Abstract
In the budding yeast Saccharomyces cerevisiae, mother cells switch mating types between a and α forms, whereas daughter cells do not. This developmental asymmetry arises because the expression of the HO endonuclease, which initiates the interconversion of a and α mating type cassettes, is extinguished by the daughter-specific Ash1 transcriptional repressor. When daughters become mothers in the subsequent cell cycle, Ash1 must be eliminated to enable a new developmental state. Here, we report that the ubiquitin ligase SCFCdc4 mediates the phosphorylation-dependent elimination of Ash1. The inactivation of SCFCdc4 stabilizes Ash1 in vivo, and consistently, Ash1 binds to and is ubiquitinated by SCFCdc4 in a phosphorylation-dependent manner in vitro. The mutation of a critical in vivo cyclin-dependent kinase (CDK) phosphorylation site (Thr290) on Ash1 reduces its ubiquitination and rate of degradation in vivo and decreases the frequency of mating type switching. Ash1 associates with active Cdc28 kinase in vivo and is targeted to SCFCdc4 in a Cdc28-dependent fashion in vivo and in vitro. Ash1 recognition by Cdc4 appears to be mediated by at least three phosphorylation sites that form two redundant diphosphorylated degrons. The phosphorylation-dependent elimination of Ash1 by the ubiquitin-proteasome system thus underpins developmental asymmetry in budding yeast.
The conversion of one cell type into another, whether the asymmetric division of a stem cell, differentiation of a committed precursor, or transition from one cell cycle phase to the next, requires that the proteins that define the prior state be eliminated or otherwise inactivated. While changes in the factors that determine cell fate alterations are well documented at the transcriptional level, the elimination of critical regulatory proteins that define a prior cell state is less well understood. Because biological networks are often exquisitely sensitive to the abundance of such regulatory factors, their timely elimination may be essential for developmental decisions (37). The best-understood example of the developmental elimination of cell fate determinants occurs in budding yeast, where the degradation of the mating type specificity factor α2 via the E3 ubiquitin ligases Doa10 and Slx5-Slx8 is required for the developmental switch between a and α cell types (38, 58, 81). The ubiquitin-dependent elimination of cell type determinants plays an important role in metazoan development and stem cell fate specification/renewal (8, 26, 75).
The ubiquitin-proteasome system mediates the selective intracellular degradation of proteins in all eukaryotes (30). Substrate proteins are conjugated to ubiquitin through a series of enzymatic steps mediated by E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes; the reiteration of this cascade results in substrate polyubiquitination, which leads to substrate recognition and rapid degradation by the 26S proteasome. The E3 enzymes are the critical factors that confer substrate specificity. Two main classes of E3 exist, as characterized by the presence of either a HECT domain or a RING domain (57). The SCF complexes are the archetypal RING domain E3 enzymes and are composed of the subunits Skp1, Cdc53, Rbx1, and any one of a number of F-box proteins that act as substrate-specific adapters (1, 56, 77). The large number of F-box proteins—21 in budding yeast and over 70 in humans—enables the core SCF complex to target a diverse array of substrates and processes (78). A superfamily of cullin-RING-based ligases (CRLs) based on the SCF architecture targets many additional regulatory factors in development and disease (23, 47, 56, 73). F-box proteins and related CRL adapters contain protein interaction domains that bind substrates, often in a phosphorylation-dependent manner (1, 54, 62). The yeast F-box protein Cdc4 recognizes its substrates via a phosphorylated motif termed the Cdc4 phosphodegron (CPD), which has the optimal consensus sequence Ile/Leu-Ile/Leu/Pro-pSer/pThr-Pro-<Lys/Arg>4, where <> indicates a disfavored basic residue (48, 52). The core pSer/pThr-Pro site in this motif matches the preferred phosphorylation site consensus for the cyclin-dependent kinase (CDK) enzymes that typically target substrates to Cdc4. The archetypal Cdc4 substrate, the CDK inhibitor Sic1, contains multiple CDK sites that bear mismatches to flanking residues in the consensus CPD (48). Biochemical evidence and nuclear magnetic resonance (NMR) evidence suggest that the multiple weak CPDs in Sic1 combine to generate a high-affinity multisite phosphodegron (44, 45, 48, 52). The phosphorylation of a second Ser/Thr residue at the +3 or +4 position can substantially increase the affinity of CPD phosphopeptides, suggesting further complexity in the recognition of multisite phosphorylated substrates (28). The multisite requirement for Sic1 recognition by Cdc4 renders substrate degradation ultrasensitive to CDK activity (21, 48). Other SCF substrates, including Far1, Gcn4, and Cln2 in yeast and cyclin E, Myc, Klf5, and Mdm2 in metazoan cells, may also be recognized in a multisite phosphorylation-dependent manner (4, 13, 29, 33, 39, 43, 46, 74).
Like multicellular organisms, unicellular eukaryotes undergo differentiation into different cell types (31). Haploid budding yeast cells exist as one of two distinct mating types, a and α, as determined by specific gene expression programs (27). When haploid cells of opposite mating types are brought into close proximity, a gametogenesis program is triggered, which enables conjugation and the formation of a diploid cell. In wild-type haploid cells, the cell cycle-dependent expression of the HO endonuclease gene in the G1 phase causes cells to switch from one mating type to the other. Ho-mediated DNA cleavage at the MAT locus initiates a gene conversion event, in which DNA encoding α mating type information is replaced with a information (or vice versa), as provided by silent a or α mating type cassettes (27).
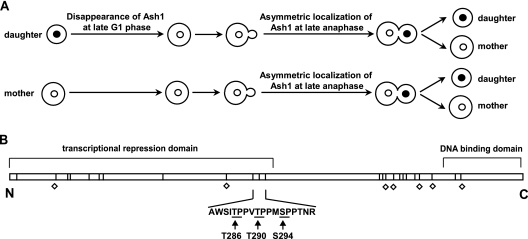
Importantly, the mating type switching program is also controlled in a cell cycle-dependent manner. The mother and daughter cells produced at each cell division are nonequivalent: while approximately 70% of mother cells switch mating type, daughter cells do not switch (64). This differential mating type frequency is imposed by the GATA-1 family Ash1 transcriptional repressor (7, 16, 61), which prevents the expression of the HO gene in daughter cells (Fig. 1A). Ash1 is recruited to the HO promoter in early G1 phase shortly after the Swi5 transcriptional activator (17, 41). ASH1 transcription and protein abundance are strongly cell cycle periodic, with a peak in late mitosis/early G1 phase (7, 18). At the onset of anaphase, ASH1 is transcribed in both mother and daughter nuclei and then asymmetrically distributed to the daughter cell by several mechanisms. ASH1 mRNA from the mother nucleus is transported and anchored to the bud tip of the daughter cell (5, 40, 66) in a manner dependent on cis-acting RNA sequences, the actin cytoskeleton, and the SHE genes (16, 53). The inhibition of ASH1 mRNA translation in the mother cell also contributes to the asymmetric distribution of Ash1 (53). However, the process whereby Ash1 is eliminated from daughter cells as they become mothers in the subsequent cell cycle is not well understood. The CDK Pho85 has been implicated in Ash1 degradation (42), but the mechanism of Ash1 elimination remains unknown. Here, we show that the Cdc28 kinase, which is the predominant cell cycle CDK, targets Ash1 to the SCFCdc4 ubiquitin ligase to enforce the developmental asymmetry between mother and daughter cells.
FIG. 1.
(A) Regulation of Ash1 during the cell cycle. In early G1 phase, daughter cells contain Ash1 in the nucleus (filled circles), whereas mother cells usually do not (open circles). Ash1 in the daughter cell disappears at late G1 phase. At late anaphase, Ash1 is expressed and preferentially localized to the daughter nucleus. (B) Schematic of domains and phosphorylation site motifs in Ash1 (588 residues). Twenty-four minimal Ser/Thr-Pro CDK sites are represented by vertical lines. Perfect consensus phosphorylation sites for Cdc28 (Thr/Ser-Pro-X-Arg/Lys) are indicated by open diamonds. The sequence of a critical Cdc4-targeting region containing residues Thr286, Thr290, and Ser294 is shown.
MATERIALS AND METHODS
Yeast strain construction and culture.
The strains and plasmids used in this study are listed in Tables 1 and 2. Yeast strains were constructed and cultured according to standard methods (69). All strains were derived from W303. Mutant ASH1 alleles were generated by QuikChange site-directed mutagenesis (Stratagene) and confirmed by sequencing. Details of oligonucleotides used for plasmid constructions are available upon request. Pedigree analysis for mating type switching frequency was performed as described previously (7). Chitin-rich bud scars were stained with 100 ng/ml calcofluor white and imaged by UV fluorescence. Green fluorescent protein (GFP) fluorescence was imaged by using the Piston GFP filter set (Nikon). Images were captured on an ORCA-II charge-coupled-device (CCD) camera (Hamamatsu) and processed with MetaMorph software or captured and processed with an Opera HCS confocal imaging system (Perkin-Elmer).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| MT235 | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL1 psi+ KN699 | K. Nasmyth |
| MT668 | MATacdc4-1 | This study |
| MT2079 | MATa/MATα ASH1 HO (KN4709) | K. Nasmyth |
| MT2189 | MATa/MATα cdc4-1 HO | This study |
| MT2192 | MATacdc28-as1 (JAU05) | D. Morgan |
| MT2195 | MATa/MATα ash1::ASH1T290A HO | This study |
| MT2203 | MATaash1::kanMX6-GAL1-ASH1MYC cdc4-1 | This study |
| MT2205 | MATaash1::kanMX6-GAL1-ASH1MYC | This study |
| MT2223 | MATaash1::ASH1MYC | This study |
| MT2224 | MATaash1::ASH1MYC cdc4-1 | This study |
| MT2459 | MATaash1::kanMX6-GAL1-ASH1MYC GAL1-HA-UB | This study |
| MT2526 | MATaash1::ASH1-GFP-kanMX6 | This study |
| MT2527 | MATaash1::ASH1T290A-GFP-kanMX6 | This study |
| MT2685 | MATaash1::kanMX6-GAL1-ASH1MYC pho85F82G | This study |
| MT2772 | MATaash1::ASH1MYC cdc15-2 | This study |
| MT2905 | MATacdc15-2 ash1::ASH1MYC cdc28-as1 | This study |
| MT2908 | MATaash1::kanMX6-GAL1-ASH1MYC cdc28-as1 | This study |
| MT3008 | MATaash1::ASH1-GFP-kanMX6 cdc4-1 | This study |
| MT3059 | MATaash1::ASH1MYCT290A cdc15-2 | This study |
| MT3169 | MATacdc28-as1 cdc4-1 | This study |
| MT4256 | MATa/MATα ash1::ASH1-GFP-kanMX6 HO | This study |
| MT4257 | MATa/MATα ash1::ASH1T290A-GFP-kanMX6 HO | This study |
| MT4258 | MATaGAL1-HA-UB | This study |
| MT4259 | MATaash1::kanMX6-GAL1-ASH1MYCT290A GAL1-HA-UB | This study |
| MT4208 | MATaash1::ASH1-GFP-kanMX6 pho85::PHO85F82G | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic | Source |
|---|---|---|
| pMT962 | pGEX 4T1 | Pharmacia |
| pMT2005 | pRS426 GAL1 URA3 2μm | ATCC 87333 |
| pMT2289 | pGAL1-ASH1 TRP1 2μm | This study |
| pMT3275 | pGEX-ASH1 | This study |
| pMT3276 | pGAL1-ASH1FLAG URA3 2μm | This study |
| pMT4217 | pGAL1-ASH1HA URA3 2μm | This study |
| pMT4326 | pGAL1-ASH1HAT286A URA3 2μm | This study |
| pMT4327 | pGAL1-ASH1HAT290A URA3 2μm | This study |
| pMT4328 | pGAL1-ASH1HAS294A URA3 2μm | This study |
| pMT4329 | pGEX-ASH1T290A | This study |
| pMT4498 | pGAL1-ASH1HAT286A/S294A URA3 2μm | This study |
| pMT4499 | pGAL1-ASH1HAT286A/T290A/S294A URA3 2μm | This study |
Immunoblot and immunoprecipitation assays.
