Abstract
SR proteins are well known to promote exon inclusion in regulated splicing through exonic splicing enhancers. SR proteins have also been reported to cause exon skipping, but little is known about the mechanism. We previously characterized SRSF1 (SF2/ASF)-dependent exon skipping of the CaMKIIδ gene during heart remodeling. By using mouse embryo fibroblasts derived from conditional SR protein knockout mice, we now show that SR protein-induced exon skipping depends on their prevalent actions on a flanking constitutive exon and requires collaboration of more than one SR protein. These findings, coupled with other established rules for SR proteins, provide a theoretical framework to understand the complex effect of SR protein-regulated splicing in mammalian cells. We further demonstrate that heart-specific CaMKIIδ splicing can be reconstituted in fibroblasts by downregulating SR proteins and upregulating a RBFOX protein and that SR protein overexpression impairs regulated CaMKIIδ splicing and neuronal differentiation in P19 cells, illustrating that SR protein-dependent exon skipping may constitute a key strategy for synergism with other splicing regulators in establishing tissue-specific alternative splicing critical for cell differentiation programs.
The splicing machinery is largely conserved in eukaryotic cells. However, compared to budding yeast, where critical splicing signals are nearly invariant, higher eukaryotic cells rely on auxiliary factors to help define functional splice sites that are only loosely conserved. Most genes in higher eukaryotic cells also undergo alternative splicing, which is subject to regulation by a variety of RNA binding proteins (2). SR proteins are unique to higher eukaryotes and are among the best-characterized RNA binding proteins involved in both constitutive and regulated splicing (29, 31, 48). Intensive biochemical analysis in the past 2 decades has established that the RNA recognition motifs (RRMs) of SR proteins are responsible for sequence-specific binding to the pre-mRNA, whereas the RS domain appears to mediate both protein-protein and protein-RNA interactions during the splicing reaction (17, 39).
Individual SR proteins exhibit distinct RNA binding specificities for various exonic splicing enhancers (ESEs), a second code in higher eukaryotic genomes that is critical for defining functional splice sites. In many cases, multiple SR proteins bind to several ESEs within the same exon, which is thought to provide redundant functions to ensure constitutive splicing against variation of SR proteins in different cell types and tissues. However, it has become abundantly clear that individual SR proteins are not functionally redundant in vivo (1, 4, 30). Because exons are short whereas introns are highly variable in length, functional splice sites in most mammalian genes are initially recognized by the exon definition mechanism, in which ESE-bound SR proteins promote U2AF recognition of the 3′ splice site and U1 binding to the downstream 5′ splice site across the exon (16). Initial exon definition complexes are then switched to intron definition complexes to facilitate spliceosome assembly events across the upstream intron, and SR proteins have been shown to contribute directly to both exon definition and the subsequent intron definition process (8).
In regulated splicing, SR proteins are best known for their ability to promote exon inclusion by binding to ESEs and enhancing the communication between the two most proximal 5′ and 3′ splice sites, thus ensuring that multiexon-containing genes are spliced in a linear order (20). However, a number of studies have also documented SR protein-dependent exon skipping events (3, 9, 22, 27, 40, 41). The best-characterized mechanism for SR protein-induced exon skipping is through an intronic binding site, which triggers the formation of splicing-like complexes on a decoy exon, thereby interfering with the recognition of nearby functional splice sites (3, 22). In other cases, SR protein-dependent exon skipping appears to be still dependent on ESEs, and different SR proteins show opposite activities in promoting exon inclusion or skipping on the same genes (9, 10, 27, 35, 40). These findings raise a general question of whether SR protein-mediated exon inclusion or skipping events utilize fundamentally distinct mechanisms. Further adding to the complexity of the problem is the recently documented effect of SR proteins on transcriptional elongation (28), which may influence kinetic coupling between transcription and cotranscriptional RNA splicing (25). For example, reduction of an SR protein may impair the selection of an alternative exon through its splicing activity, and this effect may be counteracted by reduced polymerase II processivity on the same gene, therefore affording a certain kinetic advantage on the selection of the upstream alternative exon.
Here, we used the CaMKIIδ gene as a model to understand the mechanism underlying SR protein-mediated exon inclusion or skipping. The CaMKIIδ gene is well characterized as undergoing tissue-specific alternative splicing, involving three alternative exons (exon 14, 15, and 16) (see Fig. 1A, below), to generate several kinase isoforms that are differentially targeted to different cellular compartments in the heart and neurons (19). Our previous work demonstrated that in vivo depletion of the SR protein SRSF1 induced the inclusion of exons 15 and 16, and the resultant CaMKIIδ isoforms were targeted to myocardial tubes, triggering a hypercontraction phenotype in the heart (45). By analyzing mouse embryo fibroblasts (MEFs) derived from conditional knockout mice, we found that transient withdrawal of either SRSF1 (SF2/ASF) or SRSF2 (SC35) induced exon 15 and/or 16 inclusion. Functional dissection of this model now reveals a new rule for SR protein-mediated exon inclusion or skipping, which depends on where a specific SR protein binds to the alternative or a flanking constitutive exon and on collaboration with at least one other SR protein.
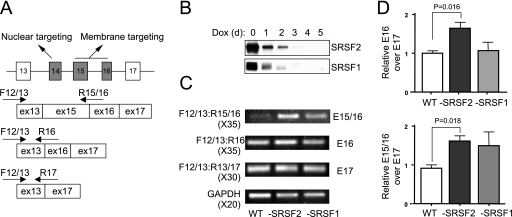
FIG. 1.
