Abstract
Alpha interferon (IFN-α) controls homeostasis of hematopoietic stem cells, regulates antiviral resistance, inhibits angiogenesis, and suppresses tumor growth. This cytokine is often used to treat cancers and chronic viral infections. The extent of cellular responses to IFN-α is limited by the IFN-induced ubiquitination and degradation of the IFN-α/β receptor chain 1 (IFNAR1) chain of the cognate receptor. IFNAR1 ubiquitination is facilitated by the βTrcp E3 ubiquitin ligase that is recruited to IFNAR1 upon its degron phosphorylation, which is induced by the ligand. Here we report identification of protein kinase D2 (PKD2) as a kinase that mediates the ligand-inducible phosphorylation of IFNAR1 degron and enables binding of βTrcp to the receptor. Treatment of cells with IFN-α induces catalytic activity of PKD2 and stimulates its interaction with IFNAR1. Expression and kinase activity of PKD2 are required for the ligand-inducible stimulation of IFNAR1 ubiquitination and endocytosis and for accelerated proteolytic turnover of IFNAR1. Furthermore, inhibition or knockdown of PKD2 robustly augments intracellular signaling induced by IFN-α and increases the efficacy of its antiviral effects. The mechanisms of the ligand-inducible elimination of IFNAR1 are discussed, along with the potential medical significance of this regulation.
Cells respond to a milieu of extracellular regulators by timely activation of diverse intracellular signaling cascades. Cells also restrict the magnitude and duration of these signaling events by ligand-inducible degradation of the cognate receptors (reviewed in references 8 and 19). Delineating the mechanisms that underlie specific proteolytic elimination of receptors is important for understanding the pathogenesis of numerous human disorders elicited by an unabated signaling. Conversely, an intimate knowledge of targets that can be used to interfere with such “eliminative signaling” should expand our abilities to augment the therapeutic efficacy of the ligands of medical importance.
Among such ligands are type I interferons (IFNs), including alpha and beta interferons (IFN-α and IFN-β), which exhibit potent antitumor, antiviral, and immunomodulatory activities and which are widely used in therapy of human tumors (25), chronic viral infections (5), and multiple sclerosis (24). These cytokines trigger their signaling via activating the cognate cell surface receptor assembled by the IFN-α/β receptor chain 1 (IFNAR1) and IFNAR2. This event is followed by activation of Janus tyrosine kinases (JAK) JAK1 and Tyk2, tyrosine phosphorylation of receptors, and recruitment of the signal transducers and activators of transcription (STAT1 and STAT2) that induce gene expression through binding to the IFN-stimulated response element (ISRE) within the promoters of IFN-stimulated genes (reviewed in references 1, 29, 39, and 46).
This pathway is under the control of several mechanisms of negative regulation (including the effects of tyrosine dephosphorylation, JAK inhibition and degradation, and STAT sumoylation) that are common to numerous JAK-STAT-activating cytokines and polypeptide hormones (reviewed in reference 17). Conversely, the ligand-specific rapid termination of IFN-α/β signaling is mediated by elimination of its receptor that depends on endocytosis and subsequent lysosomal degradation of the IFNAR1 chain (4).
Degradation of IFNAR1 is stimulated by its ubiquitination, which is facilitated by the SCFβTrcp E3 ubiquitin ligase. This ligase is recruited to IFNAR1 in a manner that depends upon phosphorylation of specific serine residues within a well-defined degron (26-28). Serine phosphorylation of the IFNAR1 degron on Ser535 is essential for IFNAR1 ubiquitination and degradation. The physiologic IFN-α- or IFN-β-inducible pathway requires catalytic activity of Tyk2 to stimulate phosphorylation of the IFNAR1 degron, recruitment of βTrcp, and IFNAR1 ubiquitination and degradation (27, 32, 34). Alternatively, the basal ligand- and JAK-independent phosphorylation (32) is mediated by casein kinase 1α (CK1α) (30) in a manner that is regulated by the priming phosphorylation of IFNAR1 (2). This priming phosphorylation can be further stimulated by inducers of unfolded protein response such as thapsigargin (TG) (31).
Potent catalytic activity and abundance of CK1α hindered the efforts to identify the long-sought serine kinase(s) that mediates IFNAR1 degron phosphorylation within the ligand-induced pathway. Using the cell lysates devoid of CK1α in an in vitro phosphorylation-binding assay helped to obviate this problem. Here we report identification of protein kinase D2 (PKD2) as a type I IFN-inducible kinase that can be activated by IFN-α/β and, in turn, is capable of phosphorylating the serines within the degron of IFNAR1. PKD2 regulates ubiquitination and degradation of IFNAR1 and contributes to the control of IFN-α signaling and antiviral defenses.
MATERIALS AND METHODS
Plasmids, oligonucleotides, cells, and gene transfer.
Vectors for mammalian expression of Flag-IFNAR1 and bacterial expression of glutathione S-transferase (GST)-IFNAR1 (28), βTrcp2/HOS (14), and the 5× ISRE-luciferase reporter (a gift from C. Horvath [38]) have been described elsewhere. Vectors for mammalian expression of human GST-tagged PKD1-3 species (wild-type or kinase-dead mutants [54]) were kindly provided by V. Malhotra. Mutations were generated by site-directed mutagenesis. All resulting mutants were verified by dideoxy sequencing. Short hairpin RNA (shRNA) against PKD constructs based on pLKO.1-puro was purchased from Sigma (MISSION shRNA, e.g., SHGLY-NM_016457). Control shRNA and small interfering RNA (siRNA) were targeted against green fluorescent protein (GFP) (23) and luciferase (28), respectively. siRNA oligonucleotides, including siPKD1 (Hs_PRKCM_2_HP Validated siRNA, SI00301350), siPKD2 (Hs_PRKD2_5_HP Validated siRNA, SI02224768), and siPKD3 (Hs_PRKCN_1_HP Validated siRNA, SI00301357), were purchased from Qiagen and transfected into cells using HiPerFect transfection reagent (Qiagen).
Human embryo kidney 293T cells and epithelial HeLa cells obtained from ATCC were maintained and transfected as described elsewhere (31). Human fibrosarcoma 2fTGH cells and their STAT1-deficient U3A derivatives (35) or Tyk2-deficient 11.1 derivatives (50) were kindly provided by G. Stark and S. Pellegrini. All these cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (HyClone).
Transient transfections of 293T cells or 2fTGH cells and their derivatives using Lipofectamine Plus (Invitrogen) and of HeLa cells using Lipofectamine-2000 (Invitrogen) were carried out according to the manufacturer's recommendations. For stable transfection, replication-deficient lentiviral particles encoding shRNA against PKD2 or vector control were prepared by cotransfecting 293T cells with three other helper vectors as described previously (12). Viral supernatants were concentrated by polyethylene glycol 8000 (PEG 8000) precipitation and used to infect HeLa cells or 2fTGH cells in the presence of Polybrene (3 μg/ml; Sigma). Cells were selected and maintained in the presence of puromycin (2 μg/ml).
