Abstract
Drosophila microRNAs (miRNAs) and small interfering RNAs (siRNAs) are generally produced by different Dicer enzymes (Dcr-1 and Dcr-2) and sorted to functionally distinct Argonaute effectors (AGO1 and AGO2). However, there is cross talk between these pathways, as highlighted by the recognition that Drosophila miRNA* strands (the partner strands of mature miRNAs) are generated by Dcr-1 but are preferentially sorted to AGO2. Here, we show that a component of the siRNA loading complex, R2D2, is essential both to load endogenously encoded siRNAs (endo-siRNAs) into AGO2 and to prevent endo-siRNAs from binding to AGO1. Northern blot analysis and deep sequencing showed that in the r2d2 mutant, all classes of endo-siRNAs were unable to load AGO2 and instead accumulated in the AGO1 complex. Such redirection was specific to endo-siRNAs and was not observed with miRNA* strands. We observed functional consequences of altered sorting in RNA interference (RNAi) mutants, since endo-siRNAs generated from cis-natural antisense transcripts (cis-NAT-siRNA) exhibited evidence for biased maturation as single strands in AGO1 according to thermodynamic asymmetry and a hairpin-derived endo-siRNA formed cleavage-competent complexes with AGO1 upon mutation of r2d2. Finally, we demonstrated a direct role for the R2D2/Dcr-2 heterodimer in sensing central mismatch positions that direct miRNA* strands to AGO2. Together, these data reveal new roles of R2D2 in organizing small RNA networks in Drosophila.
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are conserved families of small regulatory RNAs (11, 60) that operate within related molecular pathways (16, 45). In both cases, cytoplasmic Dicer class RNase III enzymes metabolize double-stranded RNA (dsRNA) precursors into small RNA duplexes. These mature into single-stranded RNA associated with an Argonaute (AGO) protein in an RNA-induced silencing complex (RISC), which is guided by the small RNA to target transcripts. Animal miRNAs have an additional preceding processing step in which primary miRNA transcripts are cleaved by the nuclear RNase III enzyme Drosha, yielding ∼60- to 70-nucleotide (nt) pre-miRNA hairpins that serve as Dicer substrates (29).
RNase III enzymes often require dsRNA binding protein (dsRBP) cofactors. Drosophila Drosha binds directly to the dsRBP Pasha, which is orthologous to mammalian DGCR8; this family is essential for pre-miRNA generation in all animals (57). Drosophila encodes two Dicers, of which the miRNA processing enzyme Dcr-1 binds the dsRBP Loquacious (Loqs)-PB isoform to generate miRNA duplexes (29). Curiously, a distinct isoform, Loqs-PD, binds the siRNA-generating enzyme Dcr-2 and is required for endogenously encoded siRNA (endo-siRNA) biogenesis (19, 65). Another dsRBP, R2D2, also binds Dcr-2 and is required to load siRNAs into AGO2. The R2D2/Dcr-2 heterodimer is the key constituent of the RISC loading complex (RLC), which determines dominant guide strands by sensing thermodynamic asymmetry of highly paired small RNA duplexes (35, 56). In particular, the duplex strand whose 5′ end resides in the more unstable end is usually selected as the mature guide strand (28, 51). The single mammalian Dicer binds the dsRBPs TRBP and PACT, which are also involved in RISC loading (29).
Despite their parallels, the miRNA and siRNA pathways are substantially separated in Drosophila cells (32, 44). Their small RNAs are not only generated by different Dicers but also sorted into functionally distinct Argonautes. miRNAs accumulate preferentially in AGO1, which is specialized for its capacity to repress targets with limited complementarity, as little as 6 to 7 nucleotides of base pairing to miRNA 5′ ends (positions 2 to 7 or 2 to 8). siRNAs accumulate preferentially in AGO2, which exhibits greater enzymatic cleavage activity toward highly or perfectly complementary targets. Since the siRNA/AGO2 pathway is specifically exploited when the RNA interference (RNAi) technique is used, it is relevant to elucidate AGO2 cargoes and understand mechanisms that ensure correct siRNA loading and function.
The first known endogenous function of Drosophila RNAi was to defend against RNA viruses (1). In addition, endo-siRNAs are generated from transposable elements (TEs), overlapping transcripts (cis-natural antisense transcripts [cis-NATs]), and structured loci (hairpin RNAs [hpRNAs]) to regulate expression of TEs and mRNA targets (8, 9, 14, 26, 42, 43). Recently, miRNA* strands, the partner strands of mature miRNAs, were recognized as a substantial source of AGO2-loaded small RNAs (10, 15, 46). Building upon earlier mechanistic work (13, 55), these studies documented that AGO1 and AGO2 have distinct strand preferences according to 5′ nucleotide identity and central structure in the precursor duplex. In particular, AGO2 strongly prefers the strand whose 9th and 10th nucleotides from the 5′ end are paired, and this structural preference can override thermodynamic asymmetry.
Because mutants of siRNA pathway components seem to exhibit little or no RNAi activity (32, 35, 44), it is usually assumed that the miRNA pathway cannot incorporate siRNAs as guide molecules. Although there is competition between AGO1 and AGO2 loading machineries (13, 20, 55), a UV cross-linking assay showed that AGO1 can largely reject synthetic siRNAs even in the absence of AGO2 loading machinery (55). On the other hand, genetics suggested possible interactions between these pathways, since double mutants of miRNA and siRNA pathway genes exhibited synthetic phenotypes (24, 40). Moreover, the sorting of miR-277, whose mature strand resides in both AGO1 and AGO2 complexes, is altered by the availability of the two Argonautes (13, 20). Taken together, it is not clear how broadly such flexible sorting applies. In addition, a mechanism to sense central duplex mismatch positions, postulated to underlie the distinct strand preference of AGO2 and AGO1 for miRNA/miRNA* duplex strands, has not been elucidated.
In this study, we focused our attention on the roles of R2D2 in small RNA sorting. Analysis of AGO2-associated small RNAs showed an essential role of R2D2 in endogenous small RNA loading to AGO2. Furthermore, all classes of endo-siRNAs, but specifically not miRNA* species, were bound more efficiently by AGO1 in r2d2 and ago2 mutants than in the wild type. Functional consequences for endo-siRNA misdirection into AGO1 in r2d2 mutants were demonstrated, and in vitro assays demonstrated that the R2D2/Dcr-2 complex is directly involved in sensing central mismatch positions to determine the strand for AGO2 loading. Together, these results establish new roles of R2D2 in small RNA-mediated gene regulatory networks.
MATERIALS AND METHODS
Fly stocks and reagents.
We used hen-1[f00810] (20, 50), r2d2[1] (24), ago2[414] (44), and dcr-2[L811fsX] (32) mutants and FLAG-AGO2 transgenic flies (9). The FLAG-AGO2 transgenic flies were used to obtain FLAG-AGO2, r2d2[1] and FLAG-AGO2, dcr-2[L811fsX] recombinant chromosomes.
Small RNA Northern blot analyses and ß-elimination.
Small RNA Northern blot analyses were performed as described previously (44). Probes used in this study are listed in Table S7 in the supplemental material. ß-Elimination was described previously (20). Small RNA enriched samples were prepared using a Mirvana miRNA isolation kit (Ambion) according to manufacturer's instruction.
Photo-cross-linking assay.
A photo-cross-linking assay was done as described previously (56). Lysates were prepared from 0- to 14-h embryos. 5-Iodouridine (5-IU)-modified oligonucleotides were purchased from Dharmacon (see Table S7 in the supplemental material). Other oligonucleotides were described previously (46). One strand of the duplexes was labeled with 32P and annealed with the phosphorylated partner strand. Oligonucleotide RNA duplex (∼50 fmol) was incubated in 20 μl of a standard in vitro RNAi reaction mixture containing 5 mg/ml lysate for 60 min at room temperature. Recombinant R2D2/Dcr-2 complex (80 fmol) was added to 20 μl of the in vitro RNAi reaction mixture containing 2.5 mg/ml bovine serum albumin (BSA). UV irradiation at 312 nm using a handheld UV illuminator (UVP) was done for 15 min on a 96-well plate with a polystyrene lid. The proteins were separated on a two-layered (12% and 8%) SDS-polyacrylamide gel.
Argonaute immunoprecipitations.