Cultures were grown to early log phase in 2% raffinose medium, arrested as indicated, induced with 2% galactose for 1 h, and then washed and cultured in medium containing 2% glucose and 1 mg/ml cycloheximide. For temperature-sensitive strains, cultures were immediately transferred into a 37°C water bath shaker after the addition of galactose. Samples were processed for immunoblot analysis as described previously (71), using either antihemagglutinin (anti-HA) or anti-MYC monoclonal antibody; equal protein loading was confirmed by Ponceau S staining and/or anti-Pgk1 immunoblotting. Signal intensities were quantified by using Quantity One software (Bio-Rad). Covalent ubiquitin conjugates were detected by the reimmunoprecipitation of denatured immunopurified complexes (71). Immunoprecipitation-kinase assays were performed as described previously (71), in the presence or absence of 1NMPP1.
Ubiquitination assays.
E1, Cdc34, SCFCdc4, and Cln2-Cdc28-Cks1 were produced as described previously (68). Ash1FLAG was purified from a wild-type strain bearing a GAL1-ASH1FLAG plasmid (pMT3276) after a 1-h induction in 2% galactose medium. Cells were disrupted by glass bead lysis in a solution containing 50 mM Tris HCl (pH 7.5), 150 mM NaCl, 50 mM NaF, 5 mM EDTA, and 0.1% NP-40 containing a phosphatase inhibitor cocktail (10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, and 0.1 mM sodium orthovanadate) and protease inhibitors. Ash1FLAG was captured on anti-FLAG M2 agarose resin (Sigma) and eluted with Tris-buffered saline (TBS) buffer containing 100 μg/ml FLAG peptide and protease inhibitors. Ash1FLAG-HA was similarly purified from a cdc28-as1 strain bearing a GAL1-ASH1FLAG-HA plasmid (pMT4217) grown in the presence of 5 μM 1NMPP1 for 30 min prior to galactose induction; buffers used during purification contained 2.5 μM 1NMPP1 and no phosphatase inhibitors. For Ash1FLAG ubiquitination, 6 μl of Ash1FLAG was incubated at 30°C for 1 h with 10 μl ubiquitination mix (0.2 μg E1, 0.4 μg Cdc34, 0.2 μg SCFCdc4, 1 μg ubiquitin, and 1.6 μl 10 × ubiquitination buffer [250 mM Tris HCl {pH 7.5}, 100 mM MgCl2, 20 mM ATP, and 0.5 mM dithiothreitol {DTT}]). Ash1FLAG-HA was phosphorylated in vitro by a Cln2-Cdc28-Cks1 kinase complex prior to incubation with ubiquitination reagents and detection of reaction products by anti-FLAG immunoblotting.
Mass spectrometry.
Ash1FLAG was purified from a 2-liter culture using anti-FLAG agarose resin as described above. Purified protein was eluted from the resin at pH 2, concentrated on an SCX column (PolyLC), reduced with DTT, alkylated with iodoacetamide, digested with trypsin, and eluted for liquid chromatography (LC)-mass spectrometry (MS) analysis. Samples were analyzed with a Thermo FT-LTQ instrument using a 75-μm by 10-cm C18 column and an Agilent 1100 CapLC chromatography system. Data were acquired with 3 data-dependent tandem MS (MS/MS) scans or with 1 data-dependent MS/MS scan and 3 MS/MS/MS scans and analyzed with Mascot and X! Tandem search engines. All phosphorylation sites detected were manually verified. MS/MS/MS spectra were also verified manually and used to validate site assignments of triply phosphorylated peptide species.
Time-lapse microscopy and microfluidics.
Exponentially growing cells were concentrated in fresh synthetic complete (SC) glucose medium to an optical density at 600 nm (OD600) of 1.5 and mixed at a ratio of 1:1 with a 3% gel of low-melting agarose in SC glucose medium at 30°C. The suspension was vortexed to homogeneity and transferred into a microfluidic device (20, 70), which was then cooled to 4°C for 3 min in order to immobilize the suspension. Individual cell imaging chambers were in diffusive contact with perfusion channels to allow for constant growth conditions throughout the experiment. Cells were perfused for 10 h with SC glucose medium, and bright-field and fluoroscent images of ash1::ASH1-GFP and ash1::ASH1T290A-GFP strains were acquired every 14 min on a Leica DMIRE2 inverted fluorescent microscope using a 63× oil immersion objective and an ORCA-ER CCD camera (Hamamatsu). GFP fluorescence was collected by using an EL6000 external light source, a Leica L5 filter cube, and a 1.5-s exposure time. Parallel bright-field images were acquired to ascertain bud emergence and cell cycle position. The automated operation of the chip and the microscope was controlled through LabVIEW (National Instruments Corporation). A custom MATLAB (Mathworks) image analysis pipeline was used to perform cell segmentation and tracking on bright-field images and to extract the GFP signal from the nucleus (20). Daughter cells were recognized by the segmentation algorithm as separate from the mother once a new bud appeared. Segmentation and bud emergence were manually verified before GFP intensity was calculated. In each cell, pixels in the nucleus above a threshold were identified using Otsu's method for thresholding as implemented in MATLAB. The relative Ash1-GFP intensity was calculated by subtracting the average intensity of nonnuclear pixels from that of nuclear pixels. MATLAB was used to construct image videos.
Fluorescence polarization.
Peptides were labeled at the N terminus with fluorescein with a free carboxylate at the C terminus and suspended in a solution containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM DTT, and 0.1 mg/ml bovine serum albumin (BSA) at a concentration of 5 to 10 nM. Skp1-Cdc4263-744 was purified as described previously (52) and titrated into a peptide solution, and fluorescence polarization was measured at 22°C with an Analyst HT instrument (Molecular Devices, CA). Data were analyzed by using GraphPad Prism 4 software.
RESULTS
Ash1 elimination depends on Cdc4.
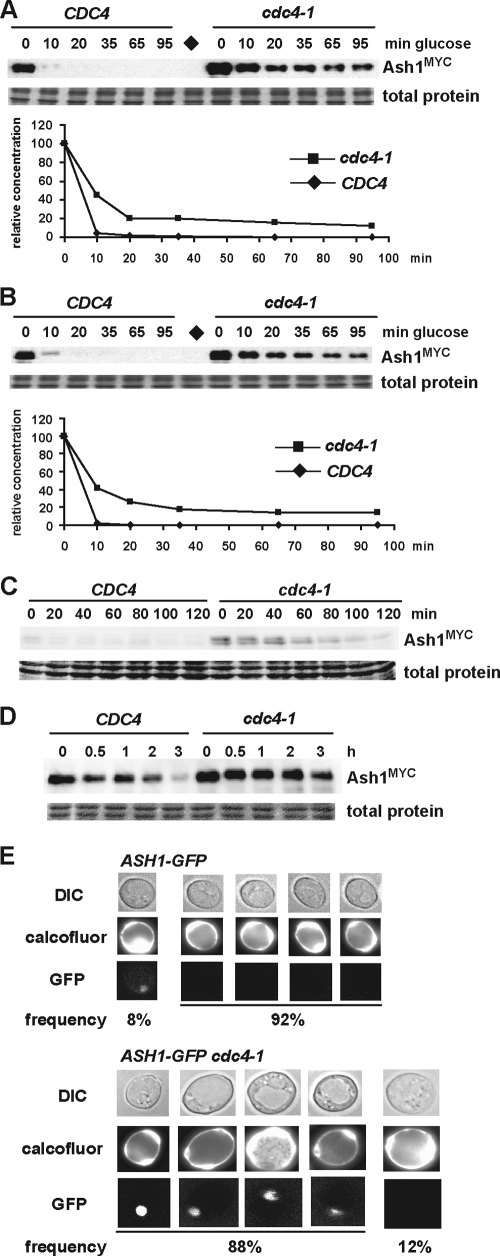
Ash1 contains 24 Ser/Thr-Pro minimal CDK sites that could form potential CPD motifs (Fig. 1B). This high density of potential CPD motifs in Ash1 prompted us to test whether Ash1 might be degraded in a Cdc4-dependent manner. We transiently expressed a MYC epitope-tagged allele of ASH1 from the GAL1 promoter followed by rapid repression to monitor Ash1 protein stability; this approach eliminated the confounding variable of cell cycle-regulated ASH1 expression (7, 61). In order to examine Ash1 at the cell cycle phase when it is likely to be unstable (7, 61), ash1::GAL1-ASH1MYC and ash1::GAL1-ASH1MYC cdc4-1 strains were arrested in mitosis with nocodazole, shifted into galactose medium at 37°C for 1 h, and then repressed for Ash1 production by transfer into glucose medium containing cycloheximide. In contrast to the virtually complete elimination of Ash1 after 10 min in a wild-type strain, Ash1 was markedly stabilized in the cdc4-1 strain and persisted for the duration of the 90-min time course (Fig. 2A). The inactivation of the core SCF subunit Skp1 or the cognate E2 enzyme for SCFCdc4, Cdc34, also stabilized Ash1 (data not shown). To examine the potential cell cycle position effects on Ash1 stability, we arrested cells in late G1 phase with the mating pheromone α-factor and again found that Ash1 was stabilized in the absence of Cdc4 function (Fig. 2B). Although limited by the resolution of the time points taken, over several independent experiments we estimated an Ash1 half-life of <3 min for the wild-type strain, versus >10 min for the cdc4-1 strain. In wild-type cells, Ash1 is thus an exceedingly unstable protein.