Chemicals, antibodies, and immunotechniques.
Antibodies against Flag, GST, and β-actin (Sigma), hemagglutinin (HA) (12CA; Roche), CK1α, PKD1/2 (PKCμ), PKD3 (PKCν), intracellular domain of hIFNAR1 and PKR (Santa Cruz), anti-pan-phospho-tyrosine (4G10), phospho-STAT1 and STAT1 (Cell Signaling), phospho-S710 of PKD2 (Biosource), PKD2 (Bethyl Laboratories), mIFNAR1 (Leinco), and ubiquitin (FK2; Biomol) were purchased. AA3, GB8, and EA12 antibodies which recognize endogenous IFNAR1 (15) and antibodies against IFNAR1 phosphorylated on Ser535 (pS535) (27) were described previously. Antibodies against βTrcp were described elsewhere (45). Secondary antibodies conjugated to horseradish peroxidase were purchased from Chemicon and LI-COR. Immunoprecipitation and immunoblot procedures are described elsewhere (14). Protein degradation was carried out by cycloheximide (CHX) chase in the presence of CHX (20 μg/ml). Immunoblot detection and quantification were carried out using LI-COR's Odyssey Infrared Imaging System.
The IFNAR1 internalization assay was carried out using the fluorescence-based assay that determines the internalization of IFNAR1 by measuring the loss of cell-surface immunoreactivity of endogenous receptor using AA3 antibody as described elsewhere (26). The same antibody in combination with anti-mouse-biotin (Jackson Laboratory) and streptavidin-PE (e-Bioscience) was used for analysis of cell surface human IFNAR1 levels using a FACSCalibur flow cytometer (BD Pharmingen). Levels of mIFNAR1 were determined using an anti-mIFNAR1 antibody (Leinco).
Recombinant human IFN-α2 (Roferon) was purchased from Roche. Thapsigargin (TG), cycloheximide (CHX), and methylamine HCl were purchased from Sigma. H89, bisindolylmaleimide (Bis-I), Gö6976, SP600125, SB203580, LY294002, and D4476 were from Calbiochem. BAY 43-9006 was a kind gift of M. Herlyn. CID755673 was purchased from Tocris Bioscience.
Phosphorylation-binding and kinase assays.
For binding assays, the lysates from 293T cells (treated for various times with IFN-α, 2,000 IU/ml) underwent two rounds of immunodepletion of CK1α, as described elsewhere (30, 32). GST-IFNAR1 proteins (wild type or S535 539A mutant) were expressed and purified from bacterial cells using glutathione-Sepharose. Purified bacterial proteins (2 μg) were incubated in the presence of CK1α-depleted cell lysates (5 μg) and unlabeled ATP (0.5 mM) in a total volume of 20 μl (containing 25 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM KCl, 1 mM EGTA, 1 mM Na3VO4, 0.1 mM dithiothreitol [DTT], and 3.5% glycerol) for 30 min at 30°C. When indicated, the following kinase inhibitors were added to the reaction in 1 μl of dimethyl sulfoxide (DMSO) to the final concentration indicated: H89 (200 nM), BAY43-9006 (50 nM), Bis-I (25 nM), Gö6976 (25 nM), LY294002 (1.5 μM), SP600125 (100 nM), D4476 (400 nM), and SB203580 (100 nM). The reactions were stopped by placing the tubes on ice and adding 150 μl of ice-cold binding buffer (phosphate-buffered saline [PBS] supplemented with 0.1% NP-40 and 50 mM NaF) and incubated with glutathione-Sepharose beads (20 μl) for 2 h at 4°C. The beads were then washed three times with 0.5 ml of binding buffer and incubated with 2 μl of in vitro-translated and 35S-methionine-labeled βTrcp2 for 1 h at 4°C. Beads were then washed three times with 1 ml of binding buffer, and the proteins were eluted using Laemmli buffer, resolved by SDS-PAGE, and analyzed by autoradiography and Coomassie staining.
In vitro phosphorylation of GST-IFNAR1 by cell extracts or PKD preparations (via immunopurification or GST pulldown) was carried out in kinase buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, and 2 mM DTT) with either 0.2 mM unlabeled ATP or 10 μCi of [γ-32P]ATP for 10 to 20 min at 30°C. The products of this reaction were separated by SDS-PAGE and analyzed either by immunoblotting using anti-pS535 antibody or by autoradiography. An in vitro kinase assay using commercially available GST-PKD2 (0.8 pmol, catalog number 7692; Cell Signaling) was carried out using 4 μg of GST-IFNAR1 in kinase buffer (5 mM MOPS, pH 7.2, 2.5 mM glycerol 2-phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, and 50 μM cold ATP) with 10 μCi of [γ-32P]ATP for 30 min at 30°C.
Virus and viral infection.
The antiviral effect of IFN-α was determined by pretreating cells overnight prior to infection with vesicular stomatitis virus (VSV) (Indiana serotype, a gift from R. Harty; propagated in HeLa cells) at a multiplicity of infection (MOI) of 0.1 or 0.5 for 1 h. After the virus inocula were removed, cells were fed with fresh medium and incubated for 20 h. The culture supernatant was harvested and viral titer was determined in HeLa cells overlaid with methylcellulose as described elsewhere (43), and PFU (PFU/ml) numbers were calculated. Cells were observed to determine the cytopathic effect, and expression of VSV-M protein was analyzed by immunoblotting.
RESULTS
PKD2 mediates ligand-inducible phosphorylation of the IFNAR1 degron.