Immunoprecipitation (IP) was done according to a published protocol, with some modifications (46). We used RIPA buffer (0.5% NP-40, 0.5% deoxycholate, 0.1% SDS, 1× Complete EDTA free [Roche] in 1× phosphate-buffered saline [PBS]) for experiments with results shown in Fig. 2 and 4 or HEPES-NP-40 buffer {30 mM HEPES-KOH [pH 7.3], 150 mM potassium acetate [KOAc], 2 mM magnesium acetate [Mg(OAc)2], 5 mM dithiothreitol [DTT], 0.1% NP-40, 1× Complete EDTA free} for AGO1-IP libraries (libraries from small RNAs coimmunoprecipitated with AGO1) to lyse cells and to wash beads. AGO1 IPs were done with rabbit anti-AGO1 (AbCam) bound by GammaBind G (GE Healthcare) or Dynabeads (Invitrogen). FLAG-M2 beads (Sigma) or FLAG-M2 antibody bound by Dynabeads was used for FLAG-AGO2 IPs.
Small RNA library analysis.
Small RNA (18 to 28 nt) libraries were constructed as described previously (9), using samples recovered from wild-type (WT) or mutant ovaries and/or AGO1 immunoprecipitates via phenol-chloroform extraction. Each library was subjected to a single lane of sequencing on an Illumina 1G platform (BC Genome Sciences Centre). We also analyzed published ß-elimination libraries and their untreated controls (14, 15).
The raw reads were clipped of 3′ linkers, and perfect matches to the fly genome (FlyBase r5.22 without Uextra) were mapped using Bowtie (30). We then extracted reads to known miRNA and miRNA* reads (49), TEs (31), hpRNAs (43), and 3′ cis-NATs (42). We considered only 21-mer reads mapped to canonical transposon sequences from Repbase as TE siRNAs (23). Only uniquely mapping 21-mers were considered cis-NAT-siRNAs. For the hpRNA-derived siRNA (hp-siRNA) 5′ position analysis, we grouped the reads that share the same 5′ ends and plotted read counts normalized against genomic locations. To study mononucleotide additions, we trimmed one nucleotide at the 3′ end of the unmapped reads and mapped them to each element (miRNA, miRNA*, TEs, hpRNAs, and 3′ cis-NATs).
Target cleavage assay.
We performed the cleavage reaction of the hp-CG4068 target using previously described conditions (43), with the following modifications. We purified AGO1 and AGO2 using Dynabeads (Invitrogen) from adult female flies in RIPA buffer and washed them 5 times each with RIPA buffer and in vitro RNAi lysis buffer (30 mM HEPES-KOH [pH 7.4], 2 mM magnesium acetate, 5 mM DTT) before use. AGO2 immunoprecipitates were incubated with 5′ labeled target RNA in reaction buffer (30 mM HEPES-KOH [pH 7.4], 2 mM magnesium acetate, 5 mM DTT, 0.4 mM ATP, 25 μg/ml yeast tRNA) in the presence of 100 nM antisense oligonucleotide inhibitor at room temperature for 2 h. The oligonucleotide sequences for the templates and inhibitors have been described previously (43).
Nucleotide sequence accession numbers.
The complete small RNA data are available at the modENCODE Data Coordination Center and/or the NCBI Gene Expression Omnibus: Canton S ovary AGO1-IP (modENCODE_2744, GSE24310); ago2[414] ovary AGO1-IP (modENCODE_2746, GSE24311); dcr-2[L811fsX] ovary AGO1-IP (modENCODE_2833, GSE24316); r2d2[1] ovary AGO1-IP (modENCODE_2749, GSE24313); r2d2[1] ovary total RNA (GSM628271); ago2[414] ovary total RNA (GSM628272); Canton S adult ovaries (modENCODE_2976, GSM609246); and dcr-2[L811fsX] ovary small RNA (modENCODE_2975; GSM609245).
RESULTS
R2D2 is essential for efficient loading of endogenous small RNAs into AGO2.
Dcr-2 binds to the dsRBPs R2D2 and Loqs-PD. Loqs-PD is strongly required for endo-siRNA accumulation and function (9, 19, 42, 43, 65). In contrast, R2D2 is dispensable for converting exogenous dsRNA into siRNAs (35), and r2d2 mutant flies accumulate mature endo-siRNAs abundantly (38, 65). Nevertheless, R2D2 is essential for loading and function of exogenous siRNAs (exo-siRNAs) (35, 56) and is genetically required for regulatory processes controlled by endo-siRNAs (38). Altogether, R2D2 is clearly an essential RNAi factor, but its roles in endogenous small RNA pathways are not completely understood.
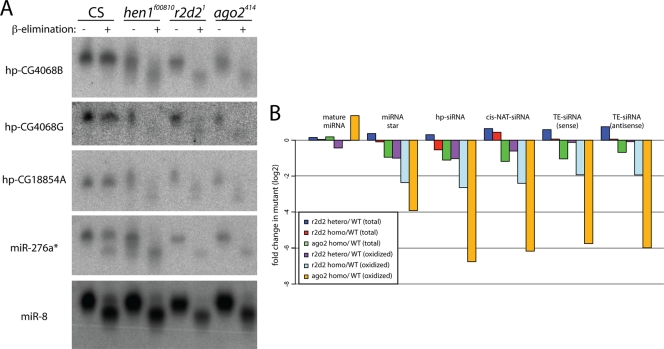
We sought to clarify the influence of siRNA loading factors on endo-siRNA biogenesis by examining the 3′-end structures of endo-siRNAs in null mutants of AGO2 (44) or r2d2 (35). Small RNA duplexes or AGO1-loaded single-stranded small RNAs have free 2′ and 3′ OH groups at their 3′ ends and react to this treatment, thereby increasing their mobility on acrylamide gels. On the other hand, single-stranded small RNAs matured in the AGO2 complex carry 2′ O-methyl modification at their 3′ ends and hence are resistant to ß-elimination. Consistent with previous reports (14, 26, 43), endo-siRNAs were resistant to ß-elimination in a Hen1-dependent manner, while mature miR-8 exhibited increased mobility after ß-elimination (Fig. 1A). A recent study showed that the endo-siRNA hp-CG4068B (also known as esi-2.3) was sensitive to ß-elimination in the r2d2 mutant (38). We assayed other hp-siRNAs and observed that all species tested, hp-CG4068B, hp-CG4068G, and hp-CG18854A, were fully sensitive to ß-elimination in the r2d2[1] mutant. Moreover, mutation of AGO2 had similar effects on hp-siRNA modification (Fig. 1A).
FIG. 1.
R2D2 and AGO2 are essential for 3′ modification of endo-siRNAs. (A) 3′ structures of different small RNAs in RNAi pathway mutants. Total RNAs (hp-CG4068B, miR-276*, and miR-8) or small RNA enriched samples (hp-CG4068G and hp-CG18854A) were isolated from wild-type (Canton S [CS]) and hen1, r2d2, and ago2 mutant male flies, ß-eliminated, and analyzed by Northern blotting. In the wild type, miR-8 was sensitive to this reaction while the endo-siRNAs hp-CG4068B, hp-CG4068G, and hp-CG18854A were resistant; this was dependent on Hen1 methyltransferase, indicating their authentic modification. endo-siRNAs present in r2d2 and ago2 mutants were fully sensitive to ß-elimination. A majority of miR-276* was ß-elimination resistant in the wild type, consistent with sorting to the AGO2 complex. However, its ß-elimination-sensitive population was not increased in r2d2 or ago2 mutants, in contrast to those of endo-siRNAs. (B) Genome-wide analysis of the 3′ modification of endo-siRNAs. We reanalyzed published small RNA libraries from r2d2 and ago2 mutants (14, 15) and plotted normalized ratios between the mutant and wild-type samples in log2 scale. miRNA* species and all classes of endo-siRNAs were relatively constant in total RNA from the wild type and these mutants. In contrast, all of these classes were reduced in oxidized samples from the mutants, demonstrating essential roles for R2D2 and AGO2 in 3′ modification.
We extended this analysis to other small RNA species by using published small RNA sequencing data from oxidized RNA samples (15). Small RNAs that bear 2′,3′ hydroxyl termini are oxidized at their 3′ termini upon treatment with NaIO4; this blocks adaptor ligation and depletes them from the cloned library. As a result, libraries made from oxidized samples are enriched with 3′ modified endo-siRNAs (14). We calculated changes in abundance of small RNAs by dividing normalized read counts in the mutant libraries by those in the wild-type libraries. This analysis confirmed the finding that miRNA* species are modified in an R2D2- and AGO2-dependent manner (14, 15) (Fig. 1B; also see Fig. S1 and Tables S1 to S5 in the supplemental material). We similarly observed that other classes of endo-siRNAs, including TE-siRNAs, cis-NAT-siRNAs, and hp-siRNAs, were all reduced in oxidized libraries from r2d2 or ago2 mutant heads relative to wild-type levels (Fig. 1B; also see Fig. S1 in the supplemental material). Therefore, the 3′ modification of all known endo-siRNAs depends on R2D2 and AGO2. Since Drosophila Hen1 can modify only 3′ ends of properly loaded small RNAs, the simplest interpretation is that R2D2 is required to load endo-siRNAs into AGO2, as with exogenous siRNAs.