FIG. 2.
Inactivation of Cdc4 stabilizes Ash1. (A) ash1::GAL1-ASH1MYC and ash1::GAL1-ASH1MYC cdc4-1 strains were arrested with nocodazole, transferred into galactose medium at 37°C for 1 h, and then repressed for GAL1-ASH1MYC expression (t = 0 min). Ash1MYC was detected by anti-MYC immunoblotting and quantified. The diamond indicates a negative-control strain that lacks Ash1MYC. (B) An experiment similar to that in A was performed with α-factor-arrested G1-phase cells. (C) ash1::ASH1MYC and ash1::ASH1MYC cdc4-1 strains were synchronized in S phase by hydroxyurea, washed and resuspended at 37°C in medium containing nocodazole, and analyzed at the indicated time points by anti-MYC immunoblotting. (D) Early-G1-phase daughter cells from ash1::ASH1MYC and ash1::ASH1MYC cdc4-1 strains were isolated by centrifugal elutriation, resuspended in 37°C medium containing α-factor (t = 0 min), and analyzed for Ash1MYC at the time points indicated. (E) ash1::ASH1-GFP and ash1::ASH1-GFP cdc4-1 strains were cultured at the semipermissive temperature of 30°C, and GFP signals were counted for 100 mother cells from each strain, as identified by calcofluor white staining for bud scars. Five representative mother cells from each strain are shown; the frequency of each Ash1-GFP signal is shown below. DIC, differential interference contrast.
To rule out possible effects caused by the transient expression of Ash1 from the GAL1 promoter, we examined the behavior of Ash1 expressed from its endogenous chromosomal locus. Consistent with the above-described half-life effects, endogenous Ash1MYC accumulated to higher levels in a cdc4-1 strain and persisted in a nocodazole block at the restrictive temperature (Fig. 2C), even though ASH1 is only weakly expressed at this cell cycle position (7, 61). We then addressed whether the turnover of endogenous Ash1 in daughter cells requires Cdc4 function. Early-G1-phase daughter cells were isolated by centrifugal elutriation from wild-type and cdc4-1 strains bearing an integrated ash1::ASH1MYC allele. In order to rule out effects of cell cycle positions between wild-type and cdc4-1 strains, elutriated G1-phase fractions were incubated at 37°C in medium containing α-factor to arrest cells in late G1 phase, followed by analysis of the Ash1MYC abundance. The inactivation of Cdc4 attenuated the gradual loss of Ash1 in daughter cells (Fig. 2D).
To determine whether endogenous Ash1 accumulated in mother cells when Cdc4 was inactivated, we examined the levels of Ash1-GFP expressed from the endogenous ASH1 locus in wild-type and cdc4-1 strains. Calcofluor white, which stains bud scars, was used to identify mother cells in random fields of asynchronous cells cultured at 30°C, the semipermissive temperature for the cdc4-1 allele. In wild-type cells, only 8% of mother cells had a visible Ash1-GFP signal in their nuclei, whereas in the cdc4-1 strain, 88% of mother cells exhibited a nuclear Ash1-GFP signal (P < 0.0001, chi-square test) (Fig. 2E). We conclude that Ash1 degradation is dependent on Cdc4 in various cycle stages and that the inactivation of Cdc4 causes an accumulation of endogenous Ash1 in both mother and daughter cells.
Ubiquitination of Ash1 by SCFCdc4 in vitro.
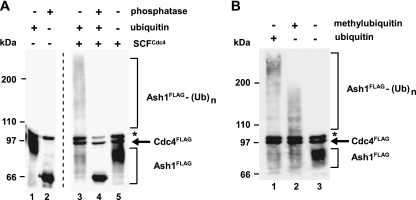
To corroborate the dependence of Ash1 degradation on Cdc4 in vivo, we assessed whether Ash1 could be ubiquitinated by recombinant SCFCdc4 in an in vitro assay (Fig. 3A). Ash1FLAG was purified from wild-type yeast cells on anti-FLAG antibody resin, eluted using FLAG peptide, and then incubated with recombinant E1, E2, ubiquitin, and SCFCdc4 in solution. SCFCdc4 efficiently converted the purified Ash1 into high-Mr species, consistent with the covalent attachment of polyubiquitin chains (Fig. 3A). This conversion was dependent on the phosphorylation of Ash1 by one or more kinases in vivo, since the treatment of Ash1 with lambda phosphatase both increased Ash1 mobility and prevented Ash1 ubiquitination (Fig. 3A, lanes 2 and 4). The substitution of the chain-terminating derivative methylubiquitin for ubiquitin in the reaction mix caused a marked decrease in the Mr of Ash1 species, demonstrating that Ash1 was indeed polyubiquitinated by SCFCdc4 (Fig. 3B). These results indicate that Ash1 isolated from yeast is sufficiently phosphorylated to allow its recognition and ubiquitination by SCFCdc4.
FIG. 3.
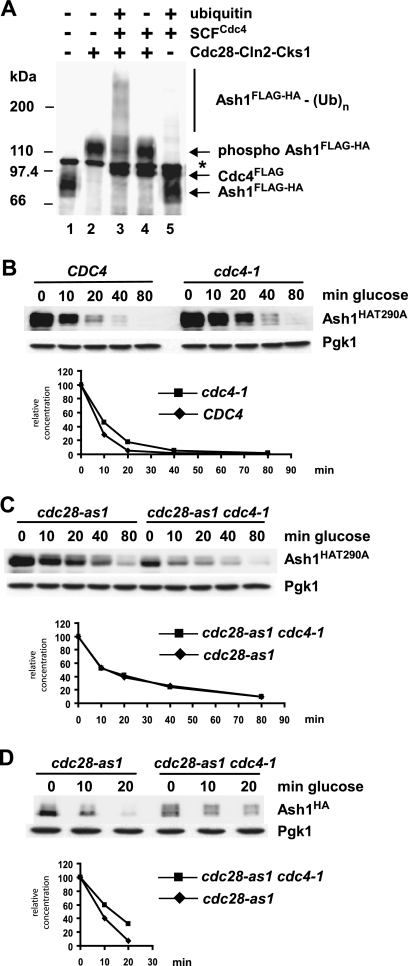
Ash1 is ubiquitinated by SCFCdc4 in vitro. (A) Ash1FLAG purified from yeast was incubated with E1, E2, SCFCdc4, and ubiquitin, as indicated, and reaction products were analyzed by anti-FLAG immunoblotting. In lane 2, Ash1FLAG was treated with lambda phosphatase prior to the ubiquitination reaction. Recombinant Cdc4FLAG was also detected on the immunoblot. Asterisks indicate a nonspecific band. An irrelevant lane between lanes 2 and 3 was cropped from the final image. (B) Same reactions as in panel A in the presence of either ubiquitin (lane 1), methylubiquitin (lane 2), or neither (lane 3). (Ub)n, polyubiquitin conjugates.
Ash1 phosphorylation and ubiquitination in vivo.
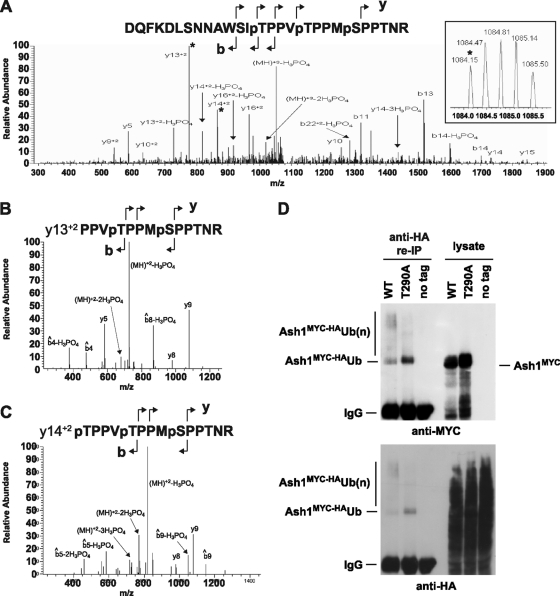
We used mass spectrometry to identify candidate phosphorylation sites on Ash1 that might mediate its recognition and ubiquitination by Cdc4. Ash1FLAG was purified from wild-type cells as described above, digested with trypsin, and analyzed by LC-MS/MS. Thr286, Thr290, and Ser294, each of which is followed by a Pro residue, were identified as in vivo-phosphorylated residues on Ash1 (Fig. 4A, B, and C). Data-dependent LC-MS revealed that the single tryptic peptide that encompasses residues Thr286, Thr290, and Ser294 occurred as unphosphorylated, singly phosphorylated, doubly phosphorylated, and triply phosphorylated species. Analysis of the spectrum corresponding to the singly modified peptide confirmed that Thr290 was phosphorylated, while residues Thr290 and Thr286 were phosphorylated in the doubly modified form (Fig. 4B). We also detected a peptide that corresponded to the correct mass and relative elution time for a triply phosphorylated species. An LC-MS experiment that targeted this peptide exclusively generated MS/MS and MS/MS/MS data that supported the phosphorylation of residues Thr286, Thr290, and Ser294 of Ash1 in vivo (Fig. 4C).
FIG. 4.
