Abstract
The mobilization of nucleosomes by the ATP-dependent remodeler INO80 is quite different from another remodeler (SWI/SNF) that is also involved in gene activation. Unlike that recently shown for SWI/SNF, INO80 is unable to disassemble nucleosomes when remodeling short nucleosomal arrays. Instead, INO80 more closely resembles, although with notable exceptions, the nucleosome spacing activity of ISW2 and ISW1a, which are generally involved in transcription repression. INO80 required a minimum of 33 to 43 bp of extranucleosomal DNA for mobilizing nucleosomes, with 70 bp being optimal. INO80 prefers to move mononucleosomes to the center of DNA, like ISW2 and ISW1a, but does so with higher precision. Unlike ISW2/1a, INO80 does not require the H4 tail for nucleosome mobilization; instead, the H2A histone tail negatively regulates nucleosome movement by INO80. INO80 moved arrays of two or three nucleosomes with 50 or 79 bp of linker DNA closer together, with a final length of ∼30 bp of linker DNA or a repeat length of ∼177 bp. A minimum length of >30 bp of linker DNA was required for nucleosome movement and spacing by INO80 in arrays.
Nucleosomes, the fundamental unit of chromatin, are formed by wrapping 147 bp of DNA around the histone octamer, and this further condenses into higher-order structures (29). The compaction of eukaryotic DNA into chromatin obstructs much of the DNA surface, making it poorly accessible to transcription, replication, and DNA repair. Cells employ two major classes of multiprotein enzymes to counteract the obstacles imposed by chromatin. One class covalently modifies the nucleosome core histones through acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (13, 45). The second class catalyzes nucleosome reorganization in an ATP-dependent manner (4, 16). ATP-dependent chromatin remodelers can mobilize histone octamers on DNA, which is sometimes referred to as nucleosome sliding, or they can be involved in exchange of histones and complete histone eviction from DNA (16). Even though these mechanisms are distinct, they are functionally interconnected inside the cell. In some cases these two functions coexist in the same complex (19).
ATP-dependent chromatin remodelers belong to the SWI2/SNF2 (switching/sucrose nonfermenting) superfamily and can be divided into several subfamilies on the basis of their ATPase domain structure and protein motifs outside the ATPase domain (14). The four most prominent subfamilies are the SNF2, ISWI, CHD, and INO80 subfamilies. INO80 was first identified as a coactivator of genes in inositol metabolism (12). DNA microarray studies have shown that INO80 regulates about 20% of Saccharomyces cerevisiae genes both positively and negatively (26, 32, 49). The INO80 subfamily has the unique and distinctive feature of the ATPase domain being split by insertion of an additional 300 amino acids between the two lobes of the ATPase (1).
Ino80 is the largest subunit of the complex and contains the conserved ATPase/helicase domain (38). The N-terminal part of Ino80 has a TELY motif that is conserved in human, Drosophila melanogaster, and yeast Ino80 proteins and is believed to be an interacting domain for actin, Arp4, Arp8, and Taf14/Anc1 (2, 38, 39). The helicase-SANT-associated (HSA) domain, which is conserved in remodelers containing actin-related proteins, is also present in Ino80 within the N-terminal region and binds to Arp4 and Arp8 (46). Arp8 is a unique subunit of INO80, while the Arp4 subunit is also part of the SWR1 and NuA4 complexes (32, 38, 44). Arp5 is another unique subunit of INO80 that interacts with the region inserted in the ATPase domain (26, 39). Rvb1 and Rvb2 are highly conserved and essential AAA+ helicases that are part of the INO80 and SWR1 complexes. Rvb1 and 2 are closely related to the bacterial RuvB helicase involved in branch migration and Holliday junction resolution (24, 36). Rvbs are essential for the chromatin remodeling activity of INO80, and they recruit Arp5 to form a functional INO80 complex (26).
INO80 is important for transcription activation of such genes as INO1 and PHO5. Both INO80 and SWI/SNF have been shown to act on the PHO5 and PHO8 promoters (3, 17, 18, 53). INO80 mostly recently was found to be involved in DNA repair (33, 49). The Nhp10 subunit of INO80 has been shown to interact with phosphorylated gamma H2AX, thus helping to recruit the complex to double-strand breaks (6, 33, 34, 49). INO80 is recruited to replication origins, and the chromatin remodeling activity of INO80 regulates the efficient progression of replication forks (52). INO80 plays a crucial role in stabilizing a stalled replisome to ensure the proper restart of DNA replication (35) and promotes the recovery of stalled replication forks (41, 47). Subunits of the INO80 complex have also been shown to be involved in regulation of telomere structure (54).
