Abstract
The expression of the histone genes is regulated during the cell cycle to provide histones for nucleosome assembly during DNA replication. In budding yeast, histones H2A and H2B are expressed from divergent promoters at the HTA1-HTB1 and HTA2-HTB2 loci. Here, we show that the major activator of HTA1-HTB1 is Spt10, a sequence-specific DNA binding protein with a putative histone acetyltransferase (HAT) domain. Spt10 binds to two pairs of upstream activation sequence (UAS) elements in the HTA1-HTB1 promoter: UAS1 and UAS2 drive HTA1 expression, and UAS3 and UAS4 drive HTB1 expression. UAS3 and UAS4 also contain binding sites for the cell cycle regulator SBF (an Swi4-Swi6 heterodimer), which overlap the Spt10 binding sites. The binding of Spt10 and binding of SBF to UAS3 and UAS4 are mutually exclusive in vitro. Both SBF and Spt10 are bound in cells arrested with α-factor, apparently awaiting a signal to activate transcription. Soon after the removal of α-factor, SBF initiates a small, early peak of HTA1 and HTB1 transcription, which is followed by a much larger peak due to Spt10. Both activators dissociate from the HTA1-HTB1 promoter after expression has been activated. Thus, SBF and Spt10 cooperate to control the timing of HTA1-HTB1 expression.
The basic structural unit of chromatin is the nucleosome, which is composed of two molecules each of the four core histones, H2A, H2B, H3, and H4, formed into an octamer, around which is wrapped ∼147 bp of DNA in 1.75 superhelical turns (34). Nucleosomes are formed into regularly spaced arrays along the DNA and can be mobilized by various ATP-dependent remodeling machines, such as SWI/SNF and RSC. The histones are subject to numerous posttranslational modifications, including acetylation and methylation (65). Chromatin structure plays a central role in gene regulation and other nuclear processes, including DNA replication (23). In the latter case, the cell must replicate not only its DNA during S phase but also its chromatin. Consequently, the histone genes are activated at the beginning of S phase to provide sufficient core histones to assemble all of the replicated DNA into chromatin. The inhibition of DNA synthesis results in a rapid repression of the histone genes, indicating that it is tightly coupled to DNA replication (48). Some histone gene expression is independent of replication, providing histones for chromatin assembly occurring outside S phase (61). In general, though, the regulation of the histone genes is an important aspect of eukaryotic cell cycle control (4).
The major core histone genes of the yeast Saccharomyces cerevisiae are organized into four loci, each containing two histone genes divergently transcribed from a central promoter: two loci encode H2A and H2B (HTA1-HTB1 and HTA2-HTB2) (27), and the other two loci encode H3 and H4 (HHT1-HHF1 and HHT2-HHF2) (57). The regulation of the yeast histone genes has been studied extensively (24, 48). Our focus is on the HTA1-HTB1 locus, in which several elements that are important for cell cycle-dependent regulation have been identified.
First, there are four upstream activation sequences (UASs) (histone UAS elements), which direct correct cell cycle regulation when transposed onto a heterologous promoter (49). Histone UAS elements are found in all of the major core histone promoters (16). We have shown that they are bound by Spt10, a sequence-specific DNA binding protein (16), which contains a histone acetyltransferase (HAT) domain (43). This HAT domain has not yet been shown to have HAT activity, but the putative catalytic residues are required for the transcriptional activation of a reporter (28). Although there is indirect evidence that Spt10 might acetylate H3-K56 at the histone gene promoters (68), it has since been shown that Rtt109 is the HAT responsible for most, if not all, H3-K56 acetylation in yeast (15, 25). Spt10 is a dimer that binds specifically to pairs of histone UAS elements, each having the bipartite sequence RTTC-N7-TTCNC, which are found only in the histone promoters (16, 38, 39). Although SPT10 is not an essential gene, the null mutant is very sick (14, 41, 42). It displays global changes in gene regulation, which we have attributed to a defective chromatin structure arising from insufficient histone synthesis (16).
Second, the HTA1-HTB1 promoter contains an ∼67-bp negative regulatory region, referred to as the CCR or NEG region (48). The NEG region presumably contains one or more binding sites for sequence-specific factors, but these have not yet been identified. A short consensus sequence that is present in all histone loci except HTA2-HTB2 has been termed the NEG or NRS element (20, 35, 48). The NEG element overlaps another element, called CCR′, which has some palindromic character. CCR′ confers cell cycle-dependent expression on a heterologous promoter, although the peak is early relative to that of normal histone gene expression (49). The NEG region is required for the rapid shutdown of histone gene transcription that occurs when cells are treated with hydroxyurea to inhibit DNA replication. This shutdown is dependent on the HIR complex (54, 58, 67), which contains Hir1, Hir2, Hir3, and Hpc2 (22, 51). The HIR complex has additional functions in transcript elongation (19, 46) and silencing (53), probably involving its histone chaperone activity (22). The NEG region is required for the recruitment of the histone chaperones Asf1 (59) and Rtt106 (18), the SWI/SNF complex (11), and the RSC complex (44). In addition, the NEG region and the HIR complex are required to mediate a compensatory response to variations in copy number of HTA1-HTB1 (40). Thus, the NEG region plays a major role in the cell cycle control of HTA1-HTB1. However, it is important to note that multiple copies of UAS elements from HTA1-HTB1 or from HHT1-HHF1 are by themselves sufficient to direct correct cell cycle activation (20, 48, 49).
Third, the 3′-untranslated region (UTR) of HTB1, when attached to a reporter gene, also directs cell cycle-dependent expression, although the peak is slightly later than the peak of endogenous HTB1 expression (7, 66).
Another sequence-specific protein with a role in HTA1-HTB1 regulation is the SBF transcription factor, a well-studied regulator of G1/S-specific genes (5, 6, 12). SBF is a complex between Swi4 and Swi6 (1). The activity of Swi4, which contains the DNA binding domain (DBD) (Swi4-DBD), is regulated by the cell cycle-dependent nuclear import of Swi6 (2, 55). Swi6 also binds Mbp1 to form MBF, the other major G1/S-phase regulator in yeast. Although cells can survive without Swi4, Swi6, or Mbp1, they are rather sick, and an swi4Δ mbp1Δ double mutant is inviable (33). The evidence that SBF plays a role at HTA1-HTB1 includes (i) genetic studies showing that both the swi4Δ and mbp1Δ mutations are synthetically lethal with the spt10Δ mutation (29), (ii) SBF mutants exhibiting a modest reduction in HTA1 and HTB1 mRNA levels (29), (iii) chromatin immunoprecipitation (ChIP) data indicating that Swi4 is present at HTA1-HTB1 (56), and (iv) predicted SBF binding sites in the HTA1-HTB1 promoter (discussed below). SBF activity has been correlated with the dimethylation of H3-K79, suggesting a link between this histone mark and the regulation of the G1-to-S-phase transition (37, 52).
Here, we address the roles of Spt10 and SBF in the cell cycle-dependent regulation of HTA1-HTB1. We show that these proteins bind to overlapping sites within two of the four histone UAS elements in a mutually exclusive manner. Together, these two activators can account for the cell cycle dependence of HTA1-HTB1 transcription: SBF is responsible for a small early peak of transcription, while Spt10 contributes a later, much larger peak. Although Spt10 and SBF both activate HTA1-HTB1 expression, the difference in the timing of this activation suggests that their functions are not redundant. In their absence, there is little residual expression. We conclude that SBF and Spt10 together control the timing of HTA1-HTB1 expression.
MATERIALS AND METHODS
All oligonucleotides used in this work are listed in Table 1.