In vivo phosphorylation and ubiquitination of Ash1. (A) MS/MS spectrum from the triply phosphorylated peptide D273-R299 (DQFKDLSNNAWSITPPVTPPMSPPTNR). The inset shows the precursor mass from the MS spectrum. The transition ions Y132+ and Y142+, which correspond to the peaks fragmented in panels B and C, are marked with asterisks. (B) MS/MS/MS from the Y132+ fragment (PPVTPPMSPPTNR) shows neutral losses of only 2 phosphate groups, with similar fragmentation patterns observed for both MS/MS/MS spectra. (C) MS/MS/MS from the Y142+ fragment (TPPVTPPMSPPTNR) shows neutral loss of all 3 phosphate groups. (D) Immunopurified anti-MYC complexes from strains bearing an integrated HA-tagged allele of ubiquitin alone or in combination with either wild-type (WT) ash1::GAL1-ASH1MYC or ash1::GAL1-ASH1MYCT290A were denatured by boiling in SDS-PAGE sample buffer, diluted in nondenaturing buffer, recaptured on anti-HA resin, and then probed with either anti-MYC or anti-HA antibody as indicated. Input lysates were probed in parallel.
Low-mobility polyubiquitinated species of wild-type Ash1 could be detected in vivo, as shown by the coprecipitation of endogenous levels of Ash1MYC with an HA-tagged version of ubiquitin under harsh denaturing conditions (Fig. 4D). In contrast, a version of Ash1MYC bearing a Thr290Ala phosphorylation site mutation exhibited a lower degree of polyubiquitination (Fig. 4D). Notably, the Ash1MYCT290A mutant appeared to be preferentially modified by a single ubiquitin moiety, based on the small mobility shift of the ubiquitinated form compared to unmodified Ash1. These results demonstrate that an in vivo phosphorylation site on Ash1 is necessary for its efficient polyubiquitination.
Phospho-Thr290 is necessary for efficient Ash1 degradation in vivo.
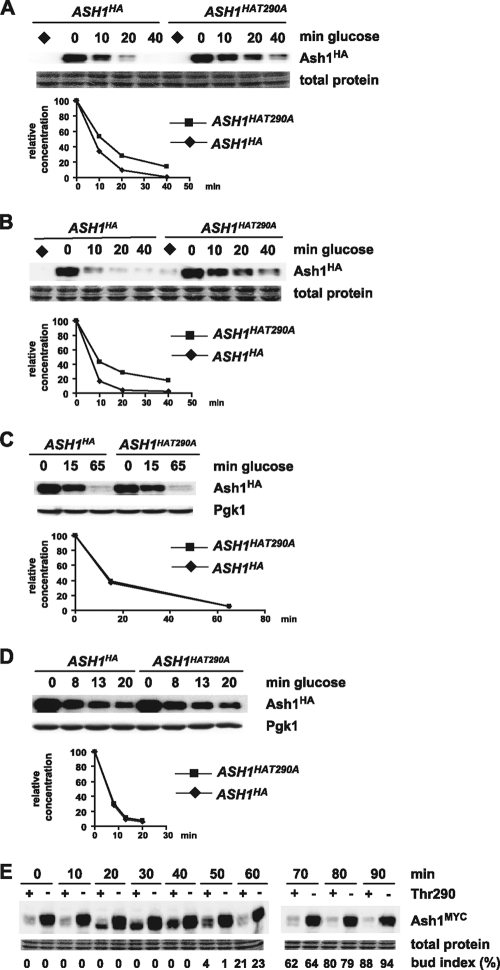
To test the potential roles of each of the above-described phosphorylated residues in Ash1 degradation, we individually mutated Thr286, Thr290, and Ser294 and assessed the stability of each residue in a GAL1 promoter induction/repression assay. Consistent with its defect in polyubiquitination in vivo, Ash1T290A was substantially more stable than wild-type Ash1 in a nocodazole-arrested wild-type strain (Fig. 5A). In contrast, Ash1T286A and Ash1S294A were not overtly stabilized (but see below). Ash1T290A was also stabilized in an α-factor-arrested wild-type strain (Fig. 5B), suggesting that Thr290 phosphorylation is important for Ash1 degradation in different cell cycle stages. To test whether Thr290 phosphorylation acts in the same pathway as Cdc4, cdc4-1 strains bearing GAL1-ASH1HA or GAL1-ASH1HAT290A were arrested with either nocodazole or α-factor, and Ash1 stability was assessed at 37°C. As expected, the cdc4-1 defect caused a stabilization of wild-type Ash1; notably, however, the Thr290Ala mutation did not further stabilize Ash1 when Cdc4 was inactivated (Fig. 5C and D). Additional experiments with a minimal fragment of Ash1 that exhibits Cdc4-dependent degradation (residues 135 to 300) confirmed that the phosphorylation of Thr290 lies in the same Ash1 degradation pathway as Cdc4 (data not shown).
FIG. 5.
Thr290 is required for Ash1 instability in vivo. (A) Wild-type strains transformed with either GAL1-ASH1HA or GAL1-ASH1HAT290A plasmids were arrested by nocodazole and subjected to promoter induction/repression for the indicated times, and Ash1HA or Ash1HAT290A was detected by anti-HA immunoblotting and quantified. Diamonds indicate negative-control samples not induced with galactose. (B) The same experiment as in panel A was performed with α-factor-arrested strains. (C) The same experiment as in panel A was performed, except with nocodazole-arrested cdc4-1 strains and with induction/repression at 37°C. (D) The same experiment as in C was performed with α-factor-arrested strains. (E) ash1::ASH1MYC cdc15-2 and ash1::ASH1MYCT290A cdc15-2 were arrested at 37°C (t = 0) and released at 23°C for the indicated times, and Ash1MYC or Ash1MYCT290A was detected by anti-MYC immunoblotting. Ash1MYC is displayed in “+” lanes, and Ash1MYCT290A is displayed in “−” lanes. The budding index at each time point is indicated.
We then determined the effect of the Thr290 mutation on endogenous levels of Ash1 expressed from its chromosomal locus. Conditional cdc15-2 strains bearing either an ash1::ASH1MYC or an ash1::ASH1MYCT290A allele were arrested in late anaphase at 37°C and released to 23°C to initiate synchronous cell cycle progression, and the Ash1 protein level and bud index were assessed at various times after release (Fig. 5E). As expected, wild-type Ash1 protein levels were low in the late mitotic block (0 min), peaked in G1 phase (40 min), and then rapidly declined as cells budded and entered a new cell cycle (60 min). In marked contrast, Ash1T290A protein levels were elevated at all time points following release from the cdc15-2 arrest (Fig. 5E, lanes with a minus sign), consistent with a role for phosphorylation in the elimination of endogenous levels of Ash1.
Phospho-Thr290 is needed for the elimination of Ash1 in mother cells and efficient mating type switching.
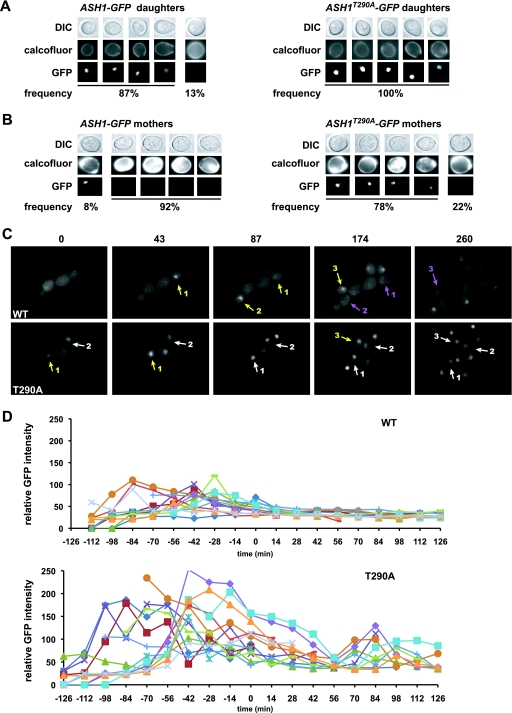
To ascertain the role of Thr290 on Ash1 at the single-cell level, we examined Ash1-GFP in daughter or mother cells in ash1::ASH1-GFP and ash1::ASH1T290A-GFP strains, using calcofluor white staining to distinguish mothers and daughters. As expected, an Ash1-GFP signal was present in most daughter cell nuclei; correspondingly, Ash1T290A-GFP was also present in all daughter cell nuclei, although the signal was noticeably more intense (Fig. 6A). Strikingly, and in contrast to wild-type Ash1-GFP, Ash1T290A-GFP was present in the vast majority of mother cells (Fig. 6B).
FIG. 6.
Thr290 is required for cell cycle regulation of Ash1. (A) ash1::ASH1-GFP and ash1::ASH1T290A-GFP strains were grown to log phase, and 100 daughter cells from each strain were identified by staining with calcofluor white and assessed for Ash1-GFP signals. Five representative daughter cells from each strain are shown; the frequency of occurrence is indicated below. (B) An experiment similar to that in A was conducted with mother cells. (C) Real-time analysis of ash1::ASH1-GFP and ash1::ASH1T290A-GFP strains in a microfluidic device. Yellow arrows indicate the GFP signal that appears in a daughter cell, purple arrows indicate the GFP signal that is lost in daughter cells as they bud to become new mother cells, and white arrows indicate GFP signals that persist in mother cells. Numbers indicate the same cell across a series of frames. For clarity, not all GFP-positive cells are labeled. Due to random nuclear motion relative to the focal plane variation, some cells with a persistent Ash1T290A-GFP signal displayed a rapid variation in intensity over the experimental time course. Panels are taken from GFP movies shown in Videos S1 and S2 in the supplemental material. (D) Quantitative analysis of Ash1-GFP and Ash1T290A-GFP intensities in the nuclei of 13 individual single cells of each strain plotted as a function of cell cycle position, as determined by the time from bud emergence.
