Abstract
The interaction of architectural proteins such as the linker histone H1 and high-mobility-group (HMG) proteins with nucleosomes leads to changes in chromatin structure and histone modifications and alters the cellular transcription profile. The interaction of HMG proteins with chromatin is dynamic. However, it is not clear whether the proteins are constantly and randomly redistributed among all the nucleosomes or whether they preferentially associate with, and turn over at, specific regions in chromatin. To address this question, we examined the genome-wide distribution of the nucleosome binding protein HMGN1 and compared it to that of regulatory chromatin marks. We find that HMGN1 is not randomly distributed throughout the genome. Instead, the protein preferentially localizes to DNase I hypersensitive (HS) sites, promoters, functional enhancers, and transcription factor binding sites. Our results suggest that HMGN1 is part of the cellular machinery that modulates transcriptional fidelity by generating, maintaining, or preferentially interacting with specific sites in chromatin.
In eukaryotic cells, DNA is wrapped around the four core histone proteins H2A, H2B, H3, and H4 to form the fundamental structural unit of chromatin, the nucleosome. In addition to core histone proteins, linker histone H1 plays a significant role in organizing chromatin into higher-order structures (15, 29, 56). These higher-order structures are inhibitory to chromatin-related processes, such as transcription, replication, and DNA repair. Therefore, alterations to the structure of the chromatin fiber are critical to control cellular processes and regulate the expression fidelity of a distinct battery of genes in particular cell types (39). Such regulation is realized by a combination of several factors, including posttranslational modifications of histone tails and chromatin remodeling enzymes. Among the many factors that contribute to this process are the architectural proteins, such as histone H1 and members of the high-mobility-group (HMG) superfamily (5, 12). Histone H1 reduces the accessibility of chromatin to regulatory factors by promoting and stabilizing condensed chromatin structures, while HMG proteins compete with histone H1 and make the chromatin more accessible (12, 16). The HMG superfamily consists of three families (HMGA, HMGB, and HMGN) that have distinct functional motifs and cellular functions (5, 9). Changes in the levels of these proteins can result in developmental defects leading to various diseases (for a review, see the article by Hock et al. [31]).
Among the HMG proteins, the HMGN family is unique in its ability to bind directly to the nucleosome core particles. The HMGN family consists of five members: HMGN1, HMGN2, HMGN3, HMGN4, and HMGN5 (41). These proteins display no specificity to the underlying DNA sequence and bind chromatin through their nucleosomal binding domains (8, 44, 46). Interestingly, HMGN proteins have been implicated in the generation and maintenance of open chromatin regions due to their association with transcriptionally active genes (54). While stabilization of nucleosomal core particles by HMGN1 and HMGN2 has been suggested by several earlier studies (19, 26, 43), the exact role of HMGN proteins in this process is unclear. Likewise, their effect on the chromatin remodeling activity of ATP-dependent enzymes is controversial (30, 43). Moreover, while studies have demonstrated a role for HMGN proteins in relaxing chromatin and maintaining active chromatin conformation, contradictory data regarding their association with gene activity exist (8, 10, 21, 42, 46, 51, 54). This lack of consensus is primarily due to most of the previous studies relying on in vitro systems, which has resulted in an incomplete understanding of the mechanism of action of this important group of proteins.
It is clear that HMGN proteins bind dynamically to nucleosomes and alter the architecture of their binding sites. However, it remains unresolved whether the proteins are uniformly distributed throughout the genome, randomly and continuously affecting the structure of all the nucleosomes, or whether they preferentially bind to selected sites at which they are continuously turning over. To gain insights into this question, we used ChIP-Seq to identify genome-wide localization profiles of HMGN1 in human CD4+ T cells and compared the distribution to that of other features known to affect chromatin structure and activity.
MATERIALS AND METHODS
CD4+ T-cell purification, ChIP-Seq, and scoring profiles.
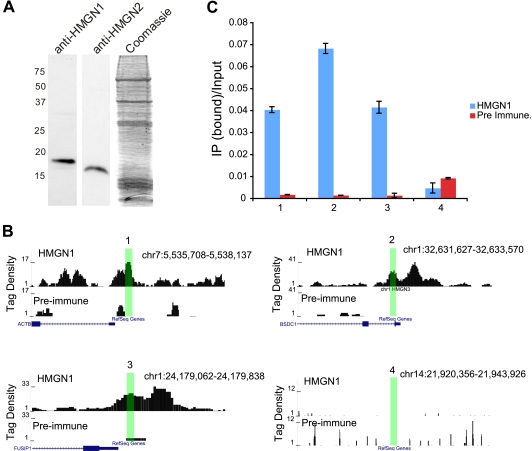
Human CD4+ T cells were isolated as previously described (3). For chromatin immunoprecipitation (ChIP), the formaldehyde-cross-linked cells were sonicated to obtain DNA fragments ranging in size from 150 to 200 bp. The antibodies used for ChIP specifically recognized the HMGN1 protein (Fig. 1A). Cluster generation and sequencing were performed as previously described (3).
FIG. 1.