To test this directly, we introduced a FLAG-AGO2 genomic transgene (9) into the dcr-2 and r2d2 mutants. We found that the tagged AGO2 transgene provided full rescue of the RNAi defect in ago2 mutant embryos (see Fig. S2A in the supplemental material), indicating its functionality. Unexpectedly, although R2D2 was previously described to be important for fertility (24), r2d2[1] flies homozygous for FLAG-AGO2 (i.e., bearing 4 copies of AGO2) could be maintained as a stable homozygous stock. Extra copies of AGO2 may partially compensate for the absence of R2D2; alternatively, meiotic recombination might have removed background mutations from the parental r2d2[1] chromosome.
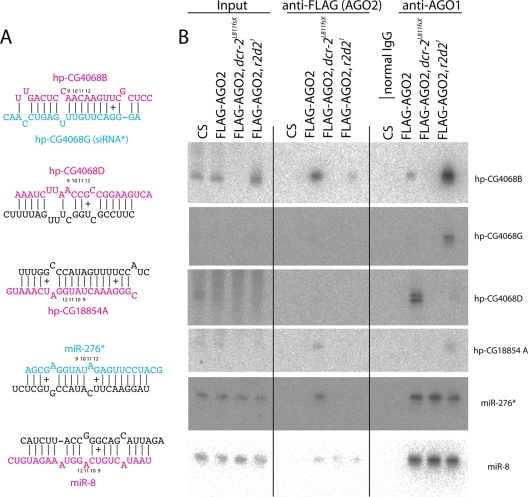
Using these flies, we sequentially purified AGO2 and AGO1 complexes from embryos or male adult flies with anti-FLAG or anti-AGO1 antibody and analyzed the copurified RNAs by Northern blotting (Fig. 2 B; also see Fig. S3 in the supplemental material). We verified that the precipitation efficiency and expression levels of AGO1 and FLAG-AGO2 were not strongly affected in the mutants (see Fig. S4 in the supplemental material). Consistent with the ß-elimination experiment, coprecipitation of endo-siRNAs (hp-CG4068B, -D, and -G and hp-CG18854A) and a miRNA* species (miR-276a*) with AGO2 was dramatically reduced in the r2d2 mutant. These results provided direct evidence that R2D2 is required for proper loading of endogenous small RNAs in the AGO2 complex.
FIG. 2.
hp-siRNAs but not miRNA* strands are misdirected to AGO1 in the r2d2 mutant. (A) Structures of the small RNA duplexes analyzed. (B) FLAG-AGO2 and AGO1 complexes were purified from wild-type and dcr-2 and r2d2 mutant embryos (hp-CG4068B) or male adults (others); all genotypes carry the FLAG-AGO2 transgene. Canton S was used as a control for FLAG-AGO2 IP, and normal rabbit IgG was used as a control for AGO1 IP; inputs contain ∼5% of IP reactions. In the wild type, mature miR-8 was sorted exclusively to AGO1, while the endo-siRNAs hp-CG4068B, hp-CG4068G, hp-CG18854A, and miR-276* were highly enriched in AGO2 (also see Fig. S3 in the supplemental material for a longer exposure of hp-CG4068G, which more clearly illustrates this rare siRNA in the AGO2 complex). In contrast, endo-siRNAs were loaded into AGO1 in the r2d2 mutant. Endo-siRNAs were not detected in the dcr-2 mutant, consistent with the essential role for Dcr-2 in siRNA biogenesis. miR-276* was not loaded in AGO2 in the dcr-2 and r2d2 mutants; however, loading of miR-276* in AGO1 was not changed in r2d2 and dcr-2 mutants. hp-CG4068D was enriched in AGO1 in the wild type (10); this siRNA was reduced in AGO1 in the r2d2 mutant, suggesting that R2D2 may also be involved in hp-siRNA processing.
endo-siRNAs but not miRNA*s are redirected to AGO1 in RNAi pathway mutants.
Since hpRNA-derived siRNAs detected in r2d2[1] flies were sensitive to ß-elimination, we hypothesized that they were either not unwound or incorporated into another complex in this mutant background. We tested whether they might associate with AGO1 using Northern blotting. As reported previously, the endo-siRNAs hp-CG4068B, hp-CG4068G, and hp-CG18854A preferentially sort to AGO2 in the wild type (9, 14, 26, 43). In contrast, we observed efficient incorporation of all of these endo-siRNAs in AGO1 in the r2d2 mutant, indicating that endo-siRNAs can efficiently be sorted to the AGO1 complex in the absence of R2D2 activity (Fig. 2B). This re-sorting accounts for the accumulation of endo-siRNAs in r2d2 mutants.
The exceptional hpRNA-derived siRNA hp-CG4068D (esi-2.3) accumulated preferentially in AGO1 in the wild type (10, 41), consistent with the unstable base pairing in the central region of the hp-CG4068D/E duplex (Fig. 2A). We note that this endo-siRNA was decreased in AGO1 when R2D2 was mutated. Since AGO1 loading is believed to be independent of R2D2 activity, we infer that R2D2 has a minor role in hpRNA processing, as suggested recently (38). Finally, we note that miR-276a* did not accumulate further in AGO1 in the r2d2 mutant. This was consistent with our observation that a portion of miR-276* is normally sensitive to ß-elimination in the wild type and that this population was not further increased in r2d2 and ago2 mutants (Fig. 1A). Therefore, not all AGO2-loaded species are redirected into AGO1 in the absence of the RNAi pathway.
Genome-wide analysis of small RNA sorting in RNAi-defective mutants.
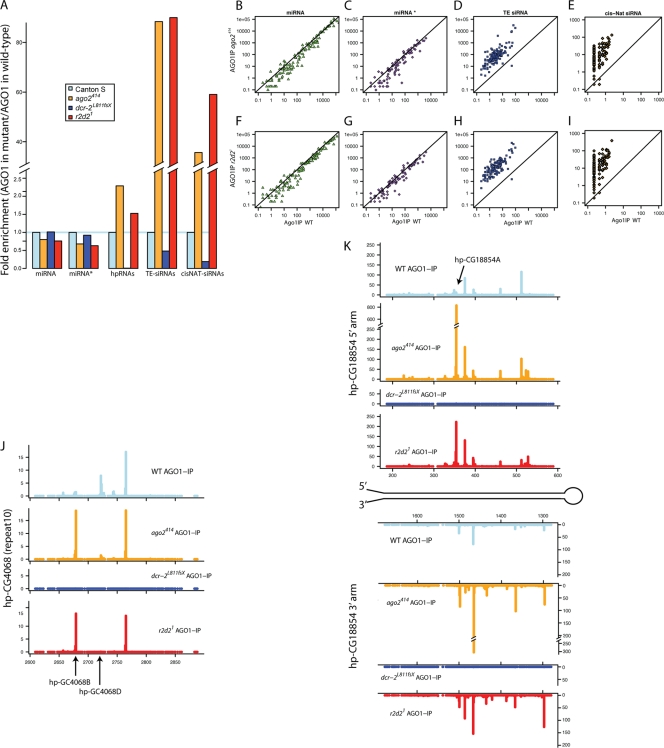
Since various hp-siRNAs, but not miR-276a*, were re-sorted to AGO1 in r2d2 mutant flies, we were interested to determine the breadth of changes in the sorting profiles in RNAi-defective mutants. To do so, we prepared libraries from small RNAs coprecipitated with AGO1 from wild-type and ago2, dcr-2, and r2d2 mutant ovaries. Illumina sequencing of these libraries yielded 4,438,146, 5,178,786, 5,137,283, and 5,206,992 mapped reads, respectively. In order to obtain an appropriate baseline for interpreting any changes in sorting, we also prepared libraries from total ovary RNA samples from the same genetic backgrounds, yielding 17,913,478, 23,077,842, 18,807,309, and 22,521,947 mapped reads, respectively. We extracted reads that mapped to mature miRNA strands, miRNA* strands, transposable elements (TEs), and hp-siRNA and 3′ cis-NAT-siRNA loci. We normalized read counts by the total mapped reads in each data set and calculated the ratio between the normalized reads in the mutants and those in the wild type (Fig. 3A; also see Fig. S5A and Table S1 in the supplemental material).