We used a microfluidic device to monitor the kinetics of Ash1 elimination in live cells for ash1::ASH1-GFP and ash1::ASH1T290A-GFP strains. In the wild-type strain, Ash1-GFP accumulated in the incipient daughter cell after the completion of mitosis and was eliminated in almost all new mother cells shortly after bud emergence (Fig. 6C, top, and see Video S1 in the supplemental material for the full movie). In contrast, Ash1T290A-GFP persisted in the mother cell, often through several divisions (Fig. 6C, bottom, and see Video S2 in the supplemental material for the full movie). Quantification of the GFP signals along individual cell trajectories revealed that Ash1T290A-GFP was on average at least 2-fold more intense than the wild type. Moreover, Ash1T290A-GFP persisted across the entire cell cycle, whereas the wild-type protein was eliminated in daughter cells shortly after bud emergence (Fig. 6D). The decrease in levels of Ash1T290A-GFP after bud emergence and the buildup of Ash1T290A-GFP during G1 phase parallel the known cell cycle-regulated transcription of ASH1 (7, 61). These results demonstrate that the phosphorylation of Thr290 is necessary for the elimination of Ash1 in new mother cells shortly after the G1/S-phase transition.
To determine the consequences of Ash1 stabilization on mating type switching frequency, we performed a pedigree analysis using ASH1 HO, ash1::ASH1T290A HO, and cdc4-1 HO strains. For each strain, 100 mother-daughter cell pairs were microdissected and assessed for mating type by placing them next to a barrier wall of α cells; MATa cells respond morphologically to the secreted α-factor, whereas MATα cells do not (7, 61). As expected, daughter cells did not switch mating type, regardless of the genotype. In the wild-type ASH1 HO strain, 72% of mother cells switched mating type, consistent with previous results (7, 61). In contrast, in the ash1::ASH1T290A HO or cdc4-1 HO strain, the switching rates of mother cells decreased to 51% and 50%, respectively (P < 0. 002, chi-square test). The incomplete suppression of mating type switching by the Thr290Ala mutation was less marked than expected from the persistence of Ash1T290A-GFP evident in most mother cells. The GFP tag itself appeared to slightly weaken the repressive activity of Ash1, as 80% of mother cells switched mating type in an ash1::ASH1-GFP HO strain and 60% switched in an ash1::ASH1T290A-GFP HO strain; consistently, large epitope tags can interfere with Ash1 function in mother cells (7). As a strong overexpression of ASH1 is sufficient to eliminate all switching in mother cells (7, 61), it is possible that Ash1T290A simply does not accumulate to a sufficient level to repress HO in all mother cells. Alternatively, as ubiquitination can attenuate the activity of other transcription factors (22, 76), the monoubiquitinated form of Ash1T290A that appears to accumulate in vivo may be compromised for HO repression. Despite these mitigating effects, the stabilization of Ash1 by the elimination of Thr290 phosphorylation still significantly impairs the rate of mating type switching in mother cells, as predicted.
CDK-mediated targeting of Ash1 to Cdc4.
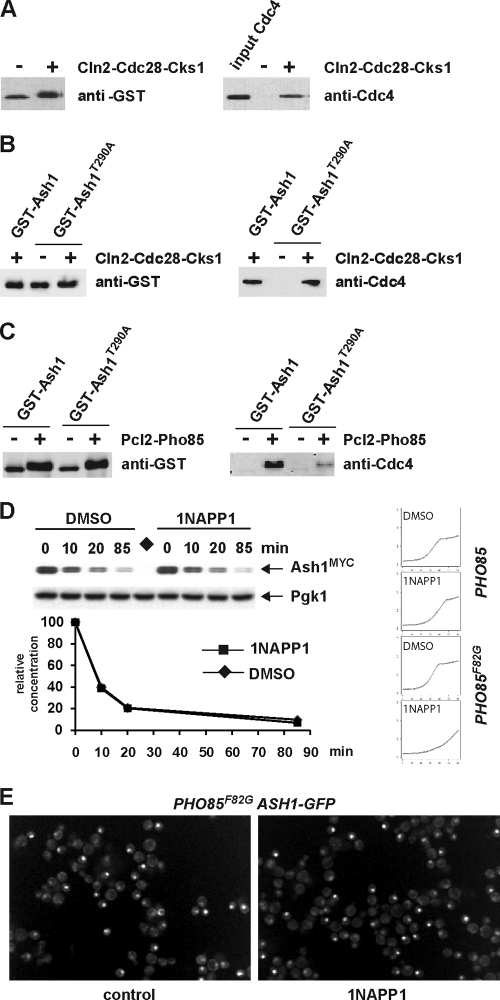
We next sought to identify the kinase(s) that might target Ash1 to Cdc4. Based on known Cdc4 substrates (78), the proline-directed kinases Cdc28 and Pho85 were likely candidates, especially as Pho85 was previously implicated in Ash1 degradation (42). To examine the phosphorylation dependence of the Ash1-Cdc4 interaction in vitro, we monitored the capture of Cdc4 from solution onto glutathione resin loaded with a full-length glutathione S-transferase (GST)-Ash1 fusion protein produced in bacteria that was either unphosphorylated or phosphorylated with either recombinant Cln2-Cdc28-Cks1 or Pcl2-Pho85 kinase complexes. The phosphorylation of GST-Ash1 by Cln2-Cdc28-Cks1 enabled the efficient capture of Cdc4 onto the resin (Fig. 7A), although the interaction was largely independent of Thr290 in this context (Fig. 7B). In the same format, Pcl2-Pho85 was also able to drive the capture of Cdc4, but in this case, the interaction was strongly dependent on Thr290 (Fig. 7C). These results indicate that both Cdc28 and Pho85 are capable of targeting Ash1 to SCFCdc4 in vitro but that Cdc28 can apparently bypass the requirement for Thr290, at least with the high concentrations achieved in an in vitro capture assay.
FIG. 7.
Phosphorylation-dependent interactions of Ash1 with Cdc4 in vitro. (A) Full-length recombinant Ash1 was purified from bacteria as a GST fusion protein on glutathione resin, incubated with or without the recombinant Cln2-Cdc28-Cks1 kinase complex, washed, and then incubated with recombinant SCFCdc4 and rewashed. SCFCdc4 captured on GST-Ash1 resin was detected by anti-Cdc4 immunoblotting; input Cdc4 is also shown (right). Input GST-Ash1 was detected with anti-GST antibody (left). (B) The same experiment as that in A was performed with full-length recombinant Ash1T290A, also purified from bacteria. Full-length GST-Ash1 was included in the assay as a positive control. (C) Experiments similar to those in A and B were performed except with GST-Ash1 phosphorylated by the Pcl2-Pho85 kinase complex. GST-Ash1 and GST-Ash1T290A were both treated with or without Pcl2-Pho85 and analyzed by anti-GST immunoblotting (left). SCFCdc4 captured by either GST-Ash1 or GST-Ash1T290A resin was detected by anti-Cdc4 immunoblotting (right). (D) An ash1::GAL1-ASH1MYC pho85::PHO85F82G strain was arrested in late G1 phase with α-factor and then incubated with 10 μM 1NAPP1 or dimethyl sulfoxide (DMSO) control solvent for 20 min, followed by GAL1-ASH1MYC induction/repression and analysis by anti-MYC immunoblotting. Growth curves in the presence and absence of 5 μM 1NAPP1 (left) indicated that the long-term inhibition of Pho85F82G impairs cell growth, as expected (9). (E) An ash1::ASH1-GFP PHO85::pho85F82G strain was incubated for 60 min in the absence or presence of 5 μM 1NAPP1, followed by image acquisition. The fractions of cells that exhibited a nuclear Ash1-GFP signal were 29% (294/1,009 counted) in the control culture and 28% (307/1,093 counted) in the 1NAPP1-treated culture.
Our in vitro binding data suggested that the phosphorylation of Ash1 by Pho85 might stimulate Ash1 recognition by Cdc4, as was suggested previously (42). However, previous studies with a pho85Δ strain revealed only long-term effects on Ash1 stability, which are confounded by the severe growth defect in pho85Δ strains (42). We thus reexamined the role of Pho85 using a PHO85F82G allele that is specifically susceptible to rapid and reversible chemical inactivation by the 1NAPP1 inhibitor (9). The acute inhibition of Pho85F82G had little effect on Ash1 stability in a GAL1-ASH1MYC repression assay (Fig. 7D), unlike the pronounced effect of the Thr290Ala mutation. We also examined the consequences of transient Pho85F82G inactivation on Ash1-GFP abundance in single cells and again observed no obvious accumulation of Ash1 (Fig. 7E). We conclude from this analysis that Pho85 does not overtly contribute to Ash1 degradation in vivo.
Cdc28 targets Ash1 to Cdc4 in vivo.
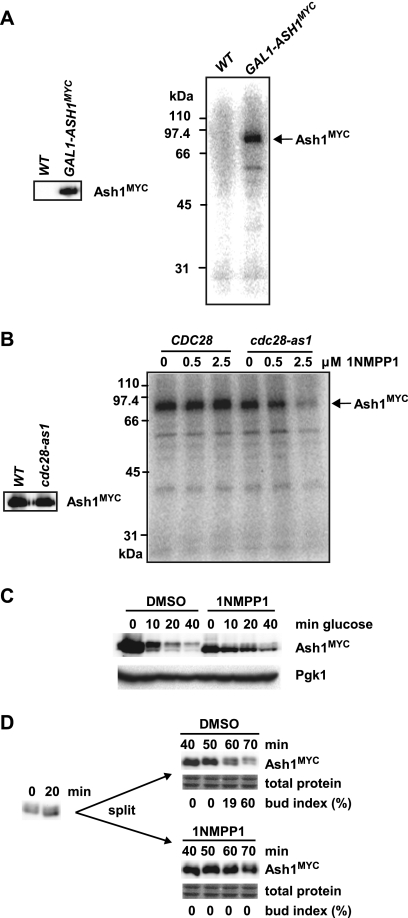
To investigate the potential role of Cdc28 in targeting Ash1 in vivo, we first examined whether Ash1 isolated from cell lysates was associated with a kinase activity; this sensitive method can often detect weak substrate-kinase interactions (71). Ash1MYC was immunopurified from either control or GAL1-ASH1MYC cultures and incubated with [γ-32P]ATP, and the reaction products were resolved by SDS-PAGE. A radioactive species of the same mobility as that of the Ash1MYC protein (82 kDa) was detected specifically in complexes from the GAL1-ASH1MYC strain (Fig. 8A). To determine if the Ash1-associated kinase might correspond to Cdc28, we repeated the experiment using strains bearing either wild-type CDC28 or the cdc28-as1 allele (6). Ash1MYC immune complexes were isolated from each strain and incubated with [γ-32P]ATP in the presence of increasing concentrations of 1NMPP1, which specifically inhibits the Cdc28-as1 kinase (Fig. 8B). Ash1 isolated from the wild-type strain was phosphorylated in vitro in both the absence and presence of 1NMPP1. In contrast, the Ash1 derived from the cdc28-as1 strain was efficiently phosphorylated only in the absence of 1NMPP1. These results indicate that the kinase activity that is associated with Ash1 in vivo and phosphorylates Ash1 in vitro is attributable mainly to Cdc28.
