (14). For those 21 genes, we examined the effect of spt6-1004 on nucleosome levels and found that most of them did not show significant nucleosome loss (Table 3). Consistent with this finding, our ChIP-chip results showed that most of these genes had low levels of RNAP II and Spt6 associated. These findings are not surprising, as our method for detection of cryptic initiation was biased toward genes transcribed at low levels (14, 46). Nevertheless, the results show that cryptic initiation occurs at genes without significant nucleosome loss in an spt6-1004 mutant.
Spt6 is required for repression of CHA1 under low-serine conditions by remodeling the +1 nucleosome.
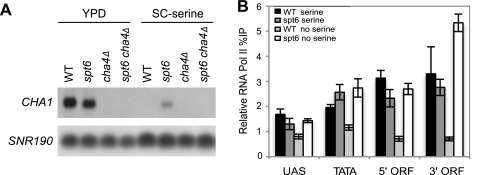
To further understand the role of Spt6 in the regulation of chromatin structure and how it relates to its role in transcriptional regulation, we decided to focus on CHA1, a gene at which Spt6 exerts strong effects on nucleosome levels across its coding region and, as described below, on transcriptional regulation under particular growth conditions. CHA1 encodes a catabolic l-serine/l-threonine deaminase, and its transcription responds to serine and threonine levels: CHA1 transcription is tightly repressed when serine or threonine is absent, and its transcription is strongly induced under high-serine or -threonine conditions (58). An spt6-1004 mutation has no detectable effect on the induced levels of CHA1 mRNA when cells are grown in rich medium (Fig. 5), even though the spt6-1004 mutation causes significant nucleosome loss across the CHA1 coding region. However, in the course of our studies, we discovered that CHA1 repression is defective in an spt6-1004 mutant when cells are grown in the absence of serine (examples are shown in Fig. 6A and 7D). Thus, Spt6 is required for repression of CHA1 transcription.
FIG. 6.
Spt6 is required for the repression of CHA1 at low serine levels. (A) CHA1 mRNA levels were measured by Northern blot analysis. RNA was isolated from the indicated strains grown under activating (YPD) or repressive (SC-serine) conditions at 30°C and then shifted to 37°C for 80 min. SNR190 was used as a loading control. (B) RNAP II levels at CHA1 were measured by ChIP in the wild type and spt6 mutants that were grown either in SC-serine or SC-serine supplemented with 0.1 mg/ml serine. The relative percent IP was determined as described in Materials and Methods. Each bar represents the mean percent immunoprecipitation value ± SEM (n = 6). The DNA sequences for each primer pair used in the real-time analysis are shown in Table S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm.
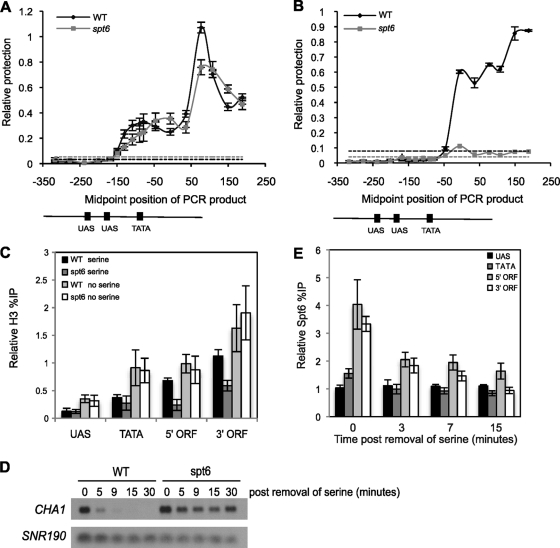
FIG. 7.