FIG. 3.
Global changes in small RNA sorting in ago2 and r2d2 mutants. (A) Enrichment of endo-siRNAs in the AGO1 complex in RNAi mutants. Reads matching TEs, 3′ cis-NATs, hpRNAs, and mature or star strands of miRNAs in AGO1-IP from RNAi mutants were normalized by their counts in those in AGO1-IP from the wild type. As expected, endo-siRNAs were strongly reduced in the dcr-2 mutant. TE- and hp-siRNAs were enriched in AGO1 in ago2 and r2d2 mutants, suggesting that sorting of endo-siRNAs is globally affected in these mutants. However, none of these mutants showed significant changes in miRNA or miRNA* sorting, even though miRNA* species are substantially incorporated in AGO2 in the wild-type background (10, 15, 46). (B to I) Small RNAs matching to mature miRNAs (B and F), miRNA* strands (C and G), TEs (D and H), and 3′ overlapping transcripts (cis-NATs) (E and I) in the wild type and RNAi mutants. Individual points represent single miRNA or miRNA* species, reads summed across single 3′ cis-NAT regions, or reads summed across the canonical sequences of individual TE families. (J and K) hpRNA analysis. Normalized small RNA density (reads/million mapped reads; y axes) was mapped on the genomic regions corresponding to 1 of 20 repeats in the hp-CG4068 locus (J) and the hairpin region of the hp-CG18854 gene (K). The positions of 5′ ends are shown (x axes; nt). The majority of peaks are higher in the r2d2 and ago2 mutants than in the wild type in the AGO1-IP libraries, with a small number of exceptions, such as hp-CG4068D.
The relative abundance of mature miRNA reads was largely unchanged in these mutants, in both AGO1-IP and total RNA libraries (Fig. 3A, B, and F; also see Fig. S5A and Table S2 in the supplemental material). This was consistent with the fact that RNAi factors are not essential for normal miRNA regulation. As predicted by a previous study (15) and our Northern blot analysis (Fig. 2B), the overall abundance of miRNA* species in the AGO1 complex was not changed in any of the RNAi-defective mutants (Fig. 3A, C, and G; also see Table S2 in the supplemental material). In contrast, hp-siRNAs were enriched in the AGO1 complex in r2d2 and ago2 mutant ovaries compared to levels in the wild type (Fig. 3A), confirming our Northern blotting data (Fig. 2B). hp-siRNAs were essentially eliminated in the dcr-2 mutant, as expected from its essential role in siRNA production (Fig. 3A; also see Fig. S5A in the supplemental material).
We further analyzed changes in AGO1-sorted hp-endo-siRNAs by examining individual duplexes grouped by their 5′ ends (Fig. 3J and K; also see Table S5 in the supplemental material). Endo-siRNAs from most hpRNA duplexes showed strong enrichment in AGO1 complex in r2d2 and ago2 mutants, indicating that the aggregate enrichment was not skewed by individual outliers. However, some hp-siRNAs were reduced in AGO1-sorted population in the r2d2 mutant (Fig. 3J), including hp-CG4068D (Fig. 2B). The hp-siRNAs reduced in the r2d2 mutant appear to have mismatches in their duplex central regions and were relatively abundant in AGO1 in the wild-type background. The reduction of some hp-siRNAs supports the notion that R2D2 plays a role in endo-siRNA processing.
More compellingly, we observed 40 to 80 times enrichment of TE-siRNAs and cis-NAT-siRNAs in AGO1 complexes from r2d2 and ago2 mutants (Fig. 3A). This was not a consequence of the biased behavior of a subset of loci, since siRNAs from essentially all the different families of TEs (Fig. 3D and H) and from all the different cis-NAT regions (Fig. 3E and I) exhibited these characteristic re-sorting patterns. The representation of TE-siRNAs in AGO1 in these mutants was particularly remarkable. About 9% of AGO1-bound small RNAs in r2d2 and ago2 mutants were TE-siRNAs (452,267 and 463,001 reads, respectively), whereas these comprised only ∼0.1% (4,447 reads) in wild-type AGO1-IP (see Table S1 in the supplemental material).
We note that TE-siRNAs and cis-NAT-siRNAs were slightly overrepresented in the total RNA libraries prepared from the r2d2 and ago2 mutants (see Table S1 and Fig. S5 in the supplemental material). Since RNAi restricts TEs, it is conceivable that a mild increase in TE substrates leads to increased siRNA production; however, this does not satisfactorily explain increased cis-NAT-siRNAs in AGO1. Another possibility is that 3′ modified and unmodified species may have differential cloning efficiencies, potentially at the 3′ linker ligation step (59). However, the 2-fold increase in 3′ OH availability does not account for the proportionally stronger enrichments of all endo-siRNAs in the AGO1 complex from r2d2 and ago2 mutant ovaries (Fig. 3A; also see Tables S3 and S4 in the supplemental material).
We hypothesize that the differences in re-sorting patterns between TE- or cis-NAT-siRNAs and hp-siRNAs are a consequence of their distinct precursor structures. hp-siRNA duplexes usually have mismatches between the two strands, whereas TE-siRNAs are believed to derive from perfect/nearly perfect dsRNA molecules. The mismatches in hpRNA precursors may allow loading to AGO1 to some extent, while perfectly duplexed siRNAs almost never enter AGO1 in the wild type. In summary, our data demonstrate that in vivo, AGO1 is capable of binding to all classes of endo-siRNAs: TE-siRNAs, hp-siRNAs, and cis-NAT-siRNAs. However, the activity of the R2D2-driven loading pathway into AGO2 efficiently depletes endo-siRNAs from the miRNA pathway in wild-type flies.
Role for R2D2 in endo-siRNA fidelity.
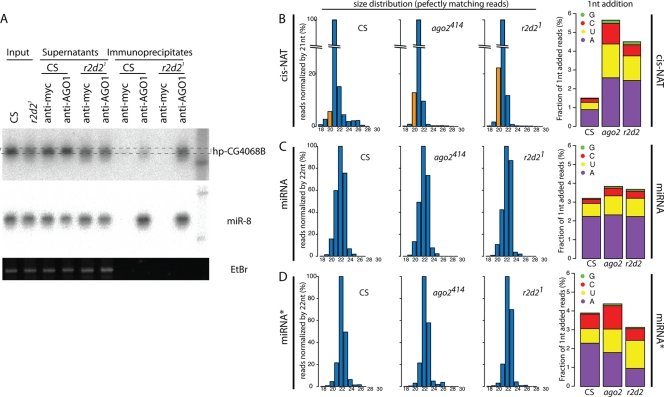
We noticed that hp-siRNAs that were misdirected into AGO1 in r2d2 mutants exhibited a broader size distribution in wild-type total RNA, occupying a size range extended by ±1 to 2 nt on Northern blots (Fig. 4A). We examined the heterogeneity of other endo-siRNA species misloaded to AGO1, with respect to size distribution and 3′ untemplated additions (Fig. 4B). In general, endo-siRNAs from perfect or nearly perfect dsRNAs exhibit a strong preference for 21 nt, defined by Dcr-2 cleavage (8, 9, 14, 42). To avoid the possibility that the size distribution of TE-siRNAs was affected by the breakdown of abundant TE-piwi interacting RNAs (TE-piRNAs) present in ovaries, we focused our analysis on uniquely mapping cis-NAT-siRNAs.
FIG. 4.
Variability of endo-siRNA sequences in the AGO1 complex. (A) Heterogeneity of siRNA hp-CG4068B in the AGO1 complex. AGO1 complexes were purified from the wild type and the r2d2 mutant. RNA samples prepared from the input, supernatant, and immunoprecipitate were analyzed on a denaturing gel. Anti-myc antibody was used as a negative control. The hp-CG4068B signal in the r2d2 mutant shows heterogeneity in the migration, whereas miR-8 and ethidium bromide (EtBr) staining did not show evidence of RNA degradation. (B to D) Size distribution and 3′ mononucleotide additions of cis-NAT-siRNAs and miRNAs. Size distribution is shown by normalizing the read counts with 21-nt reads for cis-NATs (B) or with 22-nt reads for miRNAs (C) and miRNA* species (D). 3′ addition was analyzed by calculating the fractions of mononucleotide-added read counts compared to the total number of reads.