Some of the fundamental biochemical properties of INO80 are not known, such as the requirements of extranucleosomal DNA and histone tails, directional preference for nucleosome movement, and how it reorganizes short nucleosomal arrays. This study determined that INO80 spaces nucleosomes in vitro and has several properties in common with the known nucleosome spacing factors ISW2 and ISW1a in yeast.
MATERIALS AND METHODS
Purification of INO80 complex.
The INO80 complex was purified from Saccharomyces cerevisiae by FLAG immunoaffinity chromatography as described previously (40), with two copies of the FLAG tag attached to the C terminus of Ino80. The genotype of the strain was MATaINO80-FLAG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 and was received from X. Shen (38).
DNA probe synthesis.
DNA probes containing the 601 positioning sequence and various lengths of flanking DNA were synthesized by PCR using pGEM-3Z/601 or p199-1 plasmids as templates (31). One of the two primers used for probe synthesis was radiolabeled at its 5′ end with Optikinase (US Biological) and [γ-32P]ATP (6000 Ci/mmol) prior to PCR. PCR probes were purified with the QIAquick PCR purification kit (Qiagen).
Nucleosome reconstitutions.
Mononucleosomes were assembled at 37°C by salt dilution with 7 to 10 μg of recombinant Xenopus laevis octamers (wild-type, Cys-mut, or histone tail-negative octamers), 100 to 200 fmol of 32P-labeled 601 DNA probes, 5 to 10 μg of either sheared salmon sperm DNA or PCR-amplified 601 DNA, and 1.8 M NaCl in a starting volume of 10 μl. The reaction mixture was stepwise diluted to 1.2 M, 790 mM, and 300 mM NaCl by addition of buffer containing 25 mM Tris-HCl [pH 8.0], 1 mM 2-mercaptoethanol at 15-min intervals (30). Nucleosome assemblies were analyzed on a 4% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 36:1) in 0.5× TBE (45 mM boric acid, 45 mM Tris, 1 mM EDTA) at 4°C.
Nucleosome binding and sliding assays.
Reaction conditions for INO80 binding assays were 10 mM Na-HEPES (pH 7.8), 4 mM MgCl2, 60 mM NaCl, 0.2 mM EGTA, 0.04 mM EDTA, 8% glycerol, and 0.1 μg/μl of bovine serum albumin in a 15-μl volume. Reaction mixtures were incubated for 30 min at 30°C. Four microliters of the reaction mixture was analyzed by electrophoretic mobility gel shift assay on a 4% native polyacrylamide gel (4% acrylamide; 36:1 acrylamide:bisacrylamide ratio; 20 mM Tris-HCl [pH 8], 2 mM Na-EDTA, 5% glycerol) in 1× TE buffer (20 mM Tris-HCl [pH 8], 2 mM EDTA) at room temperature.
Nucleosome mobilization assays were conducted in the same manner, except that ATP was added to a final concentration of 800 μM. Reactions were stopped and INO80 competed from nucleosomes by the addition of γ-S-ATP and sonicated salmon sperm DNA to a final concentration of 1.5 mM and 300 ng/μl. Four microliters was loaded on a 5% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 60:1) in 0.2× TBE with buffer recirculation at 4°C.
INO80 was prebound with nucleosomes at 30°C for 30 min in time course experiments before addition of ATP (800 μM) and incubation for the desired times. Reactions were stopped as described previously.
High-resolution site-directed mapping.
Serine 53 in histone H2B was replaced with cysteine by site-directed mutagenesis and introduced into the recombinant octamer by refolding with wild-type H2A, H3, and H4 as described previously (28). These octamers were assembled into nucleosomes with PCR-amplified, end-labeled DNA, which was purified using QIAquick (Qiagen), followed by additional purification with DEAE-Sephadex A-25. The unique cysteine residue of H2B was coupled with p-azidophenacyl bromide (APB) by incubating the nucleosomes for 3 h with APB. Nucleosome mobilization assays were performed using APB-modified nucleosomes, INO80, and ATP and were analyzed by 5% native PAGE as described above. Samples were cross-linked using a UV transilluminator at 312 nM for 3 min, denatured by adding SDS to a final concentration of 0.1%, and heated at 70°C for 20 min. Histone-DNA conjugates were extracted with phenol-chloroform (4:1). DNA at the site of cross-linking was cleaved under alkaline conditions. DNA was ethanol precipitated and analyzed on a 6.5% PAGE gel containing 8 M urea, along with sequencing ladders of the same starting point and DNA strand.