Confirmation of HMGN1 signals obtained by ChIP-Seq. (A) Western blot showing the specificity of HMGN1 antibodies. The Coomassie blue staining shows the whole protein extract from cells, which was used for Western blotting with antibodies specific for either human HMGN1 or HMGN2. (B and C) Confirmation of HMGN1 peaks using TaqMan quantitative real-time PCR. (B) Boxes 1 to 3, HMGN1 ChIP-Seq positive regions; box 4, HMGN1 ChIP-Seq negative region. Highlighted regions were tested for HMGN1 enrichment using quantitative PCR analysis. (C) Quantitative PCR validation of ChIP-Seq results. HMGN1 binding is enriched in regions 1 to 3 compared to IgG control, whereas region 4 shows no HMGN1 enrichment.
The ChIP-Seq reads were aligned to the March 2006 build (NCBI Build 36.1; hg18) of the human genome. All reads mapping uniquely with zero, one, or two mismatches were retained for further analysis. To reduce the effects of amplification biases, redundant reads were filtered to remove any more than three copies at the same location. A total of 18,666,896 nonredundant uniquely mapped reads were obtained for HMGN1. A replicate library consisting of 24,632,415 nonredundant uniquely mapped reads was also created through a similar process.
For visualization of ChIP-Seq data on a genome browser, score profiles for the HMGN1 data were created in a manner similar to that described previously (45). Briefly, the genome was scanned with 10-bp windows, and any read aligning to the sense (antisense) strand within 80 bp upstream (downstream) of a given window contributes an equal amount to the score of the given window. The data displayed in Fig. 2 consist of ChIP-Seq data for CTCF (20), H3K27me3 (3), H3K36me3 (3), H3K4me3 (3), H3K9ac (53), and RNA Pol II (3).
FIG. 2.
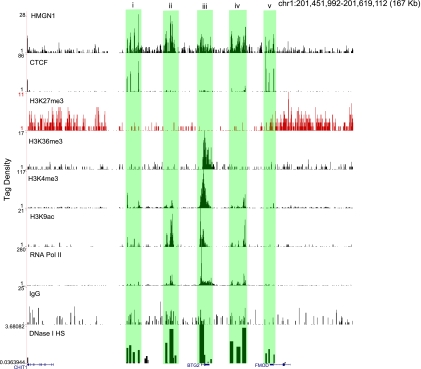
HMGN1 binding profile at BTG2 locus. Shown are the binding profiles of HMGN1 along with CTCF, H3K27me3, H3K36me3, H3K4me3, H3K9ac, RNA Pol II, IgG, and DNase I HS sites at a 167-kb region surrounding the BTG2 gene (see Materials and Methods for details on scoring profiles). Boxes i to v, colocalization of HMGN1 with DNase I HS sites. Note that boxes i and v also contain CTCF binding sites. The IgG control does not shown any enrichment.
HMGN1 binding site detection with SICER.
Binding sites for HMGN1 were identified using the spatial clustering approach for the identification of ChIP-enriched regions (SICER) algorithm (57) to identify domains enriched for HMGN1 ChIP-Seq signals to be considered binding sites. The parameters used to run SICER were as follows: window size, 10 bp; gap size, 50 bp; E value, 1; q value, 10−3; and an effective genome size of 74% of the entire human genome (corresponding to 25-bp reads). This procedure produces 146,127 HMGN1 binding regions in the human genome.
ChIP-Seq validation.
For validation of the ChIP-Seq signals we chose several regions (Fig. 1B, regions 1 to 3), which we found to be highly enriched in the HMGN1-precipitated chromatin. We chose region 4, which did not show any HMGN1 enrichment, as a control. Real-time TaqMan quantitative PCR analysis of these regions validated the ChIP-Seq results (Fig. 1C) using the following primers: 1F, CGCACAGTGCAGCATTTTTT; 1R, CTCCCTCCTCCTCTTCCTCAA; 1-Probe, ACCCCCTCTCCCCTCCTTTTGCG; 2F, AACGCCTAGGAGCAAAACGA; 2R, TCAGTAAATAGTGGTGATGTCATGCA; 2-Probe, AAGCTCACCCTTCCGCCATCTTGC; 3F, AGCCTCAGACACACACAGCTAGAG; 3R, CTTTGCGCGGAGGTAGGA; 3-Probe, CTCTGCTACGGCAACCGACTCTTCTCG; 4F, CCTGAGTGCAAACAGAGATTGAAA; 4R, GGACATGCTTCACCTATGATCAAC; 4-Probe, AACATTTTTCTGTCACTGCCCAATGGGA.
DNase I hypersensitive site data.
The DNase I hypersensitive site (HS) data from Boyle et al. (6) were obtained for the May 2004 (NCBI Build 35) build of the human genome from the UCSC table browser (34). Coordinates were converted to those of the March 2006 (NCBI Build 36.1) build using the liftOver tool provided by UCSC (34).
High-resolution analysis of HMGN1 and nucleosome positions at promoter regions, binding sites, and DNase I HS site boundaries.
The binding profiles of HMGN1 at promoter regions were created in a manner similar to that described previously (45). Briefly, all UCSC genes (32) were filtered so that if any genes overlapped, only a single gene was retained for further analysis. Then a stalling index was calculated using polymerase II (Pol II) ChIP-Seq data as the ratio of the number of Pol II ChIP-Seq reads found in a 1-kb region centered on the transcription start site (TSS) over the mean number of Pol II ChIP-Seq reads found in nonoverlapping 1-kb windows throughout the gene. Genes were classified as expressed if the stalling index was between 1 and 3 and the average number of Pol II ChIP-Seq reads in 1-kb windows throughout the gene was at least 5. A total of 8,936 expressed genes were identified through this process. All genes in this set were aligned with respect to their TSS. ChIP-Seq reads were separated by strands and read starts for reads on both strands were counted in 5-bp windows from 1 kb upstream to 1 kb downstream of the TSS.