CaMKIIδ alternative splicing in response to SR protein depletion in MEFs. (A) Alternative splicing of CaMKIIδ and detection of its major isoforms by splice junction-specific primers. Constitutive exons are shown as white boxes, and alternative exons are shown as gray boxes. The function of the alternative exons in mediating differential intracellular targeting of the kinase is also indicted. (B) Induction of SRSF2 and SRSF1 depletion in conditional knockout MEFs. (C) Analysis of CaMKIIδ isoforms in wild-type or SRSF2- or SRSF1-deficient MEFs by RT-PCR. The number of PCR cycles is indicated in parentheses for each experiment. (D) Quantification of the induced CaMKIIδ isoform containing exon 16 (E16) by quantitative RT-PCR. Because of the noise during the detection of the CaMKIIδ isoform containing both exons 15 and 16 (E15/16) by real-time PCR, quantification of the isoform was based on the specific band on the gel, which was first normalized against glyceradehyde-3-phosphate dehydrogenase (GAPDH) and then against the major skipped isoform (E13 to E17). Error bars are based on three independent experiments. Statistical significance was determined by a two-tailed t test, and the significant changes are labeled.
We also utilized the CaMKIIδ gene to learn how SR proteins contribute to tissue-specific alternative splicing in collaboration with other tissue-specific RNA binding splicing regulators. We found that overexpression of a Fox protein (now renamed RBFOX) or depletion of an SR protein each partially induced CaMKIIδ alternative splicing in MEFs, but a combined treatment fully reconstituted the splicing pattern of the CaMKIIδ gene, as observed in developing heart where RBFOX1/RBFOX2 are highly expressed and SR proteins are in low abundance. Consistently, overexpression of SRSF1 in neuroblast P19 cells impaired the isoform switch of the CaMKIIδ gene induced by retinoic acid (RA). Significantly, the RA-induced cell differentiation program was also severely compromised by augmented expression of the SR protein, likely due to multiple other altered splicing events. Collectively, these findings suggest that synergistic actions of SR proteins and other splicing regulators play critical roles in establishing tissue-specific splicing programs in development.
MATERIALS AND METHODS
Cell culture.
SRSF2- and SRSF1-deficient MEFs derived from conditional SRSF2 and SRSF1 knockout mice were engineered to express an exogenous SR protein from a tet-off promoter as described previously (30, 44). The MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% tet-free fetal bovine serum (FBS) and antibiotics. Depletion of exogenous SR proteins was performed by addition of 10 μg/ml of doxycycline (Dox) for 5 days.
Plasmid construction and mutagenesis.
All DNA constructs used in this study were confirmed by sequencing. The chimeric minigene, DUP4CX, was derived from DUP4-1 (32), in which human β-globin exons were fused to the cytomegalovirus (CMV) promoter. Mouse CaMKIIδ exons 13, 14, 15, 16, or 17 were amplified by PCR and inserted into ApaI-BglII site between human β-globin exon 1 and exon 2 of DUP4-1. pcDNA3 (Invitrogen) was used as a parent plasmid to construct CAM15/16 plasmids. The insert encoding the exons and the truncated introns of the CaMKIIδ gene was constructed without disturbing the endogenous splice sites and fused to pcDNA3. The CaMKIIδ minigene was expressed by a CMV promoter. To make the CAM16 plasmid, CAM15/16 plasmid was digested with EcoRV and ApaI and then the EcoRV-ApaI fragment was purified with a gel extraction kit (Qiagen). CAM15/16 plasmid was digested with NheI, followed by treatment with Klenow fragment and digestion with ApaI. The linearlized plasmid was ligated with the gel-purifed EcoRV-ApaI fragment. Mutagenesis of CAM16 and CAM15/16 was performed with the QuikChange site-directed mutagenesis kit (Stratagene). The mutant plasmids were also confirmed by sequencing.
To construct the splicing reporters that carry two MS2 binding sites in each exon of DUP4-16, an AgeI site was introduced into each exon by site-directed mutagenesis. Two sets of oligonucleotide pairs, each carrying two wild-type (wt; 5′-CCGGT cgtaca ccatca gggtac gctgca gtcgac ttcgcg tacacc atcagg gtacgA-3′ and 5′-Tcgtac cctgat ggtgta cgcgaa gtcgac tgcagc gtaccc tgatgg tgtacg ACCGG-3′) or mutant (5′-CCGGT cgtacc catcag ggtacg ctgcag tcgact tcgcgt acccat cagggt acgA-3′ and 5′-Tcgtac cctgatg ggtacg cgaagt cgactg cagcgt accctg atgggta cgACCG-3′) MS2 binding sites were annealed and ligated into the AgeI site in each parental plasmid. Sequence portions shown in lowercase letters are MS2 sequences, and portions in uppercase letters are AgeI restriction sites. All constructs were confirmed by sequencing.
Transfection and reverse transcription-PCR (RT-PCR).
The MEFs were cultured in 60-mm plates to 50 to 60% confluence in the presence or absence of Dox for 3 days and transfected with 2 μg of a minigene reporter in the absence of FBS at 37°C for 4 h. At 4 h posttransfection, the medium was replaced by DMEM supplemented with 10% tet-free FBS and antibiotic, and the MEFs were cultured for 48 h in the presence or absence of Dox. Total RNAs were extracted from the MEFs by using TRIzol, and 1 μg of the total RNA was used to synthesize cDNA with SuperScript III (Invitrogen) according to the manufacturer's instructions. PCR was performed using 1 μl of the cDNA in 50-μl reaction mixtures with TaqGold (Roche) and analyzed using 2% agarose gel eletrophoresis.
Nuclear extract preparation and UV cross-linking assay.
Preparation of MEF nuclear extracts was performed as described previously (18). Individual CaMKIIδ exons and intron were in vitro transcribed and internally labeled with [32P]UTP. Each labeled RNA was incubated with the nuclear extract under splicing conditions for 10 min and UV-cross-linked at 800 kJ for 10 min on ice, followed by anti-hemagglutinin (HA) immunoprecipitation and analyzed by SDS-PAGE.
Neuronal differentiation of P19 cells.