Spt6 regulates CHA1 expression by remodeling the +1 nucleosome under repressive conditions. (A and B) Nucleosome scanning of the CHA1 promoter and 5′ coding region under repressive (SC-serine) (A) and activating (SC-serine plus 1 mg/ml serine) (B) conditions. DNA from wild-type and spt6-1004 cells was digested with micrococcal nuclease and analyzed by quantitative PCR as described in Materials and Methods. The relative protection of the template for each PCR product was calculated as the ratio of the level of each PCR product to that for a nucleosome-bound template in the GAL1 promoter and graphed using the midpoint position of the PCR product. The dashed lines represent the relative protection of the nonnucleosomal GAL1 template for each sample. The data represent the average ± SEM of six independent experiments. The numbers along the x axis represent the position relative to the ATG of the CHA1 open reading frame, where the A is +1. UAS, upstream activation sequence. (C) Histone levels in SC-serine or SC-serine supplemented with 1 mg/ml serine were determined as described in Materials and Methods. Each bar represents the mean percent immunoprecipitation value ± SEM (n = 7). (D) Northern blot analysis of CHA1 mRNA levels in wild-type and spt6 cells following a shift to repressive conditions. RNA was isolated from cells grown at 30°C in SC-serine supplemented with 0.1 mg/ml serine and then shifted to SC-serine at 25°C for the indicated time. SNR190 was used as a loading control. The blot is representative of three independent experiments. (E) Spt6 levels over CHA1 following a shift to repressive conditions were determined as described in Materials and Methods. The background percent immunoprecipitations obtained from ChIP analysis of the corresponding untagged strains are shown in Fig. S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm. Each bar represents the mean percent immunoprecipitation value ± SEM (n = 4).
Since the induction of CHA1 in rich medium is dependent upon the serine-responsive activator Cha4, which is constitutively bound to the CHA1 promoter (28, 64), we tested whether the aberrant transcription of CHA1 in an spt6-1004 mutant under normally repressing conditions requires Cha4. To do this, we compared CHA1 mRNA levels in spt6-1004 and spt6-1004 cha4Δ strains and found that Cha4 is indeed required for the aberrant CHA1 transcription in an spt6-1004 mutant (Fig. 6A). To examine transcription by another method, we also did RNAP II ChIP at CHA1 and found that the results agreed with the Northern analysis (Fig. 6B). These results show that Spt6 is required for the repression of CHA1 transcription initiation at low serine levels and that CHA1 transcription in the spt6-1004 mutant likely occurs using the same mechanism during normal CHA1 induction.
To test whether Spt6 represses CHA1 transcription initiation by controlling chromatin structure over the CHA1 5′ regulatory region, we mapped the positions of nucleosomes over this region at high resolution in both wild-type and spt6-1004 strains. To do this, we performed nucleosome scanning (67) as described in Materials and Methods. Briefly, we purified micrococcal nuclease-protected mononucleosomal DNA from wild-type and spt6-1004 cells grown in the presence or absence of serine. We then used this DNA as a template in quantitative PCRs using 17 different primer pairs to amplify overlapping 95- to 115-bp fragments spanning 600 bp covering the CHA1 promoter and 5′ coding region. Previous studies have shown that, under repressive conditions (in the absence of serine), the CHA1 promoter contains a well-positioned nucleosome that occludes the TATA box and transcription start site; upon activation by the addition of serine, this nucleosome undergoes a Cha4-dependent displacement (53). When we examined CHA1 under repressing conditions by nucleosome scanning, similar to these previous results, we detected a well-positioned nucleosome over the TATA box in wild-type cells (Fig. 7A). In the spt6-1004 mutant, however, this nucleosome is shifted to a different position, approximately 100 bp 3′ of its position in the wild type. This change would expose the TATA box to a more accessible site on the nucleosome, explaining the partial derepression of CHA1 observed in spt6-1004 mutants (Fig. 6A and 7D). We also examined CHA1 by nucleosome scanning under activating conditions and showed that, in wild-type cells, the promoter region is nucleosome free while there are nucleosomes present over the 5′ coding region (Fig. 7B). In the spt6-1004 mutant, the CHA1 locus was completely devoid of nucleosomes, in agreement with our ChIP results (compare Fig. 7B and 2A to C).
To confirm the nucleosome-scanning results, we performed ChIP analysis of histone H3 at the CHA1 promoter and coding regions in wild-type and spt6-1004 cells under both repressing and inducing conditions. Consistent with the nucleosome-scanning data, under repressive conditions, histone H3 is present over the TATA region in both the wild type and spt6-1004 mutant, coinciding with the presence of the +1 nucleosome in this region in a repressive state (Fig. 7C). In addition, we see H3 present over the CHA1 coding region in both wild-type and spt6-1004 strains, even though CHA1 is transcribed in the spt6-1004 mutant. When CHA1 is induced, H3 levels are reduced over both the CHA1 promoter region and the coding region in the spt6-1004 mutant compared to wild-type cells, similar to what was previously described (compare Fig. 7C and 2A). Taken together, our data suggest that under repressing conditions, Spt6 is required to position a nucleosome over the CHA1 TATA, suggesting that it inhibits TATA-binding protein (TBP) binding to its site and that the effect of Spt6 on nucleosome levels over the coding region has little if anything to do with CHA1 regulation.