TABLE 1.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| 667 | CATTAGCGCTGTTCCAAAATTTTCGCCTCACTGTGCGAAGCTATTGGAATGGAGTGTATT |
| 668 | AAATACACTCCATTCCAATAGCTTCGCACAGTGAGGCGAAAATTTTGGAACAGCGCTAAT |
| 698 | ATCGATTGCTTCATTCTTTT |
| 711 | CTGAATCTTTCGTTACCAAT |
| 986 | GGCGCGCTGCAGGGATCCCATTTTATATTTTATATGTATGAAATTTG |
| 987 | GGCGCGACTAGTGCTAGCCATTGTATGTGTGTATGGTTT |
| 988 | GGCGCGGGATCCGCTTCTTCTGAAGATGTCATCAC |
| 989 | GGCGCGCTGCAGTTACAAGAACAAGTGGTGTCTACC |
| 990 | GGCGCGGCTAGCATTCTGTCTAAAGGTGAAGAATTATTC |
| 991 | GGCGCGACTAGTTTAAGATCTTTTGTACAATTCATCCATACCATGG |
| 992 | GGCGCGACTAGTTAATGAAATCACTTCCTTTGGTTAT |
| 993 | GACGTTGCGGCCGCATTGTTCACACGAGCCAATG |
| 994 | GGCGCGCTGCAGTAAGATCGGTTCTGGTATTTTAAAGAAG |
| 995 | GGCGCGCTCGAGGACAGAAGCCGCGGGCT |
| 996 | GGCGCGAGATCTGCATCCAACTTGAACATTTCG |
| 997 | GGCGCGAGATCTCTATATTACTTGGGTATTGCC |
| 1004 | GGCGCGCATATGCCATTTGATGTTTTGATATCAAACC |
| 1005 | GGCGCGCTCGAGTTACTGGTTGATCGATGCATTTTTC |
| 1010 | P-GCTGACTCAAAATTCCTATCTCACTGTACAGGGCTATTGAGGTGGAGTGTATTTGGTGGCTC |
| 1034 | P-CCCTTTCTTCCGCGCGCAAATGATCCAGATGGATTTAAAATCAAGAGAATTG |
| 1035 | P-TGCAACGCGCTCCAAAGTGACTTAAGCCTTCCCTTTCGGGC |
| 1036 | CTACCAAGGCTTCTCAAGAATTAT |
| 1037 | GCAACAGAGAAAACAGGAAGACAC |
| 1151 | GGCGGCCCGCGGCTATGCCTCGGTAATGATTTTC |
| 1152 | GGCGGCCCGCGGCCGTTCAGAATGACACGTATAG |
| 1192 | CTGCTAAAGCCGAAAAGAAAC |
| 1193 | TTAGCCAATTCACCTGGTAAG |
| 1223 | CTCTTTAATTAAGTACAATCTTGATCCGGAGCT |
| 1224 | CTCTTTAATTAAGCAGGCAAGTGCACAAACAATA |
| 1229 | GGCGCGTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCATGCCATTTGATGTTTTGATATC |
| 1230 | GGCGCGGGTACCGTGATGATGATGATGATGTGAGCCCTGGTTGATCGATGCATTTTTC |
| 1252 | CATCGCGAAATTTGTCTCAAC |
| 1253 | GAAACATAACGGTTTTTGGCG |
| 1254 | CTCAATGTCGCCCGAAAG |
| 1255 | CTCTTGATTTTAAATCCATCGTTC |
| 1280 | TTCTCATTTCCCTTCTGTCCCTCTTGCGTAGTTCTAAAAGGTTGATTTATTCGAGAAATCCGGATCCCCGGGTTAATTAA |
| 1281 | AAAAAAATTTAAAAACTCTGATAATATAGTAAAAATTATTGGTACATTGTGAATTAAAATGAATTCGAGCTCGTTTAAAC |
| 1282 | GATCGAATTCATTTTAATTCACAATGTACCAATAAT |
| 1283 | ACATCTCAACTTGGATGATTCATTAC |
| 1292 | CAAATAAACCATACACACATACAATG |
| 1293 | P-ATCGTTAACGAAAGAGTTCAAG |
| 1294 | CTAGTATTTACGATCCACTGG |
| 1295 | P-GTATTCAGAAACCGATGTATACG |
| 1296 | GGCGCGGGTACCGTGATGATGATGATGATGTGAGCCTGC |
| 1358 | CGGTGAACAAAAGCTTATCTCCGAGGAAGACTTGAACTCTTAAC |
| 1359 | TCGAGTTAAGAGTTCAAGTCTTCCTCGGAGATAAGCTTTTGTTCACCGGTAC |
| 1360 | GGCGCGCATATGGGTACAGATTATAAAGATGACGATGACAAGGGTACCGCGTTGGAAGAAGTGGTAC |
| 1361 | GGCGCGGGATCCTCATGTACCAGCGTAATCTGGAAC |
| 1409 | CTATGGTTAGACGCTCAATGTCG |
| 1410 | CACTACTAAGGCCAATTCTCTTG |
| 1413 | GAACCTCAAAACTGCGTGTTC |
| 1420 | GTGCTCTTCGCTAGGTATCC |
| 1425 | TGGCGAAACCAATTTGTTCTCTCAA |
| 1426 | GGAGGATATATTACCTTTGATGTT |
| 1501 | GTAGCACGTCGCGTTTATGG |
| 1502 | GATGGCGCTCGTGATCCTG |
| 1527 | CCCGTATGGGACTCGTCAT |
| 1528 | AGTTGGCGAGGGGAACATT |
Reporter plasmids.
A reporter plasmid (p532) containing the wild-type (WT) divergent HTA1-HTB1 promoter with the HTA1 promoter driving RedStar-2 (RDS2) and the HTB1 promoter driving green fluorescent protein (GFP) was constructed as follows. The HTA1-HTB1 promoter (from start codon to start codon) was obtained as an 841-bp PstI-SpeI PCR fragment using p474 (16) as a template with primers 986 and 987, which also introduced BamHI and NheI sites, and inserted into pRS415 (Stratagene) cut with PstI and SpeI to obtain p526. The RDS2 open reading frame (ORF) was obtained by PCR as a 681-bp BamHI-PstI fragment using pYM43 (p524) (30) as a template and primers 988 and 989 and inserted at these sites into p526 to give p528. The 3′-UTR of HTA1 was obtained as a 378-bp PstI-XhoI fragment by PCR using p474 as a template and primers 994 and 995 and inserted after the RDS2 ORF in p528 to give p529. The GFP ORF with a BglII site inserted just prior to the stop codon was obtained as a 732-bp NheI-SpeI fragment by PCR using pYM44 (p525) (30) as a template and primers 990 and 991 and inserted at these sites in p529 to yield p530. The 3′-UTR of HTB1 was obtained as a 310-bp SpeI-NotI fragment by PCR using p474 as template with primers 992 and 993 and inserted after the GFP ORF in p530 to give p531. A 543-bp BglII fragment encoding PEST sequences from CLN2 (36) was obtained by PCR using yeast genomic DNA as a template and primers 996 and 997 and was inserted at the BglII site at the C terminus of GFP to give p532. For reporter plasmids with mutations in both UAS1 and UAS2 (p533), the 441-bp BamHI-AfeI HTA1 promoter fragment in p532 was replaced with a mutated version obtained by PCR using p531 as a template and primers 986 and 1010 (mutations in UAS1 are marked with a BsrGI site; those in UAS2 are marked with a PleI site). For mutations in both UAS3 and UAS4 (p541), the 829-bp BamHI-NheI HTA1-HTB1 promoter fragment in p532 was replaced with a mutated version obtained by ligating two PCR fragments together, made using p532 as a template and primers 986 and 1035, and primers 987 and 1034 (mutations in UAS3 are marked with an AflII site; those in UAS4 are marked with a BssHII site). For all four fully mutated UAS elements (p545), the 441-bp BamHI-AfeI HTA1 promoter fragment in p541 was replaced with a mutated version made by using primers 986 and 1010. Mutations in all four Spt10 half-sites (p598) and point mutations in the two Swi4 sites (p652) were introduced into p532 using synthesized mutated HTA1-HTB1 promoter fragments. All inserts were sequenced.
Integration plasmids.
An integration plasmid containing the wild-type HTA1-HTB1 locus with HIS3 inserted downstream of HTA1 (p592) was constructed as follows. The 2,903-bp MscI-BamHI fragment from pCC67 (8), containing the HTA1-HTB1 locus, was inserted into pNEB193 (New England Biolabs) cut with SmaI and BamHI to form p577. p592 was obtained by the insertion of a 984-bp BtgI fragment containing the HIS3 gene (obtained by PCR using pGEM-TAHIS3 [31] as a template with primers 1151 and 1152) at the BtgI site downstream of HTA1 in p577, such that HIS3 and HTA1 are transcribed in the same direction. Mutations in all four Spt10 half-sites were introduced into p592 by replacing the PflMI-EcoRV fragment with mutated versions made by PCR (p656): a megaprimer was made by using p598 as a template with primers 1292 and 1293 and then paired with primer 986; the final product was obtained by using primers 1293 and 1294. Point mutations in the two Swi4 sites were introduced into p592 in the same way, using p652 as a template, to obtain p657. Yeast was transformed with an EcoRI-BamHI digest of the integration plasmid. An integration plasmid for tagging SWI4 with 3 Flag tags at its C terminus was constructed by gene synthesis (p623) followed by the insertion of an 880-bp PacI fragment containing TRP1 (made by PCR using oligonucleotides 1223 and 1224 as primers and pRS414 [Stratagene] as a template) at a PacI site introduced into the 3′-UTR of SWI4 to obtain p626. Yeast was transformed with a PmeI digest.
Expression plasmids for Swi4 and Swi6.
A plasmid for expressing the first 202 residues of Swi4 followed by a 6-histidine tag and a single Flag tag (p618), based on pET28a(+) (Novagen), was constructed by replacing the entire XbaI-KpnI SPT10 insert in p348 (38) with a 671-bp XbaI-KpnI SWI4 fragment obtained by PCR: an initial PCR using yeast genomic DNA as a template and primers 1004 and 1005 was used to generate a template for use with primers 1229 and 1230. The N terminus of the Swi4-DBD is native. A plasmid for expressing full-length Swi4 with a native N terminus and C-terminal 6-histidine and Myc tags (p660) was constructed by replacing the SnaBI-KpnI fragment in p618 with a 3,186-bp SnaBI-KpnI fragment containing most of the SWI4 ORF, made using yeast genomic DNA as a template and primers 1295 and 1296 to obtain p648 and then replacing the single Flag tag with a single Myc tag, inserted as a synthetic KpnI-XhoI double-stranded oligonucleotide (primers 1358 and 1359). An expression plasmid for full-length Swi6 with an N-terminal Flag tag and a C-terminal hemagglutinin (HA) tag (p659) was constructed by the insertion of the SWI6 ORF obtained as a 2,490-bp NdeI-BamHI fragment using primers 1360 and 1361 into pET11a (Novagen).
Yeast strains.
YPE250 (MATa can1-100 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 GAL+ ADE2+ SPT10-3HA::URA3) was used for the reporter studies and was constructed by transforming a W303 wild-type strain with a SacI-HindIII digest of p355 (16) to tag SPT10. ChIP studies were performed with YPE334, containing both SPT10-3HA and SWI4-3FLAG, made by transforming YPE250 with a PmeI digest of p626 (SWI4-3FLAG::TRP1). The binding-site mutants were constructed by transforming YPE334 with an EcoRI-BamHI digest of p592 (WT), p656 (Spt10 half-site mutations), or p657 (Swi4 site mutations) to obtain YDC360, YDC361, or YDC362, respectively. In these strains, HIS3 was integrated downstream of HTA1. The mutations were verified by the sequencing of PCR fragments containing the HTA1-HTB1 promoter, obtained by PCR using genomic DNA from these strains. An swi4Δ strain (YPE352 [MATa can1-100 ade2-1 his3-11 leu2-3,112 LYS2+ trp1-1 ura3-1 GAL+ swi4Δ::KanMX]) was constructed by transforming a diploid W303 strain with an swi4Δ::KanMX fragment made by ligating two PCR fragments together (an swi4Δ::KanMX fragment was made by using primer pair 1280/1281 with pFA6a-KanMX6 [64] as a template, and a fragment containing 3′ SWI4 sequences was made by using primer pair 1282/1283; both were digested with EcoRI, ligated together, and amplified by using primer pair 1280/1283). The germination efficiency of swi4Δ spores was low; the resulting haploid cells were enlarged and tended to clump.
Reporter assay.