ATPase assays.
All ATPase assays were carried out with reconstitutions that contained only PCR-generated DNA and no carrier DNA. Each binding reaction mixture contained 33 nM nucleosomes and 6.7 nM INO80. After binding for 30 min at 30°C, [γ-32P]ATP (1 μl [γ-32P] ATP; 6,000 Ci/mmol, 10 mCi/ml [Perkin-Elmer] plus 18 μl 1 mM ATP) was added to a final concentration of 80 μM. Reactions were stopped by adding SDS and EDTA to final concentrations of 2% and 100 mM, respectively, at 5, 10, 20, 40, 80, 160, 320, and 640 s. Reaction mixtures were spotted on a polyethyleneimine cellulose thin-layer chromatography (TLC) plate (JT Baker) and developed with 0.5 M LiCl and 0.5 M formic acid. TLC plates were visualized by phosphorimaging.
Restriction enzyme accessibility assay.
Nucleosome mobilization assays were carried out and stopped with γ-S-ATP and sonicated salmon sperm DNA as described above. Restriction endonuclease assays were performed with NotI and PmlI added to a final concentration of 7 U/μl and incubation for 30 min at 37°C and stopped by adding to a final concentration 15 mM Tris-HCl (pH 7.5), 50 mM EDTA, 1.5% SDS, 10% glycerol. Proteinase K (Roche) was added to 3.3 mg/ml and incubated for 1 h at 37°C, and then a 4-μl was loaded on a 6% native PAGE gel. The gel was dried and analyzed by phosphorimaging.
RESULTS
INO80 preferentially binds to nucleosomes with extranucleosomal DNA.
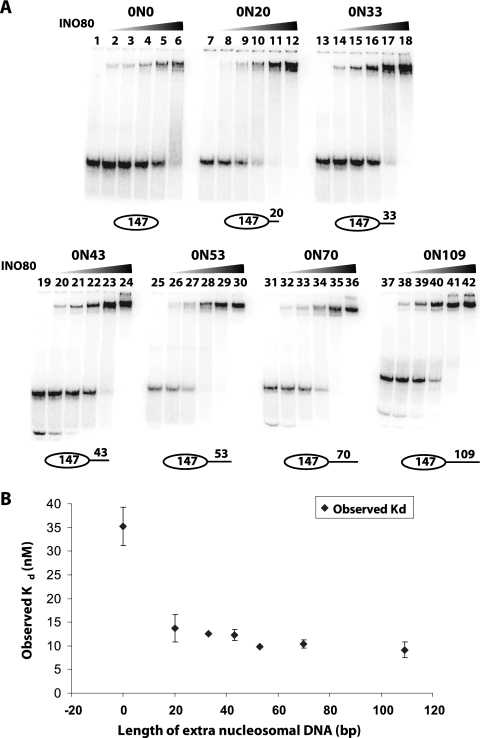
Nucleosomes were assembled with 601 nucleosome positioning DNA (31), and the relative dissociation constant (Kd) of INO80 for nucleosomes was measured with different lengths of extranucleosomal DNA (Fig. 1A and B; see also Table S1 in the supplemental material). The affinity of INO80 for nucleosomes was about 3-fold lower with no linker DNA (0N0), compared to nucleosomes with 20 bp of extranucleosomal DNA (0N20). INO80 affinity for nucleosomes was not significantly increased with longer lengths of extranucleosomal DNA. Previously, the affinity of ISW2 for nucleosomes was found to increase 5-fold with the addition of 20 bp of extranucleosomal DNA compared to the nucleosome core particle (27), and similar increases in affinity of ISW1a for nucleosomes with the addition of ∼30 bp of DNA were observed (20). These results showed that INO80 behaves in a similar manner to ISW2 and ISW1a in terms of its dependence on extranucleosomal DNA for efficient binding to nucleosomes. However, unlike ISW1a, the affinity of INO80 did not further increase when extranucleosomal DNA was on both sides of the nucleosome core particle (see Table S1 and Fig. S2 in the supplemental material).