CTCF binding sites were obtained from the work of Cuddapah et al. (20) and aligned relative to their midpoints. YY1 binding sites were identified using the model-based analysis for ChIP-Seq (MACS) algorithm (58) with a P value threshold of 10−10. All promoter-bound (±1 kb surrounding annotated TSSs) sites for CTCF and YY1 were removed from further analysis. Profiles of HMGN1 and nucleosomes were created as described above.
Histone modification score profiles at DNase I HS sites.
The coordinates for DNase I HS sites were obtained from the work of Boyle et al. (6). ChIP-Seq reads were adjusted to “center” them on the sequenced fragment by moving reads one-half of the average fragment size along the strand to which each read aligned. The score profiles shown in Fig. 3E to G were obtained by summing adjusted read starts in windows from 2 kb upstream to 2 kb downstream of each DNase I HS site where the window size is 5% of the total hypersensitive site within the HS site and 100 bp outside the site. The score in each window was normalized to the number of total base pairs within the window and the number of reads in each ChIP-Seq library.
FIG. 3.
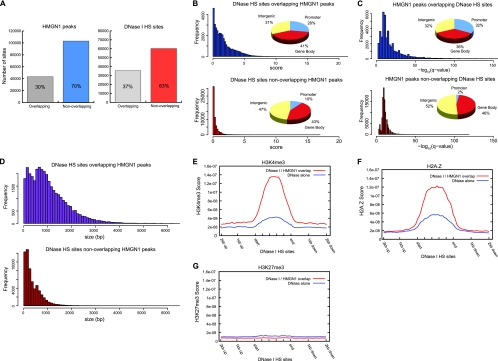
HMGN1 binding sites and DNase I HS sites overlap. (A) The overlap between HMGN1 and DNase I HS sites in the genome. (B) DNase I HS sites that overlap HMGN1 sites are more hypersensitive and more likely to occur in promoters than DNase I HS sites that do not overlap HMGN1 sites. (C) HMGN1 sites that overlap DNase I HS sites have stronger HMGN1 signals and are more likely to occur in promoters than HMGN1 sites that do not overlap DNase I HS sites. (D) DNase I HS sites that overlap HMGN1 sites tend to be larger than those sites that do not overlap HMGN1 sites. (E) H3K4me3 is enriched at DNase I HS sites overlapping HMGN1 sites relative to DNase I HS sites that are not overlapping HMGN1 sites (see Materials and Methods for details on modification score profiles). (F) H2A.Z is enriched at DNase I HS sites overlapping HMGN1 sites relative to DNase I HS sites that are not overlapping HMGN1 sites. (G) H3K27me3 is depleted at the HMGN1 binding sites.
Plotting HMGN1 signals across intron-exon boundaries.
The coordinates of exon starts and ends were obtained from the UCSC gene coordinates for all genes classified as expressed and nonexpressed genes from the work of Schones et al. (45). The score profiles shown (see Fig. 5) were obtained by summing adjusted read starts in 5-bp windows and normalizing to the total number of genes in each set.
Plotting HMGN1 signals across active and inactive genes.
Lists of expressed and nonexpressed genes were obtained from the work of Schones et al. (45). For each gene in a list, the HMGN1 reads were summed in 3-kb windows from 60 kb upstream to the transcription start site (TxStart) and from the transcription end site (TxEnd) to 60 kb downstream. Within the gene bodies, reads were summed in windows equal to 5% of the length of the given gene. For all regions, the total counts were averaged over all genes in each gene set to obtain the read densities.
Data accession numbers.
We deposited sequence reads for HMGN1, YY1, and IgG, files containing all aligned reads, and files containing statistically significant peaks identified in this study in NCBI's Gene Expression Omnibus (GEO series accession number GSE25674).
RESULTS
HMGN1 is enriched in active chromatin regions.
We found that HMGN1 is enriched in transcriptionally active domains, as exemplified by the BTG2 locus (Fig. 2). The BTG2 locus shows a clear domain structure: (i) an enrichment of active chromatin marks, such as H3K4me3 and H3K9ac, is seen close to the gene; (ii) the gene is actively transcribed in resting CD4+ T cells with RNA Pol II across the promoter and gene body and H3K36me3 marking the gene body; (iii) the regions immediately surrounding the gene locus contain strong signals for H3K27me3, a known repressive mark; and (iv) the insulator binding and chromatin domain protein CTCF is found at the edges of the repressive H3K27me3 domains, consistent with previous findings (20).
We find that HMGN1 signals are enriched in the active domains but not in the transcriptionally inactive chromatin regions marked by H3K27me3. The BTG2 locus contains several DNase I HS sites (6), which indicate the presence of potential regulatory elements (18). Strikingly, each of the HS sites is marked by strong signals of HMGN1 (Fig. 2, boxes i through v). Furthermore, CTCF binding sites at the domain boundaries are also marked by HMGN1 binding (Fig. 2, boxes i and v).