Cultures (500 ml) were grown in synthetic medium without leucine to an A600 of ∼0.2 and arrested using 10 μg/ml α-factor (a synthetic peptide corresponding to residues 104 to 120) for 2 h at 30°C. Cells were released from arrest by rapid filtration (0.45 μm) (Millipore HA), washed briefly with prewarmed medium, and resuspended in 500 ml prewarmed medium. Aliquots of cells were removed at various times, washed in cold water, and stored frozen. RNA was prepared by using the hot-phenol method, concentrations were determined spectrophotometrically, and RNA integrity was confirmed by ethidium staining of agarose gels. Northern blots of denaturing gels containing 2.5% formaldehyde were probed either with both 3′ HTA1 and ACT1 probes or with the HTB1 coding region and GFP probes. Northern probes were PCR fragments radiolabeled by random priming: for 3′ HTA1, ACT1, and CLN2 ORFs, primer pairs 1036/1037, 698/711, and 1527/1528, respectively, were used with yeast genomic DNA as a template; for the HTB1 ORF, primer pair 1192/1193 was used with pCC67 as a template; and for GFP, primer pair 990/991 was used with pYM44 as a template. An aliquot of the same RNA sample was included on each blot for the normalization of GFP/HTB1 signals. Phosphorimages were quantified by using ImageQuant software.
Purification of Spt10, the Swi4-DBD, and SBF.
The purification of Spt10 using baculoviral expression was described previously (16). The Swi4-DBD was purified as follows. A total of 450 ml Escherichia coli BL21(DE3) cells transformed with p618 was grown to an A600 of ∼0.6 in LB medium with 60 μg kanamycin/ml at 30°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. The cell pellet was resuspended in TI buffer (25 ml 50 mM Tris HCl [pH 8.0], 0.5 M NaCl, 0.1% Triton X-100, 5 mM 2-mercaptoethanol, 2 mM imidazole, and protease inhibitors without EDTA [catalog number 05056489001; Roche]) and lysed by 1 passage through a French press. The debris was removed by spinning at 10,000 rpm for 20 min in a Sorvall SA600 rotor. The supernatant was syringe filtered (0.45-μm Millex HA; Millipore), added to 2.5 ml Talon resin (Clontech) equilibrated in TI buffer, and mixed on a tube rotator for 20 min at 4°C. The resin was spun out, and the supernatant was removed. The resin was washed once with 10 ml TI buffer (10 min on rotator). An equal volume of TI buffer was added to the resin. The resuspension was transferred into a disposable gravity feed column. The column was washed with 5 ml TI buffer and eluted with a solution containing 150 mM imidazole, 20 mM Tris HCl (pH 8.0), 0.5 M NaCl, and 0.1% Triton X-100 containing protease inhibitors; 0.9-ml fractions were collected. The highly purified Swi4-DBD was obtained in an excellent yield (∼10 mg). Its concentration was measured by using the Bradford assay (3a) with IgG as a standard. Swi4-6His-Myc and Flag-Swi6-HA were coexpressed in E. coli Rosetta 2(DE3)pLysS cells (EMD Biosciences) transformed with p659 (Kanr) and p660 (Ampr). Cells were grown to an A600 of ∼1 in 1 liter LB medium containing 34 μg chloramphenicol/ml, 100 μg ampicillin/ml, and 40 μg kanamycin/ml at 30°C and induced for 4 h with 1 mM IPTG. The cells were lysed by sonication in 10 ml TI buffer. The extract was spun to remove debris and filtered, and Swi4 (with associated Swi6) was purified by using Talon resin as described above. To remove excess Swi4, Swi6 (with associated Swi4) was purified by affinity chromatography: the Talon peak fractions were pooled and mixed with 0.5 ml anti-Flag resin (catalog number A2220; Sigma) equilibrated in LS/LT buffer (0.15 M NaCl, 50 mM HEPES-K [pH 7.5], 0.2% Triton X-100, 1 mM Na-EDTA, and 10 μM pepstatin A with protease inhibitors) for 1 h at 4°C on a tube rotator. The resin was washed twice with LS/LT buffer containing 0.5 M NaCl-1% Triton X-100 and once with LS/LT buffer. SBF was eluted with 250 μl 1 mg 3-Flag peptide/ml (catalog number F4799; Sigma) in LS/LT buffer (the pH was adjusted to ∼7 with 0.1 M HEPES-K [pH 7.5]). Elution was done for 60 min at 4°C on a tube rotator. The eluate was stored in aliquots at −80°C.
Gel shift experiments.
The following probes were used: annealed oligonucleotides 667 and 668 (60 bp) for UAS1/UAS2, a 97-bp PCR fragment made using oligonucleotides 1254 and 1255 as primers and p526 as a template (mutated versions of this probe were obtained by using the same primers with plasmids containing the required mutations as templates) for UAS3/UAS4, and a 101-bp PCR fragment made using oligonucleotides 1252 and 1253 as primers and yeast genomic DNA as a template for the CLN2 promoter. The PCR fragments were gel purified, and their concentrations were determined by measuring the A260. Binding reactions were performed by using 5 nM radioactive probe in 15 μl of a solution containing 20 mM HEPES-K (pH 7.5), 70 mM NaCl, 5 mM MgCl2, 10 μM zinc acetate, 0.1 mg/ml bovine serum albumin (BSA) (IgG-free) (catalog number A6918; Sigma), 0.05% Triton X-100, 10% glycerol, and 0.05 mg/ml poly(dI-dC) (catalog number P4929; Sigma) with protease inhibitors (as described above). After incubation at room temperature for 20 min, the reactions were analyzed in 6% (19:1) polyacrylamide gels containing 40 mM Tris-acetate (pH 8.3) and 5% glycerol. The binding of SBF was analyzed with 3.5% (50:1) polyacrylamide gels. Gels were electrophoresed at 150 V for ∼2 h at room temperature using 40 mM Tris-acetate (pH 8.3) as a running buffer. Phosphorimages were quantified by using ImageQuant software.
ChIP experiments.
ChIP experiments were performed as described previously (32), except for the following modifications. Cells (750 ml) were grown in synthetic complete (SC) medium to an optical density at 600 nm (OD600) of ∼0.5. Cultures were synchronized with α-factor (10 μg/ml) for 2 h at 30°C; 125 ml was removed for cross-linking. The remaining cells were washed to remove α-factor, resuspended in fresh medium (625 ml), and shaken at 30°C. Aliquots (125 ml) were removed at specific time points and fixed with 1% formaldehyde for 15 min at room temperature with shaking. Fixation was terminated by the addition of 100 ml 2.5 M glycine to the mixture and shaking for 5 min. Fixed cells were collected by filtration. Cross-linked chromatin was fragmented by using a Misonix Sonicator 3000 (96 pulses of 10 s separated by 10-s pauses for cooling), resulting in DNA fragments of ∼250 bp on average (17). For immunoprecipitation reactions, 0.8 μg cross-linked chromatin (measured using a Hoechst assay) was mixed with protein G magnetic beads (catalog number 100.04; Dynal) carrying prebound M2-anti-Flag (catalog number F3165; Sigma) or anti-HA F7 (sc7392; Santa Cruz) monoclonal antibody. Elutions were performed by using specific peptides (3-Flag peptide) (catalog number F4799; Sigma) or HA peptide (catalog number 11666975001; Roche). Multiplex PCR was performed as described previously (32), using primer pair 1409/1410 to amplify UAS3/UAS4 as a 127-bp fragment, primer pair 1413/1420 for the CLN2 promoter (82 bp), primer pair 1425/1426 for the YLR454 ORF (105 bp), and primer pair 1501/1502 for the HTA2-HTB2 promoter (163 bp).
RESULTS
Reporter assay for the effects of UAS mutations on HTA1-HTB1 expression.
A reporter plasmid was constructed in which all sequence elements important for HTA1 and HTB1 expression were retained (Fig. 1A). These are the four UAS elements, the NEG region located between UAS2 and UAS3, and the 3′-UTRs. In particular, the divergent promoter organization was preserved. In effect, the open reading frames encoding H2A (HTA1) and H2B (HTB1) were replaced with those encoding the fluorescent proteins RedStar-2 (RDS2) (30) and GFP, respectively. The modified HTA1-HTB1 locus was inserted into a single-copy CEN ARS LEU2 plasmid.
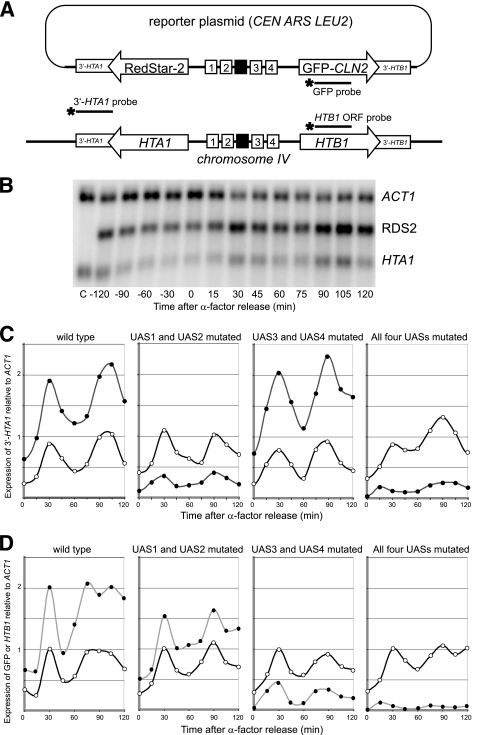
FIG. 1.
The UAS elements direct the cell cycle-dependent activation of HTA1-HTB1. (A) Reporter plasmid for assaying mutations in the HTA1-HTB1 promoter. The coding regions of HTA1 and HTB1 were replaced with RDS2 and GFP fused to PEST sequences from CLN2, respectively. Numbered boxes, UAS elements; black box, NEG region. RDS2 and HTA1 mRNAs were detected by using a single probe (3′-UTR of HTA1). Separate GFP and HTB1 probes were used for HTB1. (B) Cells were synchronized by using α-factor. Shown is a typical Northern blot probed for 3′ HTA1 and ACT1. C, RNA from a strain lacking a reporter plasmid. (C) Expression of RDS2 (filled circles) and HTA1 (open circles) in cells carrying the wild-type reporter or reporters with UAS1 and UAS2 mutated, UAS3 and UAS4 mutated, or all four UAS elements mutated. Relative levels of RDS2 and HTA1 may be compared because the same probe was used to detect both transcripts. (D) Expression of GFP (filled circles) and HTB1 (open circles). All four blots were hybridized at the same time with the same GFP/HTB1 probe mix to allow a direct comparison of GFP signals in each strain.