FIG. 1.
INO80 preferentially binds to nucleosomes with extranucleosomal DNA. (A) The affinities of INO80 for end-positioned nucleosomes with various lengths of extranucleosomal DNA were measured by gel shift analysis on a 4% native PAGE with 1× TE. The binding reaction mixtures had 30 nM nucleosomes with end-labeled PCR DNA and increasing amounts of INO80 (2.5, 5, 10, 20, and 40 nM). These reaction mixtures did not contain any salmon sperm DNA. (B) The observed Kd values of INO80 for nucleosomes with different extranucleosomal DNA lengths was determined from the data shown in panel A and are plotted with respect to the length of extranucleosomal DNA. These data are from three independent binding titration experiments, and the standard deviations are shown.
INO80 remodels nucleosomes toward the center of DNA.
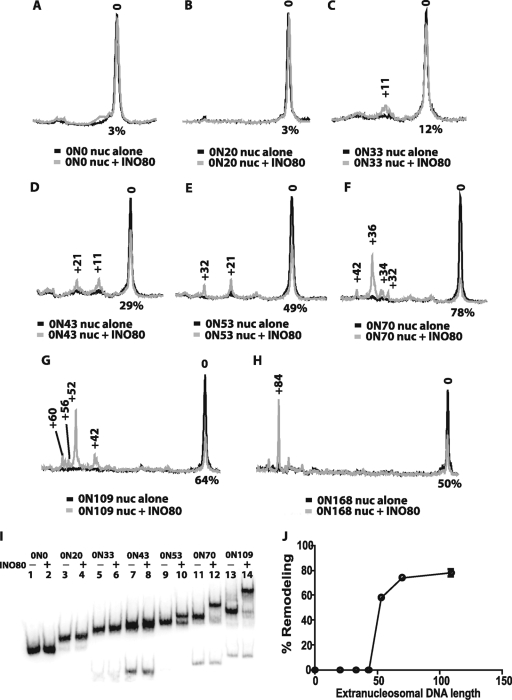
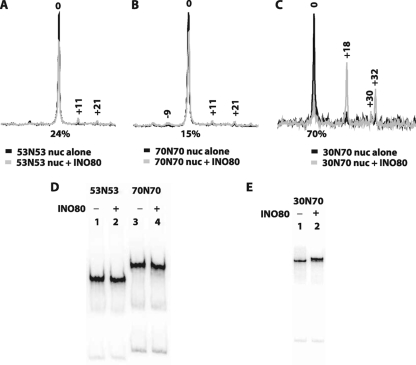
The effect of extranucleosomal DNA length on nucleosome mobilization by INO80 was examined by gel shift analysis and high-resolution site-directed mapping of nucleosomal translational positions with ∼1-bp resolution. Site-directed mapping of nucleosome positions was done with nucleosomes containing histone H2B, in which serine 53 was changed to cysteine (H2B S53C), and coupled to a photo-cross-linker or an aryl azide. Nucleosomes were photo-cross-linked before or after remodeling in order to map changes in the histone-DNA contacts upon INO80 remodeling (see Materials and Methods). Nucleosomes were mobilized with saturating amounts of INO80 and ATP. Even though INO80 can bind to nucleosomes with 20 bp of extranucleosomal DNA, INO80 was unable to mobilize these nucleosomes (Fig. 2B). Analysis of end-positioned nucleosomes with increasing lengths of extranucleosomal DNA showed that INO80 required ∼33 bp to minimally mobilize nucleosomes. INO80 remodeled nucleosomes with 33 bp of extranucleosomal DNA at only one entry site (referred to as to 0N33, with 0 and 33 indicating the length of extranucleosomal DNA at the two entry sites) by moving nucleosomes ∼11 bp from the original position, with a low efficiency of only 12% of the nucleosomes being moved (Fig. 2C). As the length of extranucleosomal DNA was increased, remodeling efficiency increased, with the optimal length being 70 bp or longer (Fig. 2C to H and I, lanes 1 to 14). The distance of nucleosome movement was also governed by the length of extranucleosomal DNA. As more extranucleosomal DNA was added, the distance nucleosomes moved increased from 11 (0N33) to 21 (0N43) and 32 bp (0N53), with progressively fewer of the shorter distances present. Although the efficiency of nucleosome movement did not change from 70 to 168 bp of extranucleosomal DNA, the major remodeled nucleosomes were moved to dramatically distinct positions and were, respectively, 36, 52, and 84 bp from the original translocation positions for 0N70, 0N109, and 0N168 nucleosomes and generally placed nucleosomes in the center of the DNA (Fig. 2F to H). It is particularly significant that the bulk of remodeled nucleosomes were at one primary translational position, with a smaller fraction offset by about 10 bp rather than a wider distribution of translational positions. These data indicate that INO80 appears to be adept at sensing the center of DNA, whether the length of the DNA is 180 or 315 bp, as in the case for 0N33 and 0N168 nucleosomes, and places nucleosomes at the center of DNA. Similarly, INO80 was unable to mobilize nucleosomes that started in a central position on the DNA, such as with the 53N53 and 70N70 nucleosomes (Fig. 3A, B, and D, lanes 1 to 4).