HMGN1 binding colocalizes with DNase I hypersensitive sites.
Hypersensitivity to DNase I digestion is a hallmark of regulatory regions in chromatin. Several different gene regulatory elements have been shown to be DNase I hypersensitive (6, 22, 28, 47). However, DNase I hypersensitive sites are also found in transcriptionally silent regions of the genome. A possible link between the presence of HMGN proteins and DNase I HS sites was previously detected in transcriptionally active chromatin regions (54). However, this link has not been rigorously tested, largely due to the lack of information on HMGN binding sites in chromatin. Boyle et al. recently profiled DNase I HS sites in human CD4+ T cells, identifying 94,925 sites across the genome (6). To investigate the relationship between HMGN1 and DNase I hypersensitivity, we obtained the DNase I HS sites identified by Boyle et al. (6) (see Materials and Methods for details) and analyzed their overlap with HMGN1 binding sites.
Analysis of the HMGN1 and DNase I binding sites revealed that 30% of HMGN1 sites overlapped DNase I HS sites and 37% of DNase I HS sites overlapped HMGN1 sites (Fig. 3A). Considering that the binding sites of HMGN1 and DNase I HS sites constitute only 1.5% and 2% of the genome, respectively (assessed by counting the total number of base pairs in each set of enriched regions), the overlap between HMGN1 and DNase I HS sites is extensive.
In addition, Boyle et al. demonstrated that a range of hypersensitivity exists across identified DNase I HS sites and they developed a method to quantify the degree of hypersensitivity (6, 7). To investigate the relationship between HMGN1 binding and DNase I hypersensitivity, we compared the hypersensitivity of the DNase I HS sites that overlap HMGN1 binding sites with that of those DNase I HS sites that do not. We found that the hypersensitivity of the DNase I HS sites that overlapped HMGN1 sites was substantially higher than that of those HS sites that did not overlap HMGN1 sites (Fig. 3B, bar graph). Further analysis revealed that while 28% of DNase I HS sites that overlap HMGN1 sites were located in promoter regions, only 10% of the nonoverlapping sites were located in promoters (Fig. 3B, pie charts). Similarly, HMGN1 signal strength was much higher in the HMGN1 binding sites that overlapped DNase I HS sites. Interestingly, while 32% of the HMGN1 peaks that overlap with the DNase I HS sites were located in promoter regions, only 2% of the HMGN1 sites that did not overlap with DNase I HS sites were located in promoters. Moreover, DNase I HS sites overlapping HMGN1 sites are also considerably larger (Fig. 3D). These findings indicate that the chromatin regions containing HMGN1 are more accessible to nuclease digestion, perhaps reflecting a less-compacted chromatin structure, a feature that is characteristic of regulatory regions across the genome.
Indeed, we observed a significant enrichment of active chromatin marks such as histone H3K4me3 and histone variant H2A.Z (Fig. 3E and F) at the DNase I HS sites overlapping HMGN1 sites relative to the nonoverlapping sites. In contrast, the significant enrichment of neither HMGN1 nor DNase I HS sites was detected in chromatin regions containing H3K27me3, a modification associated with repressed regions of the genome (Fig. 3G). Taken together, our findings indicate that HMGN1 is preferentially bound to regulatory sites in chromatin.
HMGN1 is colocalized with CTCF binding sites.
To further investigate the relationship between HMGN1 binding and regulatory sites, we compared the genome-wide chromatin binding of HMGN1 to that of the zinc finger protein CCCTC-binding factor (CTCF). CTCF is a multifunctional protein, which demarcates active and inactive chromatin domains and affects chromatin-related processes such as the cellular transcription profile, imprinting, and X chromosome inactivation (20, 24, 40, 55). We previously reported the profile of CTCF sites in CD4+ T cells, identifying 28,661 CTCF binding sites across the genome. Genome-wide analysis of the overlap between CTCF and HMGN1 binding sites revealed that 51% of CTCF sites in the genome overlapped with sites of HMGN1 binding (as exemplified in Fig. 4A).
FIG. 4.