Interestingly, when the read numbers were normalized by the number of 21-nt cis-NAT (i.e., “siRNA”) reads, 20-nt cis-NAT small RNAs were slightly more frequent in the ago2 or r2d2 mutant than in the wild type (Fig. 4B). We also counted reads containing single nucleotide mismatches to the reference genome at their 3′ ends, which are generally considered to derive from untemplated nucleotide addition (3, 5, 7). We observed that the mutant libraries contained a higher proportion of reads with terminal mismatches than the wild type (Fig. 4B, right), with additions of adenosine or uridine being much more frequent than guanosine or cytosine additions. In contrast, the size distribution and untemplated addition of miRNAs and miRNA* strands were not altered in these RNAi mutants. This suggested that variability of small RNA sequences in the mutants was specific for endo-siRNAs misloaded to AGO1 (Fig. 4B, C, and D).
These observations mirror recently reported trimming and tailing effects on miRNAs in the presence of artificial perfectly complementary targets (3). Since endo-siRNAs usually have endogenous perfectly complementary targets, unlike most miRNAs, such tailing and trimming processes may predict that endo-siRNAs in the AGO1 complex would be unstable. However, although our analysis detected increased 3′ variation of endo-siRNAs in ago2 and r2d2 mutants, this did not effectively deplete bulk endo-siRNAs from AGO1, the large majority of which remained 21 nt in length. We were therefore interested to address whether re-sorted endo-siRNAs were functional as regulatory species in the AGO1 complex.
Functionality of AGO1-sorted endo-siRNAs.
It was not a given that endo-siRNAs could program active AGO1 complexes, since AGO1 is poorly capable of unwinding perfectly base paired siRNA duplexes in vitro (25, 55). We confirmed using lysate preparations that AGO1 could bind artificial siRNA duplexes weakly but did not unwind them efficiently in vitro (see Fig. S6 in the supplemental material). In this system, 3′ overhangs were required for efficient binding to AGO1, arguing against the possibility of nonspecific associations stemming from a high concentration of synthetic small RNA duplexes in the reactions (see Fig. S6). These in vitro tests suggested that a strong proportion of perfectly complementary endo-siRNA duplexes might be arrested for AGO1-RISC maturation.
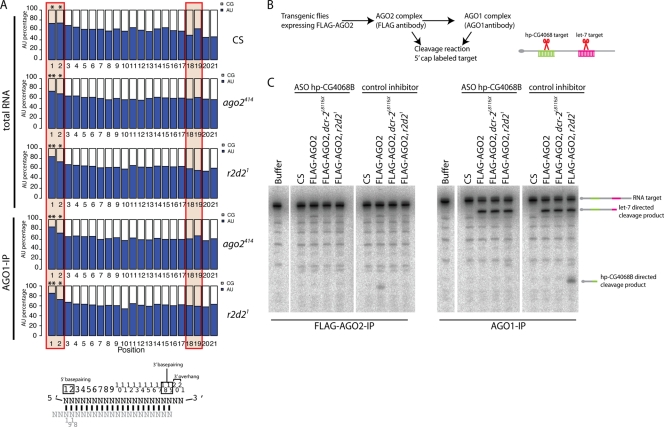
Still, we sought to assay whether any endo-siRNAs from perfectly base paired cis-NAT regions could be unwound in vivo. As cis-NAT-siRNAs were too rare to detect by Northern blotting (data not shown), we could not perform native gel analysis to distinguish duplex and single-stranded cis-NAT-siRNAs. However, we could address this issue using the deep sequencing data, by taking advantage of the fact that strand selection by active RISC is influenced by duplex thermodynamic asymmetry. In particular, AGO1 preferentially selects the strand whose 5′ end resides in the less stable duplex end. In the total RNA sample from the wild type, the 5′ ends of cis-NAT-siRNAs (i.e., precisely 21-nt reads) exhibited a strong bias for A and U nucleotides, consistent with the thermodynamic asymmetry rule (Fig. 5A). Interestingly, we observed significantly higher (P < 0.05) A-U contents only at positions 1 and 2 of cis-NAT-siRNAs in the total RNA and AGO1-IP libraries from ago2 and r2d2 mutants (Fig. 5A; also see Table S6 in the supplemental material).
FIG. 5.
Functionality of endo-siRNAs misloaded into AGO1. (A) cis-NAT-siRNAs in the RNAi mutants exhibit single-stranded siRNA features. The nucleotide composition of 21-nt cis-NAT-siRNAs was analyzed using the total RNA and AGO1-IP libraries from Canton S and the r2d2 and ago2 mutants. The average A-U content across all cis-NAT regions was ∼66%. In all libraries, the first two nucleotides were enriched with A and U nucleotide, especially at the first position (**, P < 1E−6; *, P < 0.05) (also see Table S6 in the supplemental material). On the other hand, nucleotides 18 and 19, which are predicted to base pair with the 5′ 2 nucleotides of their partner strand, exhibit lower A-U contents than the first two nucleotides. The asymmetric distribution of A-U nucleotides argues that the cis-NAT-siRNA population in the mutants contains a significant fraction of unwound siRNAs, as opposed to arrested siRNA duplexes. (B) In vitro cleavage assay. FLAG-AGO2 and AGO1 complexes were purified from wild-type and dcr-2 and r2d2 mutant female flies and subjected to an on-bead cleavage assay using a cap-labeled RNA target. The RNA target contains sequences perfectly complementary to hp-CG4068B and let-7. Thus, the cleavage activity by an endo-siRNA (hp-CG4068B) and a mature miRNA (let-7) can be monitored in the same reaction. (C) In vitro cleavage assays were carried out in the presence of antisense oligonucleotide (ASO) inhibitors against hp-CG4068B or an unrelated sequence (hp-CG18854A) to demonstrate assay specificity. In the wild type, hpRNA-directed cleavage activity was associated exclusively with AGO2. As expected, AGO1 complexes from different genotypes exhibited similar cleavage activities against the let-7 target sequence. In the dcr-2 mutant, neither Argonaute complex showed hp-CG4068-directed target cleavage activity. In the r2d2 mutant, CG4068B-mediated cleavage activity was not detected in the AGO2 complex but was instead robustly detected in the AGO1 complex.
If cis-NAT-siRNAs were double stranded, as seen from in vitro tests, then their 5′ A-U enrichment should be mirrored by A-U enrichment at positions 18 and 19, which base pair with positions 1 and 2. Such complementary features of small RNA pairs are seen, for example, between sense/antisense piRNAs with a 10-nt overlap. In this setting, the 5′ U bias of Aub-loaded piRNAs is mirrored by an A10 bias of AGO3-loaded piRNAs, indicating their guide-target relationships (4, 18). However, positions 18 and 19 of endo-siRNAs exhibited lower A-U bias than their corresponding 5′ nucleotides, and this result was observed consistently among total RNA pools and AGO1-IP pools from the wild type and the RNAi mutants (Fig. 5A). These data were consistent with the notion that cis-NAT-siRNA populations were biased toward single-stranded species, as a result of duplex thermodynamic asymmetry and/or 5′ U preference of AGO1 for mature guide strands. This in vivo analysis suggested that a substantial proportion of cis-NAT-siRNAs in AGO1 complexes from RNAi mutants were unwound and available to serve as regulatory molecules.
Because a mismatch in the seed region of the guide or passenger strand can strongly facilitate unwinding by AGO1 (25, 55), we hypothesized that the activity of siRNAs derived from imperfect duplexes, such as hp-siRNAs, might be even more readily detectable in the AGO1 complex. To test whether hp-siRNAs were active in AGO1 in the r2d2 mutant background, we challenged FLAG-AGO2 and AGO1 complexes from wild-type and dcr-2 and r2d2 mutant female adult flies to cleave cap-labeled targets (Fig. 5B). Consistent with previous reports, miRNA- and endo-siRNA-mediated cleavage activities were associated with AGO1 and AGO2, respectively, in wild-type flies (26) (Fig. 5C). Neither complex showed endo-siRNA-directed cleavage activity in the dcr-2 mutant, because Dcr-2 is essential for endo-siRNA production. In the r2d2 mutant, AGO2 was not capable of cleaving the hpRNA target, confirming the importance of R2D2 for efficient AGO2 loading. On the other hand, the AGO1 complex purified from the r2d2 mutant could efficiently cleave the hp-siRNA target. let-7-directed cleavage activities were similar among the AGO1-IP samples, a control that excluded artifacts such as changes in IP efficiency.