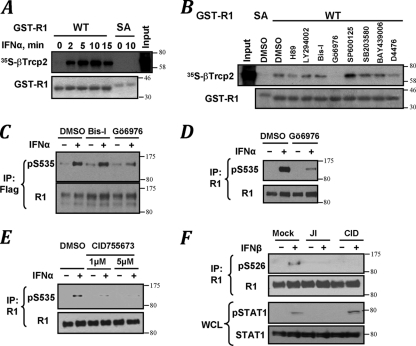
Serine kinases known to be activated by IFN include the members of the protein kinase C family, PKA, PI-3K-Akt-IKK, and MAPK (JNK, p38, Erk, and their downstream kinases; reviewed in references 11, 29, and 39). However, a major basal IFNAR1 kinase activity in cell lysates that often masks the ligand-inducible activity is conferred by CK1α (30). We assessed an in vitro binding of 35S-labeled βTrcp2 to the bacterially expressed GST-tagged intracellular domain of IFNAR1 upon its phosphorylation by the cell lysates that were immunodepleted of CK1α. A greater binding of βTrcp2 to wild-type GST-IFNAR (GST-IFNAR1WT) (but not to its S535 539A mutant) was mediated by the lysates from IFN-α-treated cells (Fig. 1A; see Fig. S1 in the supplemental material). When various kinase inhibitors were tested in this phosphorylation reaction, an opposite effect of two PKC inhibitors was observed. Whereas a dramatic inhibition was achieved using Gö6976 compound, a pan-PKC inhibitor bisindolylmaleimide I (Bis I) did not affect βTrcp2 recruitment (Fig. 1B and Fig. S1). Pretreatment of cells with Gö6976 also inhibited IFN-α-induced Ser535 phosphorylation of either exogenous receptor expressed in human fibrosarcoma 2fTGH-derived U3A cells (Flag-IFNAR1; Fig. 1C) or endogenous IFNAR1 in 293T cells (Fig. 1D). Given that, besides some forms of PKC, Gö6976 was shown to inhibit other kinases, including protein kinase D (PKD) (16), we used a recently identified benzoxoloazepinolone (CID755673) shown to exhibit high specificity against PKD in vitro and in cells (42). Pretreatment of 293T cells with this compound led to a significant inhibition of Ser535 phosphorylation of endogenous IFNAR1 in response to IFN-α (Fig. 1E). Furthermore, similar to JAK inhibitor I, this PKD inhibitor, CID755673, also decreased the efficiency of IFN-β-induced phosphorylation of Ser526 (mouse analogue of human Ser535) in mouse embryo fibroblasts (Fig. 1F). Given that CID755673 treatment did not suppress STAT1 activation, it is plausible that this inhibitor acted downstream of Janus kinases. Taken together, these results suggest that phosphorylation of the degron of IFNAR1 in response to type I IFN might be regulated by a protein kinase sensitive to CID755673, such as PKD.
FIG. 1.
Pharmacological analyses of IFN-induced phosphorylation of IFNAR1 degron. (A) Binding of 35S-βTrcp2 to GST-IFNAR1 (wild type [WT] or S535 539A mutant [SA]) upon their phosphorylation using CK1α-depleted lysates from 293T cells treated with IFN-α as indicated. Levels of GST-IFNAR1 are analyzed by Coomassie blue staining. The SA mutant migrates more slowly in SDS-PAGE due to the presence of four additional amino acids in the linker (31). (B) The effects of various pharmacologic kinase inhibitors (whose activities were verified in kinase-specific assays; data not shown) on phosphorylation of GST-IFNAR1 by the CK1α-depleted lysate from IFN-α-treated (for 10 min) cells were analyzed by subsequent binding of 35S-labeled βTrcp2, as described for panel A. (C) Immunoblot analysis of Flag-IFNAR1 immunopurified from U3A human cells pretreated with kinase inhibitors and then treated with IFN-α as indicated. (D and E) Endogenous IFNAR1 immunoprecipitated from the lysates from 293T cells (treated as indicated) was analyzed as in panel C. (F) Immunoblot analysis of IFNAR1 and STAT1 in mouse embryo fibroblasts not treated (Mock) or treated with IFN-β (500 IU/ml) with or without pretreatment with JAK inhibitor I (JI) or PKD inhibitor CID755673 (CID) as indicated.
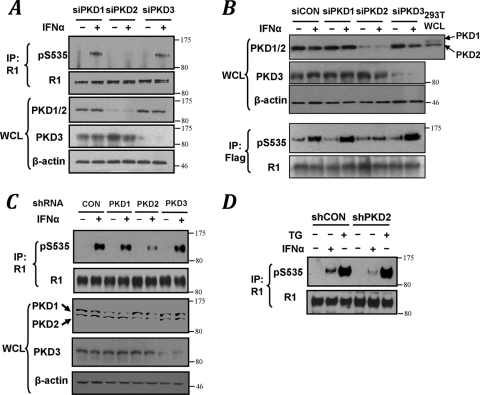
PKD represents a family of serine/threonine protein kinases that is comprised of three members (PKD1/PKCμ, PKD2, and PKD3/PKCν; reviewed in reference 40). In HeLa cells that do not express PKD1 (3), siRNA specific against PKD2 (but not PKD1 or PKD3) markedly inhibited IFN-α-stimulated IFNAR1 phosphorylation on Ser535 (Fig. 2A). Similar data were obtained on exogenous Flag-IFNAR1 stably expressed in U3A cells (Fig. 2B). In 293T cells that express both PKD1 and PKD2, a specific and efficient knockdown of PKD1 did not influence IFNAR1 degron phosphorylation (see Fig. S2 in the supplemental material). Furthermore, experiments that used transduction of these cells with constructs for expressing the hairpins against various PKD species demonstrated that knockdown of PKD2 (but not PKD1 or PKD3) markedly decreased the extent of the ligand-stimulated phosphorylation of Ser535 of IFNAR1 (Fig. 2C). These results implicate PKD2 in mediating IFN-α-inducible phosphorylation of the IFNAR1 degron. Furthermore, given that knockdown of PKD2 affected neither basal IFNAR1 degron phosphorylation (Fig. 2B) nor its induction by an inducer of the unfolded protein response thapsigargin (Fig. 2D), which is known to stimulate IFNAR1 phosphorylation and degradation via the ligand-independent pathway (2, 31), it appears that the role of PKD2 might be limited to the ligand-inducible pathway.
FIG. 2.
Ligand-induced phosphorylation of the IFNAR1 degron is mediated by PKD2. (A) Immunoblot analysis of endogenous IFNAR1 immunopurified from HeLa cells that received indicated siRNA oligonucleotides. Levels of PKD species and β-actin in whole-cell lysates (WCL) were also analyzed. (B) Immunoblot analysis of exogenous Flag-IFNAR1 immunopurified from U3A cells that received indicated siRNA oligonucleotides. Levels of PKD species and β-actin in WCL were also analyzed. WCL from 293T cells is used as a control of mobility of PKD2 species. (C) Immunoblot analysis of endogenous IFNAR1 immunopurified from 293T cells transduced with shRNA against GFP (CON) or PKD species (as indicated) and then treated with IFN-α was carried out as in panel A. Levels of PKD species and β-actin in WCL were also analyzed. (D) Immunoblot analysis of endogenous IFNAR1 immunopurified from HeLa cells stably transduced with shRNA against GFP (shCON) or PKD2 (shPKD2) and then treated with IFN-α or thapsigargin (TG).