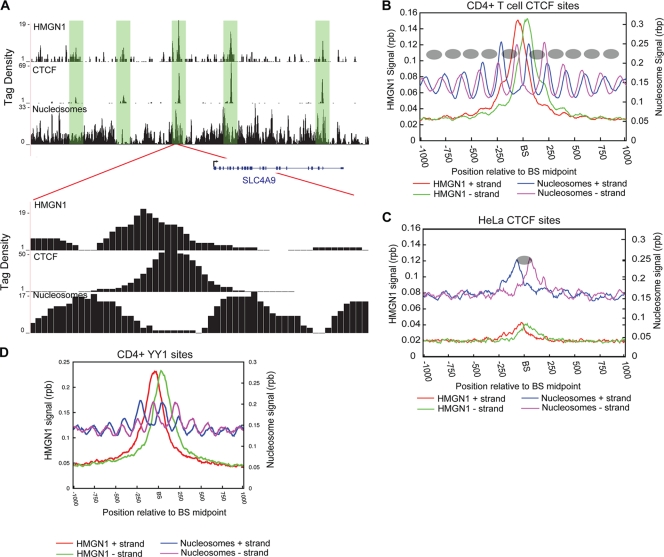
CTCF and YY1 binding sites are associated with HMGN1. (A) Nucleosome profiles along with HMGN1 and CTCF binding profiles in a segment of chromosome 5 near the SLC4A9 locus. A substantial overlap between HMGN1 and CTCF binding is observed. Lower panel, enlargement of a 500-bp region. CTCF binds to the linker region, and HMGN1 appears to bind to the nucleosome adjacent to the CTCF binding site. Tag density indicates the number of adjusted reads in 10-bp windows. (B) In CD4+ T cells CTCF binds to linker regions surrounded by well-positioned nucleosomes and displays an enrichment of HMGN1 at the binding sites. Blue and purple lines indicate nucleosome reads from sense and antisense strands, respectively. Red and green lines indicate HMGN1 reads from sense and antisense strands, respectively. rpb, reads per bp. The inferred nucleosome positions are represented as ovals. BS, CTCF binding site. (C) In CD4+ T cells, the HeLa cell-specific CTCF sites are occluded by nucleosomes and HMGN1 binding levels are low. Blue and purple lines indicate nucleosome reads from sense and antisense strands, respectively. Red and green lines indicate HMGN1 reads from sense and antisense strands, respectively. BS, CTCF binding site. (D) YY1 binds to linker regions bracketed by well-positioned nucleosomes and HMGN1 is enriched surrounding the binding sites. BS, YY1 binding site.
CTCF binds to linker regions surrounded by well-positioned nucleosomes (20, 23). HMGN proteins are the only nuclear proteins that specifically recognize the generic structure of the nucleosomal core particle (CP), the building block of the chromatin fiber. Although the interaction of HMGN with nucleosomes reduces the compaction of the 30-nm chromatin fiber, it also counteracts the action of nucleosome remodeling factors and stabilizes the structure of the CP (8, 43). Utilizing nucleosome profiling data that we obtained in a previous study (45), we investigated the relationship between nucleosome positioning at CTCF sites and the binding of HMGN1. Examination of HMGN1 and nucleosome profiles at specific CTCF binding sites indicated that HMGN1 preferentially bound to the borders of the two well-positioned nucleosomes (Fig. 4A, lower panel). To examine this at a genome-wide level, we plotted the HMGN1 and nucleosome signals at all CTCF binding sites in CD4+ T cells. The results indicate that, indeed, HMGN1 bound to the well-positioned nucleosomes bracketing CTCF binding sites (Fig. 4B).
We demonstrated in an earlier study that while CD4+ T cells and HeLa cells shared a majority of the CTCF binding sites, about 20% of the binding sites were cell type specific (20). In CD4+ T cells, the HeLa-specific CTCF binding sites are occluded by nucleosomes, blocking the sites from CTCF binding (20). Given that we identified an enrichment of HMGN1 at CTCF binding sites, we examined the HMGN1 occupancy of the HeLa-specific sites in CD4+ T cells. We found a reduction in the enrichment of HMGN1 at HeLa-specific sites compared to that of the global CTCF sites bound in CD4+ T cells (Fig. 4C, compare with HMGN1 signals in Fig. 4B). The results suggest that HMGN1 might play a role in generating or maintaining the positioning of nucleosomes around functional CTCF binding sites in chromatin.
HMGN1 is colocalized with YY1 binding sites.
To further test the preferential localization of HMGN1 with active regulatory regions, we examined the transcription factor YY1 in CD4+ T cells. YY1 is a ubiquitous transcription factor which can function either as a transcriptional activator or a repressor, depending on its interacting partner (27, 33, 48). Using ChIP-Seq, we identified 10,958 YY1 binding sites in CD4+ T cells (see Materials and Methods for details). Among these, 9,470 binding sites (90%) colocalized with HMGN1. Analysis of the nucleosome positioning around YY1 binding sites revealed that similarly to CTCF, YY1 bound the linker DNA with regular-positioned nucleosomes on either side of the binding site (Fig. 4D).
The binding patterns of HMGN1 around CTCF and YY1 binding sites thus suggest that HMGN1 is part of the cellular mechanism that generates or maintains regulatory regions of chromatin by affecting nucleosome positioning and perhaps by enhancing access to linker DNA. HMGN1 may affect nucleosome positioning either by binding to the DNA at its nucleosomal entry/exit points and stabilizing the structure of the core particle (38) or by interfering with the action of chromatin remodelers (43).
HMNG1 is enriched at intron-exon boundaries.
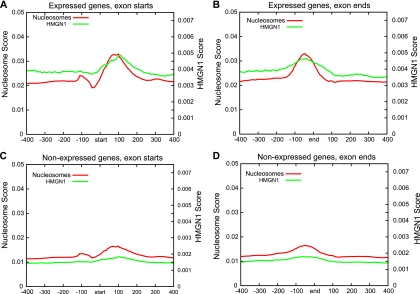
Recent studies have demonstrated that nucleosomes are well positioned relative to intron-exon boundaries in the human genome and that these nucleosomes contain characteristic modifications (2, 49). These results combined with our observations that HMGN1 binds to the borders of well-positioned nucleosomes prompted us to examine the profiles of HMGN1 at intron-exon boundaries. We aligned all internal exon boundaries for expressed and nonexpressed genes separately and plotted the HMGN1 and nucleosome profiles relative to these boundaries. Consistent with previous studies, we found that nucleosomes at the intron-exon boundaries of expressed genes are more strongly positioned than those at boundaries for nonexpressed genes. Furthermore, we determined that HMGN1 binding was enriched at the intron-exon boundaries of expressed genes compared to that at these boundaries for nonexpressed genes (Fig. 5).