FIG. 2.
INO80 optimally requires 70 bp of extranucleosomal DNA to move nucleosomes to the center of DNA. (A to H) Nucleosome movement was tracked by site-directed mapping, with nucleosomes having a photoreactive group attached to amino acid residue 53 of histone H2B. Nucleosomes with various lengths of extranucleosomal DNA were used as described for Fig. 1, except that in these reaction mixtures salmon sperm DNA was added in place of unlabeled PCR DNA. The reaction mixtures contained 33 nM nucleosomes (based on octamer concentration), 40 nM INO80, and 800 μM ATP. The samples were analyzed as described in Materials and Methods, and the phosphorimages before (black) and after (gray) INO80 remodeling are overlaid to illustrate changes in nucleosome position. The number 0 refers to the original nucleosome position, and the other numbers to how many base pairs the nucleosome has shifted after INO80 remodeling. The lengths of extranucleosomal DNA in these experiments were as follows: no extranucleosomal DNA (0N0) (A), 20 bp DNA (0N20) (B), 33 bp (0N33) (C), 43 bp (0N43) (D), 53 bp (0N53) (E), 70 bp (0N70) (F), 109 bp (0N109) (G), and 168 bp (ON168) (H). The extent of nucleosomes shifted from the original position is indicated below each plot as a percentage of the initial nucleosomes that were moved. (I) The same remodeling reactions were stopped with γ-S-ATP and competitor DNA before loading on to a 5% PAGE gel (60:1, high resolution). The − and + symbols above each lane indicate whether INO80 was added, and the nucleosome nomenclature is the same as for panels A to H. (J) The extent of remodeling, based on changes in electrophoretic mobility indicated in panel I, was quantified and plotted relative to the length of extranucleosomal DNA.
FIG. 3.
INO80 does not remodel centrally positioned nucleosomes. (A to C) Site-directed mapping of nucleosomes with extranucleosomal DNA at both entry sites before and after INO80 remodeling was performed as described for Fig. 2A to H. In panels A and B, the nucleosomes start in the center of DNA with either 53 or 70 bp of extranucleosomal DNA, respectively, at both entry sites. In panel C nucleosomes are asymmetrically placed on DNA with 30 and 70 bp of extranucleosomal DNA at either entry site. (D and E) The samples shown in panels A to C were analyzed by gel shift as described for Fig. 2I.
If nucleosomes were placed at an intermediate position between the center and end of the DNA with 30 and 70 bp of extranucleosomal DNA (30N70), then INO80 preferentially slid nucleosomes 18 to 30/32 bp toward the longer extranucleosomal DNA, thereby giving rise to a more central position (Fig. 3C and E). As shown in previous studies, both ISW2 (27) and ISW1a (20) also moved nucleosomes toward the center of DNA, further confirming the similarity between INO80 and ISW2 and ISW1a.
INO80 bound to centrally positioned nucleosomes is not catalytically inactive in terms of ATPase activity.