Our original plan was to measure the output of the reporters by measuring RDS2 and GFP fluorescence. However, this did not work well, perhaps because of slow protein maturation or turnover (3). Instead, reporter expression was measured by hybridizing Northern blots with probes corresponding to the 3′-UTRs of HTA1 and HTB1. This had the advantage that the probe detects both the reporter and the endogenous transcripts, facilitating quantitative comparisons of reporter expression relative to the endogenous HTA1-HTB1 locus. Although this approach worked well for RDS2/HTA1, it was not quantitatively reliable for GFP/HTB1, probably because the 3′-HTB1 probe is extremely AT rich. For HTB1 and GFP, separate coding-region probes were used; the relative amounts were determined by probing all blots with the same probe preparations or by including a reference sample.
Yeast cells transformed with the reporter plasmid were synchronized using α-factor, which arrests cells in the G1 phase of the cell cycle. The cell cycle dependence of RDS2 and HTA1 expression in cells carrying a reporter plasmid containing a wild-type HTA1-HTB1 promoter is shown in Fig. 1B and quantified in Fig. 1C (left). In both cases, two peaks of expression were observed: the first peak occurred ∼30 min after α-factor release, and the second peak occurred after ∼90 to 100 min. The expression profile for RDS2 was qualitatively very similar to that of endogenous HTA1. The amount of RDS2 mRNA was approximately double that of HTA1, for reasons which are unclear. GFP and HTB1 were expressed in a qualitatively similar manner: both gave peaks at ∼30 min and ∼90 min after α-factor release (Fig. 1D). Thus, the reporter genes behaved very similarly to the endogenous genes.
UAS1 and UAS2 drive HTA1, and UAS3 and UAS4 drive HTB1, with some cross talk.
We have shown previously that Spt10 is a dimer and that it requires two UAS elements to bind with high affinity (16, 39). Therefore, the contributions of the four UAS elements to HTA1 and HTB1 expression were determined by mutating pairs of UAS elements: either UAS1 and UAS2 or UAS3 and UAS4. All but one of the bases important for Spt10 binding were mutated in each UAS. The mutation of UAS1 and UAS2 (UAS3 and UAS4 intact) reduced RDS2 expression to only ∼15% of that of the wild-type reporter (Fig. 1C). In contrast, the level of GFP expression was only mildly reduced when UAS1 and UAS2 were mutated (to ∼75% of that of the wild type) (Fig. 1D). Thus, UAS1 and UAS2 are of major importance for the expression of HTA1/RDS2 but have only a minor effect on HTB1/GFP. The mutation of UAS3 and UAS4 (UAS1 and UAS2 intact) had essentially no effect on RDS2 expression but reduced GFP expression to just ∼25% of that of the wild type (Fig. 1C and D). Thus, UAS3 and UAS4 are of major importance for the expression of HTB1/GFP but have no effect on HTA1/RDS2, the opposite effect of the UAS1 and UAS2 mutations. Mutations in all four UAS elements resulted in just a small amount of residual expression of both reporter genes, although this residual expression was clearly cell cycle dependent and perhaps slightly early, peaking at 15 min. The residual expression was at levels significantly below those observed for mutations in two of the four UAS elements, revealing that UAS1 and UAS2 have a small but significant effect on HTB1/GFP and confirming that UAS3 and UAS4 have a relatively minor effect on HTA1/RDS2. In conclusion, UAS1 and UAS2 primarily drive HTA1 and UAS3 and UAS4 primarily drive HTB1, but each pair of UAS elements has a small but significant stimulatory effect on the more distant gene.
Swi4 binding sites in UAS3 and UAS4 overlap the Spt10 binding sites.
Initially, we attributed the effects of mutations in the UAS elements to Spt10. However, there are predicted binding sites for Swi4 (SBF) that are almost entirely contained within the UAS elements (Fig. 2A). Two closely related Swi4 consensus sites have been suggested: CACGAAAA (60) and CGC(C/G)AAA (26). There are no exact matches to the former consensus in the HTA1-HTB1 promoter, but UAS3 and UAS4 both contain the sequence CGCGAAA, an exact match to the latter consensus. In addition, UAS1 and UAS2 contain related sequences (TGCGAAG and GGCGAAA, respectively).
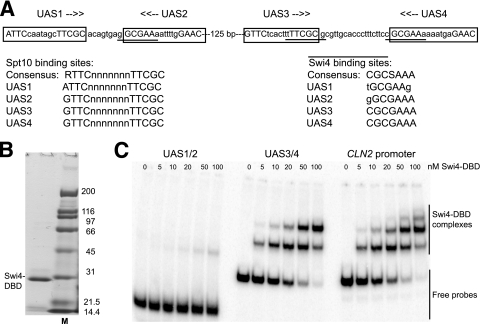
FIG. 2.
The DNA binding domain of Swi4 binds specifically and with high affinity to UAS3 and UAS4 but not to UAS1 and UAS2. (A) Spt10 and Swi4 binding sites in the HTA1-HTB1 promoter. Uppercase type indicates bases important for Spt10 binding. Putative Swi4 binding sites are underlined. Spt10 and Swi4 sites are listed (lowercase type indicates deviations from the consensus). (B) Purified Swi4 DNA binding domain analyzed in a protein gel stained with Coomassie blue. M, marker. (C) Gel shift assay for binding of the Swi4-DBD using a 67-bp probe containing UAS1 and UAS2, a 97-bp probe containing UAS3 and UAS4, and a 101-bp probe with part of the CLN2 promoter (positive control). The Swi4-DBD gives two complexes with UAS3/UAS4, corresponding to the occupation of one or both binding sites. The CLN2 probe contains at least three Swi4 sites.
To determine whether the predicted Swi4 binding sites are actual sites, the Swi4-DBD, corresponding to the N-terminal 202 residues (63), was prepared for gel shift analysis (Fig. 2B). A 101-bp DNA fragment corresponding to part of the CLN2 promoter containing three Swi4 binding sites (47) was used as a positive control (Fig. 2C). As expected, the Swi4-DBD bound with high affinity to the CLN2 promoter: three complexes were observed, corresponding to the binding of one, two, and three Swi4-DBD molecules to the same DNA molecule. Analysis of the binding data revealed that there are two high-affinity sites in the CLN2 promoter with a Kd (dissociation constant) of 15 ± 6 nM (n = 2) and a Hill coefficient close to 1, indicating that these sites are essentially independent. The third site appeared to have a significantly lower affinity. A much weaker binding (Kd of ∼800 nM) was reported for N-terminal fragments of Swi4 to an oligonucleotide containing one of the CLN2 promoter sites (63); it is unclear why our Swi4-DBD binds with a much higher affinity.
The Swi4-DBD bound with similar affinity to a 97-bp fragment containing both UAS3 and UAS4, yielding two complexes corresponding to binding to one or both UAS elements (Kd = 25 ± 6 nM; n = 4). Analysis of the binding data for UAS3 and UAS4 yielded a Hill coefficient close to 1, indicating that the Swi4-DBD binds independently to UAS3 and UAS4; i.e., it binds noncooperatively. This was consistent with a Kd of ∼18 nM measured for UAS3 in a DNA fragment carrying mutations in UAS4. In contrast, the Swi4-DBD bound extremely weakly to a 67-bp fragment containing UAS1 and UAS2 (Fig. 2C), indicating that these are very-low-affinity sites (Kd of >500 nM), presumably reflecting the mismatches to the consensus sequence. In conclusion, the fragment containing UAS3 and UAS4 contains two high-affinity binding sites for the Swi4-DBD, whereas the binding of the Swi4-DBD to the fragment containing UAS1 and UAS2 is so weak that it is unlikely to be biologically significant.
Spt10 and SBF compete for binding to UAS3 and UAS4.
There is extensive overlap between the binding sites for Spt10 and Swi4 in UAS3 and UAS4: both Swi4 sites (TTTCGCG on the opposite strand) overlap the downstream half of the bipartite site recognized by Spt10 (GTTC-N7-TTCGC) (Fig. 2A). This overlap is so extensive that it seemed likely that the binding of Spt10 and binding of Swi4 would be mutually exclusive; i.e., they would compete for binding to UAS3 and UAS4. However, in a competition gel shift experiment in which increasing amounts of Spt10 were mixed with sufficient Swi4-DBD to bind almost all of the probe, additional complexes were observed (Fig. 3A). This indicated that, surprisingly, both Spt10 and the Swi4-DBD could bind to the same DNA molecule, presumably by binding on opposite sides of the DNA helix. However, the Swi4-DBD is much smaller than the SBF complex (Swi4 has 1,093 residues, and Swi6 has 803). Therefore, SBF might inhibit the binding of Spt10 through much increased steric hindrance. To test this hypothesis, recombinant SBF was prepared by the coexpression of Swi4 (with C-terminal 6-histidine and Myc tags) and Swi6 (with an N-terminal Flag tag and a C-terminal HA tag) in E. coli. The Swi4-Swi6 complex was affinity purified first by immobilized metal affinity chromatography to obtain Swi4 and then by using anti-Flag resin to obtain Swi4 bound to Swi6. Although both of these large proteins were susceptible to degradation, reasonably pure recombinant SBF was obtained (Fig. 3B).
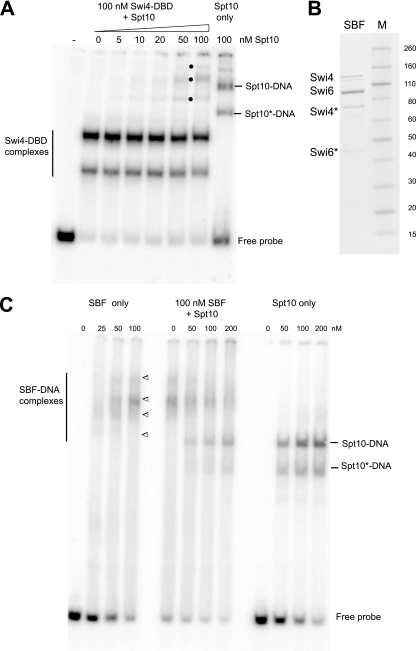
FIG. 3.