FIG. 5.
HMGN1 is enriched at intron-exon boundaries. (A and B) HMGN1 and nucleosome profiles at the intron-exon junctions of expressed genes. (C and D) HMGN1 and nucleosome profiles at the intron-exon junctions of nonexpressed genes. In the x axis “start” denotes exon start and “end” denotes exon end regions. The numbers denote base pairs from these reference points. The “Nucleosome Score” and “HMGN1 Score” on the y axis denote the score as determined by the adjusted sense (antisense) read contributions in 5-bp windows normalized by the number of genes in each set (see Materials and Methods for details).
HMGN1 preferentially binds active promoters.
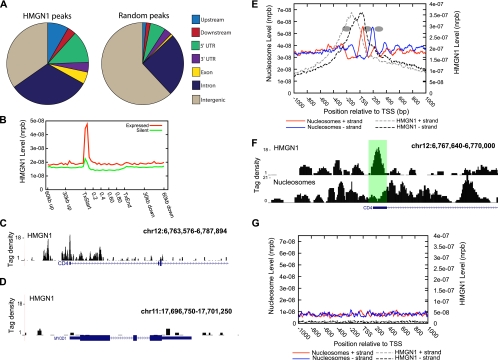
Given that our initial analysis suggested that HMGN1 binds to regulatory regions such as DNase I HS sites and distal transcription factor binding sites, we wanted to assess HMGN1 binding profile at a genomic level. We counted HMGN1 peaks identified using our approach (see Materials and Methods for details) in upstream (2 kb upstream of an annotated TSS) and downstream (2 kb downstream of an annotated TSS) regions, 5′ and 3′ untranslated regions (UTR), and exon, intron, and general intergenic regions and compared the results to a similar number of randomly generated peaks. This analysis revealed an association of HMGN1 with genic regions with respect to randomly generated peaks (Fig. 6A). Extending this analysis further revealed an enrichment of HMGN1 in promoter regions of expressed genes relative to those of nonexpressed genes (Fig. 6B to D). Periodic positioning of nucleosomes relative to the start sites of promoters of active genes in the human genome is well known (37, 45). We investigated the relationship between HMGN1 and nucleosomes in promoter regions by examining the distribution of reads representing HMGN1 binding and reads representing nucleosome positions at the promoters of genes being actively transcribed (see Materials and Methods for details). This analysis revealed preferential localization of HMGN1 between the two well-positioned nucleosomes surrounding the nucleosome-depleted region (Fig. 6E and F), again suggesting a role for HMGN1 in anchoring nucleosomes at regulatory regions, as postulated for the role of HMGN1 at distal CTCF and YY1 binding sites (Fig. 4). Silent gene promoters exhibited no periodic positioning of nucleosomes and very low levels of HMGN1 binding (Fig. 6G).
FIG. 6.
HMGN1 marks promoters of active genes. (A) Distribution of HMGN1 binding sites in CD4+ T cells. The distribution of “Random” peaks is shown as a control. “Upstream” and “Downstream” regions are defined as 2 kb upstream of the TSS and 2 kb downstream of the transcription end site, respectively. 5′ and 3′ UTR, Exon, and Intron coordinates were obtained from UCSC Gene coordinates. Only internal (not first or last) exons were considered. “Intergenic” regions were classified as all regions not within 2 kb of an annotated gene. (B) HMGN1 binding profiles across the gene bodies of expressed and nonexpressed genes. The x axis scale between the transcription start (TxStart) and end (TxEnd) sites denotes the percentage of gene body length. The y axis indicates the normalized read count per base pair (nrpb). (C) HMGN1 binding is significantly enriched at the promoter of the CD4 gene, which is expressed in CD4+ T cells. The “Tag density” indicates the number of adjusted reads aligning in 10-bp windows. (D) The MyoD gene, which is silent in CD4+ T cells, is devoid of HMGN1 binding. (E) Nucleosome position signals of the genes expressed in CD4+ T cells plotted with the HMGN1 signals. Gray and black dashed lines indicate the HMGN1 reads from sense and antisense strands of DNA. Red and blue lines indicate the nucleosome reads from sense and antisense strands of DNA. The inferred nucleosome positions are shown as ovals. (F) HMGN1 and nucleosome signals at the CD4 locus. (G) HMGN1 and nucleosome profiles in a 2-kb region surrounding transcription start sites of genes that are not expressed in CD4+ T cells. Red and blue lines indicate nucleosome reads from the sense and antisense strands, respectively. Gray and black dashed lines indicate HMGN1 reads from the sense and antisense strands, respectively.