Both P19 and SRSF1-overexpressing P19 cell lines were differentiated by addition of retinoic acid, and neuronal differentiation was monitored by analyzing the neuronal marker Mash1 by RT-PCR and immunofluorescence analysis with the neuron-specific anti-β-III tubulin antibody. Expression of CaMKIIδ isoforms was measured by RT-PCR with specific splice junction primers.
RESULTS
Induction of SR protein-dependent CaMKIIδ exon skipping in MEFs.
We previously established that conditional knockout of the SR protein SRSF1 induced exon inclusion of the CaMKIIδ gene in the heart (45). To determine whether this effect reflects a general function of SR proteins on the CaMKIIδ gene, we employed MEFs derived from conditional SRSF1 and SRSF2 knockout mice to test CaMKIIδ mRNA isoform switch in response to SR protein depletion, which could be accomplished by the addition of Dox to culture medium (Fig. 1B). Total RNA was extracted from Dox-treated wt and conditional knockout MEFs, and the expression of individual CaMKIIδ isoforms was measured by RT-PCR or real time PCR using specific exon junction primers. We detected the predominantly skipped isoform (exon 13 directly linked to exon 17) in both wt and conditional knockout MEFs (Fig. 1C), consistent with the splicing pattern of the CaMKIIδ gene in nonneuronal cells (38). However, by amplifying cDNAs with a higher PCR cycle, we clearly detected Dox-induced inclusion of the alternative exon 16 in SRSF2-depleted MEFs and double inclusion of exons 15 and 16 in SRSF2-depeleted cells (Fig. 1C and D). We also detected variable inclusion of exons 15 and 16 in SRSF1-depleted MEFs, but the effect was not statistically significant (Fig. 1C and D). Much more robust induction of exon inclusion was seen with CaMKIIδ-derived minigenes in transfected cells (see below), likely reflecting some chromatin-related effects on cotranscriptional RNA processing.
These data demonstrated an increase in exon inclusion in response to transient depletion of SR proteins in MEFs, as we observed in developing heart, suggesting that SR proteins are similarly involved in regulated CaMKIIδ splicing in different cell types. Interestingly, we noted that, in MEFs depletion of SRSF2 was more efficient than SRSF1 in inducing CaMKIIδ splicing, whereas depletion of SRSF1 had a much stronger effect in developing heart (45). These differences imply that different SR proteins may have quantitatively distinct activities in regulated splicing of the same genes in different cell types, perhaps due to the presence of other tissue-specific splicing regulators that render differential sensitivity of SR proteins in the regulation of different splicing events. Importantly, the current results establish an inducible model for dissecting the mechanism underlying SR protein-dependent exon skipping in mammalian cells.
Portable SR protein-dependent exon skipping events.
To approach SR protein-dependent exon skipping, we first examined potential cis-acting elements within alternative and flanking constitutive exons in the CaMKIIδ gene. Using the ESEfinder program (http://exon.cshl.edu/ESE), we identified multiple putative SR protein-responsive elements in each exon, particularly in the alternative exon 16, which appears to contain even stronger ESEs than the downstream constitutive exon 17 (Fig. 2A). Because ESE activities are well known to subject influence by the sequence context (11), we experimentally determined the relative strength of individual exons, and more importantly, tested whether the regulatory information on individual exons was transferable, by cloning each exon into a splicing reporter derived from the human β-globin gene (pDUP4), in which the inclusion of the internal exon is strictly dependent on a functional ESE (33) (Fig. 2B). The resulting chimeric constructs, which contained identical splicing signals in both upstream and downstream intronic regions, were individually transfected into wt MEFs, and their splicing patterns were analyzed by RT-PCR. Surprisingly, we detected full inclusion of exon 17, but no inclusion of exon 13, despite the fact that both of these exons are constitutively included in their native context. In comparison, we observed no, weak, and significant inclusion of exons 14, 15, and 16, respectively (Fig. 2C). As the inclusion of each test exon in the reporter altered the reading frame of the globin reporter, we examined the splicing pattern in the presence of the translational inhibitor cycloheximide and detected the same splicing pattern, indicating a lack of nonsense-mediated RNA decay (NMD) effects on the reporter system in transfected cells.
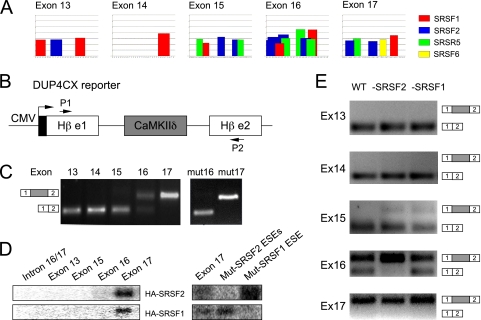
FIG. 2.
Exon strength analysis on a splicing reporter. (A) Identification of putative ESEs in the alternative and flanking constitutive exons of the CaMKIIδ gene based on the ESEfinder program. (B) The β-globin minigene (DUP4CX)-based reporter. Individual CaMKIIδ exons (exons 13 through 17) are flanked by the identical intronic splicing signals. Arrows indicate the primers for RT-PCR analysis. (C) Wild-type MEF lines were transfected by the chimeric β-globin minigenes, and the inclusion of each CaMKIIδ exon was analyzed by RT-PCR. (D) Binding levels of individual wt and mutant exons and an intron control with SRSF2 or SRSF1 were measured by UV cross-linking using MEF-derived nuclear extracts containing HA-tagged SR proteins. (E) The inclusion of individual CaMKIIδ exons induced by SR protein depletion.
To determine whether this gradient in splicing strength is correlated with SR protein binding on individual exons, we performed UV cross-linking experiments with nuclear extracts prepared from conditional knockout MEFs in which exogenous SRSF2 and SRSF1 were each expressed as an HA-tagged fusion protein (Fig. 1B). Uniformly labeled exonic sequences were incubated under splicing conditions followed by UV-induced cross-linking and anti-HA immunoprecipitation. We found that only exon 17 was bound to a detectable level by both SRSF2 and SRSF1 (Fig. 2D), suggesting that ESEs in exon 17 are, indeed, much stronger than those in exon 13 through exon 16. The lack of detectable SRSF1 or SRSF2 protein binding to exon 16 suggests that either the binding assay was not sufficiently sensitive compared to the reporter assay or the splicing activity of the reporter might be due to other SR proteins.