SBF and Spt10 compete for binding to UAS3 and UAS4. (A) Simultaneous binding of the recombinant Swi4-DBD and Spt10 to a 97-bp probe containing UAS3 and UAS4. The Swi4-DBD was fixed at 100 nM; Spt10 was added in increasing amounts. Bullets indicates complexes formed only in the presence of both proteins. Spt10* indicates degraded Spt10. (B) Recombinant SBF analyzed in a protein gel stained with Coomassie blue. Swi4* and Swi6* indicate degradation products. (C) Competition between SBF and Spt10 for binding to the UAS3/UAS4 probe. Increasing amounts of Spt10 were mixed with a fixed amount of SBF, and DNA was then added. For SBF-DNA complexes, the upper band is the probe with two SBFs bound, the upper middle complex is the probe with one SBF bound, and the lower middle and lower complexes are the degraded forms of SBF.
The binding of SBF to the probe containing both UAS3 and UAS4 was analyzed in gel shift experiments (Fig. 3C). It was necessary to use a probe with two UAS elements, because Spt10 requires two such elements for high-affinity binding (16). To resolve the complexes, very-low-percentage polyacrylamide gels were used (the Swi4-Swi6 heterodimer has a molecular weight of ∼215,000); these had the unfortunate property of causing the DNA to smear somewhat in the presence of SBF. SBF formed four complexes on the UAS3/UAS4 probe. The two lower complexes were attributed to Swi4-Swi6 degradation products, the major band was attributed to the binding of SBF to either UAS3 or UAS4, and the weak upper band was attributed to the binding of a second SBF at the second UAS. This interpretation is based on the fact that the upper complex did not appear when a probe with mutations in UAS4 was used (not shown). The binding of SBF appeared to be negatively cooperative, unlike that of the Swi4-DBD (Fig. 2C), presumably due to a steric hindrance of the binding of a second SBF by the first. The analysis of SBF binding to UAS3/UAS4 gave a Kd of ∼120 nM. This is significantly higher than that of the Swi4-DBD (Kd = 25 nM), but the smearing of DNA in the gel complicated the measurement of the Kd, and some of the SBF might be inactive.
To determine whether both SBF and Spt10 can bind to the UAS elements at the same time, or whether they compete for binding, gel shift experiments were performed in which sufficient SBF to bind almost all of the probe was mixed with increasing amounts of Spt10 prior to the addition of the DNA (Fig. 3C). As Spt10 was titrated in, complexes corresponding to the binding of Spt10 alone appeared in increasing amounts, and concomitantly, the complexes corresponding to the various SBF complexes decreased, indicating that SBF and Spt10 compete for binding to UAS3 and UAS4. The same result was obtained when SBF was mixed with DNA for 10 min before the addition of Spt10 and when Spt10 was mixed with DNA before the addition of SBF (not shown), indicating that the binding of both factors is reversible. In conclusion, the binding of SBF and binding of Spt10 to UAS3 and UAS4 are mutually exclusive.
Binding-site mutations in UAS3 and UAS4, which discriminate between Spt10 and Swi4.
The fact that UAS3 and UAS4 contain high-affinity sites for Swi4 presents a problem for the interpretation of the reporter experiments (Fig. 1), because the UAS mutations described above would prevent the binding of Swi4 as well as that of Spt10. This is in fact the case (Fig. 4). Spt10 bound with moderately high affinity to UAS3 and UAS4 (Kd = 81 ± 8 nM; n = 2), which is somewhat weaker than that for binding to UAS1 and UAS2 (Kd = 45 nM) (16). The lower affinity of Spt10 for UAS3/UAS4 than that for UAS1/UAS2 might be due to the difference in distance between the UAS elements (6 bp between UAS1 and UAS2 and 17 bp between UAS3 and UAS4). As expected (16), Spt10 did not bind at all to the mutated UAS elements used in the reporter assay (Fig. 4B). The level of binding of the Swi4-DBD to the fully mutated UAS elements was strongly reduced (Kd of ∼200 nM, corresponding to a ∼10-fold reduction in affinity) but, surprisingly, was not completely eliminated (Fig. 4C). This is probably due to the accidental creation of two weak binding sites with one mismatch to the consensus, both of which include some flanking bases and some bases within mutated UAS4 (CGCGCAA and CGCGGAA).
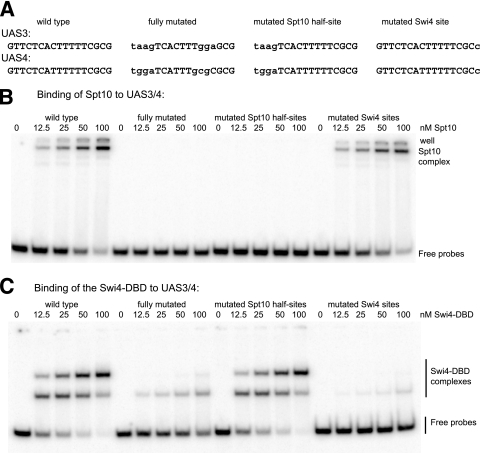
FIG. 4.
Binding-site mutations in UAS3 and UAS4 which discriminate between Spt10 and Swi4 binding in vitro. (A) Mutations (lowercase type) in UAS3 and UAS4 tested for binding by Spt10 or Swi4. Fully mutated indicates mutations tested in the reporter assay (Fig. 1). (B) The Spt10 half-site mutations in UAS3 and UAS4 abolish Spt10 binding in vitro, and the Swi4 site mutations do not affect Spt10 binding. Gel shift assays used a series of 97-bp probes. (C) Mutations in the Swi4 sites strongly reduced the binding of the Swi4-DBD, and mutations in the Spt10 half-sites do not affect Swi4-DBD binding.
New UAS mutations to discriminate between the binding of Swi4 and that of Spt10 were designed. Spt10 binding was eliminated without affecting Swi4 binding by mutating all four bases in each upstream half-site (GTTC), which does not overlap with the Swi4 site (Fig. 4A and B). The elimination of Swi4 binding without affecting Spt10 binding was more challenging, because the downstream Spt10 half-site includes all but the outermost bases of the Swi4 site: T(TTCGC)G. The mutation of both outermost bases to G(TTCGC)T eliminated the binding of the Swi4-DBD but also had a small but significant effect on Spt10 binding (not shown). However, a mutation of the downstream G to C had a major effect on the binding of the Swi4-DBD (Kd of >500 nM), a >40-fold reduction in affinity, without affecting Spt10 binding (Fig. 4B and C). The mutation of this G to A had less effect on affinity (Kd of ∼100 nM). In conclusion, the mutation of the upstream half-site recognized by Spt10 prevents Spt10 binding without affecting that of Swi4, and the mutation of the Swi4 site from CGCGAAA to GGCGAAA severely reduces the binding of Swi4 without affecting that of Spt10. Thus, these mutations can be used to discriminate between the functions of Spt10 and Swi4 in vivo.
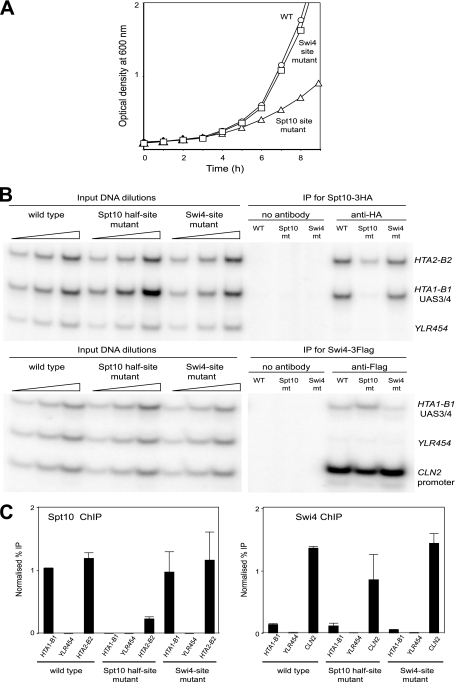
Effects of mutating the Spt10 or Swi4 binding sites in the chromosomal HTA1-HTB1 locus in vivo.
The Spt10 upstream half-site mutations were introduced into all four UAS elements in the native HTA1-HTB1 locus. This strain exhibited a marked growth phenotype: its doubling time in synthetic complete (SC) medium was 2.8 h, whereas the doubling time of the wild type was only 1.7 h (Fig. 5A). This result confirms that Spt10 plays a major role in the regulation of HTA1-HTB1 and shows that Swi4 cannot compensate for the loss of Spt10 function. Another strain, in which point mutations were introduced into UAS3 and UAS4, changing both Swi4 sites from CGCGAAA to GGCGAAA, was constructed. This strain did not have a significant growth defect (doubling time = 1.7 h) (Fig. 5A), indicating that the contribution of SBF to HTA1-HTB1 regulation is more subtle than that of Spt10.
FIG. 5.
Effects of introducing Spt10 and Swi4 binding-site mutations at the chromosomal HTA1-HTB1 locus in vivo. (A) Growth in SC medium of strains carrying Spt10 half-site or Swi4 site mutations at the chromosomal HTA1-HTB1 locus. (B) Binding of Spt10 and Swi4 in cells arrested with α-factor. Shown are data for ChIP using a strain expressing Spt10-3HA and Swi4-3Flag. Multiplex PCR analysis was done using primers for UAS3/UAS4 in HTA1-HTB1 and for the coding region of YLR454 (negative control). Positive controls were the HTA2-HTB2 promoter for Spt10 and the CLN2 promoter for Swi4. Phosphorimages are shown (in the case of Swi4, some lanes between the inputs and the test samples have been removed). (C) Quantitation of ChIP data. Error bars indicate standard deviations for two independent experiments.