Our results therefore suggest two functional consequences of the redirection of endo-siRNAs into AGO1 in the absence of r2d2 and ago2. First, endo-siRNA duplexes may occlude maturation of AGO1 complexes. Second, those endo-siRNAs that mature in AGO1 complexes are expected to have altered function as a consequence of the distinct functions of AGO2 and AGO1 (13, 21). We provided evidence that populations of both cis-NAT-siRNAs and hp-siRNAs are available in single-stranded form in AGO1 and could execute regulatory function.
R2D2/Dcr-2 complex directionally binds to miRNA duplexes.
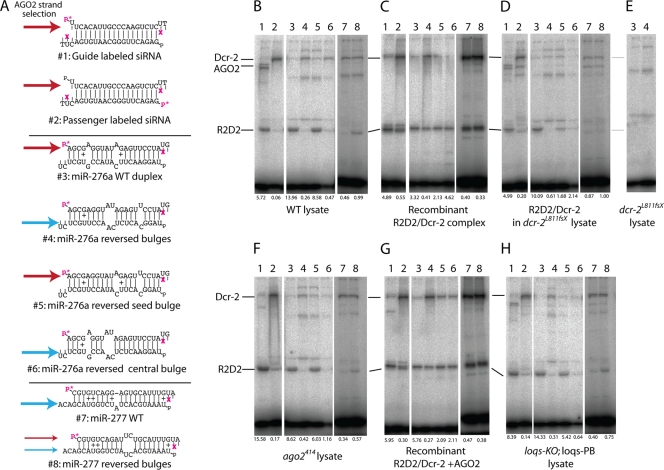
Since our results indicated that R2D2 plays a central role in small RNA loading to AGO2 (Fig. 1 and 2), we were interested to further test the role of R2D2 in strand selection. Recent studies showed that Drosophila AGO2 prefers the strand whose 9th and 10th nucleotides from the 5′ end are paired (10, 46). To study the mechanisms of sensing central mismatch positions, we took advantage of a photo-cross-linking assay (56). A photoreactive cross-linker, 5-iodouridine (5-IU), was introduced at the second nucleotide from the 3′ ends of synthetic sod1 small RNAs (Fig. 6A). As shown previously, R2D2 and Dcr-2 cross-linked with the guide and passenger strands of the thermodynamically asymmetric sod1 siRNA duplex, respectively (Fig. 6A and B, duplexes #1 and #2) (56).
FIG. 6.
R2D2/Dcr-2 complex senses central duplex mismatches. (A) Small RNA duplexes tested in photo-cross-linking assays. Duplexes #1 and #2 are thermodynamically asymmetric sod1 siRNA duplexes (56); duplexes #3 to #8 are wild-type and variant mir-276a and mir-277 duplexes characterized previously (46). The dominant guide strands known to be preferred by AGO2 are indicated by arrows. The arrows are blue when AGO2 selects the mature miRNA and red when AGO2 selects the miRNA* strands or the guide siRNA strands. Purple “x”s depict 5-iodouridine modification, and P*s represent 32P labels. Note that the second nucleotide from the 3′ end of miR-276* and miR-277* was changed from C to U to incorporate the cross-linker. Cross-linked proteins were visualized via the radiolabeled RNA and identified by their characteristic migration in SDS-PAGE. (B to H) Photo-cross-linking assays using embryo lysates and/or recombinant proteins, as indicated below each set of panels; the numbers at the bottom indicate ratios of R2D2/Dcr-2 signals. The R2D2/Dcr-2 complex orients directionally onto thermodynamically asymmetric duplexes (B, lanes 1 and 2) (54), as well as onto miRNA duplexes whose central structure determines strand preference by AGO2 (B, lanes 3 to 8). R2D2 dominantly bound the star (top) strand of the WT mir-276a duplex (B, lane 3), but reversal of its unpaired bases caused Dcr-2 to bind this strand preferentially (B, lane 4). Such directional binding was not determined by the seed bulge (B, lane 5) but instead by the duplex central bulges (B, lane 6). In the WT mir-277 duplex, whose star (top) strand is strongly rejected by AGO2, we observed no cross-linking between miR-277* and R2D2 (B, lane 7). The reversal of central duplex bulges, which allows partial loading of miR-277* to AGO2, permits detectable cross-linking with R2D2 (B, lane 8). The recombinant R2D2/Dcr-2 complex also bound directionally to asymmetric siRNA (C, lanes 1 and 2) and mir-276a (C, lanes 3 and 4) duplexes. However, asymmetric binding of recombinant proteins was less prominent than in embryo lysate, and purified R2D2/Dcr-2 did not orient directionally on some duplexes (C, lanes 5 to 8). This was rescued by supplementing recombinant R2D2/Dcr-2 with a dcr-2 mutant lysate (D) that was confirmed to lack the functional R2D2/Dcr-2 complex (E). Directional binding of the R2D2/Dcr-2 complex was normal in the ago2 mutant lysate (F), and recombinant AGO2 did not improve binding of purified R2D2/Dcr-2 (G). The loqs mutant lysate specifically lacking siRNA cofactor activity (Loqs-PD) exhibited full directional binding of R2D2/Dcr-2 (H). Functional validation of the ago2 mutant lysate, recombinant AGO2, and the loqs-KO; loqs-PB lysate is shown in Fig. S3 in the supplemental material.
We tested whether the positions of central mismatches were also sensed by the R2D2/Dcr-2 complex. We used the mir-276a duplex as a model because its central structure is known to direct AGO2 to select its star strand (Fig. 6A, duplex #3) (10, 46). When embryo lysate was incubated with the synthetic mir-276a duplex, in which miR-276a* was radioactively labeled and also contained 5-IU at its second-to-last position, we observed that R2D2 cross-linked strongly to the miR-276a* strand (Fig. 6B, lane 3). We previously showed that the strand selection of AGO2 was strongly changed by moving the mismatch positions (46). We modified the sequence of the bottom strand to reverse the relative positions of internal mismatches, without changing the first 4 nucleotides of either of the ends (Fig. 6A, duplex #4). The binding direction of the R2D2/Dcr-2 complex on this substrate was remarkably altered (Fig. 6B, lanes 3 and 4). These results indicate that the R2D2/Dcr-2 complex possesses the ability to sense internal duplex mismatches and that the asymmetric binding determines the dominant strand selected by AGO2.
To further dissect the effect of mismatch positions on the binding direction, we prepared another set of mutant duplexes. When the mismatch in the seed region of the star strand was moved to the opposite side, AGO2 selected the star strand (Fig. 6A, duplex #5). Consistently, R2D2 was more efficiently cross-linked than Dcr-2 (Fig. 6B, lane 5). When only the central mismatches were moved, AGO2 selected the mature strand (Fig. 6A, duplex #6), and correspondingly the star strand was cross-linked with Dcr-2 (Fig. 6B, lane 6).
Because the end energies of the miR-276a duplex are relatively similar, we wished to confirm our results with a duplex with greater thermodynamic asymmetry. In our previous report, we showed that changes in the central structure of a terminally asymmetric duplex, the mir-277 duplex, could detectably alter AGO2 strand selection (46). Although AGO2 selects exclusively the mature strand from the WT mir-277 duplex, we were able to change the central structure of the miR-277 duplex to load a significant amount of miR-277* strand to AGO2 in our previous report (Fig. 6A, duplexes #7 and #8). Using the 5-IU-modified oligonucleotides, we observed the expected cross-linking patterns. With the WT mir-277 duplex, we could not detect cross-linking with R2D2, consistent with the complete rejection of the star strand from AGO2 loading (Fig. 6B, lane 7). In contrast, we observed substantial cross-linking of R2D2 when the centrally mutated mir-277 duplex was used (Fig. 6B, lane 8).
These results indicate that the R2D2/Dcr-2 complex is involved in sensing the positions in the internal structures. These aspects mirrored previous observations made using perfectly duplexed siRNAs (56) and suggested that the mechanisms of sensing terminal free energy and internal mismatch positions are tightly linked processes.
R2D2/Dcr-2 is directly involved in central mismatch sensing.