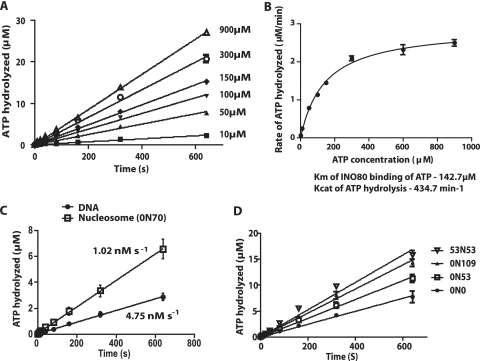
The coupling of ATP hydrolysis to nucleosome movement was examined by first determining the rates of ATP hydrolysis with different nucleosome substrates. The Km and Kcat values of INO80 were then determined by nonlinear fitting of the data to the Michaelis-Menten equation, using a range of ATP concentrations (2 to 900 μM) in the presence of excess 0N70 nucleosomes. The 0N70 nucleosomes were used because, as shown earlier, they are efficiently mobilized by INO80. The Km of INO80 was found to be 143 ± 12 μM (mean ± standard deviation) and the Kcat was 435 ± 12 s−1 (Fig. 4B), and these values are comparable to the values reported for yeast SWI/SNF (43). Nucleosomes stimulated the ATPase activity of INO80 more than free DNA, as shown when the rate of ATP hydrolysis was stimulated 2-fold more by 0N70 nucleosomes than by the same free DNA used to reconstitute these nucleosomes (Fig. 4C). The nucleosome-dependent stimulation of INO80 is thus similar to that observed with ISW2 and ISW1a (20, 55).
FIG. 4.
The ATPase activity of INO80 is enhanced with increasing lengths of extranucleosomal DNA. (A) The rate of ATP hydrolysis by INO80 with 0N70 nucleosomes was measured with different concentrations of ATP (10 to 900 μM). Nucleosomes (33 nM) were prebound with INO80 (6.67 nM) for 15 min at 30°C before addition of ATP. The amount of ATP hydrolyzed at different time points was determined using thin-layer chromatography, and ATP hydrolyzed (in μM) versus time (in seconds) was plotted for the different ATP concentrations. (B) The Km and Kcat values for INO80 were determined by plotting the rate of ATP hydrolyzed (in μM min−1) versus the concentration of ATP, with nonlinear fitting to the Michaelis-Menten equation using GraphPad. (C) The rates of ATP hydrolysis of INO80 with DNA or nucleosomes were determined as for panel A with 80 μM ATP. The free DNA used was the same DNA used to reconstitute the 0N70 nucleosomes. (D) The effects of extranucleosomal DNA length on the rate of ATP hydrolysis by INO80 were examined using nucleosome core particle (0N0), or 53 or 109 bp of extranucleosomal DNA at only one entry site (0N53 and 0N109), or with 53 bp of extranucleosomal DNA at both entry sites (53N53). The assays were performed as described for panel A, except with a fixed concentration of 80 μM ATP. In all of these reactions, only PCR-generated DNA was used.
The effect of extranucleosomal DNA length and nucleosome placement (end versus centered position) on the ATPase activity of INO80 was determined using nucleosomes with extranucleosomal DNA of 0 to 109 bp. The rate of ATP hydrolysis progressively increased as extranucleosomal DNA was changed from 0 (12.4 nM s−1) to 53 (18.1 nM s−1) to 109 bp (23.2 nM s−1) with end-positioned nucleosomes, which paralleled the changes in efficiency of nucleosome movement (Fig. 4D). The rate of ATP hydrolysis almost doubled from core nucleosome with no extranucleosomal DNA to nucleosomes with 109 bp of extranucleosomal DNA. Nucleosome movement by INO80 is slow enough that the time interval used to determine the rate of ATP hydrolysis in these experiments reflects the rate of ATP hydrolysis during nucleosome mobilization and not that of its terminal, steady-state position on DNA. This is shown by 0N70 nucleosomes being completely remodeled in 10 min with 800 μM ATP and only 50% of 0N70 nucleosomes being moved in 40 min with 80 μM ATP, the latter being the same conditions as for the ATPase assay (data not shown).
Somewhat surprisingly, the centrally positioned 53N53 nucleosomes, although unable to be moved by INO80, nonetheless stimulated the ATPase activity of INO80 as much as 0N109 nucleosomes (Fig. 4D and 3A and D, lanes 1 and 2). The inability of INO80 to mobilize 53N53 nucleosomes is therefore not likely due to an inability to hydrolyze ATP. In contrast, the ATPase activity and ability to mobilize nucleosomes were both negatively impacted with ISW1a and centrally positioned nucleosomes (20). Similarly, 0N0 nucleosomes stimulate the ATPase activity of INO80 more than the free DNA used to reconstitute 0N70 nucleosomes, even though these nucleosomes cannot be mobilized by INO80. INO80 appears to stall in its ability to mobilize nucleosomes after reaching the center of DNA, even though its ATPase activity is not diminished at this stage of the process, and it is distinct in this regard from ISW1a.