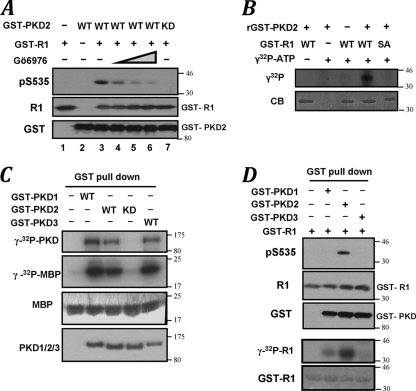
Incubation of GST-tagged PKD2 purified from IFN-α-treated cells with GST-IFNAR1 and ATP enabled phosphorylation of the latter protein on Ser535 detected by phosphospecific antibody. This in vitro reaction could be inhibited by either adding Gö6976 to this reaction (Fig. 3A, lanes 3 to 6) or using a kinase-dead (KD) PKD2K580N mutant (Fig. 3A, lane 3 versus 7), indicating that PKD2 catalytic activity is important for this phosphorylation. Furthermore, commercially available recombinant PKD2 incubated with a 100-fold molar excess of GST-IFNAR1 resulted in phosphorylation of this substrate; a markedly lesser phosphorylation was seen when the GST-IFNAR1SA mutant was used (Fig. 3B). In addition, phosphorylation of GST-IFNAR1 by immunoprecipitated endogenous PKD2 was also detected (Fig. 4B). Together, these results suggest that PKD2 is capable of directly phosphorylating Ser535 of IFNAR1.
FIG. 3.
PKD2 is capable of phosphorylating the IFNAR1 degron in vitro. (A) Effect of Gö6976 (20 to 200 nM) on in vitro phosphorylation of GST-IFNAR1 on Ser535 by purified GST-PKD2 (wild type or kinase dead [KD]) was analyzed by immunoblotting. (B) In vitro phosphorylation of wild-type or SA mutant GST-IFNAR1 (96.0 pmol) by recombinant PKD2 (0.8 pmol) in the presence of γ-32P-labeled ATP was analyzed by autoradiography and Coomassie blue staining. (C) Indicated GST-tagged PKD proteins expressed in 293T cells and purified by pulldown with glutathione beads were incubated with myelin basic protein (MBP) and [γ-32P]ATP. Incorporation of labeled phosphate into PKD and MBP was analyzed by autoradiography. Levels of MBP (Coomassie blue staining) and GST-tagged PKD species (immunoblot with antibody against GST) are also shown. (D) In vitro phosphorylation of GST-IFNAR1 on Ser535 by purified GST-PKD species was carried out using either unlabeled (top two panels) or labeled (bottom two panels) ATP and analyzed by immunoblotting (as in panel A) or autoradiography (as in panel B).
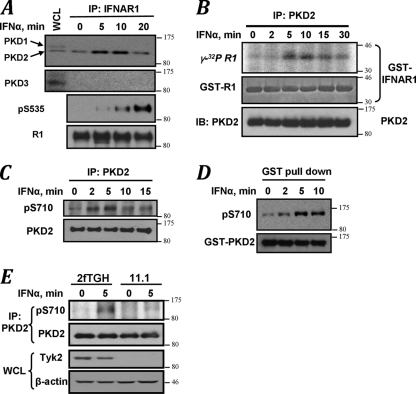
FIG. 4.
Ligand-induced recruitment of PKD2 to IFNAR1 and stimulation of PKD2 kinase activity. (A) IFNAR1 was immunoprecipitated from the lysates of 293T cells treated with IFN-α for the indicated times, and the reaction was analyzed by immunoblotting using the indicated antibodies. The leftmost lane of the upper panel represents the whole-cell lysates from untreated cells. (B) In vitro immunokinase activity of endogenous PKD2 purified from HeLa cells (treated with IFN-α as indicated) toward GST-IFNAR1 as a substrate in the presence of [γ-32P]ATP as analyzed by autoradiography (upper panel), CB staining (middle panel), and immunoblotting with an antibody against PKD (lower panel). (C) Immunoblot analysis of phosphorylation of Ser710 within the activation loop of PKD2 carried out on endogenous PKD2 immunoprecipitated from HeLa cells treated as indicated. (D) Immunoblot analysis on GST-PKD2 purified from HeLa cells was carried out as outlined for panel C. (E) Human 2fTGH fibrosarcoma cells or their isogenic Tyk2-deficient 11.1 derivatives were treated with IFN-α as indicated. Endogenous PKD2 was immunoprecipitated and analyzed as in panel C.
Whereas biochemical properties of PKD2 remain largely uncharacterized, published studies on peptide substrates in vitro have suggested that related PKD1/PKCμ exhibits a preference for a consensus site, L/I/V-X-R/K-XX-s/t (9, 20, 37), which is not congruent with the sequence proximal to the IFNAR1 degron (QTSQDs535) (residues that undergo phosphorylation are underlined). We next sought to compare the activities of various PKD forms (expressed and purified from human cells) toward Ser535 in vitro. All tested GST-tagged PKD species displayed comparable activities in autophosphorylation or phosphorylation of an artificial substrate, myelinic basic protein (Fig. 3C). Despite that, either overall phosphorylation of GST-IFNAR1 (assessed by incorporation of radiolabeled phosphate) or specific phosphorylation on Ser535 (assessed using immunoblot analysis) in the presence of either PKD1 or PKD3 was noticeably inferior to phosphorylation mediated by PKD2 (Fig. 3D). These results indicate that PKD2 possesses a greater ability to phosphorylate Ser535 within the degron of IFNAR1 than other PKD species.
We next sought to determine how IFN-α-induced signaling stimulates phosphorylation of IFNAR1 on Ser535 by PKD2. Interaction between endogenous IFNAR1 and PKD2 (but not PKD1 or PKD3) detected in untreated 293T cells (Fig. 4A; see Fig. S3 in the supplemental material) was further stimulated upon treatment with the ligand (Fig. 4A). An increase in catalytic activity of endogenous PKD2 in HeLa cells treated with IFN-α was detected either by immunokinase assay with GST-IFNAR1 as a substrate (Fig. 4B) or by phosphorylation of Ser710 within the activation loop in either endogenous (Fig. 4C) or exogenous (Fig. 4D) PKD2.
Ligand-inducible phosphorylation of the IFNAR1 degron requires Tyk2 (34). Indeed, this phosphorylation was impaired in cells treated with JAK inhibitor (32) (Fig. 1F). Stimulation of PKD2 (assessed by Ser710 phosphorylation) in response to IFN-α was evident in 2fTGH cells but noticeably less pronounced in the 2fTGH-derived 11.1 cells that lacked Tyk2 expression (Fig. 4E). These data collectively support a hypothesis that a Tyk2-dependent PKD2 activation along with a specific recruitment of PKD2 to IFNAR1 may contribute to an IFN-α-stimulated increase in phosphorylation of the IFNAR1 degron.
PKD2 mediates ligand-inducible phosphorylation, ubiquitination, and degradation of IFNAR1.