To establish the function of the deduced ESEs in mediating exon inclusion in the reporter assay, we mutated the ESE for SRSF1 in exon 16 or two ESEs for SRSF2 in exon 17 (see Fig. 3D, below), and we observed that inactivation of the SRSF1 ESE in exon 16 (mut 16) impaired exon inclusion, while mutations in exon 17 had no effect (Fig. 2C, right panel). These results indicate that the ESE for SRSF1 in exon 16 may have measurable activity in the reporter assay, while multiple functional ESEs in exon 17, as indicated by the UV cross-linking result (Fig. 2D, right panel), may provide redundant functions to ensure efficient inclusion of this constitutive exon (this was further established by mutagenesis on the CaMKIIδ-derived minigenes, as shown below in Fig. 3). Importantly, the result of the reporter assay fully agrees with the well-established role of ESE-dependent exon inclusion, thus suggesting that there is no unusual property associated with the alternative exons, particularly exon 16, in the CaMKIIδ gene.
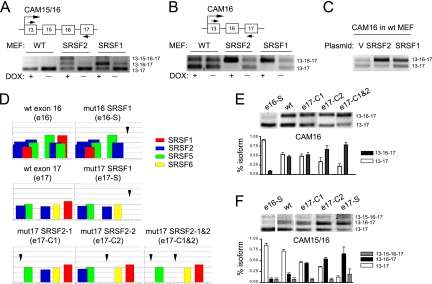
FIG. 3.
Mutational analysis of CaMKIIδ alternative splicing. (A and B) The CaMKIIδ-based minigenes (CAM16 and CAM15/16) and their splicing patterns in transfected wt and conditional knockout MEFs before and after induced depletion of the specific SR proteins indicated. (C) Induction of exon 16 inclusion by overexpression of individual SR proteins. (D) Mutations of ESEs in exons 16 and 17. Arrows show the ESE scores on the mutated regions. (E and F) RT-PCR analysis of the wt and mutant CaMKIIδ minigenes in transfected MEFs. The results are quantified at the bottom in each panel. Error bars are based on three independent experiments.
We next asked whether the effects of ESE mutations on exon inclusion mirror the effect of SR protein depletion on the same reporter system. Paradoxically, we detected a modest enhancement in exon 15 inclusion in response to depletion of both SRSF2 and SRSF1 and a strong induction of exon 16 inclusion in SRSF2-depleted MEFs (Fig. 2E). Thus, the effect of SR protein depletion is opposite to that with individual ESE mutants, as seen in Fig. 2C. These data suggest that the SR proteins under investigation may not solely act on the ESEs on exon 16, and the functional consequence of SR protein depletion may result from a more complex action mechanism, perhaps through ESEs on the globin exons, as previously elucidated (37).
Mechanistic dissection of SR protein-dependent exon skipping.
The ability to transfer the regulatory information by the alternative exons permitted us to focus on exonic sequences to dissect potential cis-acting elements in the CaMKIIδ gene. For this purpose, we constructed two CaMKIIδ minigenes, one containing the alternative exons 15 and 16 (CAM15/16) and the other carrying only the alternative exon 16 (CAM16). Compared to the endogenous gene, the transfected minigenes responded robustly to SR protein depletion to include both alternative exons (Fig. 3A and B). In the case of CAM16, the induction of exon 16 was almost complete upon in vivo withdrawal of SRSF2 or SRSF1 (Fig. 3B). We performed a converse experiment on one of these minigenes (CAM16) by overexpression, and again, we observed enhancement of exon 16 inclusion upon SRSF2 and SRSF1 overexpression in transfected MEFs (Fig. 3C). To our knowledge, this is the first report that depletion and overexpression of SR proteins can induce a splicing switch in the same direction, although it is important to keep in mind potential indirect effects in these in vivo analyses.
To understand why knockdown and overexpression of SR proteins both enhanced exon 16 inclusion, we initially attempted to use in vitro splicing with the CaMKIIδ minigene to investigate the underlying mechanism, but we could not detect any spliced products even though the globin pre-mRNA control spliced well under the same conditions. The poor splicing efficiency in cell extracts might be due to the weak exon 13, as indicated in the reporter assay. We therefore employed a mutagenesis approach (Fig. 3D) to determine specific ESEs responsible for SR protein-mediated exon skipping in the context of CAM16 and CAM15/16 minigenes in wt MEFs, which also avoided potential compensatory effects that might be induced by in vivo depletion or overexpression of SR proteins (26). Consistent with the splicing reporter assay, we found that the SRSR1 ESE mutation (e16-S) on exon 16 impaired its inclusion (Fig. 3E and F), but this mutant continued to respond to SRSF1 depletion, resulting in full inclusion of the exon, as we observed with the wt reporter in SRSF1-depleted cells. This prompted us to consider the potential role of ESEs in the flanking constitutive exon 17 in mediating SR protein-dependent skipping of exon 16.
Based on ESE predictions, we introduced a mutation in the ESE for SRSF1 and two mutations, either alone or in combination, in the ESEs for SRSF2 on exon 17. According to ESEfinder, the mutations selectively diminished the ESE we intended to disrupt (Fig. 3D). We tested all these mutations in the CAM16 and CAM15/16 reporters. Indeed, opposite to the effect of the ESE mutation on exon 16, we detected enhanced inclusion of the alternative exons when the ESEs in exon 17 were inactivated (Fig. 3E and F). Individual mutations in the SRSF2 ESEs had a small but detectable effect, but a combination of the mutations dramatically promoted exon 16 inclusion (Fig. 3E). These findings reveal that, while the ESE in exon 16 mediates exon inclusion, the ESEs in exon 17 are responsible for SR protein-dependent exon skipping.