The effectiveness of these mutations in preventing the binding of Spt10 or Swi4 in vivo was determined by ChIP using a yeast strain expressing Spt10 with three C-terminal HA tags and Swi4 with three C-terminal Flag tags (Fig. 5B and C). The presence of Spt10-3HA at the HTA1-HTB1 locus in wild-type cells was confirmed by a strong ChIP signal. Spt10-3HA was not detectable in the middle of the YLR454 gene, which served as a negative control. The Spt10 half-site mutations eliminated the binding of Spt10-3HA at the HTA1-HTB1 promoter, as expected. Surprisingly, the binding of Spt10 at the HTA2-HTB2 promoter, used as a positive control, was also strongly reduced in this mutant (∼4-fold). This might indicate that the nuclear Spt10 concentration is lower in the half-site mutant cells, presumably as an indirect result of the poor growth of this strain. Clearly, the loss of the Spt10-dependent activation of HTA1-HTB1 is not compensated for by increased Spt10 binding at HTA2-HTB2. The mutation of the Swi4 sites in UAS3 and UAS4 did not have a significant effect on the binding of Spt10 at HTA1-HTB1 (Fig. 5C). This finding suggests that Swi4 does not compete strongly with Spt10 in arrested cells, because mutations preventing Swi4 binding might be expected to increase Spt10 binding, if that were the case. However, competition between Spt10 and Swi4 at UAS3/UAS4 might be masked by Spt10 binding at UAS1/UAS2, since the distance between UAS2 and UAS3 is only 125 bp, and the average size of the chromatin fragments used in these ChIP experiments was ∼250 bp.
As expected, Swi4 was also clearly detectable at the HTA1-HTB1 promoter. However, the Swi4 signal at the CLN2 promoter, used as a positive control, was ∼10-fold higher than that at HTA1-HTB1. This large quantitative difference might indicate that the occupancy of the Swi4 sites in the HTA1-HTB1 promoter is much lower than that of the Swi4 sites in the CLN2 promoter, even though these sites have very similar affinities for Swi4 (Fig. 2C). However, the fact that the CLN2 promoter contains five predicted high-affinity Swi4 sites (located between positions −605 and −473 relative to the CLN2 start codon), whereas HTA1-HTB1 has only two such sites, must also be taken into account. The point mutations in UAS3 and UAS4 reduced the Swi4 signal by ∼2-fold and had no significant effect on Swi4 binding at the CLN2 promoter (Fig. 5C). The Swi4 site mutations were less effective than expected, given the large decrease in affinity observed in vitro (Fig. 4C), but it should be noted that the level of the signal observed for the Swi4 site mutant was close to background levels (Fig. 5B). The mutation of the Spt10 sites did not result in an increased binding of Swi4 at UAS3/UAS4, as would be expected for a simple competition. However, the binding of Swi4 at the CLN2 promoter was significantly reduced relative to that of wild type cells (Fig. 5C), as observed for Spt10 binding at HTA2-HTB2, suggesting that cell cycle regulation is generally compromised in these slow-growing cells.
In conclusion, UAS mutations that discriminate between Spt10 and Swi4 binding at the HTA1-HTB1 promoter have been described.
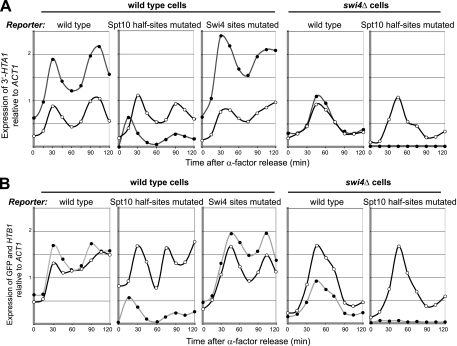
Spt10 is responsible for the major cell cycle-dependent peak in HTA1-HTB1 expression, whereas Swi4 drives a small, early peak of expression.
The effects of the Spt10 upstream half-site and Swi4 site mutations on reporter expression were determined (Fig. 6). In the case of HTA1/RDS2, the mutation of the Spt10 half-sites resulted in a large reduction in expression and a shift in the peak of expression: a small, early, peak was observed 15 min after α-factor release, instead of a major peak at ∼30 min (Fig. 6A). The same was observed for HTB1/GFP (Fig. 6B). The difference between the wild type and the Spt10 half-site mutant represents the contribution of Spt10 to the regulation of HTA1 and HTB1. Thus, Spt10 is required for most of the cell cycle-dependent expression of HTA1 and HTB1. The implication is that the small, early peak is due to Swi4. To determine whether this peak is dependent on Swi4, the reporter carrying the Spt10 half-site mutations was assayed in an swi4Δ strain (Fig. 6). The expression of the endogenous HTA1 and HTB1 genes was delayed relative to that of wild-type cells, peaking 15 min later, at ∼45 min after α-factor release. This is consistent with the slow-growth phenotype exhibited by the swi4Δ strain, which represents an extension of the cell cycle. In contrast, the Spt10 site mutations abolished the expression of RDS2, and GFP expression was reduced to very low levels, confirming that Swi4 is indeed required for the small, early peak of expression.
FIG. 6.
Mutations preventing the binding of Spt10 result in a small, early, Swi4-dependent peak of HTA1 and HTB1 expression. (A) Effects of mutations in the Spt10 half-sites in all four UAS elements, or the Swi4 sites in UAS3/UAS4, on the cell cycle-dependent expression of RDS2 (filled circles) and HTA1 (open circles) in synchronized WT or swi4Δ cells. (B) Effects of the same mutations on GFP (filled circles) and HTB1 (open circles) expression. GFP levels in each of the strains may be compared directly, because the signals were normalized to a common RNA sample used as a standard in all of the blots. The same RNA samples were used for panels A and B.
In wild-type cells, the level of expression of RDS2 and GFP from a reporter plasmid carrying the Swi4 site mutations in UAS3 and UAS4 was somewhat higher than that for the wild-type reporter (Fig. 6), but this difference is probably within experimental error. There was no obvious effect on the shape of the peak at early times, but the expected loss of the small peak in expression due to Swi4 would be difficult to measure in the presence of the major peak due to Spt10. However, the Swi4 site mutations did have an effect on the timing of the GFP peak, which shifted from ∼30 min to ∼45 min, and there might also be a small shift in the RDS2 peak to later times, although this might be within experimental error. Remarkably, this shift was also observed for endogenous HTB1 expression, which has a wild-type promoter, suggesting an effect of the reporter mutations in trans. There was also an effect on the timing of endogenous HTB1 expression in the Spt10 half-site mutant reporter, such that the second cycle was earlier than that for the wild-type reporter. These observations are being investigated further. In conclusion, Swi4 appears to initiate the cell cycle-dependent expression of HTA1 and HTB1, driving a small, early peak in expression, which is followed soon after by an overlapping and much larger peak of expression driven by Spt10.
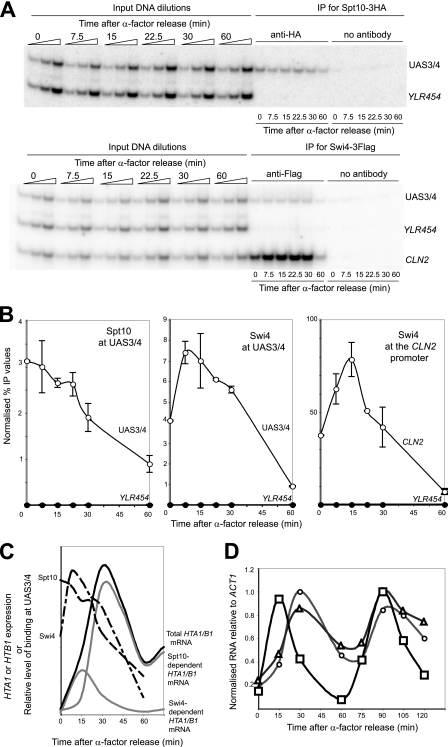
Cell cycle-dependent binding of Spt10 and Swi4 at the HTA1-HTB1 promoter.
To determine whether the peaks of HTA1 and HTB1 expression correlate with the binding of Spt10 and Swi4, their presence at the HTA1-HTB1 promoter was measured by ChIP as a function of time (Fig. 7). Cells containing both Spt10-3HA and Swi4-3Flag were synchronized with α-factor in the same way as described above for the reporter assays. Spt10 was bound at UAS3/UAS4 at relatively high levels in cells arrested with α-factor, eventually declining to just ∼20% of the initial level ∼60 min after α-factor release (Fig. 7A and B), coinciding with the low point of HTA1-HTB1 expression. Thus, the amount of Spt10 bound at the HTA1-HTB1 promoter was highest within 15 min of α-factor release, coinciding with increasing expression levels of the genes, but there was no obvious peak in Spt10 binding. This was also the case for Spt10 binding at UAS1/UAS2 (not shown). Thus, the correlation between Spt10 binding and HTA1/HTB1 expression is not exact, since Spt10 was bound at maximal levels in arrested cells, when the level of expression was low (Fig. 7C).
FIG. 7.
The binding of Spt10 and Swi4 at the HTA1-HTB1 promoter is cell cycle dependent. ChIP experiments used a yeast strain carrying SPT10-3HA and SWI4-3Flag. (A) Binding of Spt10 and Swi4 at the HTA1-HTB1 promoter (phosphorimages). Multiplex PCR was done with primers for UAS3/UAS4, the coding region of YLR454 (negative control), and the CLN2 promoter (positive control for Swi4). The PCR products were 127 bp, 105 bp, and 82 bp, respectively. (B) Quantitation of Spt10 and Swi4 binding. Error bars indicate standard deviations for two independent experiments. The Swi4 data for the CLN2 promoter are plotted separately because the signal was so strong. (C) Timing of Spt10 and Swi4 binding and HTA1/HTB1 transcription (idealized). The relative amounts of Spt10 and Swi4 binding (dashed lines) should not be compared. The idealized expression of HTA1 or HTB1 is shown with the deduced contributions of Swi4 and Spt10 (gray lines) (see text). (D) Expression of HTA1 (circles), HTB1 (triangles), and CLN2 (squares) over two cell cycles. Northern blot data were normalized to ACT1 mRNA and to the highest value in each data set.
The time course of the binding of Swi4 at UAS3/UAS4 and the CLN2 promoter indicated that it was bound at high levels in cells arrested with α-factor, like Spt10 (Fig. 7A and B). However, unlike Spt10, the Swi4 signal increased after α-factor release, showing a clear peak in binding at HTA1-HTB1 between 7.5 and 15 min (this peak appeared slightly later at the CLN2 promoter, at 15 min) and then declining steeply to very low levels after 60 min. The binding profile for Swi4 at HTA1-HTB1 is consistent with expression due to Swi4 in that it peaks concurrently with, or just before, the small, early expression peak at 15 min (Fig. 7C). However, as for Spt10, the level of binding of Swi4 at HTA1-HTB1 and CLN2 was rather high in arrested cells, even though they expressed HTA1 and HTB1 at low levels. This implies that both Spt10 and Swi4 are bound in a poised state in G1 phase, awaiting a signal to activate expression.