FIG. 8.
Cdc28 physically associates with Ash1 and mediates Ash1 degradation in vivo. (A) Anti-MYC immunoprecipitates from wild-type cells and GAL1-ASH1MYC cells were analyzed by anti-MYC immunoblotting (left) and by incubation with [γ-32P]ATP followed by SDS-PAGE (right). (B) Ash1MYC immunoprecipitates isolated from GAL1-ASH1MYC and GAL1-ASH1MYC cdc28-as1 strains were analyzed by anti-MYC immunoblotting (left) and by incubation with [γ-32P]ATP in the presence of various concentrations of 1NMPP1 (right). (C) A nocodazole-arrested GAL1-ASH1MYC cdc28-as1 strain was treated with either 5 μM 1NMPP1 or DMSO solvent control for 20 min, followed by GAL1-ASH1MYC induction/repression and analysis by anti-MYC immunoblotting. (D) A cdc15-2 ash1::ASH1MYC cdc28-as1 strain was arrested at 37°C, transferred to 23°C (t = 0 min) for 20 min, and then divided into two, treated with either 1NMPP1 or DMSO, and analyzed by anti-MYC immunoblotting. The bud index was determined for each time point to monitor cell cycle progression.
Given that the activity of Cdc28 appeared to be the predominant kinase activity associated with Ash1, we next asked whether Ash1 is stabilized in vivo when Cdc28 is inactivated. A GAL1-ASH1MYC cdc28-as1 strain was arrested with nocodazole and treated with either 1NMPP1 or solvent control for 20 min, and Ash1 stability was assessed as described above. Ash1 protein levels decreased rapidly in the control culture but persisted in the 1NMPP1-treated culture (Fig. 8C). 1NMPP1 had no effect on Ash1 stability in the absence of the cdc28-as1 mutation (data not shown). We note that Ash1 levels were reduced in the cdc28-as1 strain, probably because the GAL1 promoter is less effective when Cdc28 is inactivated (82). In addition, the inactivation of Cdc28-as1 in vivo by 1NMPP1 also increased the mobility of Ash1 (Fig. 8C), consistent with the ability of Cdc28 to readily phosphorylate Ash1 in vitro.
To ensure that the observed effects were not specific to ectopically expressed Ash1, we also determined whether the inactivation of Cdc28 influenced the stability of endogenous Ash1. An ash1::ASH1MYC cdc28-as1 cdc15-2 strain was arrested in late mitosis at 37°C, allowed to resume cell cycle progression at 23°C for 30 min, and treated with either 1NMPP1 or solvent control, and endogenous Ash1 levels at various time points were assessed by immunoblotting. The inhibition of Cdc28-as1 by 1NMPP1 caused a marked delay in the disappearance of Ash1MYC as cells progressed through G1 phase (Fig. 8D). Taken together, these results indicate that Ash1 degradation depends heavily on Cdc28 activity in vivo.
We then determined whether Cdc28 was able to stimulate Ash1 ubiquitination by SCFCdc4 in vitro. Ash1FLAG-HA was purified from a 1NMPP1-treated cdc28-as1 cell culture and phosphorylated in vitro by recombinant Cln2-Cdc28-Cks1 kinase prior to incubation with recombinant SCFCdc4 in a ubiquitination assay. In contrast to Ash1 isolated from wild-type cells (Fig. 3), Ash1 isolated from cells devoid of Cdc28 activity could be efficiently ubiquitinated in vitro only if it was subsequently phosphorylated by Cln2-Cdc28-Cks1 (Fig. 9A). This result demonstrated the critical role of Cdc28-dependent phosphorylation in Ash1 recognition and ubiquitination by SCFCdc4.
FIG. 9.
Cdc28 targets Ash1 for SCFCdc4-dependent ubiquitination in vitro and Cdc4-dependent degradation in vivo. (A) Ash1FLAG-HA was purified from a cdc28-as1 strain bearing a GAL1-ASH1FLAG-HA plasmid, which was grown in the presence of 1NMPP1 for 30 min prior to galactose induction. Purified Ash1FLAG-HA was treated with or without Cln2-Cdc28-Cks1 and then incubated in an SCFCdc4 ubiquitination reaction. Reaction products were analyzed by anti-FLAG immunoblotting; Cdc4FLAG present in the reaction was also detected. The asterisk indicates a nonspecific cross-reactive species. (B) Wild-type and cdc4-1 strains bearing a GAL1-ASH1HAT290A plasmid were arrested by nocodazole followed by GAL1-ASH1HAT290A induction/repression at 37°C and analysis by anti-HA immunoblotting. (C) cdc28-as1 and cdc28-as1 cdc4-1 strains bearing a GAL1-ASH1HAT290A plasmid were arrested by nocodazole, followed by 1NMPP1 treatment for 20 min, GAL1-ASH1HAT290A induction/repression at 37°C in the continued presence of 1NMPP1, and analysis by anti-HA immunoblotting. (D) An experiment similar to that in panel C was performed with a wild-type GAL1-ASH1HA plasmid.
To determine the interrelationship between Thr290 phosphorylation, Cdc28 activity, and Cdc4-mediated Ash1 degradation, we examined the stability of the Ash1T290A protein in nocodazole-arrested CDC4 and cdc4-1 strains. Ash1T290A was still partially dependent on Cdc4 for its residual instability (Fig. 9B), suggesting that Thr290 is not the only residue of Ash1 that contributes to its Cdc4-dependent degradation. This observation is consistent with the results showing that the Thr290Ala mutation did not further stabilize Ash1 in a cdc4-1 strain (Fig. 5) and that the phosphorylation of Ash1T290A by Cln2-Cdc28-Cks1 allowed the recognition of Ash1 by SCFCdc4 (Fig. 7B).
We then asked whether the Cdc4-dependent degradation of Ash1T290A requires Cdc28 function by examining the stability of Ash1T290A in cdc28-as1 and cdc28-as1 cdc4-1 strains. Each strain was arrested with nocodazole and treated with 1NMPP1 for 20 min, following which the cultures were shifted to 37°C and induced/repressed for GAL1-ASH1HAT290A, followed by immunoblot detection of Ash1HAT290A. The inactivation of Cdc4 did not appear to increase the stability of Ash1T290A beyond that caused by the inhibition of Cdc28 inactivation (Fig. 9C). This result suggested that, aside from Thr290, the Cdc4-dependent degradation of Ash1 depends entirely on Cdc28. In contrast, when a similar experiment was performed with wild-type Ash1, the inactivation of both Cdc28 and Cdc4 caused a further stabilization of Ash1 (Fig. 9D), suggesting that an additional as-yet-unidentified kinase may target Thr290 in the absence of Cdc28.
Multisite phosphodegron in Ash1.
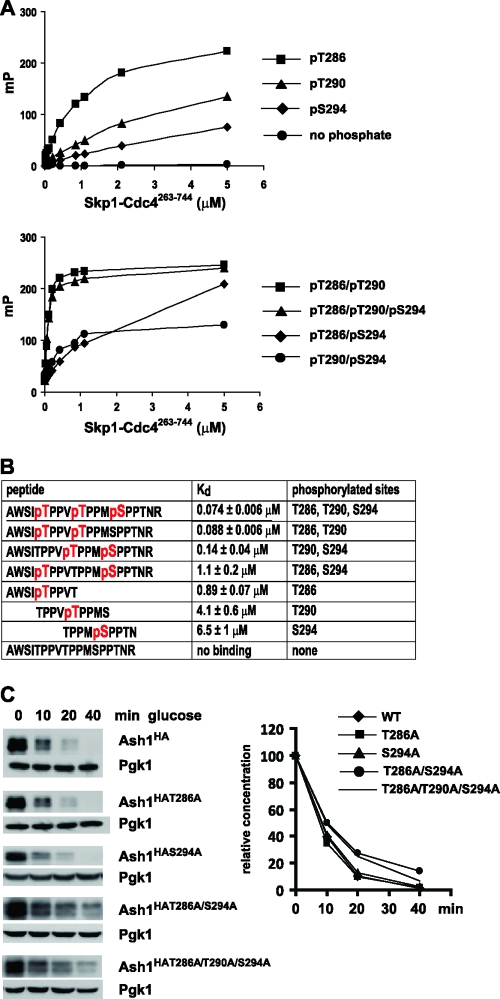
The above-described results suggested that Cdc28, perhaps in conjunction with another kinase(s), might generate multiple phosphodegrons to mediate the Cdc4-dependent degradation of Ash1. Given that the Thr286, Thr290, and Ser294 residues of Ash1 are phosphorylated in vivo (Fig. 4), we assessed the binding interactions of recombinant Cdc4 with synthetic peptides in which each site was phosphorylated individually and in all possible double and triple combinations. Quantitative affinities were determined in a fluorescence polarization assay using fluorescein-labeled phosphopeptides (48). Each of the individual pThr286, pThr290, and pSer294 peptides interacted with Cdc4 in this assay (Fig. 10). The highest-affinity interaction occurred with the pThr286 peptide (Kd [dissociation constant] = 0.89 μM), followed by the pThr290 peptide (Kd = 4.1 μM) and the pSer294 peptide (Kd = 6.5 μM). However, when pThr290 was combined with either of the two adjacent phosphorylated sites, compared to the highest affinity of the single-site peptides, the interaction affinity was increased by 10-fold for the pThr286/pThr290 diphosphopeptide (Kd = 0.088 μM) or 30-fold for the pThr290/pSer294 diphosphopeptide (Kd = 0.14 μM). The triply phosphorylated pThr286/pThr290/pSer294 peptide also bound with a similarly high affinity (Kd = 0.074 μM).
FIG. 10.