We sought to determine whether the R2D2/Dcr-2 complex itself or other cofactors determine the binding direction, since it is known that the RISC loading complex contains additional factors (48). We tested whether the known Dcr-2 binding factors AGO2 and Loqs-PD isoform could contribute to directional binding. We used the ago2[414] mutant or the loqs[KO] mutant complemented by the loqs-PB transgene (47, 65) (see Fig. S2C in the supplemental material). As described previously, loqs[KO]; P[loqs-PB] flies are rescued for miRNA processing and are consequently viable, but this genotype is highly compromised for endo-siRNA accumulation due to the absence of Loqs-PD (19, 65); we therefore refer to this genotype as the loqs-PD mutant. Loqs-PD appears to be dispensable for siRNA function, judging from relatively normal siRNA-initiated target cleavage activity in this background (see Fig. S2A in the supplemental material). Mutant lysates from ago2 or loqs-PD mutant flies did not reveal obvious defects in asymmetric binding, indicating that these factors are not essential for strand selection (Fig. 6F and H).
In reciprocal experiments, we used a purified protein complex of R2D2/Dcr-2. Notably, a substantial switch in binding direction was also observed when a purified, recombinant R2D2/Dcr-2 complex was used (Fig. 6C, lanes 3 and 4). We also added functional recombinant AGO2 protein (see Fig. S2B in the supplemental material) to the purified R2D2/Dcr-2 heterodimer, but this also did not improve binding asymmetry (Fig. 6G), arguing against involvement of AGO2 in this step. Although we demonstrated a new activity for R2D2/Dcr-2 in sensing central mismatch positions, the asymmetry of the binding was more prominent in lysate than in purified complex (Fig. 6B and C); some substrates exhibiting asymmetric binding in the lysate were not similarly bound asymmetrically by recombinant proteins (e.g., duplexes 5 and 6). Indeed, it was noted previously that directional binding of R2D2/Dcr-2 onto thermodynamically asymmetric duplexes was better in lysate than in purified proteins (56). This could suggest the existence of other auxiliary factors for asymmetric binding. In fact, the directional binding was partially improved by adding dcr-2 mutant lysate (Fig. 6D), which lacks signals for both Dcr-2 and R2D2 (Fig. 6E). While the results with recombinant proteins and complex cell extracts are not directly comparable, they may suggest an additional factor(s) that promotes directional binding of the RISC loading machinery.
DISCUSSION
The nearly complete lack of RNAi activity in mutants of core RNAi components led to the conclusion that the Drosophila miRNA pathway is not capable of incorporating siRNAs as guide molecules (32, 35, 44). In fact, in vitro studies using dcr-2 or ago2 mutant lysates showed that AGO1 still prefers mismatched duplexes in RNAi-defective mutants (25, 55). However, our results not only demonstrate that R2D2 is essential for endogenous small RNA loading to AGO2 but also reveal that the RNAi pathway prevents endo-siRNA duplexes from binding to AGO1. All classes of endo-siRNAs were enriched in AGO1 in the r2d2 mutant, whereas the abundance of miRNA* species in AGO1 remained unchanged (Fig. 2 and 3). The different behaviors of endo-siRNAs and miRNA* species in the r2d2 mutant emphasize the sophistication of sorting machineries.
Distinct behaviors of endo-siRNAs and miRNA* species in r2d2 and ago2 mutants suggested that all classes of endo-siRNAs are sorted primarily to AGO2 by the degree of base pairing in the central region. The dramatic change of sorting in the r2d2 and ago2 mutants fits well with this hypothesis. In contrast, miRNA* species were not further accumulated in AGO1 in RNAi-deficient mutants, consistent with the notion that miRNA* species are sorted to AGO2 by the distinct strand selection mechanisms of the two Argonautes (10, 15, 46). Although additional miRNA/miRNA* duplexes become available for AGO1 in RNAi mutants, AGO1 continues to incorporate the mature strands preferentially. Reciprocally, AGO1 knockdown can increase the amount of miRNA* species (10, 15), suggesting that loss of AGO1 can increase the pool of miRNA duplexes available to AGO2 for miRNA* strand incorporation.
R2D2, together with Dcr-2, directly mediates strand selection according to the thermodynamic stabilities of strongly paired siRNA duplexes (56). It was proposed that the dsRNA binding domains of R2D2 have greater affinity for the more stably base paired end, thereby orienting Dcr-2 at the less stable duplex end. We analyzed the role of R2D2 in strand selection from imperfectly paired duplexes and found that the direction of R2D2/Dcr-2 binding was strongly influenced by characteristic positions of internal mismatches (Fig. 6). The capacity of R2D2/Dcr-2 to sense central mismatch positions was not predicted by the existing models. We note that the purified R2D2/Dcr-2 complex showed the weakest affinity, across a panel of variant duplexes, to a duplex with a mismatch at the 9th nucleotide (55). Because position 9 is not exactly in the middle of 22-mer duplexes, this asymmetry might contribute to orienting the RLC according to positions of internal mismatch.
Although the biological requirement for RNAi as an endogenous defense system against viruses or selfish genetic elements is now well documented, it remains unclear why the RNAi pathway has to be separated from the miRNA pathway. Nevertheless, the factors for both pathways are well conserved among insect species (6, 58). Moreover, while the siRNA factors of Caenorhabditis elegans are not orthologous to those in insects, the miRNA pathway is also separated from endo-/exo-siRNA pathways in the nematode (17, 63). These observations suggest that separation of these pathways is of biological utility. Because viral infection can induce the production of large quantities of viral siRNAs (2, 12, 33, 62), the lack of R2D2 or AGO2 may conceivably saturate the miRNA pathway upon viral infection. In addition to the occlusion of AGO1 complexes by highly complementary endo-siRNA duplexes (e.g., from TEs and cis-NATs), we provide explicit evidence for activity of an endo-siRNA redirected to AGO1 in the absence of R2D2 (Fig. 5C). Our bioinformatics analysis also suggests that at least some fraction of endo-siRNAs from perfectly double-stranded RNAs is unwound in the AGO1 complex, implicating activity of endo-siRNAs (Fig. 5A). Because AGO1 is a more potent repressor against seed-matched target mRNAs than AGO2 (13, 21), the residence of normally AGO2-loaded siRNAs in AGO1 predicts potential misregulation of targets bearing seed matches. We propose that the loss of developmental robustness in RNAi mutants may potentially be due to the off-target effects of endo-siRNAs in AGO1 (36), in addition to the direct loss of endo-siRNA-mediated regulation.
Off-target effects of siRNAs on seed-matched targets are a major problem in mammalian cells (22). In addition, transfection of synthetic siRNAs in mammalian cells can saturate the miRNA pathway, yielding derepression of endogenous miRNA targets (27). More strikingly, injection of siRNAs into zebrafish embryos induces non-sequence-specific effects, possibly by saturating the miRNA pathway (64). In contrast to invertebrate model systems, siRNAs and miRNAs seem to be broadly incorporated into all Argonautes in mammals (34, 39), perhaps suggesting that there are inherent limitations to our ability to completely control the regulatory capacity of mammalian siRNAs. Intriguingly, though, the mammalian endogenous siRNA and miRNA pathways appear to be temporally segregated by keeping miRNAs inactive in oocytes and very early embryonic stages (37, 52) and by allowing the majority of siRNA production only in oocytes (53, 61). Our results highlight the central role of R2D2 in organizing the Drosophila siRNA and miRNA pathways by loading duplexes to the correct complex and selecting the correct strands. In principle, such a factor could provide selectivity for mammalian small RNA sorting.
Supplementary Material
Acknowledgments
We thank Greg Hannon, Haru Siomi, and Richard Carthew for fly stocks. We also thank Stefan Ameres for his suggestion about AU contents of cis-NAT-siRNAs. K.O. was supported by the Japan Society for the Promotion of Science.
Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Alfred Bressler Scholars Fund, and the NIH (R01-GM083300 and U01-HG004261). Q.L. was supported by the Welch Foundation (I-1608) and the NIH (GM084010).