INO80 spaces nucleosomes ∼30 bp apart on di- and trinucleosomal substrates.
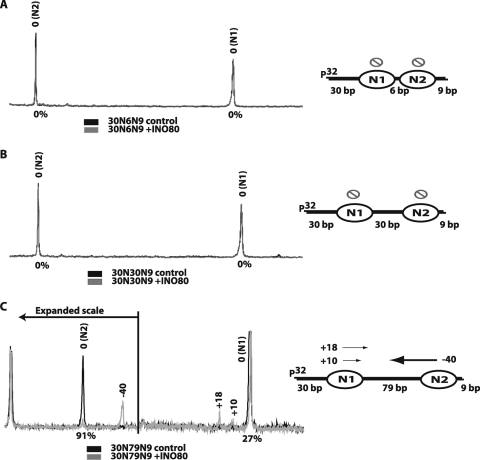
The rationale for ATP-dependent remodelers sensing extranucleosomal DNA length is better understood in the context of nucleosomal arrays in which, rather than encountering the end of DNA, INO80 will encounter an adjacent nucleosome. For this reason nucleosome mobilization by INO80 was studied with well-positioned di- and trinucleosome substrates. In these experiments, three dinucleosome constructs were used with 6, 30, and 79 bp of linker DNA between the two nucleosomes, and the flanking extranucleosomal DNA was 30 and 9 bp on either side of the dinucleosome (N1 and N2) (Fig. 5). INO80 was unable to remodel dinucleosomes with 6 and 30 bp of linker DNA. Sliding of these dinucleosomal substrates by INO80 was not observed by either gel shift analysis (data not shown) or high-resolution site-directed mapping (Fig. 5A and B). INO80, however, moved practically all of the second nucleosome (N2) 40 bp toward the first nucleosome (N1) when there was 79 bp of linker DNA (Fig. 5C). A lesser amount of N1 nucleosomes was moved 10 and18 bp toward the N2 nucleosome (Fig. 5C). INO80 preferentially spaced nucleosomes 21 to 39 bp apart and required more than 30 bp of linker DNA to mobilize arrays of nucleosomes. INO80 remodeling on short arrays suggests that INO80 can space nucleosomes in a manner similar to the ISWI family of remodelers.
FIG. 5.
INO80 cannot mobilize dinucleosomes with only 6 or 30 bp of linker DNA, but it can with 79 bp. (A to C) Dinucleosomal substrates containing 6 (A), 30 (B), or 79 bp (C) of linker DNA between the two nucleosomes were remodeled with INO80. All three nucleosomes contained 9 and 30 bp of flanking DNA on either side. Dinucleosome positions were mapped as for Fig. 2, before and after INO80 remodeling, and are displayed in the same way. On the right side is the schematic view of the nucleosomes moved by INO80. The direction of nucleosome movement and the number of bp moved are indicated by arrows and numbers, respectively. The thickness of the arrow indicates the relative amount of nucleosomes moved. In panel C, the scale on the left side has been expanded to allow better visualization of the location of the remodeled nucleosome positions.
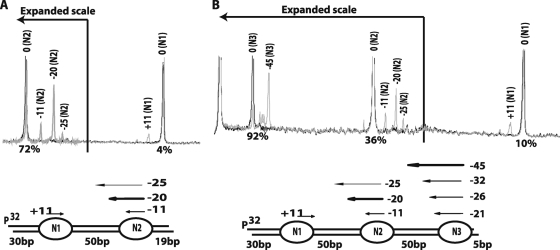
Next, a different version of dinucleosomes was used in which the linker DNA was shortened to 50 bp and the one flanking extranucleosomal DNA was increased from 9 to 19 bp (30N50N19). INO80 mobilized 30N50N19 dinucleosomes by moving nucleosomes away from the DNA ends. As before, the nucleosome (N1) that had 30 bp of flanking DNA tended to remain at its starting position, with only 4% being moved (Fig. 6A; compare to Fig. 5C). The other nucleosome (N2) with 19 bp of flanking DNA was readily moved away from the DNA end. The N2 nucleosome was moved primarily 20 bp toward N1 and changed the spacing between the nucleosomes from 50 to 30 bp when N1 did not move (Fig. 6A). Other minor changes were observed with N2 nucleosomes moving 11 or 25 bp toward N1 nucleosomes, as well as with N1 nucleosomes moving 11 bp toward N2 nucleosomes. The two nucleosomes could be separated by either 39 or 19 bp in the extreme situation of either only one nucleosome moving 11 bp or both nucleosomes moving 11 and 25 bp toward each other. In general, these observations suggest that INO80 tends to space dinucleosomes ∼30 bp apart and provide the clue that INO80 may be involved in nucleosome spacing like ISWI family members, such as ISW2 and ISW1a (48, 51). One general rule for nucleosome spacing by INO80 derived from these two sets of experiments is that 50 and 79 bp of linker DNA is sufficient to promote nucleosome movement, while with only 30 bp or less of linker DNA the two nucleosomes will tend to be immobile. Another rule appears to be that the length of flanking DNA inherently dictates which of the two nucleosomes are moved and that when the flanking DNA next to the nucleosome is 30 bp it will tend to remain in its original position but will be moved when the flanking DNA is shorter (i.e., 9 or 19 bp).