Demonstration of a location-dependent effect of SR proteins by MS2 tethering.
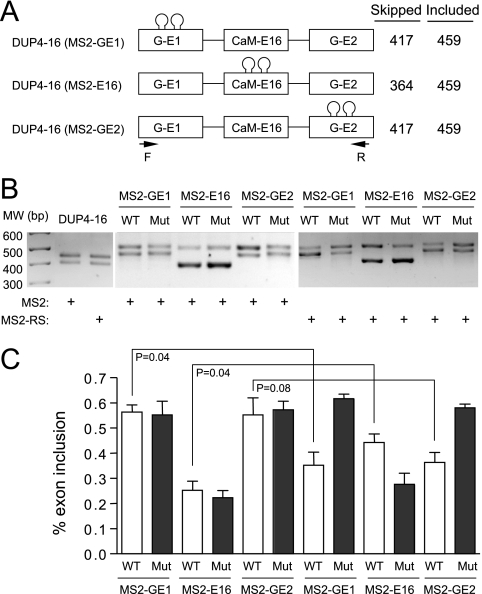
To directly demonstrate the position-dependent effect of SR proteins, we employed the MS2 tethering strategy (13) by inserting two MS2 binding sites in the flanking constitutive exons or the internal alternative exon in the DUP4-16 reporter (Fig. 4A). In each case, a mutant MS2 site was similarly introduced in all three locations as a control. The parental splicing reporter did not respond to the MS2 protein or the MS2-RS fusion protein, which carries the RS domain of the SR protein 9G8 (Fig. 4B). Insertion of the MS2 binding sites in various locations in the reporter altered the size of the spliced products, but they did not respond to MS2 binding. In contrast, insertion of the MS2 binding sites in the constitutive exons induced skipping of the internal alternative splicing exon in response to the MS2-RS fusion protein, while the presence of the MS2 binding sites in the internal exon enhanced its inclusion (Fig. 4B). The quantified results showed that the MS2-RS-induced exon skipping and inclusion were both dependent on the functional MS2 binding sites (Fig. 4C).
FIG. 4.
Induction of exon skipping or inclusion by a tethered RS domain. (A) DUP4-16-based constructs. The CaMKIIδ exon 16-containing splicing reporter was used to individually engineer wt and mutant MS2 binding sites in each exon. The expected sizes of pre-mRNA and spliced mRNA are indicated on the right. (B) The parental reporter and individual MS2-containing constructs labeled on the top were transfected into wt MEFs with either MS2 or the MS2-RS fusion gene as indicated at the bottom. The gel shows the results of RT-PCR analysis. (C) Quantification of the results in panel B. Error bars are based on three independent experiments, and statistical significance was determined by a two-tailed t test.
Based on the results of the mutagenesis studies (Fig. 3) and the MS2 tethering assay (Fig. 4), we concluded that SR protein-dependent exon skipping is mediated by the strong ESEs in the flanking constitutive exon(s). This finding is consistent with the recent CLIP analysis of SRSF1 binding in the human genome, where SRSF1 binding to downstream constitutive exons appears to correlate with exon skipping events in response to SRSF1 knockdown (36). Together, these results suggest a unifying rule for SR protein-dependent exon inclusion or skipping, which depends on preferential SR protein binding on the internal alternative exon or on a flanking constitutive exon.
Reconstitution of CaMKIIδ tissue-specific alternative splicing in MEFs.
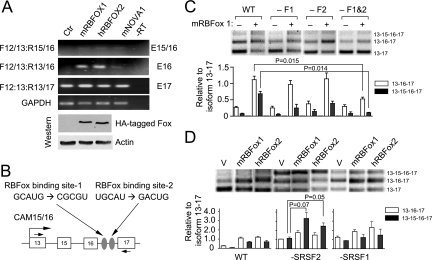
The CaMKIIδ gene is known to undergo alternative splicing in the heart and neurons. Recent global analyses of regulated splicing point to a prevalent role of the RNA binding Fox family of splicing regulators (RBFOX) in tissue-specific regulation of alternative splicing (43, 46, 47). To determine whether any tissue-specific factor(s) was sufficient to confer neuronal specific splicing in nonneuronal cells, we individually transfected the expression plasmid for mouse RBFOX1, human RBFOX2, and mouse Nova1 into MEFs. By taking advantage of HA-tagged RBFOX1 and -2 in the expression units, we confirmed equal expression of the factors in transfected MEFs and detected induced inclusion of exon 16 and to some extent exons 15 and 16, to which Nova1 showed a weaker effect (Fig. 5A). We therefore focused on further characterization of the putative RBFOX binding sites in mediating the induced exon inclusion.
FIG. 5.
Induction of CaMKIIδ alternative splicing by ectopic expression of tissue-specific splicing regulators and their synergy with SR protein depletion. (A) Effects of ectopic expression of tissue-specific splicing regulators on CaMKIIδ alternative splicing. The plasmids encoding each tissue-specific splicing regulator were transfected into wild-type MEFs, and alternative splicing of the endogenous CaMKIIδ gene was analyzed by RT-PCR. HA-tagged RBFOX proteins and endogenous β-actin were quantified by Western blotting. (B) The putative RBFOX binding sites in the CaMKIIδ minigene and the mutations introduced to the binding sites. (C) RBFOX binding site-mediated splicing response of the CAM15/16 minigene. Individual wild-type and mutant minigene reporters were cotransfected with pcDNA3 (−) or with a plasmid (+) expressing the mouse RBFOX1 gene. Quantified results are shown by the bar graph on the bottom. (D) Synergy between SR protein depletion and overexpression of tissue-specific alternative splicing regulators in the regulation of CaMKIIδ alternative splicing. Individual RBFOX expression plasmids were transfected into wild-type or SRSF2- or SRSF1-deficient MEFs, followed by RT-PCR analysis of the CaMKIIδ isoforms expressed from the CAM15/16 minigene. Quantified results are shown by the bar graph on the bottom, error bars are based on three independent experiments, and statistical significance was determined by a two-tailed t test.