Interactions between Ash1-derived peptides and Cdc4. (A) Fluorescence polarization of the indicated fluorescein-labeled peptides in the presence of increasing concentrations of the recombinant Skp1-Cdc4263-744 complex. mP, milli-polarization unit. (B) Calculated dissociation constant (Kd) values from fluorescence polarization measurements in panel A. Phosphorylated residues in each peptide sequence are indicated in red. (C) Strains bearing the indicated GAL1-ASH1HA alleles were arrested with nocodazole, induced with galactose, and then assessed for Ash1 stability upon glucose repression.
Prompted by the multiple Ash1 phosphopeptide interactions detected in vitro, we examined the effect of single and multiple phosphorylation site mutations on Ash1 stability in vivo (Fig. 10C). Unlike the dominant Thr290 site, the mutation of either Thr286 or Ser294 alone had no observable effect on Ash1 stability as measured by a GAL1-ASH1HA repression assay. However, the Thr286Ala/Ser294Ala double mutation caused a marked stabilization of Ash1. This double mutant was not further stabilized upon an additional mutation of Thr290, indicating that these sites are epistatic to each other. Collectively, these results suggest that a redundant diphosphodegron composed of pThr286 pThr290 and pThr290 pSer294 underpins the recognition of Ash1 by Cdc4 in vitro and the elimination of Ash1 in vivo.
DISCUSSION
Our results demonstrate that the asymmetric cell division factor Ash1 is targeted to SCFCdc4 by Cdc28, which is the primary CDK kinase that controls cell division. The Cdc4- and phosphorylation-dependent degradation of Ash1 serves to restrict Ash1 accumulation to a G1-phase window in daughter cells and is necessary for efficient mating type switching to occur in mother cells. When combined with other mechanisms that enforce the daughter-specific localization of Ash1, this G1-phase restriction ensures that Ash1 cannot perdure as daughters become mothers (7, 61). These findings illustrate the crucial role of protein degradation during developmental state transitions.
Kinase requirements for Ash1 degradation.
Several lines of evidence demonstrate that Cdc28 targets Ash1 for Cdc4-dependent elimination: (i) the rapid chemical inhibition of Cdc28 stabilizes Ash1 in vivo, (ii) a robust Cdc28-dependent kinase activity associates with Ash1 isolated from cell lysates, (iii) the in vitro binding and ubiquitination of Ash1 by SCFCdc4 are heavily stimulated by Cdc28, and (iv) an Ash1T290A mutant protein is further impaired for Cdc4-dependent degradation when Cdc28 is inhibited. Taken together, our genetic and biochemical data demonstrate that Cdc28 is limiting for efficient Ash1 degradation in vivo and Ash1 ubiquitination in vitro. Consistently, Ash1 is eliminated during the cell cycle phases when Cdc28 is active (7, 61) and has been identified as a potential Cdc28 substrate in a large-scale phosphorylation-based screen (72). Interestingly, it was recently demonstrated that Ash1 negatively regulates the expression of the G1 cyclin CLN3 and thereby delays cell cycle commitment in daughter cells (18). Our finding that Cdc28 mediates Ash1 degradation suggests a potential feedback loop in which Cln-Cdc28 activity may help stimulate CLN3 expression through the elimination of Ash1.
Genetic evidence implicates Pho85, which is activated in G1 phase by a variety of Pcl cyclins, as an activator of mating type switching and, by implication, an antagonist of Ash1 (42). While it was suggested that Ash1 is stabilized in the absence of Pho85, the half-life of Ash1 in a pho85Δ strain is identical to that in a wild-type strain for at least the first 60 min of a repression time course, with partial stabilization manifest only after 120 min (42). In contrast to the substantial stabilization of Ash1 that we observed upon the conditional inactivation of the cdc28-as1 allele, the chemical inactivation of the PHO85F82G allele appeared have no effect on the Ash1 half-life or abundance in vivo. Our results do, however, suggest that Pho85 can drive the recognition of Ash1 by Cdc4 in vitro in a Thr290-dependent manner. Given the lack of evidence supporting a primary role for Pho85 in Ash1 elimination in vivo, we speculate that the main requirement for Pho85 in mating type switching may be explained by the Pho85-dependent inhibition of the transcriptional repression functions of Ash1, which is mediated by the Ash1 association with histone deacetylase (HDAC) complexes (10). Pcl-Pho85 kinases are known to control the association of HDACs with the G1/S transcriptional inhibitor Whi5 (32) and may thus exert similar effects on other G1/S regulators. The Pho85 dependency of Ash1 elimination may also be complicated by the pleiotropic functions of Pho85 in metabolism, which result in a severe growth defect of the pho85Δ strain (9).
Other kinases may also contribute to Ash1 degradation under different conditions. The Cdc4-dependent Ash1 instability that we observed for mating pheromone-arrested cells seems unlikely to be explained by the low levels of Cln-Cdc28 activity at this arrest point (71). It is possible that this role is played by the Fus3 mitogen-activated protein (MAP) kinase, which is known to target the transcription factor Tec1 to Cdc4 upon pheromone stimulation (2, 14). As Ash1 also functions in filamentous growth (12) and the regulation of the G1/S transition (18), its stability may be controlled by yet other proline-directed kinases, such as Srb10, the CDK that is associated with the core transcriptional machinery (13, 49). Other SCF substrates are also targeted by multiple kinases, including Sic1 (19, 50, 80), Gcn4 (13), Clb6 (34), and human cyclin E (74). Such multiple regulatory inputs may allow signal integration and/or targeted degradation in different cellular states.
Multisite phosphodegron in Ash1.
Substrate recognition by E3 enzymes is the basis for specificity in the ubiquitin system (30). In many instances, substrate-E3 interactions depend on substrate phosphorylation, other modifications, or small-molecule cofactors (56). The theme of the phosphorylation-dependent recognition of substrates by F-box proteins has many variations. In the simplest instance, recognition entails the facile docking of a single linear phospho-epitope, such as the interaction between a single CPD phosphopeptide and Cdc4 (52) or between diphosphorylated peptides and β-TrCP (79). The CPD motif has been extended to include diphosphodegrons that depend on a secondary contact between a phosphorylated residue at the +3 or +4 position and a polar patch adjacent to the main pSer/pThr-Pro binding pocket on the WD40 domain of Cdc4; through additive thermodynamic effects, this second engagement site serves to increase the overall affinity for Cdc4 by approximately 10-fold (28). A number of other hCdc4/Fbw7 substrates appear to include diphosphodegrons (3, 73). Finally, a complex recognition mode that depends on the generation of multiple, widely separated, weak CPD sites has been posited for the Cdc4 substrate Sic1 (48). A recent NMR analysis of the interaction between Sic1 phosphorylated on six sites and Cdc4 supports this model (44, 45). Other CRL substrates, including d-Myc, Klf5, Ci, and Mdm2, also have been reported to interact in a multisite-dependent fashion (33, 39, 46, 83, 84).
Ash1 appears to represent another variation on this phosphorecognition theme. The role of the Thr290 residue in Ash1 degradation is somewhat analogous to that of the dominant Thr45 site in Sic1, the mutation of which partially stabilizes Sic1 (48). As in the case of Sic1, additional sites also appear to contribute to Ash1 recognition, as we demonstrate for the adjacent Thr286 and Ser294 sites. Notably, the interresidue distance between Thr286, Thr290, and Ser294 conforms to the +4 spacing required to generate a high-affinity diphosphodegron for Cdc4 (28). The finding that Ash1-derived phosphopeptides can generate such high-affinity sites in two different permutations (i.e., pThr286/pThr290 and pThr290/pSer294) suggests that a redundant diphosphodegron determines Ash1 recognition. The moderate interaction affinities of the pThr286 phosphopeptide and the pThr286/pSer294 diphosphopeptide may also be able to weakly target Ash1. It is possible that each individual CPD motif in Ash1 differentially facilitates the ubiquitination reaction, as suggested previously for Sic1 (55, 59). In particular, the unexpected accumulation of monoubiquitinated Ash1T290A in vivo suggests that Thr290 may be required for efficient ubiquitin chain elongation. As noted above, the monoubiquitination of Ash1 may directly attenuate its repressive activity through the remodeling of protein interactions, as observed previously for other transcription factors (22, 76). Finally, it will be of interest to determine whether the multisite phosphorylation requirement confers nonlinear properties on Ash1 degradation and/or the efficiency of mating type switching (21) and to examine the interaction dynamics of various phosphorylated isoforms of Ash1 with Cdc4 (44, 45).
Protein turnover and cell state transitions.
The transition from daughter cell to mother cell and the mating type switching of mother cells represent two simplified examples of a change in developmental state (15). In addition to Ash1 regulation, the ubiquitin-proteasome system also enforces yeast cell type identity by at least two other pathways. The SCF complex containing the F-box protein Ufo1 (SCFUfo1) targets the Ho endonuclease for ubiquitination and subsequent degradation (35). Ho accumulation is thus restricted to a narrow window during late G1 phase, which both protects genome integrity and ensures a single switch event per generation (35). Two distinct E3 ubiquitin ligases, Doa10 and the Slx5-Slx8 complex, target the α-cell-specific factor α2, which represses the transcription of a-specific genes (38, 81). Ubiquitination causes the disassembly of α2-containing complexes on DNA and the degradation of α2, both of which enable cells to rapidly transition from the α to the a cell state (38, 76). The elimination of the α1 transcriptional activator is similarly required to switch to the a cell type (51). The hierarchy that dictates the elimination of Ash1, Ho, α1, and α2 enables the precise specification of cell fate in a temporally controlled fashion. In each of these cases, however, it appears that the degradation machinery is not itself regulated. In particular, the symmetric localization of Ash1 in late anaphase in a she1Δ strain, which is defective in ASH1 mRNA transport to the daughter cell, suggests that there is no intrinsic difference in the ubiquitination machinery that eliminates Ash1 in mother cells versus daughter cells per se (7, 40, 66).