Footnotes
Published ahead of print on 6 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aliyari, R., and S. W. Ding. 2009. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 227:176-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aliyari, R., Q. Wu, H. W. Li, X. H. Wang, F. Li, L. D. Green, C. S. Han, W. X. Li, and S. W. Ding. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameres, S. L., M. D. Horwich, J. H. Hung, J. Xu, M. Ghildiyal, Z. Weng, and P. D. Zamore. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328:1534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennecke, J., A. A. Aravin, A. Stark, M. Dus, M. Kellis, R. Sachidanandam, and G. J. Hannon. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089-1103. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs, A. M., Y. Ando, M. J. de Hoon, Y. Tomaru, T. Nishibu, R. Ukekawa, T. Funakoshi, T. Kurokawa, H. Suzuki, Y. Hayashizaki, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 20:1398-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, C. L., W. C. Black IV, A. M. Hess, and B. D. Foy. 2008. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics 9:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang, H. R., L. W. Schoenfeld, J. G. Ruby, V. C. Auyeung, N. Spies, D. Baek, W. K. Johnston, C. Russ, S. Luo, J. E. Babiarz, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 24:992-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, W. J., K. Okamura, R. Martin, and E. C. Lai. 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czech, B., C. D. Malone, R. Zhou, A. Stark, C. Schlingeheyde, M. Dus, N. Perrimon, M. Kellis, J. Wohlschlegel, R. Sachidanandam, et al. 2008. An endogenous siRNA pathway in Drosophila. Nature 453:798-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czech, B., R. Zhou, Y. Erlich, J. Brennecke, R. Binari, C. Villalta, A. Gordon, N. Perrimon, and G. J. Hannon. 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell 36:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynt, A. S., and E. C. Lai. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynt, A. S., N. Liu, and E. C. Lai. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. U. S. A. 106:5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forstemann, K., M. D. Horwich, L. Wee, Y. Tomari, and P. D. Zamore. 2007. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghildiyal, M., H. Seitz, M. D. Horwich, C. Li, T. Du, S. Lee, J. Xu, E. L. Kittler, M. L. Zapp, Z. Weng, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildiyal, M., J. Xu, H. Seitz, Z. Weng, and P. D. Zamore. 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghildiyal, M., and P. D. Zamore. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishok, A., A. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 18.Gunawardane, L. S., K. Saito, K. M. Nishida, K. Miyoshi, Y. Kawamura, T. Nagami, H. Siomi, and M. C. Siomi. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315:1587-1590. [DOI] [PubMed] [Google Scholar]
- 19.Hartig, J. V., S. Esslinger, R. Bottcher, K. Saito, and K. Forstemann. 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 28:2932-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwich, M. D., C. Li, C. Matranga, V. Vagin, G. Farley, P. Wang, and P. D. Zamore. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17:1265-1272. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki, S., T. Kawamata, and Y. Tomari. 2009. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell 34:58-67. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, A. L., J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. Lim, and P. S. Linsley. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany, and J. Walichiewicz. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110:462-467. [DOI] [PubMed] [Google Scholar]
- 24.Kalidas, S., C. Sanders, X. Ye, T. Strauss, M. Kuhn, Q. Liu, and D. P. Smith. 2008. Drosophila R2D2 mediates follicle formation in somatic tissues through interactions with Dicer-1. Mech. Dev. 125:475-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamata, T., H. Seitz, and Y. Tomari. 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 16:953-960. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura, Y., K. Saito, T. Kin, Y. Ono, K. Asai, T. Sunohara, T. Okada, M. C. Siomi, and H. Siomi. 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453:793-797. [DOI] [PubMed] [Google Scholar]
- 27.Khan, A. A., D. Betel, M. L. Miller, C. Sander, C. S. Leslie, and D. S. Marks. 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209-216. [DOI] [PubMed] [Google Scholar]
- 29.Kim, V. N., J. Han, and M. C. Siomi. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126-139. [DOI] [PubMed] [Google Scholar]
- 30.Langmead, B., C. Trapnell, M. Pop, and S. L. Salzberg. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau, N., N. Robine, R. Martin, W. J. Chung, Y. Niki, E. Berezikov, and E. C. Lai. 2009. Abundant primary piRNAs, endo-siRNAs and microRNAs in a Drosophila ovary cell line. Genome Res. 19:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He, E. J. Sontheimer, and R. W. Carthew. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69-81. [DOI] [PubMed] [Google Scholar]
- 33.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437-1441. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Q., T. A. Rand, S. Kalidas, F. Du, H. E. Kim, D. P. Smith, and X. Wang. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921-1925. [DOI] [PubMed] [Google Scholar]
- 36.Lucchetta, E. M., R. W. Carthew, and R. F. Ismagilov. 2009. The endo-siRNA pathway is essential for robust development of the Drosophila embryo. PLoS One 4:e7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, J., M. Flemr, P. Stein, P. Berninger, R. Malik, M. Zavolan, P. Svoboda, and R. M. Schultz. 2010. MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 20:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques, J. T., K. Kim, P. H. Wu, T. M. Alleyne, N. Jafari, and R. W. Carthew. 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 17:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meister, G., M. Landthaler, A. Patkaniowska, Y. Dorsett, G. Teng, and T. Tuschl. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15:185-197. [DOI] [PubMed] [Google Scholar]
- 40.Meyer, W. J., S. Schreiber, Y. Guo, T. Volkmann, M. A. Welte, and H. A. Muller. 2006. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi, K., T. Miyoshi, J. V. Hartig, H. Siomi, and M. C. Siomi. 2010. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA 16:506-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamura, K., S. Balla, R. Martin, N. Liu, and E. C. Lai. 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila. Nat. Struct. Mol. Biol. 15:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamura, K., W.-J. Chung, J. G. Ruby, H. Guo, D. P. Bartel, and E. C. Lai. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamura, K., A. Ishizuka, H. Siomi, and M. C. Siomi. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18:1655-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamura, K., and E. C. Lai. 2008. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 9:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamura, K., N. Liu, and E. C. Lai. 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell 36:431-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, J. K., X. Liu, T. J. Strauss, D. M. McKearin, and Q. Liu. 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17:533-538. [DOI] [PubMed] [Google Scholar]
- 48.Pham, J. W., and E. J. Sontheimer. 2005. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J. Biol. Chem. 280:39278-39283. [DOI] [PubMed] [Google Scholar]
- 49.Ruby, J. G., A. Stark, W. K. Johnston, M. Kellis, D. P. Bartel, and E. C. Lai. 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17:1850-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito, K., Y. Sakaguchi, T. Suzuki, T. Suzuki, H. Siomi, and M. C. Siomi. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 21:1603-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199-208. [DOI] [PubMed] [Google Scholar]
- 52.Suh, N., L. Baehner, F. Moltzahn, C. Melton, A. Shenoy, J. Chen, and R. Blelloch. 2010. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 20:271-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam, O. H., A. A. Aravin, P. Stein, A. Girard, E. P. Murchison, S. Cheloufi, E. Hodges, M. Anger, R. Sachidanandam, R. M. Schultz, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453:534-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomari, Y., T. Du, B. Haley, D. S. Schwarz, R. Bennett, H. A. Cook, B. S. Koppetsch, W. E. Theurkauf, and P. D. Zamore. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116:831-841. [DOI] [PubMed] [Google Scholar]
- 55.Tomari, Y., T. Du, and P. D. Zamore. 2007. Sorting of Drosophila small silencing RNAs. Cell 130:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomari, Y., C. Matranga, B. Haley, N. Martinez, and P. D. Zamore. 2004. A protein sensor for siRNA asymmetry. Science 306:1377-1380. [DOI] [PubMed] [Google Scholar]
- 57.Tomari, Y., and P. D. Zamore. 2005. MicroRNA biogenesis: drosha can't cut it without a partner. Curr. Biol. 15:R61-R64. [DOI] [PubMed] [Google Scholar]
- 58.Tomoyasu, Y., S. C. Miller, S. Tomita, M. Schoppmeier, D. Grossmann, and G. Bucher. 2008. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vagin, V. V., A. Sigova, C. Li, H. Seitz, V. Gvozdev, and P. D. Zamore. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313:320-324. [DOI] [PubMed] [Google Scholar]
- 60.Voinnet, O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136:669-687. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe, T., Y. Totoki, A. Toyoda, M. Kaneda, S. Kuramochi-Miyagawa, Y. Obata, H. Chiba, Y. Kohara, T. Kono, T. Nakano, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453:539-543. [DOI] [PubMed] [Google Scholar]
- 62.Wu, Q., Y. Luo, R. Lu, N. Lau, E. C. Lai, W. X. Li, and S. W. Ding. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. U. S. A. 107:1606-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yigit, E., P. J. Batista, Y. Bei, K. M. Pang, C. C. Chen, N. H. Tolia, L. Joshua-Tor, S. Mitani, M. J. Simard, and C. C. Mello. 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127:747-757. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, X. F., A. Fjose, N. Larsen, J. V. Helvik, and O. Drivenes. 2008. Treatment with small interfering RNA affects the microRNA pathway and causes unspecific defects in zebrafish embryos. FEBS J. 275:2177-2184. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, R., B. Czech, J. Brennecke, R. Sachidanandam, J. A. Wohlschlegel, N. Perrimon, and G. J. Hannon. 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15:1886-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.