Inspection of the RBFOX binding consensus motif in the CAM15/16 minigene suggested two potential sites in intron 16 (Fig. 5B). We introduced point mutations either alone or in combination to these sites and tested their effects in transfected MEFs. As shown in Fig. 5C, mutations of individual RBFOX binding sites partially impaired the inclusion of exon 16, and combined mutations showed a significant additive effect. Importantly, both single mutants responded to cotransfected mRBFOX1, but the double mutant lost the responsiveness. These data demonstrated regulated exon 16 inclusion by overexpressed RBFOX through their specific binding sites in the minigene. Interestingly, one RBFOX binding site is near the alternative exon 16 and the other is close to the constitutive exon 17. According to the recently established positional rule for RBFOX, the upstream binding site may enhance the selection of exon 16, whereas the downstream binding site may suppress the selection of exon 17 (46). Thus, these two RBFOX binding sites may act together to promote exon 16 inclusion through strengthening the internal alternative exon and weakening the flanking constitutive exon.
Despite the fact that we could activate the heart- and brain-specific alternative splicing program for the CaMKIIδ gene by overexpressing a tissue-specific splicing regulator, we note that the activation was relatively inefficient, as we could not detect significant inclusion of exons 15 and 16, which is the major isoform in the heart and brain. This observation suggests that the expression of a tissue-specific splicing regulator alone may not be sufficient to fully reconstitute the tissue-specific splicing program of the CaMKIIδ gene in fibroblasts. We reasoned that SR protein-dependent exon skipping might be a key mechanism for full reconstitution of tissue-specific alternative splicing in combination with the action of tissue-specific alternative splicing regulators, such as the RBFOX proteins. We tested this possibility by examining the effect of RBFOX expression in combination with downregulation of SR proteins, a scenario that mimics the expression pattern for RBFOX1/2 and SR proteins in the heart (15, 34, 42). As shown in Fig. 5D, SRSF2 depletion exhibited a significant synergy with increased RBFOX expression in enhancing the selection of exons 15 and 16 (compare lanes 1 to 3 with lanes 4 to 6). Although without sufficient statistical significance, SRSF1 depletion seems to enhance the selection of exon 16, and to some extent both exon 15 and 16 (Fig. 4D, compare lanes 1 to 3 with lanes 7 to 9). Together, these results suggest SR protein-dependent exon skipping as a potential key strategy to collaborate with other splicing regulators to activate a full tissue-specific splicing program for the CaMKIIδ gene.
Effects of SR proteins in CaMKIIδ splicing during neuronal differentiation.
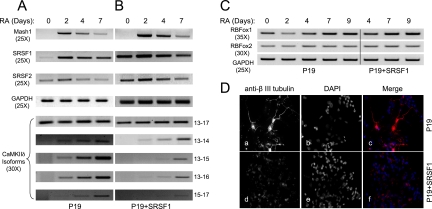
To further address SR protein-dependent exon skipping in a biological context, we took undifferentiated neuroblast P19 cells to determine the induction of CaMKIIδ splicing during neuronal differentiation and examine how over- expression of an SR protein might interfere with the development of such a neuron-specific program. For this purpose, we constructed a pair of isogenic P19 cells with or without SRSF1 overexpression. Upon the induction of neuronal differentiation by RA, we detected transient induction of the neuronal marker Mash1 in the parental P19 cells (21), and overexpression of SRSF1 did not have an apparent effect on this induction (Fig. 6A and B). Similarly, a published study has documented the induction of RBFOX1 and other neuron-specific splicing regulators (14), which were not affected by SRSF1 overexpression either (Fig. 6C). These data indicate that the RA-induced transcriptional program is largely intact in SRSF1-overexpressing P19 cells.
FIG. 6.
Effects of SRSF1 overexpression on the CaMKIIδ splicing program during RA-induced neuronal differentiation. (A) RA treatment of parental P19 cells. (B) RA treatment of SRSF1-overexpressing P19 cells. The time course is indicated on the top, and the expression of individual genes was monitored by semiquantitative RT-PCR, with the specific number of PCR cycles in parentheses in each case. Individual CaMKIIδ isoforms were detected with splice junction-specific primers. (C) RT-PCR analysis of the neuronal splicing factors RBFOX1 and RBFOX2 in P19 cells with or without SRSF1 overexpression. The data show little influence of overexpressed SRSF1 on the expression of these splicing regulators during RA-induced P19 differentiation. (D) Immunofluorescence analysis of parental and SRSF1-overexpressing P19 cells induced by RA for 7 days. Anti-β-III tubulin antibody was used to demonstrate neural differentiation.
We next used semiquantitative PCR to measure CaMKIIδ alternative splicing during neuronal differentiation, and we observed a clear induction of multiple CaMKIIδ isoforms on the parental P19 cells (Fig. 6A, lower panels). Comparison between parental and SRSF1-overexpressing P19 cells revealed the enhanced inclusion of the alternative exon 14, which is consistent with the only SRSF1-responsive ESE present in the exon (Fig. 2A), and diminished inclusion of this exon in response of SRSF1 inactivation in the heart (45). Although the initial induction of SRSF1 appeared coincident with the switch in CaMKIIδ splicing (Fig. 6A), this elevated expression alone is unlikely responsible for the induction of CaMKIIδ splicing, because of the lack of such induction when SRSF1 was expressed to a comparable level in undifferentiated P19 cells (Fig. 6B). Importantly, compared to the parental cells, the RA-induced inclusion of the alternative exons 15 and 16 was significantly delayed and overall reduced in SRSF1-overexpressing P19 cells, consistent with a role of this SR protein in suppressing the inclusion of exons 15 and 16. This observation also raises an intriguing possibility that the unexpected induction of SRSF1 may provide an additional mechanism to ensure a gradual switch in CaMKIIδ isoform expression against acute induction of cell-type-specific splicing regulators during neuronal differentiation.