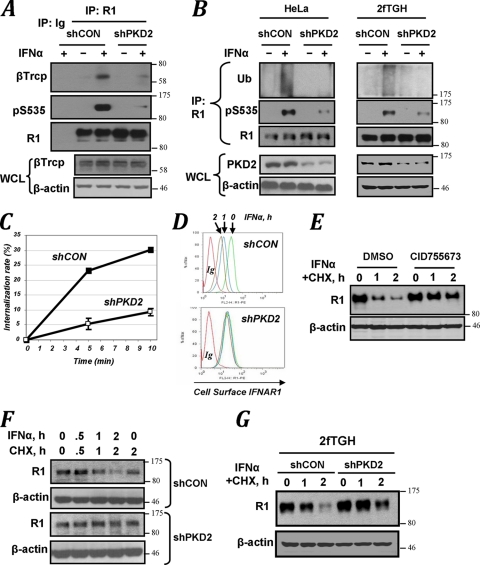
Given that IFN-α-induced Ser535 phosphorylation is required for recruitment of βTrcp and subsequent ubiquitination, endocytosis, downregulation, and degradation of IFNAR1, we next sought to investigate whether these events are mediated by PKD2. Knockdown of PKD2 in HeLa cells robustly inhibited IFN-α-stimulated interaction of IFNAR1 with βTrcp (Fig. 5A). Consistent with an important role of this E3 ubiquitin ligase in conjugation of ubiquitin to IFNAR1 (28), we observed that delivery of shRNA against PKD2 into either HeLa or 2fTGH cells markedly decreased the efficacy of IFNAR1 ubiquitination that was stimulated by IFN-α treatment (Fig. 5B). Furthermore, in line with previous reports on stimulation of IFNAR1 endocytosis by ubiquitination of this receptor, the rate of IFNAR1 internalization was decreased upon PKD2 knockdown (Fig. 5C).
FIG. 5.
PKD2 regulates ubiquitination, endocytosis, and degradation of IFNAR1. (A) Immunoblot analysis of binding of βTrcp to IFNAR1 in HeLa cells (which received indicated shRNAs) analyzed by coimmunoprecipitation and immunoblotting with indicated antibodies. Ig, reaction with irrelevant isotype antibody control. Levels of βTrcp and β-actin in whole-cell lysates (WCL) were also analyzed. (B) Immunoblot analysis of IFNAR1 immunopurified from either HeLa or 2fTGH cells that received indicated shRNA. Levels of PKD2 and β-actin in whole-cell lysates (WCL) were also analyzed. (C) Effect of PKD2 knockdown (open squares) on the rate of internalization of endogenous IFNAR1 measured by a fluorescence-based assay are presented as percentages of total cell surface IFNAR1 level (means ± standard errors of the mean). (D) Fluorescence-activated cell sorter (FACS) analysis of IFNAR1 levels on the surface of cells that received indicated shRNA. Green, blue, and brown signals represent cell surface expression of IFNAR1 after 0, 1, and 2 h of IFN-α treatment, respectively (red, isotype control). (E) Immunoblot analysis of endogenous IFNAR1 in cells untreated or pretreated with the PKD inhibitor CID755673 and then subjected to a cycloheximide (CHX) chase in the presence of IFN-α for the indicated times. (F) Degradation of IFNAR1 in HeLa cells that received indicated shRNA was assessed as for panel E. (G) Degradation of IFNAR1 in 2fTGH cells that received indicated shRNA was assessed as for panel E.
Consistent with these data, ligand-stimulated downregulation of IFNAR1 on the cell surface was noticeably impeded in cells expressing shRNA against PKD2 (Fig. 5D). Treatment of HeLa cells with the PKD inhibitor CID755673 resulted in an appreciable decrease in the rate of proteolytic turnover of IFNAR1 (Fig. 5E). Furthermore, ligand-stimulated degradation of IFNAR1 was noticeably impaired in HeLa cells that received shRNA against PKD2 (Fig. 5F). Similar results were obtained in 2fTGH cells (Fig. 5G). In all, these results suggest that PKD2 plays an important role in the ligand-inducible ubiquitination, endocytosis, and degradation of IFNAR1.
PKD2-mediated degradation of IFNAR1 restricts the extent of cellular responses to IFN-α.
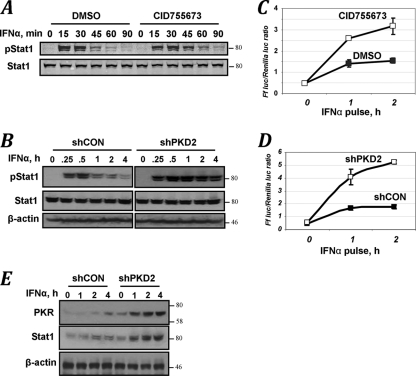
Given that knockdown of PKD2 prevented downregulation of IFNAR1 from the cell surface (Fig. 5D), it is plausible that it may enable a prolonged interaction of IFNAR1 with the ligand that could potentially result in an increase in magnitude and/or duration of IFN-α-elicited signaling. We next sought to investigate whether PKD2-mediated events indeed contribute to the regulation of cellular responses to type I IFN. Pulse treatment of HeLa cells with IFN-α led to a robust activation of STAT1 (assessed by its tyrosine phosphorylation) that peaked at 15 to 30 min and declined during the next hour. Either brief pretreatment of cells with PKD inhibitor CID755673 (Fig. 6A) or knockdown of PKD2 (Fig. 6B) noticeably increased the ligand-induced phosphorylation of STAT1. Similar augmentation of STAT1 activation by CID755673 was observed in mouse embryo fibroblasts in response to IFN-β (Fig. 1F).
FIG. 6.
PKD2 regulates the extent of cellular responses to IFN-α. (A) STAT1 Tyr phosphorylation and levels in cells untreated or pretreated with the PKD inhibitor CID755673 for 1 h and then pulse-treated with IFN-α for 15 min (followed by removal of cytokine and inhibitor and incubation of cells for the indicated times) were analyzed by immunoblotting. (B) Analysis in cells that received indicated shRNA was carried out as for panel A. (C) Relative activity of ISRE-driven firefly luciferase activity normalized to renilla luciferase activity in 2fTGH cells. Cells were pretreated with the PKD inhibitor CID755673 (for 1 h) and pulse-treated with IFN-α (for 0 to 2 h) as indicated, and the activity of luciferase was assessed 24 h later. Data from four independent experiments (each in triplicate) are presented as means ± standard errors of the mean. (D) Analysis of ISRE-driven transcription in 2fTGH cells that received the indicated shRNA and were pulse-treated with IFN-α for the indicated times was carried out as described for panel C. (E) Immunoblot analysis of PKR, STAT1, and β-actin levels in 2fTGH cells that received the indicated shRNA were pulse-treated with IFN-α for the indicated times and were analyzed 24 h thereafter.