In multicellular organisms, changes of cellular state underlie development and tissue renewal. Many gene expression programs that characterize different developmental stages have been well documented. However, the elimination of the protein cohorts that define the eclipsed states has remained less well explored. Notably, stem cells must eliminate the factors that define stemness in order to differentiate into specific cell lineages. Like budding yeast, stem cells often divide asymmetrically to renew the stem cell compartment and yield a lineage-committed daughter cell within the same division (25, 36). This change of stem cell fate is further complicated by observations that stem cells exhibit stochastic fluctuations in the abundance of critical regulators (11), which may in part occur at the posttranslational level (63). Examples of E3 components that affect the fate of multipotent cells include the Trim32/NHL proteins that target Myc during neuronal precursor differentiation (60), the elimination of tramtrack during eye development (67), and the elimination of REST by β-TrCP, also during neuronal fate specification (26, 75). Interestingly, asymmetric protein degradation between daughter cells has recently been documented for mammalian cells (24). It is likely that the ubiquitin system will emerge as a pervasive means to shape the fate of stem and progenitor cells through the elimination of key regulatory factors (65).
Supplementary Material
Acknowledgments
We thank Marcia Roy and members of the Tyers laboratory for technical assistance; Kim Nasmyth, John Diffley, and Brenda Andrews for reagents; and Brenda Andrews for helpful discussions.
This work was supported by grants to C.H., F.S., and M.T. from the Canadian Institutes of Health Research (grants MOP-93571 and MOP-57795) and by grants to M.T. from the National Cancer Institute of Canada and the Wellcome Trust. T.L.B. was supported by a grant from the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council (BB/D019621/1). C.H. was supported by a Michael Smith Foundation for Health Research Career Investigator award and a Canadian Institutes of Health Research New Investigator award (MSH-95337), F.S. was supported by a Canada Research Chair in Structural Biology, and M.T. was supported by a Scottish Universities Life Sciences Alliance research professorship and a Royal Society Wolfson Research Merit award.
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bai, C., et al. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 2.Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates III, and H. D. Madhani. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991-1000. [DOI] [PubMed] [Google Scholar]
- 3.Bao, M. Z., T. R. Shock, and H. D. Madhani. 2009. Multisite phosphorylation of the Saccharomyces cerevisiae filamentous growth regulator Tec1 is required for its recognition by the E3 ubiquitin ligase adaptor Cdc4 and its subsequent destruction in vivo. Eukaryot. Cell 9:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berset, C., et al. 2002. Transferable domain in the G1) cyclin Cln2 sufficient to switch degradation of Sic1 from the E3 ubiquitin ligase SCFCdc4 to SCFGrr1. Mol. Cell. Biol. 22:4463-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand, E., et al. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437-445. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, A. C., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. [DOI] [PubMed] [Google Scholar]
- 7.Bobola, N., R. P. Jansen, T. H. Shin, and K. Nasmyth. 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84:699-709. [DOI] [PubMed] [Google Scholar]
- 8.Bowerman, B., and T. Kurz. 2006. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development 133:773-784. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, A. S., A. C. Bishop, J. L. DeRisi, K. M. Shokat, and E. K. O'Shea. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. U. S. A. 98:12578-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrozza, M. J., et al. 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731:77-87. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, I., et al. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450:1230-1234. [DOI] [PubMed] [Google Scholar]
- 12.Chandarlapaty, S., and B. Errede. 1998. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 18:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi, Y., et al. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou, S., L. Huang, and H. Liu. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981-990. [DOI] [PubMed] [Google Scholar]
- 15.Cook, M. A., and M. Tyers. 2004. Cellular differentiation: the violin strikes up another tune. Curr. Biol. 14:R11-R13. [DOI] [PubMed] [Google Scholar]
- 16.Cosma, M. P. 2004. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 5:953-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 18.Di Talia, S., et al. 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 7:e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escote, X., M. Zapater, J. Clotet, and F. Posas. 2004. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6:997-1002. [DOI] [PubMed] [Google Scholar]
- 20.Falconnet, D., et al. 18 November 2010. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab Chip [Epub ahead of print.] doi: 10.1039/C0LC00228C. [DOI] [PMC free article] [PubMed]
- 21.Ferrell, J. E., Jr. 1996. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 21:460-466. [DOI] [PubMed] [Google Scholar]
- 22.Flick, K., et al. 2004. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 6:634-641. [DOI] [PubMed] [Google Scholar]
- 23.Frescas, D., and M. Pagano. 2008. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8:438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuentealba, L. C., E. Eivers, D. Geissert, V. Taelman, and E. M. De Robertis. 2008. Asymmetric mitosis: unequal segregation of proteins destined for degradation. Proc. Natl. Acad. Sci. U. S. A. 105:7732-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonczy, P. 2008. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9:355-366. [DOI] [PubMed] [Google Scholar]
- 26.Guardavaccaro, D., et al. 2008. Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature 452:365-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 28.Hao, B., S. Oehlmann, M. E. Sowa, J. W. Harper, and N. P. Pavletich. 2007. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26:131-143. [DOI] [PubMed] [Google Scholar]
- 29.Henchoz, S., et al. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 31.Herskowitz, I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342:749-757. [DOI] [PubMed] [Google Scholar]
- 32.Huang, D., et al. 2009. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol. 7:e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inuzuka, H., et al. 2010. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(β-TRCP) ubiquitin ligase. Cancer Cell 18:147-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, L. P., S. I. Reed, and S. B. Haase. 2006. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol. Cell. Biol. 26:2456-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. U. S. A. 97:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohlmaier, A., and B. A. Edgar. 2008. Proliferative control in Drosophila stem cells. Curr. Opin. Cell Biol. 20:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laney, J. D., and M. Hochstrasser. 2004. Ubiquitin-dependent control of development in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 7:647-654. [DOI] [PubMed] [Google Scholar]
- 38.Laney, J. D., and M. Hochstrasser. 2003. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev. 17:2259-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, N., et al. 2010. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J. Biol. Chem. 285:18858-18867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long, R. M., et al. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277:383-387. [DOI] [PubMed] [Google Scholar]
- 41.Maxon, M. E., and I. Herskowitz. 2001. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. U. S. A. 98:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride, H. J., et al. 2001. The protein kinase Pho85 is required for asymmetric accumulation of the Ash1 protein in Saccharomyces cerevisiae. Mol. Microbiol. 42:345-353. [DOI] [PubMed] [Google Scholar]
- 43.Meimoun, A., et al. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol. Biol. Cell 11:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittag, T., et al. 2010. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure 18:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mittag, T., et al. 2008. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U. S. A. 105:17772-17777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moberg, K. H., A. Mukherjee, A. Veraksa, S. Artavanis-Tsakonas, and I. K. Hariharan. 2004. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 14:965-974. [DOI] [PubMed] [Google Scholar]
- 47.Nalepa, G., M. Rolfe, and J. W. Harper. 2006. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5:596-613. [DOI] [PubMed] [Google Scholar]
- 48.Nash, P., et al. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 49.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187-190. [DOI] [PubMed] [Google Scholar]
- 50.Nishizawa, M., M. Kawasumi, M. Fujino, and A. Toh-e. 1998. Phosphorylation of Sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nixon, C. E., A. J. Wilcox, and J. D. Laney. 2010. Degradation of the Saccharomyces cerevisiae mating-type regulator α1: genetic dissection of cis-determinants and trans-acting pathways. Genetics 185:497-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243-256. [DOI] [PubMed] [Google Scholar]
- 53.Paquin, N., and P. Chartrand. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105-111. [DOI] [PubMed] [Google Scholar]
- 54.Patton, E. E., et al. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petroski, M. D., and R. J. Deshaies. 2003. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 11:1435-1444. [DOI] [PubMed] [Google Scholar]
- 56.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 57.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 58.Ravid, T., S. G. Kreft, and M. Hochstrasser. 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25:533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadowski, M., R. Suryadinata, X. Lai, J. Heierhorst, and B. Sarcevic. 2010. Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol. Cell. Biol. 30:2316-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwamborn, J. C., E. Berezikov, and J. A. Knoblich. 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136:913-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sil, A., and I. Herskowitz. 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84:711-722. [DOI] [PubMed] [Google Scholar]
- 62.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 63.Spencer, S. L., S. Gaudet, J. G. Albeck, J. M. Burke, and P. K. Sorger. 2009. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strathern, J. N., and I. Herskowitz. 1979. Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell 17:371-381. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi, K., and S. Yamanaka. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663-676. [DOI] [PubMed] [Google Scholar]
- 66.Takizawa, P. A., A. Sil, J. R. Swedlow, I. Herskowitz, and R. D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389:90-93. [DOI] [PubMed] [Google Scholar]
- 67.Tang, A. H., T. P. Neufeld, E. Kwan, and G. M. Rubin. 1997. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell 90:459-467. [DOI] [PubMed] [Google Scholar]
- 68.Tang, X., et al. 2007. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129:1165-1176. [DOI] [PubMed] [Google Scholar]
- 69.Tang, X., et al. 2005. Genome-wide surveys for phosphorylation-dependent substrates of SCF ubiquitin ligases. Methods Enzymol. 399:433-458. [DOI] [PubMed] [Google Scholar]
- 70.Taylor, R. J., et al. 2009. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc. Natl. Acad. Sci. U. S. A. 106:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tyers, M., and B. Futcher. 1993. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol. Cell. Biol. 13:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ubersax, J. A., et al. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 73.Welcker, M., and B. E. Clurman. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83-93. [DOI] [PubMed] [Google Scholar]
- 74.Welcker, M., et al. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12:381-392. [DOI] [PubMed] [Google Scholar]
- 75.Westbrook, T. F., et al. 2008. SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilcox, A. J., and J. D. Laney. 2009. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. 11:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Willems, A. R., et al. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]
- 78.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695:133-170. [DOI] [PubMed] [Google Scholar]
- 79.Wu, G., et al. 2003. Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCFβ-TrCP1 ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 80.Wysocki, R., A. Javaheri, K. Kristjansdottir, F. Sha, and S. J. Kron. 2006. CDK Pho85 targets CDK inhibitor Sic1 to relieve yeast G1 checkpoint arrest after DNA damage. Nat. Struct. Mol. Biol. 13:908-914. [DOI] [PubMed] [Google Scholar]
- 81.Xie, Y., E. M. Rubenstein, T. Matt, and M. Hochstrasser. 2010. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 24:893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu, V. P., C. Baskerville, B. Grunenfelder, and S. I. Reed. 2005. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol. Cell 17:145-151. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, Q., et al. 2009. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 106:21191-21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuang, M., et al. 2009. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.