Significantly, upon the induction of neuronal differentiation by RA, we detected neuronal cell morphology as well as the induction of expression of the neuronal cell marker β-III tubulin in the parental P19 cells (Fig. 6D). In contrast, overexpression of SRSF1 blocked proper neuronal differentiation of P19 cells, as indicated by the lack of RA-induced projections (Fig. 6D). Collectively, these data illustrate that regulated expression of certain “general” splicing regulators, such as SRSF1, is also a part of the developmental program for tissue-specific alternative splicing in mammals.
DISCUSSION
Key to the decision to include or skip a particular exon during splicing is the recognition and selection of functional splice sites by specific components of the splicing machinery. This process is controlled by a variety of cis-acting elements and trans-acting regulatory factors. One of the best-characterized trans-regulatory splicing factors is the SR family of RNA binding proteins (7, 12). While SR proteins are widely known to promote exon inclusion in both constitutive and alternative splicing, increasing evidence now suggests that SR proteins are also involved in suppressing the selection of alternative exons.
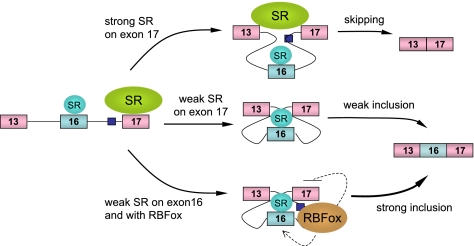
While SR protein-dependent exon inclusion has been extensively characterized in many gene models, relatively little is known about the mechanisms underlying SR protein-mediated exon skipping. In several cases, SR proteins appear to interact with specific binding sites in the intron to activate a decoy exon, thereby interfering with the selection of a real exon nearby (3, 22). In other reported cases, different SR proteins appear to show opposite effects on regulated splicing, but the mechanism has remained elusive (9, 23, 27, 40, 41). Strikingly, our current work revealed for the first time that both up- and downregulation of SR proteins may induce a similar effect on alterative splicing; this likely accounts for complex regulation of alternative splicing in mammalian cells. By characterizing SR protein-dependent exon skipping on the CaMKIIδ model, we now suggest a general model (Fig. 7) for SR protein-dependent exon inclusion or skipping dictated by different binding scenarios: (i) SR protein binding in the intron would trigger a decoy exon to induce exon skipping as previously established; (ii) SR protein binding to the alternative exon would enhance exon inclusion as observed in many well-studied models; (iii) strong SR protein binding to a flanking constitutive exon would force the communication between two constitutive exons, thereby suppressing the inclusion of the internal alternative exon. It is conceivable that different SR proteins may use a combination of these mechanisms to generate a complex pattern of alternative splicing in mammalian cells.
FIG. 7.
Model for collaboration between general and tissue-specific splicing regulators in regulated splicing. Strong SR protein interactions on the flanking constitutive exon 17 are responsible for skipping of the internal alternative exon 16. When the strong exon 17 is weakened, either by ESE mutations or by depletion of a trans-acting SR protein, the internal alternative exon 16 is selected, because of its proximity to the upstream constitutive exon 13. Inclusion of exon 16 is further enhanced by two intronic RBFOX binding events, which promote the upstream exon and inhibits the downstream exon, respectively. Downregulation of SR proteins in combination with other splicing regulators, such as RBFOX, may constitute a general strategy to achieve synergistic regulation of tissue-specific alternative splicing.
Our current work provides further mechanistic insights into several well-established rules in regulated splicing. (i) It has been generally assumed that most constitutive exons may contain strong SR protein binding sites to ensure their inclusion during the splicing reaction. Our results with exon 13 of the CaMKIIδ gene suggest that this may not be true with all constitutive exons, and other strong splicing signals may bypass the need for SR proteins in some cases. (ii) Another major accepted rule in regulated splicing is that the proximal splice sites would be paired if both of the competing splice sites were equally strong or equally weak (5, 6), implying that the activation or repression of both competing splice sites would enhance the selection of the proximal site. Our data provide further support to this rule by demonstrating the enhanced selection of the weak exon 16 when the downstream competing exon 17 is weakened by ESE mutations or by SR protein depletion. (iii) The model depicted in Fig. 7 also emphasizes the involvement of more than one SR protein in regulated splicing. For example, while SRSF2 depletion would impair its ESE activities on both exon 16 and exon 17, the impact on exon 16 may be minor relative to exon 17, thereby elevating the contribution of other SR proteins acting on exon 16. Such collaboration between different SR proteins may also underlie the observed synergy with the RBFOX family of splicing regulators, as an RBFOX bound upstream of the 3′ splice site is able to interfere with SR protein binding to the downstream exon (49). Thus, RBFOX may contribute to the repression of overall SR protein binding on the constitutive exon 17, thus creating a synergy with depletion of a particular SR protein.
Last, but not least, SR proteins have been considered general splicing factors or regulators, as they are ubiquitously expressed in most cell types, and their homeostasis appears to be maintained by autoregulation and cross-regulation loops among the SR protein-coding genes (26). Now, there is a need to modify such a static view of SR proteins, as they are clearly subjected to regulation at the transcriptional and posttranscriptional levels in different cell types and tissues. In fact, SR proteins exhibit tissue-specific expression, particularly in the heart (15), and SRSF1 has been found to vary dramatically in human cancers (24). In the present study, we noticed an intriguing induction of SRSF1 mRNA followed by a progressive decline during neuronal differentiation in P19 cells. Indeed, perturbation of SRSF1 expression in this system not only impaired CaMKIIδ splicing but also blocked RA-induced cell differentiation. These findings have thus put the regulation of SR protein expression and SR protein-mediated splice site selection in a biological context, emphasizing their contribution to tissue-specific alternative splicing through functional synergies with other tissue-specific splicing regulators in the development of the neuron-specific splicing program.