DISCUSSION
The interaction of regulatory factors with specific genomic regions constitutes a critical step in gene regulation. Most regulatory factors bind to specific sites as multiprotein complexes that affect the local structure of chromatin and recruit modifiers that alter the structure of chromatin to either enhance or repress transcriptional activity. This is well exemplified by modifications of core histone proteins. For example, acetylation of histones catalyzed by histone acetyltransferases (HATs) favors the generation of open chromatin regions, which in most instances enhances transcription. In contrast, deacetylation by histone deacetylases (HDACs) generates closed regions that are inhibitory to transcription. A balance between the two activities is essential for proper transcriptional regulation (52). Such opposing forces also control the positioning of nucleosomes, which is also an important component of transcriptional regulation. Nucleosome remodeling is an important process in the dynamic alteration of chromatin structure. Studies done in Saccharomyces cerevisiae have shown that while the ATP-dependent SWI/SNF complexes activate transcription by remodeling nucleosomes, ISWI family enzymes, especially ISW2, repress transcription by positioning nucleosomes such that regulatory regions are inaccessible (25, 50).
Our results indicate that the architectural protein HMGN1 is enriched at nucleosomes that are well positioned in regulatory regions. CTCF binding to linker DNA and the presence of well-positioned nucleosomes surrounding the binding sites have been previously demonstrated (20, 23). Our results indicate that the nucleosomes surrounding YY1 binding sites exhibit a similar profile. HMGN1 binds to the edges of nucleosomes surrounding CTCF and YY1 binding sites, a finding that is in full agreement with the known position of nucleosomes at the entry-exit points of the nucleosomal DNA (1). Furthermore, recent work by Rattner et al. demonstrated a role for HMGN1 in stabilizing nucleosomes by inhibiting the chromatin remodeling activity (43). Thus, our results suggest that HMGN1 plays a role in maintaining a nucleosome occupancy profile that enhances the accessibility of regulatory factors to their target. Consistent with this, at the HeLa-specific CTCF binding sites in CD4+ cells, nucleosomes occlude the binding site (20). The absence of HMGN1 in these sites further reinforces the idea that the protein plays a specific role in the maintenance of nucleosome positions at regulatory sites. One of the possible mechanisms through which HMGN1 could influence nucleosome positioning is through the inhibition of the SWI/SNF chromatin remodeling complexes, as Rattner and colleagues suggest (43).
However, it is also conceivable that HMGN1 associates with regulatory factors involved in the generation of open chromatin and may act to stabilize these structures. Chromatin remodeling activity of the INO80 complex has been suggested as a prerequisite for YY1 binding to its target sites (11). Further investigation is necessary to test whether HMGN1 is associated with any such complexes.
Partial loss of the canonical −1 nucleosome upstream of the TSS and precise positioning of other surrounding nucleosomes are characteristic features of promoters of genes which are being actively transcribed (45). It has been suggested that the periodically positioned nucleosomes found at the CTCF binding sites and promoters of active genes could be an outcome of one or two nucleosomes being precisely positioned and the neighboring nucleosomes being positioned relative to these nucleosomes (36). Thus, it is possible that HMGN1 stabilizes the position of the nucleosomes, perhaps anchoring them at the boundaries of the CTCF and YY1 binding sites. Our identification of a significant enrichment of HMGN1 in the nucleosome-depleted region in active promoters is consistent with previous data indicating that HMGN1 preferentially binds to “anchor” nucleosomes. Due to variation in the lengths of nucleosome-depleted regions of different promoters, the distribution of HMNG1 binding at promoters is broader than at CTCF and YY1 binding sites. Therefore, it is possible that the HMGN1 peaks that we observe at the nucleosome-depleted region of the promoter could be due to an averaging of the binding that occurs at the edges of the flanking nucleosomes. Increased rate of digestion by micrococcal nuclease at the HSP70 promoter locus is higher in Hmgn1+/+ cells than in Hmgn1−/− cells, suggesting a role for HMGN1 at the promoters of active genes (4). Consistent with this, we detected enrichment of HMGN1 at active promoters and only very low levels of HMGN1 binding at silent gene promoters.
HMGN1 may play a similar role at the DNase I HS sites, for which we found extensive colocalization. The hypersensitivity of the DNase I HS sites was also higher in regions with stronger HMGN1 signals. Conventionally, DNase I HS sites have been used in the identification of regulatory elements (6, 17, 18). Our findings indicate that HMGN1 binding data can be used in conjunction with DNase I HS site data to identify functional regulatory elements with higher confidence.
Taken together with previous analyses (12-14, 16), our results suggest that although HMGN1 moves rapidly through the nucleus, its residence time at regulatory sites is longer than at nonregulatory sites. Thus, from a functional view, HMGN1 preferentially turns over at chromatin regulatory sites, a finding that is in agreement with the observation that Hmgn1−/− mice have an altered phenotype (31). It remains to be seen whether HMGN1 is involved in the generation or maintenance of the regulatory sites, or whether it preferentially recognizes these preformed sites, perhaps targeted to specific chromatin regions as part of a metastable regulatory complex (35).
Acknowledgments
We thank Iouri Chepelev (National Institutes of Health) for helpful comments and discussions.
This work was supported by the Intramural Research Program of the National Heart, Lung and Blood Institute and of the Center for Cancer Research, National Institutes of Health.