To test whether Spt6 acts directly on the repositioning of the CHA1 +1 nucleosome, we assayed for Spt6 by ChIP during a shift of wild-type cells from activating (serine-rich) to repressing (serine-free) conditions. During this time course, CHA1 mRNA levels are greatly reduced in wild-type cells by 5 min after the removal of serine from the medium; in contrast, CHA1 mRNA levels are maintained at a significant level in the spt6-1004 mutant (Fig. 7D; ChIP signals for no-tag controls are shown in Fig. S1 at http://genepath.med.harvard.edu/∼winston/supplemental.htm). By ChIP, we were unable to detect Spt6 over the CHA1 promoter region under activating conditions or during repression (Fig. 7E). As expected, Spt6 is detectable over the CHA1 open reading frame when cells are grown in rich medium, and this level decreases during repression, correlating with CHA1 mRNA levels (Fig. 7E). These results suggest that the role of Spt6 in repositioning the nucleosomes over the CHA1 regulatory region is not the result of stable association of Spt6 with the region.
DISCUSSION
In this study, we have investigated the roles of Spt6 in chromatin structure and transcription in S. cerevisiae, leading to new insights into the roles of Spt6 in vivo. Our results show that in an spt6 mutant, there is a significant loss of nucleosomes over some, but not all, coding regions on chromosome III and that this nucleosome loss occurs primarily over genes that, in wild-type cells, are highly transcribed and have a high level of Spt6. Our results are consistent with earlier studies that showed reduced histone levels across the coding regions of FLO8 and HSP104 in an spt6 mutant (30, 34). As previous studies have shown that there are many changes in mRNAs in an spt6 mutant (14), we compared the set of genes with high levels of Spt6 in the wild type to those with transcriptional changes in an spt6 mutant and found, unexpectedly, that there was a poor correlation. This finding suggests that nucleosome loss over coding regions is not the primary cause of transcriptional changes in an spt6 mutant. Finally, at one gene, CHA1, we show that Spt6 confers transcriptional repression by positioning the +1 nucleosome over the CHA1 TATA element. These results suggest that Spt6 plays distinct roles over coding and regulatory regions and that these changes have different consequences for transcription. An extension of our results is the suggestion that nucleosome levels over coding regions may not play a prominent role in determining mRNA levels.
The effects of an spt6 mutation on nucleosome levels across coding regions are consistent with previous studies that suggested two different mechanisms for chromatin remodeling and reassembly during RNAP II transcription. These studies showed that genes with a high level of transcription have dynamic chromatin, with a significant loss, a high level of exchange, and rapid reassembly of all four core histones over transcribed regions; in contrast, for genes with more modest levels of transcription, chromatin is less dynamic, with a lower level of histone loss and exchange, particularly for histones H3 and H4 (19, 29, 31, 38, 40, 42, 44, 63, 66, 70; for reviews, see references 6, 43, and 69). Our results, taken together with these previous studies, suggest a model in which Spt6 is required for reassembly of chromatin after the passage of RNAP II over highly transcribed genes. Similar models for Spt6 have been proposed previously (23, 27, 34). By this model, nucleosomes are lost over the subset of genes that have a high level of transcription elongation, and in an spt6 mutant, nucleosomes cannot be reassembled after the passage of RNAP II. For those genes that are normally transcribed at a lower level, little or no detectable nucleosome loss occurs, bypassing any requirement for Spt6 for nucleosome reassembly. Thus, our analysis of an spt6 mutant has provided strong support in vivo for two classes of genes with respect to nucleosome loss during transcription.
An unexpected result of our studies is the finding that the reduced nucleosome levels across coding regions in an spt6 mutant do not cause transcriptional changes. This conclusion applies to two types of changes that occur in spt6 mutants: altered levels of full-length mRNAs and the activation of cryptic transcription within coding regions (14). However, there is previous evidence that Spt6 is required for normal transcription elongation, both in vitro and in vivo (5, 21). In addition, Spt6 has been shown to interact with a number of factors during transcription elongation, including RNAP II, Spn1/Iws1, Spt5, Spt2, Spt16, Elf1, and TFIIS (21, 27, 41, 47, 57, 60, 73, 78). Given these results and the significant effect of an spt6 mutation on chromatin structure across highly transcribed genes, it seems reasonable to conclude that Spt6 is required for some aspects of transcription elongation, although they may be too subtle to detect by microarray analysis.