Acknowledgments
We thank Fu lab members for cooperation and stimulating discussion during the course of the study.
This work was supported by NIH grant GM49369 to X.D.F. and by NRL grant ROA-2008-000-20053-0 from the Ministry of Education, Science and Technology of Korea to S.J.
Footnotes
Published ahead of print on 6 December 2010.
REFERENCES
- 1.Anko, M. L., L. Morales, I. Henry, A. Beyer, and K. M. Neugebauer. 2010. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat. Struct. Mol. Biol. 17:962-970. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Buratti, E., C. Stuani, G. De Prato, and F. E. Baralle. 2007. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 35:4359-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding, J. H., et al. 2004. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 23:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eperon, I. C., D. C. Ireland, R. A. Smith, A. Mayeda, and A. R. Krainer. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 12:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eperon, I. C., et al. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, X. D. 2004. Towards a splicing code. Cell 119:736-738. [DOI] [PubMed] [Google Scholar]
- 9.Gallego, M. E., R. Gattoni, J. Stevenin, J. Marie, and A. Expert-Bezancon. 1997. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 16:1772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghigna, C., et al. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20:881-890. [DOI] [PubMed] [Google Scholar]
- 11.Goren, A., et al. 2006. Comparative analysis identifies exonic splicing regulatory sequences: the complex definition of enhancers and silencers. Mol. Cell 22:769-781. [DOI] [PubMed] [Google Scholar]
- 12.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 14.Hakim, N., T. Kounishi, A. Alam, T. Tsukahara, and H. Suzuki. 2010. Alternative splicing of Mef2c promoted by Fox-1 during neuronal differentiation in P19 cells. Genes Cells 15:255-267. [DOI] [PubMed] [Google Scholar]
- 15.Hanamura, A., J. F. Caceres, A. Mayeda, B. R. Franza, Jr., and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 16.Hertel, K. J. 2008. Combinatorial control of exon recognition. J. Biol. Chem. 283:1211-1215. [DOI] [PubMed] [Google Scholar]
- 17.Hertel, K. J., and B. R. Graveley. 2005. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 30:115-118. [DOI] [PubMed] [Google Scholar]
- 18.Hicks, M. J., B. J. Lam, and K. J. Hertel. 2005. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods 37:306-313. [DOI] [PubMed] [Google Scholar]
- 19.Hudmon, A., and H. Schulman. 2002. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 71:473-510. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim el, C., T. D. Schaal, K. J. Hertel, R. Reed, and T. Maniatis. 2005. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. U. S. A. 102:5002-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh, F., T. Nakane, and S. Chiba. 1997. Expression of MASH-1, MATH-1, neuronD, and NSCL-2, basic helix-loop-helix during neuronal differentiation in p19 embryonal carcinoma cells. J. Exp. Med. 182:327-336. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Z. H., W. J. Zhang, Y. Rao, and J. Y. Wu. 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. U. S. A. 95:9155-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jumaa, H., and P. J. Nielsen. 1997. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16:5077-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karni, R., et al. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornblihtt, A. R., M. de la Mata, J. P. Fededa, M. J. Munoz, and G. Nogues. 2004. Multiple links between transcription and splicing. RNA 10:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lareau, L. F., M. Inada, R. E. Green, J. C. Wengrod, and S. E. Brenner. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446:926-929. [DOI] [PubMed] [Google Scholar]
- 27.Lemaire, R., A. Winne, M. Sarkissian, and R. Lafyatis. 1999. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur. J. Immunol. 29:823-837. [DOI] [PubMed] [Google Scholar]
- 28.Lin, S., G. Coutinho-Mansfield, D. Wang, S. Pandit, and X. D. Fu. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, S., and X. D. Fu. 2007. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 623:107-122. [DOI] [PubMed] [Google Scholar]
- 30.Lin, S., R. Xiao, P. Sun, X. Xu, and X. D. Fu. 2005. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell 20:413-425. [DOI] [PubMed] [Google Scholar]
- 31.Long, J. C., and J. F. Caceres. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417:15-27. [DOI] [PubMed] [Google Scholar]
- 32.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modafferi, E. F., and D. L. Black. 1999. Combinatorial control of a neuron-specific exon. RNA 5:687-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33:2078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanford, J. R., et al. 2008. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One 3:e3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanford, J. R., et al. 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 19:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaal, T. D., and T. Maniatis. 1999. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schworer, C. M., L. I. Rothblum, T. J. Thekkumkara, and H. A. Singer. 1993. Identification of novel isoforms of the delta subunit of Ca2+/calmodulin-dependent protein kinase II. Differential expression in rat brain and aorta. J. Biol. Chem. 268:14443-14449. [PubMed] [Google Scholar]
- 39.Shen, H., J. L. Kan, and M. R. Green. 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13:367-376. [DOI] [PubMed] [Google Scholar]
- 40.Solis, A. S., R. Peng, J. B. Crawford, J. A. Phillips III, and J. G. Patton. 2008. Growth hormone deficiency and splicing fidelity: two serine/arginine-rich proteins, ASF/SF2 and SC35, act antagonistically. J. Biol. Chem. 283:23619-23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ten Dam, G. B., et al. 2000. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J. Immunol. 164:5287-5295. [DOI] [PubMed] [Google Scholar]
- 42.Underwood, J. G., P. L. Boutz, J. D. Dougherty, P. Stoilov, and D. L. Black. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25:10005-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, E. T., et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao, R., et al. 2007. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 27:5393-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, X., et al. 2005. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59-72. [DOI] [PubMed] [Google Scholar]
- 46.Yeo, G. W., et al. 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16:130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, C., et al. 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22:2550-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong, X. Y., P. Wang, J. Han, M. G. Rosenfeld, and X. D. Fu. 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, H. L., and H. Lou. 2008. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol. Cell. Biol. 28:5507-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]