In additional ChIP experiments, the time course was extended to include a second cell cycle, but the expected second peaks for both Spt10 and Swi4 were much reduced and broadened relative to those of the first cycle (not shown). Concurrent RNA analysis indicated that HTA1 and HTB1 showed the expected periodic expression (Fig. 7D). However, CLN2 mRNA clearly peaked before the histone mRNAs in the first cycle but was coincident with them in the second cycle. This indicates a significant loss of synchrony in the second cycle and suggests an explanation for the broadened ChIP signals. This conclusion is supported by a fluorescence-activated cell sorter (FACS) analysis performed in parallel (not shown).
DISCUSSION
In summary, we have shown that the UAS elements are required for virtually all expression of HTA1 and HTB1. UAS1 and UAS2 primarily drive HTA1, whereas UAS3 and UAS4 primarily drive HTB1. Thus, each UAS pair has its greatest effect on the closest gene. There is some cross talk: UAS1 and UAS2 have a minor activating effect on HTB1, and UAS3 and UAS4 have a similar effect on HTA1. Most of the transcription is activated by Spt10, but a small peak in the expression of both genes, occurring earlier than the Spt10-dependent peak, is due to SBF, working through sites in UAS3 and UAS4. A role for SBF in HTA1-HTB1 regulation is supported by the demonstration that the predicted Swi4 sites in UAS3 and UAS4 are indeed high-affinity sites for Swi4 and SBF in vitro, that these sites are occupied in vivo, and that the small early peak is dependent on Swi4. However, the role of Spt10 in HTA1-HTB1 regulation is much more important, as shown by the fact that UAS mutations which prevent the binding of Spt10 eliminate most of the reporter expression, sparing only the small, early, Swi4-dependent peak, and result in a quite severe growth phenotype when introduced into the chromosomal HTA1-HTB1 locus (even though HTA2-HTB2, the other locus encoding H2A and H2B, remains intact). The severity of this growth phenotype is similar to that observed previously for an hta1-htb1Δ strain (45), although it is not as severe as that of an spt10Δ strain, as expected, since the latter affects the expression of all four major core histone loci.
Overlapping binding sites for Spt10 and SBF in UAS3 and UAS4.
Unlike UAS1 and UAS2, UAS3 and UAS4 contain overlapping high-affinity binding sites for Spt10 and SBF. The fact that nearly all of the critical bases in the Swi4 consensus site are also part of the downstream half-site recognized by Spt10 suggested that these proteins would be unable to bind simultaneously to the same UAS. However, the Swi4-DBD can bind at the same time as Spt10, presumably on the opposite face of the DNA helix, suggesting that the full-size proteins might perhaps bind simultaneously, if one or both were to undergo conformational changes. However, there was no sign of this in vitro, where Spt10 and SBF competed for binding to UAS3/UAS4.
Analysis of the sequences of the other major core histone gene promoters revealed only one exact match to the Swi4 consensus site, located near HHF2; this is a high-affinity site (D. J. Clark, unpublished data). Although this Swi4 site overlaps putative UAS6 (16), UAS6 is a weak match to the Spt10 consensus and does not bind Spt10 (G. Mendiratta, unpublished data). There are no high-affinity Swi4 sites in the HTA2-HTB2 or the HHT1-HHF1 promoters, but there are some weak sites that might have biological significance. Both the HTZ1 and HHO1 promoters contain a predicted high-affinity Swi4 site, suggesting that SBF also plays a significant role in the expression of H2A-Z and H1, respectively. Thus, overlapping Swi4 and Spt10 binding sites are not a general feature of histone gene regulation but appear to be specific for HTA1-HTB1, which is the most important of the two loci encoding H2A and H2B (45).
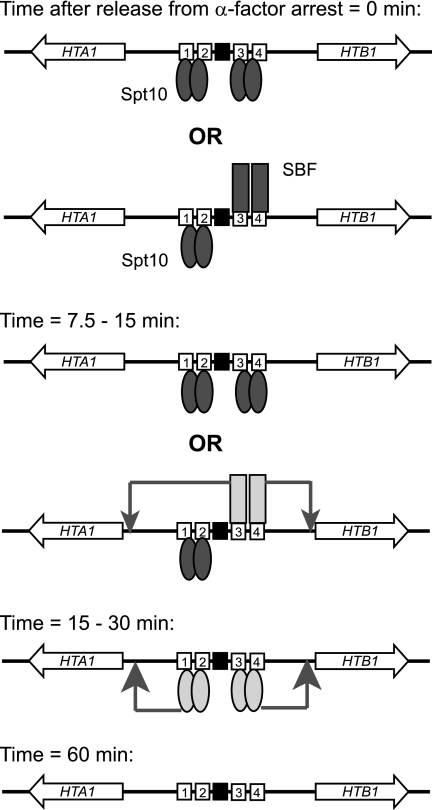
A working model for cell cycle-dependent activation of HTA1-HTB1.
Based on our data and those of others, we propose a model for the activation of HTA1-HTB1 (Fig. 8). In cells arrested with α-factor, Spt10 dimers may bind to UAS1/UAS2 and to UAS3/UAS4. There is competition between Spt10 and SBF for binding at UAS3/UAS4 but not at UAS1/UAS2. The relative occupancies of Spt10 and SBF at UAS3/UAS4 will be governed by their relative concentrations and affinities. Transcription factor binding to DNA is expected to be in dynamic equilibrium: each factor binds and dissociates, thus creating opportunities for competing factors to bind. The extent to which the UAS elements are assembled into nucleosomes will be important in determining which factors are bound and when they are bound.
FIG. 8.
Model for activation of HTA1-HTB1. At early times, there are at least two possible configurations for the HTA1-HTB1 promoter, because the binding of SBF and Spt10 at UAS3/UAS4 is mutually exclusive (indicated by “OR”). Both Spt10 and SBF bind before HTA1 and HTB1 transcription is activated and are therefore inactive in arrested cells. SBF is activated first, driving small peaks of expression of both genes. Soon afterwards, Spt10 is activated, driving the major peaks of expression of both genes. After 60 min, little Swi4 is bound, and Spt10 is reduced to ∼20% of its initial level. Numbered boxes, UASs; black box, NEG region; pair of ovals, Spt10 dimer; single rectangle, SBF. Inactive Spt10 and SBF are shaded dark; activated forms are shaded light. Arrows indicate activated promoters.
In arrested cells, Spt10 and SBF bind but do not activate HTA1-HTB1. In the case of SBF, the lack of activation is due to Whi5, a repressor which binds to SBF at its target promoters in early G1 phase (9, 10) and helps to recruit the histone deacetylase Rpd3L (62). This repression is later relieved by the phosphorylation of Whi5 by the Cln3-Cdc28 kinase (9, 10). SBF also directs the recruitment of the FACT complex (62). Nothing is known of this aspect of Spt10 regulation, but Spt10 activity might be regulated in an analogous manner, perhaps resulting in the activation of its putative HAT function, given that the HAT domain is capable of activating transcription in vivo (28). Alternatively, Spt10 might act at zero time, when its binding is maximal, leaving a memory of its presence (e.g., histone acetylation), which eventually facilitates transcription after 15 to 30 min.
Our model proposes that activated SBF stimulates HTA1 and HTB1 transcription, resulting in a small early peak of expression of both genes, ∼7.5 min after α-factor removal. At this time, Spt10 remains inactive. After 15 to 30 min, Spt10 is activated and drives much larger peaks of expression of both genes. By ∼30 min, Spt10 is fully active, but SBF contributes little. There is a precipitous decline in the binding of both Spt10 and SBF, which, in the case of SBF, reflects the phosphorylation of Swi6 and export from the nucleus (21). By 60 min, HTA1 and HTB1 are at their lowest point of expression, little or no SBF is bound, and the amount of bound Spt10 has declined to ∼20% of its maximum. It is suggested that the remaining Spt10 returns to its inactive, poised state, awaiting the next S phase.
Roles of the UAS and NEG systems in the regulation of the histone genes.
In the model (Fig. 8), it is implicit that the cell cycle regulation of HTA1-HTB1 is centered on Spt10, SBF, and the UAS elements. However, there is strong evidence for an important role for the NEG region and its associated repressive factors in the cell cycle control of HTA1-HTB1: the deletion of NEG or of NEG-associated factors results in an altered cell cycle-dependent expression of HTA1-HTB1 such that the level of expression remains high after the first peak, merging with the second (48, 50). Recently, it was proposed that the establishment of repressive chromatin domains is the primary mechanism of cell cycle regulation of the histone promoters (18). It was suggested that Rtt109 (the major HAT for H3-K56) together with the SWI/SNF and Yta7 chromatin-remodeling complexes counteract nucleosome assembly mediated by the histone chaperone Rtt106 and the HIR complex at the HTA1-HTB1 promoter. In this model, the accessibility of the HTA1-HTB1 promoter to RNA polymerase II is regulated through nucleosome assembly. However, no role for activators or the UAS elements was envisaged, even though swi4Δ and spt10Δ cells showed a reduced level of expression of an HTA1 reporter (18). Clearly, both systems play important roles in the regulation of HTA1-HTB1.
Instead, we propose that the primary mechanism of the cell cycle regulation of HTA1-HTB1 is the UAS system operating through Spt10 and SBF. This is based on several lines of evidence. First, isolated UAS elements confer cell cycle-regulated expression on a reporter with the correct timing (20, 49). Although the CCR′ element, which is located within the NEG region, can also confer cell cycle regulation on a reporter, the timing is too early (49). Second, all four of the major histone gene promoters contain histone UAS elements, which bind Spt10 (16). Spt10 requires a pair of UAS elements to bind with high affinity, and these are not found anywhere else in the genome. Third, NEG elements have not been identified in HTA2-HTB2 and so cannot account for the cell cycle regulation of this promoter. Fourth, Spt10 is not essential, but spt10Δ cells are very sick. Furthermore, the spt10Δ mutation is synthetically lethal with the swi4Δ mutation (29). This might well reflect the fact that both histone gene activators would be absent in the double mutant, although Swi4 is important for the activation of other genes at the G1-to-S-phase transition. Finally, our data here show that multiple mutations in all four UAS elements in HTA1-HTB1 almost completely eliminate reporter gene expression, even though the NEG region is intact.