For analysis of the role of PKD2 in expression of IFN-stimulated genes and antiviral defenses, we chose human 2fTGH cells. In these cells, phosphorylation, ubiquitination, and degradation of IFNAR1 were affected by PKD2 modulations similarly to HeLa cells (Fig. 2A and C; Fig. 5B, F, and G). However, these cells are highly sensitive to IFN-α/β (35) and, unlike HeLa cells, do not harbor human papillomavirus genes that may confound the analyses of events that occur downstream of the receptor. A pulse treatment with IFN-α (for 1 to 2 h) noticeably increased ISRE-driven luciferase activity in 2fTGH cells. Both pretreatment of cells with the PKD inhibitor CID755673 (Fig. 6C) and knockdown of PKD2 (Fig. 6D) robustly augmented this transcriptional induction. Furthermore, cells that harbored shRNA against PKD2 exhibited a noticeably higher expression of the products of interferon-stimulated genes, such as PKR and STAT1 (Fig. 6E), indicating that PKD2 regulates IFN-α signaling and expression of IFN-stimulated genes.
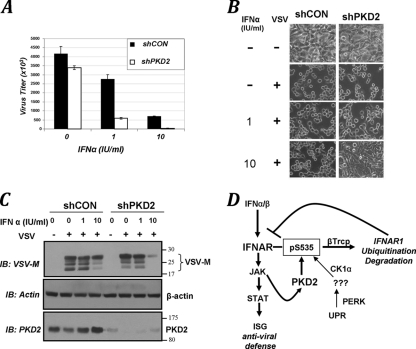
Knockdown of PKD2 in 2fTGH cells resulted in an approximately 20 to 25% lesser viral titer upon infection with vesicular stomatitis virus (VSV) (Fig. 7A), most likely in lieu of a well-characterized role of PKD in viral protein transport along the secretory pathway (22). Remarkably, pretreatment of PKD2 knockdown cells with low doses of IFN-α led to a much more robust protection against infection with VSV, as evident from the assessment of viral titer (Fig. 7A), of the cytopathogenic effects of VSV (Fig. 7B), or of the expression of virus-specific VSV-M protein (Fig. 7C). These data together suggest that PKD2 plays an important role in regulating the cellular responses to type I IFN, including the induction of antiviral defenses.
FIG. 7.
PKD2 regulates the antiviral defenses. (A) 2fTGH cells that received control shRNA (black bars) or shRNA against PKD2 (white bars) were untreated or pretreated with IFN-α (at the indicated doses) and then infected with VSV. Viral titers from three independent experiments (each in quadruplicate) are shown as means ± standard errors of the mean. (B) Cytopathogenic effect of VSV manifested in appearance of rounded, poorly attached, dying cells upon infection of 2fTGH cells that received indicated shRNAs and were pretreated with IFN-α as indicated and then infected with VSV (at an MOI of 0.5). (C) Viral load in 2fTGH cells transduced, treated, and infected with VSV as outlined for panel B was assessed by analysis of the expression of VSV-M viral protein by immunoblotting. Comparable loading was verified by analysis of β-actin (shown in the middle panel). The extent of PKD2 knockdown (lower panel) is also shown. (D) Model that outlines pathways that converge on the phosphorylation of the IFNAR1 degron and ensuing βTrcp-dependent ubiquitination and degradation of IFNAR1 and restriction of cellular responses to type I IFN. Whereas the ligand-independent “inside-out” pathway triggered by unfolded protein responses (UPR) relies on activity of PERK (2, 31) and phosphorylation of the IFNAR1 degron by CK1α (30), the IFN-α/β-inducible pathway involves JAK-mediated activation of PKD2.
DISCUSSION
PKD2 is a ligand-inducible regulator of IFNAR1 stability.
Ligand-stimulated lysosomal degradation of IFNAR1 plays an important role in limiting the magnitude and duration of type I IFN signaling. Previous studies demonstrated that this degradation is mediated by IFNAR1 ubiquitination (26), which is facilitated by the SCFβTrcp E3 ubiquitin ligase. This ligase is recruited to the receptor upon phosphorylation of its degron (27, 28). Unlike basal ligand-independent phosphorylation, ubiquitination and degradation of IFNAR1 (32) mediated by CK1α (30), and stimulation by unfolded protein response inducers via priming phosphorylation (2, 31), the mechanisms underlying the physiologic IFN-inducible events remained obscure. Presented here, data from pharmacologic, genetic, and biochemical analyses indicate a novel and important function of a previously discovered kinase, PKD2, as a serine protein kinase that is activated by IFN-α and that mediates the ligand-inducible phosphorylation of IFNAR1's degron, the recruitment of βTrcp, the ubiquitination of IFNAR1, and the proteolytic elimination of IFNAR1 (Fig. 7D).
Whereas our experiments were focused on PKD2, they did not allow us to entirely rule out a putative role of other members of the PKD family (PKD1 and PKD3) in regulation of IFNAR1 phosphorylation and stability. The primary sequence of the IFNAR1 degron does not resemble the preferred PKD1/PKCμ phosphorylation motif defined on the peptide substrates L/I/V-X-R/K-XX-s/t (9, 20, 37) and present in the majority (reviewed in reference 53) but not all (10, 18, 21, 52) of the reported substrates of PKD1, including its own autophosphorylation sites (41). Consistent with these studies, PKD1 was much less efficient in phosphorylation of IFNAR1 in vitro than PKD2 (Fig. 3D). Our data strongly suggest that PKD2 is capable of directly phosphorylating Ser535. An ability of PKD2 to phosphorylate the IFNAR1 degron was determined by diverse approaches, including the use of either immunoprecipitated endogenous kinase (Fig. 4B), exogenous GST-tagged kinase expressed in mammalian cells and purified by affinity beads (Fig. 3A and D), or commercially available baculovirus-produced recombinant kinase (Fig. 3B). However, a remote possibility that another serine/threonine kinase (which is tightly associated with PKD2 and whose activity depends on catalytic activation of PKD2 and is sensitive to PKD inhibitor) is capable of functioning downstream of PKD2 cannot be entirely excluded. Our preliminary studies that sought to investigate the putative role of other kinases reported to function downstream of PKD (such as JNK, p38 kinase, and IKK) failed to provide the evidence that these kinases are involved in IFN-α-inducible phosphorylation of IFNAR1 (data not shown). In any case, it appears that PKD2 may differ from other members of the PKD family and that some unique attributes of PKD2 enable its preferential recruitment to the vicinity of IFNAR1 (Fig. 4A) and its greater ability to phosphorylate IFNAR1 on Ser535 (Fig. 3D). Future investigation of these characteristics may lead to the development of means for specific interference with IFN-α-induced activation of PKD2. Additional studies (currently impeded by the inability of our antibody to reliably detect phosphorylated Ser539) will also address whether PKD2 also contributes to phosphorylation of this second serine within the IFNAR1 degron.