Several results suggest that the major role for Spt6 in transcriptional regulation occurs during initiation. Previous studies have analyzed in depth the role of Spt6 in the regulation of transcription initiation of two genes, SUC2 and PHO5, both of which have nucleosomes over their promoter regions (1, 10). For both genes, reassembly of these nucleosomes upon a shift to repressing conditions is impaired in an spt6 mutant, resulting in defects in transcriptional repression (1). Our analysis of CHA1 has revealed another mechanism by which Spt6 can control initiation. In contrast to SUC2 and PHO5, which have nucleosomes over their regulatory regions, CHA1 has a more typical nucleosome-free region over its regulatory sites, flanked by a +1 nucleosome over the TATA element (45, 53, 76). Our results have shown that the position of the +1 nucleosome is controlled by Spt6, as it is positioned over the TATA in wild-type cells and has an altered position in the spt6 mutant. Previous results have shown that histone H3 also controls CHA1 chromatin structure over this region (64). As Spt6 has been shown to directly interact with H3 (10), this result suggests that Spt6 may directly interact with the +1 nucleosome to control its position. However, since ChIP has not detected Spt6 at the CHA1 promoter under repressing growth conditions, it is also possible that Spt6 functions indirectly to position the +1 nucleosome. Additional studies will be necessary to elucidate the precise mechanism by which Spt6 regulates the position of the +1 nucleosome at CHA1 and whether this aspect of Spt6 function occurs at other promoters.
Supplementary Material
Acknowledgments
We thank Audrey Forest for help with the ChIP-chip experiments and Jessica Downs and Brad Cairns for providing antibodies. We are grateful to Randy Morse for many helpful discussions and for sharing plasmids and oligonucleotide sequences. We thank Christine Kiely for critical reading of the manuscript.
This work was supported by NIH grant GM032967 to F.W., NIH grant GM079205 to O.J.R., and CIHR grant 82891 to F.R.
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21:405-416. [DOI] [PubMed] [Google Scholar]
- 2.Al-Rawi, N., S. S. Laforce-Nesbitt, and J. M. Bliss. 2010. Deletion of Candida albicans SPT6 is not lethal but results in defective hyphal growth. Fungal Genet. Biol. 47:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrulis, E. D., et al. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 5.Ardehali, M. B., et al. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong, J. A. 2007. Negotiating the nucleosome: factors that allow RNA polymerase II to elongate through chromatin. Biochem. Cell Biol. 85:426-434. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., et al. (ed.). 1991. Current protocols in molecular biology. Wiley, New York, NY.
- 8.Bataille, A. R., and F. Robert. 2009. Profiling genome-wide histone modifications and variants by ChIP-chip on tiling microarrays in S. cerevisiae. Methods Mol. Biol. 543:267-279. [DOI] [PubMed] [Google Scholar]
- 9.Belotserkovskaya, R., and D. Reinberg. 2004. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 14:139-146. [DOI] [PubMed] [Google Scholar]
- 10.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 11.Brickner, D. G., et al. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos, E. I., and D. Reinberg. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559-599. [DOI] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 14.Cheung, V., et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu, Y., R. Simic, M. H. Warner, K. M. Arndt, and G. Prelich. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26:4646-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clapier, C. R., and B. R. Cairns. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273-304. [DOI] [PubMed] [Google Scholar]
- 17.Clark-Adams, C. D., and F. Winston. 1987. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dengl, S., A. Mayer, M. Sun, and P. Cramer. 2009. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J. Mol. Biol. 389:211-225. [DOI] [PubMed] [Google Scholar]
- 19.Dion, M. F., et al. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405-1408. [DOI] [PubMed] [Google Scholar]
- 20.Eitoku, M., L. Sato, T. Senda, and M. Horikoshi. 2008. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol. Life Sci. 65:414-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endoh, M., et al. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24:3324-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estruch, F., et al. 2009. A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with mex67-5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol. Genet. Genomics 281:125-134. [DOI] [PubMed] [Google Scholar]
- 23.Formosa, T., et al. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gat-Viks, I., and M. Vingron. 2009. Evidence for gene-specific rather than transcription rate-dependent histone H3 exchange in yeast coding regions. PLoS Comput. Biol. 5:e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever, G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 26.Guillemette, B., et al. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmberg, S., and P. Schjerling. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144:467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamai, A., R. M. Imoberdorf, and M. Strubin. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25:345-355. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, M. M., M. S. Christensen, B. Bonven, and T. H. Jensen. 2008. Requirements for chromatin reassembly during transcriptional downregulation of a heat shock gene in Saccharomyces cerevisiae. FEBS J. 275:2956-2964. [DOI] [PubMed] [Google Scholar]