Perhaps the most confusing aspect of the identification of Spt10 as the major activator of the histone genes has been that the levels of expression of three of the four histone loci are only modestly reduced in spt10Δ cells (HTA2-HTB2 shows a stronger reduction in levels of expression than do the others) (16, 28). However, these measurements were performed with asynchronous cells; the loss of expression is much clearer in synchronized cells (68). The relatively mild effects on histone gene expression in spt10Δ cells can be accounted for by considering the following: (i) the defective chromatin structure assembled in the absence of Spt10 might result in a derepression of the basal transcription of the histone genes (16); (ii) the activation of HTA1-HTB1 by a UAS belonging to another gene upstream or downstream of HTA1-HTB1 might occur, since a UAS is effective at a much greater distance from its target gene in spt10Δ cells (13); (iii) the very slow growth of spt10Δ cells could reflect the existence of a checkpoint system requiring cells to wait until sufficient histone mRNA is produced to allow the completion of S phase; and (iv) the fact that the Spt10 half-site mutations eliminate all but the early, Swi4-dependent, peak of HTA1 and HTB1 expression is very good evidence that Spt10, working through the UAS elements, is indeed primarily responsible for the cell cycle-dependent expression of HTA1-HTB1. Because a reporter was used in this experiment, the effects of Spt10 on transcription are divorced from the indirect and downstream effects of aberrant histone gene expression.
So what might the role of the NEG system be? The NEG system might act as a checkpoint to communicate the presence of stalled replication forks to the histone genes. This hypothesis is based on the previously demonstrated role of NEG in shutting down histone gene expression in the presence of hydroxyurea (54, 58, 67). The NEG system might temporarily shut down the UAS system, awaiting a signal to continue, probably involving a chromatin-based mechanism (18). In other words, the NEG system modulates the activity of the UAS system. In this model, the fact that the CCR′ element (which is within the NEG region) confers cell cycle-dependent expression on a reporter driven by an unrelated UAS (49) could be accounted for by a repressor that recognizes CCR′, the activity of which is also under cell cycle control, such that it is inactive during early S phase. We are currently investigating this hypothesis.
Acknowledgments
We thank Geetu Mendiratta for providing unpublished data. We thank Rohinton Kamakaka and David Stillman for helpful comments on the manuscript.
This work was supported by the Intramural Research Program of the NIH (NICHD).
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Andrews, B. J., and L. A. Moore. 1992. Interactions of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. U. S. A. 89:11852-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baetz, K., and B. Andrews. 1999. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell. Biol. 19:6729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Breeden, L. L. 2003. Periodic transcription: a cycle within a cycle. Curr. Biol. 13:R31-R38. [DOI] [PubMed] [Google Scholar]
- 5.Breeden, L., and G. E. Mikesell. 1991. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 5:1183-1190. [DOI] [PubMed] [Google Scholar]
- 6.Breeden, L., and K. Nasmyth. 1987. Cell cycle control of the yeast HO gene: cis- and trans-activating regulators. Cell 48:389-397. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, S. G., M. del Olmo, P. Beglan, and U. Bond. 2002. A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3′ processing and cell cycle regulation. Mol. Cell. Biol. 22:8415-8425. (Author's correction, 28:1873, 2008.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark-Adams, C. D., D. Norris, M. A. Osley, J. S. Fassler, and F. Winston. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2:150-159. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo, M., et al. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117:899-913. [DOI] [PubMed] [Google Scholar]
- 10.de Bruin, R. A. M., W. H. McDonald, T. I. Kalashnikova, J. Yates, and C. Wittenberg. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF-bound repressor Whi5. Cell 117:887-898. [DOI] [PubMed] [Google Scholar]
- 11.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast SWI/SNF complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 12.Dirick, L., T. Moll, H. Auer, and K. Nasmyth. 1992. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature 357:507-513. [DOI] [PubMed] [Google Scholar]
- 13.Dobi, K. C., and F. Winston. 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:5575-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollard, C., S. L. Ricupero-Hovasse, G. Natsoulis, J. D. Boeke, and F. Winston. 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:5223-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson, P. R., et al. 2005. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol. Cell. Biol. 25:9127-9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, X., N. Lamarre-Vincent, Q. Wang, and K. Struhl. 2008. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucleic Acids Res. 36:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fillingham, J., et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35:340-351. [DOI] [PubMed] [Google Scholar]
- 19.Formosa, T., et al. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman, K. B., L. R. Karns, K. A. Lutz, and M. M. Smith. 1992. Histone H3 transcription in Saccharomyces cerevisiae is controlled by multiple cell cycle activation sites and a constitutive negative regulatory element. Mol. Cell. Biol. 12:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geymonat, M., A. Spanos, G. P. Wells, S. J. Smerdon, and S. G. Sedgwick. 2004. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell. Biol. 24:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, E. M., et al. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groth, A., W. Rocha, A. Verreault, and G. Almouzni. 2007. Chromatin challenges during DNA replication and repair. Cell 128:721-733. [DOI] [PubMed] [Google Scholar]
- 24.Gunjan, A., J. Paik, and A. Verreault. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87:625-635. [DOI] [PubMed] [Google Scholar]
- 25.Han, J., et al. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653-655. [DOI] [PubMed] [Google Scholar]
- 26.Harbison, C. T., et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hereford, L., K. Fahrner, J. Woolford, M. Rosbash, and D. B. Kaback. 1979. Isolation of yeast histone genes H2A and H2B. Cell 18:1261-1271. [DOI] [PubMed] [Google Scholar]
- 28.Hess, D., B. Liu, N. R. Roan, R. Sterglanz, and F. Winston. 2004. Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 histone acetyltransferase domain and Spt21. Mol. Cell. Biol. 24:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess, D., and F. Winston. 2005. Evidence that Spt10 and Spt21 of S. cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics 170:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janke, C., et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947-962. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. U. S. A. 99:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, Y., N. McLaughlin, K. Lindstrom, T. Tsukiyama, and D. J. Clark. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 26:8607-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch, C., T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S-phase. Science 261:1551-1557. [DOI] [PubMed] [Google Scholar]
- 34.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 35.Mariño-Ramirez, L., I. K. Jordan, and D. Landsman. 2006. Multiple independent evolutionary solutions to core histone gene regulation. Genome Biol. 7:R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateus, C., and S. V. Avery. 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16:1313-1323. [DOI] [PubMed] [Google Scholar]
- 37.Mellor, J. 2009. Linking the cell cycle to histone modifications: Dot1, G1/S and cycling K79me2. Mol. Cell 35:729-730. [DOI] [PubMed] [Google Scholar]
- 38.Mendiratta, G., P. R. Eriksson, C. H. Shen, and D. J. Clark. 2006. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J. Biol. Chem. 281:7040-7048. [DOI] [PubMed] [Google Scholar]
- 39.Mendiratta, G., P. R. Eriksson, and D. J. Clark. 2007. Cooperative binding of the yeast Spt10 activator to the histone upstream activating sequences is mediated through an N-terminal dimerization domain. Nucleic Acids Res. 35:812-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moran, L., D. Norris, and M. A. Osley. 1990. A yeast H2A-H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 4:752-763. [DOI] [PubMed] [Google Scholar]
- 41.Natsoulis, G., C. Dollard, F. Winston, and J. D. Boeke. 1991. The products of the SPT10 and SPT21 genes of Saccharomyces cerevisiae increase the amplitude of transcriptional regulation at a large number of unlinked loci. New Biol. 3:1249-1259. [PubMed] [Google Scholar]
- 42.Natsoulis, G., F. Winston, and J. D. Boeke. 1994. The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics 136:93-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 44.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norris, D., and M. A. Osley. 1987. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 7:3473-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nourani, A., F. Robert, and F. Winston. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1496-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2 and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66:1015-1026. [DOI] [PubMed] [Google Scholar]
- 48.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 49.Osley, M. A., J. Gould, S. Kim, M. Kane, and L. Hereford. 1986. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45:537-545. [DOI] [PubMed] [Google Scholar]
- 50.Osley, M. A., and D. Lycan. 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prochasson, P., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulze, J. M., et al. 2009. Linking cell cycle to histone modifications: SBF and H2B mono-ubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35:626-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 54.Sherwood, P. W., and M. A. Osley. 1991. Histone regulatory (hir) mutations suppress δ insertion alleles in Saccharomyces cerevisiae. Genetics 128:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidorova, J. M., G. E. Mikesell, and L. L. Breeden. 1995. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear regulation. Mol. Biol. Cell 6:1641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon, L., et al. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 57.Smith, M. M., and K. Murray. 1983. Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J. Mol. Biol. 169:641-661. [DOI] [PubMed] [Google Scholar]
- 58.Spector, M. S., A. Raff, H. DeSilva, K. Lee, and M. A. Osley. 1997. Hir1p and Hir2p function as transcriptional co-repressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taba, M. R., I. Muroff, D. Lydall, G. Tebb, and K. Nasmyth. 1991. Changes in a SWI4/6 DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 5:2000-2013. [DOI] [PubMed] [Google Scholar]
- 61.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 62.Takahata, S., Y. Yu, and D. J. Stillman. 2009. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 28:3378-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, I. A., et al. 2000. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 39:3943-3954. [DOI] [PubMed] [Google Scholar]
- 64.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 65.Wang, G. G., C. D. Allis, and P. Chi. 2007. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 13:363-372. [DOI] [PubMed] [Google Scholar]
- 66.Xu, H., L. Johnson, and M. Grunstein. 1990. Coding and noncoding sequences at the 3′ end of yeast histone H2B mRNA confer cell cycle regulation. Mol. Cell. Biol. 10:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu, H., U. Kim, T. Schuster, and M. Grunstein. 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5249-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375-385. [DOI] [PubMed] [Google Scholar]