Mechanisms of the ligand-inducible receptor elimination.
Degradation of IFNAR1 is likely ensured once it is ubiquitinated by the SCFβTrcp E3 ligase recruited to the receptor upon degron phosphorylation stimulated by the ligand (26-28, 34). Identification of PKD2 as a protein kinase mediating Ser535 phosphorylation helps to delineate the proximal IFN-α signaling leading to IFNAR1 proteolysis. The results presented here suggest that treatment of cells with IFN-α promotes the interaction of PKD2 with IFNAR1 and stimulates the catalytic activity of PKD2.
Future studies will delineate in details how exactly IFN-α stimulates PKD2 recruitment to IFNAR1 and promotes the PKD2-mediated catalysis of phosphate transfer. Among possibilities to be explored is the IFN-α/β-stimulated activation of specific Bis-I-insensitive PKC species that can phosphorylate the PKD2 activation loop; this activation may contribute to kinase recruitment as well as increase the activity of PKD2. Whereas our data here clearly show that ligand-stimulated phosphorylation of PKD2 on Ser710 requires expression of Tyk2 (Fig. 4E), it will be important to expand these studies toward requirement of the catalytic activity of Tyk2 because this activity was demonstrated to be essential for IFN-α-stimulated phosphorylation of IFNAR1 on Ser535 (34). Our pilot experiments also indicate that PKD2 might undergo tyrosine phosphorylation in response to IFN-α/β treatment; the role of Janus kinases such as Tyk2 in this phosphorylation and the importance of this phosphorylation for PKD2 recruitment and activation are currently under investigation.
Within this proposed model, the ligand-induced immediate signaling events (i.e., activation of JAK) would give an impetus to both progressive signaling via STAT proteins, to mediate the functions of type I IFN, and eliminative signaling via PKD2, to limit the magnitude and duration of these functions (Fig. 7D). Unlike regulation of some growth factor receptors (e.g., EGFR), where phosphorylation of their Tyr residues serves as a tag for both progressive signaling and for receptor recognition by an E3 ubiquitin ligase (e.g., c-Cbl, reviewed in reference 7), IFN-α-induced eliminative signaling utilizes the additional steps of activating a different protein kinase (PKD2) that mediates phosphorylation of IFNAR1 on serines to recruit βTrcp. This apparent complexity might help to avoid the competition between positive and negative signaling regulators that utilize a similar platform of phospho-Tyr-based interaction as well as to amplify the eliminative signals and enable an additional level of regulation that could be further augmented by the control of IFNAR1-PKD2 interaction.
This mechanism may illustrate the general principle that the combination of kinase recruitment and kinase activation can parlay activation of a receptor into its phosphorylation- and ubiquitination-dependent elimination. It is anticipated that specific details (e.g., nature of a kinase, modes of its activation, characteristics of an E3 ubiquitin ligase, types of resulting ubiquitination, etc.) may vary greatly between specific ligands and their receptors (reviewed in references 8 and 19). The PKD-dependent mechanism of receptor regulation need not even be limited to events stimulated by IFN-α. Numerous stimuli shown to activate protein kinases of the PKD family include polypeptide hormones and growth factors (e.g., PDGF [55]), inducers of the G protein-coupled receptors (49), and the cross-linking of B- and T-cell receptors (44). It remains to be seen whether some of these stimuli utilize PKD to eliminate their own receptors.
PKD2 and cellular responses to type I IFN.
It is plausible that an abnormal PKD2-driven accelerated degradation of IFNAR1 could neutralize the defensive functions of IFN-α/β aimed to protect against viral infections and malignancies. Intriguingly, activation of PKD2 was reported in response to expression of the NS3 protein of hepatitis C virus (33). Furthermore, PKD2 can be activated by some proto-oncogenic proteins, including BCR-ABL (36) and Src (47, 48, 51). In addition, a single nucleotide polymorphism (SNP) within intron 3 of the Prkd2 gene was strongly associated with susceptibility to chronic lymphocytic leukemia (6). The significance of these findings for PKD2-mediated degradation of IFNAR1 and attenuation of IFN-α/β signaling (as well as for pathogenesis of infectious diseases and cancer) remains to be investigated.
Our results indicate that PKD2 plays an important role in regulating the extent of type I IFN signaling and antiviral defenses (Fig. 6 and 7). It is likely that PKD2 will also play a key role in other physiological functions of IFN-α/β, such as antitumorigenic effects, immunomodulation, and activation of dormant hematopoietic stem cells (13). Given that IFN-α/β are actively used in treatment of infections, cancers, and multiple sclerosis, advances in understanding the mechanisms of ligand-inducible activation of PKD2 and its recruitment to the receptor may be of medical importance. Generation and characterization of specific small molecules capable of inhibiting or activating PKD2 are warranted to develop potent means for modulating the IFN signaling. A synergism between such inhibitors and putative modulators of the ligand-independent pathway that involves CK1α (30) and upstream activators of priming phosphorylation of IFNAR1 (2) could be envisioned. Progress in this area is expected to provide a platform for developing a novel class of antiviral, anticancer, and immunomodulatory agents that could be used to augment or attenuate the effects of endogenous type I IFNs or to potentiate the efficacy of existing IFN-α/β-based drugs.
Supplementary Material
Acknowledgments
We thank J. A. Diehl, R. Harty, M. Herlyn, C. Horvath, J. Krolewski, V. Malhotra, S. Pellegrini and G. Stark for the reagents. We are also grateful to Z. Ronai, A. Weissman, J. A. Diehl, and V. S. Spiegelman for helpful comments and suggestions.
We all declare no conflict of interest, apart from D.P.B., who is an employee of Biogen Idec and owns shares of Biogen Idec stock.
This work was supported by Public Health Service grants CA092900 and CA142425 from the National Cancer Institute and funds from The Mari Lowe Center for Comparative Oncology Research (to S.F.).