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Alfonso, P. J., M. P. Crippa, J. J. Hayes, and M. Bustin. 1994. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J. Mol. Biol. 236:189-198. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, R., S. Enroth, A. Rada-Iglesias, C. Wadelius, and J. Komorowski. 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 19:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barski, A., et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 4.Belova, G. I., Y. V. Postnikov, T. Furusawa, Y. Birger, and M. Bustin. 2008. Chromosomal protein HMGN1 enhances the heat shock-induced remodeling of Hsp70 chromatin. J. Biol. Chem. 283:8080-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi, M. E., and A. Agresti. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15:496-506. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, A. P., et al. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle, A. P., J. Guinney, G. E. Crawford, and T. S. Furey. 2008. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24:2537-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 9.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 11.Cai, Y., et al. 2007. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14:872-874. [DOI] [PubMed] [Google Scholar]
- 12.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catez, F., and R. Hock. 2010. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta 1799:15-27. [DOI] [PubMed] [Google Scholar]
- 14.Catez, F., J. H. Lim, R. Hock, Y. V. Postnikov, and M. Bustin. 2003. HMGN dynamics and chromatin function. Biochem. Cell Biol. 81:113-122. [DOI] [PubMed] [Google Scholar]
- 15.Catez, F., T. Ueda, and M. Bustin. 2006. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 13:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catez, F., et al. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 24:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford, G. E., et al. 2006. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods 3:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford, G. E., et al. 2004. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. U. S. A. 101:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crippa, M. P., P. J. Alfonso, and M. Bustin. 1992. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 228:442-449. [DOI] [PubMed] [Google Scholar]
- 20.Cuddapah, S., et al. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Druckmann, S., E. Mendelson, D. Landsman, and M. Bustin. 1986. Immunofractionation of DNA sequences associated with HMG-17 in chromatin. Exp. Cell Res. 166:486-496. [DOI] [PubMed] [Google Scholar]
- 22.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 23.Fu, Y., M. Sinha, C. L. Peterson, and Z. Weng. 2008. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaszner, M., and G. Felsenfeld. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703-713. [DOI] [PubMed] [Google Scholar]
- 25.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, P. J., and E. Palacian. 1990. Structural and transcriptional properties of different nucleosomal particles containing high mobility group proteins 14 and 17 (HMG 14/17). J. Biol. Chem. 265:8225-8229. [PubMed] [Google Scholar]
- 27.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 28.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57:159-197. [DOI] [PubMed] [Google Scholar]
- 29.Happel, N., and D. Doenecke. 2009. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Hill, D. A., C. L. Peterson, and A. N. Imbalzano. 2005. Effects of HMGN1 on chromatin structure and SWI/SNF-mediated chromatin remodeling. J. Biol. Chem. 280:41777-41783. [DOI] [PubMed] [Google Scholar]
- 31.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 17:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu, F., W. J. Kent, H. Clawson, R. M. Kuhn, M. Diekhans, and D. Haussler. 2006. The UCSC Known Genes. Bioinformatics 22:1036-1046. [DOI] [PubMed] [Google Scholar]
- 33.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent, W. J., et al. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim, J. H., M. Bustin, V. V. Ogryzko, and Y. V. Postnikov. 2002. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J. Biol. Chem. 277:20774-20782. [DOI] [PubMed] [Google Scholar]
- 36.Mavrich, T. N., et al. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozsolak, F., J. S. Song, X. S. Liu, and D. E. Fisher. 2007. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 25:244-248. [DOI] [PubMed] [Google Scholar]
- 38.Paton, A. E., E. Wilkinson-Singley, and D. E. Olins. 1983. Nonhistone nuclear high mobility group proteins 14 and 17 stabilize nucleosome core particles. J. Biol. Chem. 258:13221-13229. [PubMed] [Google Scholar]
- 39.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips, J. E., and V. G. Corces. 2009. CTCF: master weaver of the genome. Cell 137:1194-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postnikov, Y., and M. Bustin. 2010. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta 1799:62-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postnikov, Y. V., et al. 1991. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 19:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rattner, B. P., T. Yusufzai, and J. T. Kadonaga. 2009. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell 34:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rochman, M., et al. 2009. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell 35:642-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schones, D. E., et al. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirakawa, H., J. E. Herrera, M. Bustin, and Y. Postnikov. 2000. Targeting of high mobility group-14/-17 proteins in chromatin is independent of DNA sequence. J. Biol. Chem. 275:37937-37944. [DOI] [PubMed] [Google Scholar]
- 47.Stalder, J., et al. 1980. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20:451-460. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 49.Tilgner, H., et al. 2009. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 16:996-1001. [DOI] [PubMed] [Google Scholar]
- 50.Tomar, R. S., J. N. Psathas, H. Zhang, Z. Zhang, and J. C. Reese. 2009. A novel mechanism of antagonism between ATP-dependent chromatin remodeling complexes regulates RNR3 expression. Mol. Cell. Biol. 29:3255-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vestner, B., M. Bustin, and C. Gruss. 1998. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J. Biol. Chem. 273:9409-9414. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z., et al. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z., et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisbrod, S. 1982. Active chromatin. Nature 297:289-295. [DOI] [PubMed] [Google Scholar]
- 55.Williams, A., and R. A. Flavell. 2008. The role of CTCF in regulating nuclear organization. J. Exp. Med. 205:747-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodcock, C. L., A. I. Skoultchi, and Y. Fan. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14:17-25. [DOI] [PubMed] [Google Scholar]
- 57.Zang, C., et al. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]