- 31.Jin, J., et al. 2010. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat. Struct. Mol. Biol. 17:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, S. J., et al. 2008. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J. Mol. Biol. 377:1460-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan, C. D., M. J. Holland, and F. Winston. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913-922. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keegan, B. R., et al. 2002. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development 129:1623-1632. [DOI] [PubMed] [Google Scholar]
- 37.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kok, F. O., et al. 2007. The role of the SPT6 chromatin remodeling factor in zebrafish embryogenesis. Dev. Biol. 307:214-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogan, N. J., et al. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulaeva, O. I., et al. 2009. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16:1272-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulaeva, O. I., D. A. Gaykalova, and V. M. Studitsky. 2007. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 618:116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulaeva, O. I., F. K. Hsieh, and V. M. Studitsky. 2010. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl. Acad. Sci. U. S. A. 107:11325-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, W., et al. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235-1244. [DOI] [PubMed] [Google Scholar]
- 46.Lickwar, C. R., et al. 2009. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4:e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindstrom, D. L., et al. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, C. L., S. L. Schreiber, and B. E. Bernstein. 2003. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohr, D. 1984. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 12:8457-8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longtine, M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 51.Malagón, F., and A. Aguilera. 1996. Differential intrachromosomal hyper-recombination phenotype of spt4 and spt6 mutants of S. cerevisiae. Curr. Genet. 30:101-106. [DOI] [PubMed] [Google Scholar]
- 52.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571-574. [DOI] [PubMed] [Google Scholar]
- 52a.Mayer, A., et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272-1279. [DOI] [PubMed] [Google Scholar]
- 53.Moreira, J. M., and S. Holmberg. 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 17:6028-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neigeborn, L., J. L. Celenza, and M. Carlson. 1987. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neigeborn, L., K. Rubin, and M. Carlson. 1986. Suppressors of SNF2 mutations restore invertase derepression and cause temperature-sensitive lethality in yeast. Genetics 112:741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishiwaki, K., T. Sano, and J. Miwa. 1993. emb-5, a gene required for the correct timing of gut precursor cell division during gastrulation in Caenorhabditis elegans, encodes a protein similar to the yeast nuclear protein SPT6. Mol. Gen. Genet. 239:313-322. [DOI] [PubMed] [Google Scholar]
- 57.Nourani, A., F. Robert, and F. Winston. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1496-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen, J. G., M. C. Kielland-Brandt, T. Nilsson-Tillgren, C. Bornaes, and S. Holmberg. 1988. Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics 119:527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pokholok, D. K., et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 60.Prather, D., N. J. Krogan, A. Emili, J. F. Greenblatt, and F. Winston. 2005. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:10122-10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rando, O. J. 2007. Chromatin structure in the genomics era. Trends Genet. 23:67-73. [DOI] [PubMed] [Google Scholar]
- 62.Rose, M., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual, p. 198. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Rufiange, A., P. E. Jacques, W. Bhat, F. Robert, and A. Nourani. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27:393-405. [DOI] [PubMed] [Google Scholar]
- 64.Sabet, N., et al. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. U. S. A. 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 66.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735-748. [DOI] [PubMed] [Google Scholar]
- 68.Serluca, F. C. 2008. Development of the proepicardial organ in the zebrafish. Dev. Biol. 315:18-27. [DOI] [PubMed] [Google Scholar]
- 69.Thiriet, C., and J. J. Hayes. 2006. Histone dynamics during transcription: exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation. Results Probl. Cell Differ. 41:77-90. [DOI] [PubMed] [Google Scholar]
- 70.Thiriet, C., and J. J. Hayes. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winkler, M., T. aus Dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 73.Yoh, S. M., H. Cho, L. Pickle, R. M. Evans, and K. A. Jones. 2007. The Spt6 SH2 domain binds Ser2-P RNAP II to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21:160-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoh, S. M., J. S. Lucas, and K. A. Jones. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22:3422-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Youdell, M. L., et al. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28:4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan, G. C., et al. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed] [Google Scholar]
- 77.Yuasa, T., et al. 2004. An interactive gene network for securin-separase, condensin, cohesin, Dis1/Mtc1 and histones constructed by mass transformation. Genes Cells 9:1069-1082. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, L., A. G. Fletcher, V. Cheung, F. Winston, and L. A. Stargell. 2008. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell. Biol. 28:1393-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.