Footnotes
Published ahead of print on 20 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aaronson, D. S., and C. M. Horvath. 2002. A road map for those who don't know JAK-STAT. Science 296:1653-1655. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya, S., et al. 2010. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J. Biol. Chem. 285:2318-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossard, C., D. Bresson, R. S. Polishchuk, and V. Malhotra. 2007. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 179:1123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coccia, E. M., G. Uze, and S. Pellegrini. 2006. Negative regulation of type I interferon signaling: facts and mechanisms. Cell. Mol. Biol. (Noisy-le-grand) 52:77-87. [PubMed] [Google Scholar]
- 5.Davis, G. L. 2001. Current treatment for chronic hepatitis C. Rev. Gastroenterol. Disord. 1:59-72. [PubMed] [Google Scholar]
- 6.Di Bernardo, M. C., et al. 2008. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40:1204-1210. [DOI] [PubMed] [Google Scholar]
- 7.Dikic, I. 2003. Mechanisms controlling EGF receptor endocytosis and degradation. Biochem. Soc. Trans. 31:1178-1181. [DOI] [PubMed] [Google Scholar]
- 8.Dikic, I., and S. Giordano. 2003. Negative receptor signalling. Curr. Opin. Cell Biol. 15:128-135. [DOI] [PubMed] [Google Scholar]
- 9.Doppler, H., P. Storz, J. Li, M. J. Comb, and A. Toker. 2005. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 280:15013-15019. [DOI] [PubMed] [Google Scholar]
- 10.Du, C., M. Jaggi, C. Zhang, and K. C. Balaji. 2009. Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res. 69:1117-1124. [DOI] [PubMed] [Google Scholar]
- 11.Du, Z., et al. 2007. Non-conventional signal transduction by type 1 interferons: the NF-kappaB pathway. J. Cell. Biochem. 102:1087-1094. [DOI] [PubMed] [Google Scholar]
- 12.Dull, T., et al. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essers, M. A., et al. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458:904-908. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, S. Y., A. Chen, Y. Xiong, Z. Q. Pan, and Z. Ronai. 1999. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IkappaB and beta-catenin. Oncogene 18:2039-2046. [DOI] [PubMed] [Google Scholar]
- 15.Goldman, L. A., et al. 1999. Characterization of antihuman IFNAR-1 monoclonal antibodies: epitope localization and functional analysis. J. Interferon Cytokine Res. 19:15-26. [DOI] [PubMed] [Google Scholar]
- 16.Gschwendt, M., et al. 1996. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase C isoenzymes. FEBS Lett. 392:77-80. [DOI] [PubMed] [Google Scholar]
- 17.Hilton, D. J. 1999. Negative regulators of cytokine signal transduction. Cell. Mol. Life Sci. 55:1568-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinchliffe, K. A., and R. F. Irvine. 2006. Regulation of type II PIP kinase by PKD phosphorylation. Cell. Signal. 18:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huangfu, W. C., and S. Y. Fuchs. 2010. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer 1:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutti, J. E., et al. 2004. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1:27-29. [DOI] [PubMed] [Google Scholar]
- 21.Jaggi, M., et al. 2005. E-cadherin phosphorylation by protein kinase D1/protein kinase Cμ is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 65:483-492. [PubMed] [Google Scholar]
- 22.Jamora, C., et al. 1999. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell 98:59-68. [DOI] [PubMed] [Google Scholar]
- 23.Jin, J., et al. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan, O., et al. 2002. Comparative assessment of immunomodulating therapies for relapsing-remitting multiple sclerosis. CNS Drugs 16:563-578. [DOI] [PubMed] [Google Scholar]
- 25.Kirkwood, J. 2002. Cancer immunotherapy: the interferon-alpha experience. Semin. Oncol. 29:18-26. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, K. G., et al. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J. Cell Biol. 179:935-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, K. G., J. J. Krolewski, and S. Y. Fuchs. 2004. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J. Biol. Chem. 279:46614-46620. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, K. G., et al. 2003. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 22:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larner, A., and N. C. Reich. 1996. Interferon signal transduction. Biotherapy 8:175-181. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J., et al. 2009. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol. Cell. Biol. 29:6401-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, J., et al. 2009. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5:72-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., et al. 2008. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem. Biophys. Res. Commun. 367:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, L., et al. 2008. NS3 protein of hepatitis C virus regulates cyclooxygenase-2 expression through multiple signaling pathways. Virology 371:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marijanovic, Z., J. Ragimbeau, K. G. Kumar, S. Y. Fuchs, and S. Pellegrini. 2006. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem. J. 397:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKendry, R., et al. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. U. S. A. 88:11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihailovic, T., et al. 2004. Protein kinase D2 mediates activation of nuclear factor kappaB by Bcr-Abl in Bcr-Abl+ human myeloid leukemia cells. Cancer Res. 64:8939-8944. [DOI] [PubMed] [Google Scholar]
- 37.Nishikawa, K., A. Toker, F. J. Johannes, Z. Songyang, and L. C. Cantley. 1997. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272:952-960. [DOI] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 40.Rozengurt, E., O. Rey, and R. T. Waldron. 2005. Protein kinase D signaling. J. Biol. Chem. 280:13205-13208. [DOI] [PubMed] [Google Scholar]
- 41.Rybin, V. O., J. Guo, and S. F. Steinberg. 2009. Protein kinase D1 autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J. Biol. Chem. 284:2332-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharlow, E. R., et al. 2008. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J. Biol. Chem. 283:33516-33526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma, S., et al. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 44.Sidorenko, S. P., et al. 1996. Protein kinase C mu (PKC mu) associates with the B cell antigen receptor complex and regulates lymphocyte signaling. Immunity 5:353-363. [DOI] [PubMed] [Google Scholar]
- 45.Spiegelman, V. S., et al. 2000. Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol. Cell 5:877-882. [DOI] [PubMed] [Google Scholar]
- 46.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 47.Storz, P., H. Doppler, F. J. Johannes, and A. Toker. 2003. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J. Biol. Chem. 278:17969-17976. [DOI] [PubMed] [Google Scholar]
- 48.Storz, P., and A. Toker. 2003. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 22:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturany, S., et al. 2002. Mechanism of activation of protein kinase D2(PKD2) by the CCK(B)/gastrin receptor. J. Biol. Chem. 277:29431-29436. [DOI] [PubMed] [Google Scholar]
- 50.Velazquez, L., M. Fellous, G. R. Stark, and S. Pellegrini. 1992. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70:313-322. [DOI] [PubMed] [Google Scholar]
- 51.Waldron, R. T., and E. Rozengurt. 2000. Oxidative stress induces protein kinase D activation in intact cells. Involvement of Src and dependence on protein kinase C. J. Biol. Chem. 275:17114-17121. [DOI] [PubMed] [Google Scholar]
- 52.Waldron, R. T., J. P. Whitelegge, K. F. Faull, and E. Rozengurt. 2007. Identification of a novel phosphorylation site in c-jun directly targeted in vitro by protein kinase D. Biochem. Biophys. Res. Commun. 356:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Q. J. 2006. PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol. Sci. 27:317-323. [DOI] [PubMed] [Google Scholar]
- 54.Yeaman, C., et al. 2004. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 6:106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zugaza, J. L., R. T. Waldron, J. Sinnett-Smith, and E. Rozengurt. 1997. Bombesin, vasopressin, endothelin, bradykinin, and platelet-derived growth factor rapidly activate protein kinase D through a protein kinase C-dependent signal transduction pathway. J. Biol. Chem. 272:23952-23960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.