FIG. 6.
INO80 spaces di- and trinucleosomes ∼30 bp apart. Site-directed mapping of INO80 was carried out with di- (A) and trinucleosomal (B) substrates containing 50 bp of linker DNA between the nucleosomes, and the findings are displayed as described for Fig. 5. The dinucleosomes had 19 and 30 bp of flanking DNA, while the trinucleosomes had 5 and 30 bp of flanking DNA. The scale on the left side has been expanded for ease in mapping changes in nucleosome position.
A trinucleosome substrate was used to further assess the spacing properties of INO80 which had at one end a nucleosome with 30 bp flanking DNA (Fig. 6B, N1) to make it relatively immobile. As expected, 90% of the N1 nucleosome remained at the original position after remodeling with INO80 and the other 10% moved only 11 bp from the original position (Fig. 6B). In contrast, 92% of N3 nucleosomes were moved primarily 45 bp toward the central nucleosome (N2), with a smaller fraction moving 21 to 32 bp in the same direction. The central nucleosome had moved to a lesser extent, with 36% being primarily moved 20 bp toward the N1 nucleosome. In essence, both the N2 and N3 nucleosomes shifted toward the N1 nucleosome and created an array of three nucleosomes spaced 25 to 30 bp apart. There were also other minor nucleosome movements of the N2 and N3 nucleosomes that could create some heterogeneity in nucleosome spacing, but they appear to be minor compared to the more evenly spaced array. Collectively, these data with di- and trinucleosomes point to INO80 having an intrinsic ability for spacing nucleosomes ∼30 bp apart.
Histone tails are dispensable for nucleosome binding, ATPase stimulation, and remodeling by INO80.
ISWI family members require the histone H4 tail for nucleosome remodeling in vitro (7, 8, 9, 23) and in vivo (15). The histone H4 tail is also required for stimulation of the ATPase activity of the ISWI family (7, 8, 21, 25, 48, 50). Previous studies have shown that hSWI/SNF remodels less efficiently with N-terminal tails removed by trypsin digestion than with tailed mononucleosomes (22). The requirements of histone tails for nucleosome binding, ATP hydrolysis, and mobilizing nucleosomes by INO80 were determined by systematically removing N-terminal histone tails. Recombinant histones missing their N-terminal tails were assembled into octamers to assess either the individual contribution of each histone tail or the combined effect of two or more tails. The importance of histone tails in INO80 binding was examined using gel shift assays to measure the affinity of INO80 for nucleosomes missing selected histone tails. Nucleosome binding of INO80 was not impaired in the absence of any one histone tail or in various combinations (Fig. 7A), and this suggests that histone tails are not essential for INO80 binding to nucleosomes.
FIG. 7.
INO80 does not have an H4 N-terminal histone tail requirement for binding or ATP hydrolysis. (A) The affinity of INO80 for nucleosomes with and without particular histone tails were measured by gel shift analysis. Increasing amounts of INO80 (5, 10, 20, and 40 nM) were bound with 33 nM nucleosomes for 30 min at 30°C and analyzed on a 4% native PAGE gel in 1× TE. (B) The rate of ATP hydrolysis was measured for the same nucleosome substrates as described in panel A, with INO80 (6.7 nM) prebound to nucleosomes (33 nM) for 15 min at 30°C. ATP was added to a final concentration of 80 μM and incubated for the indicated times. Reactions were stopped with SDS and EDTA as described in Materials and Methods and analyzed by thin-layer chromatography.
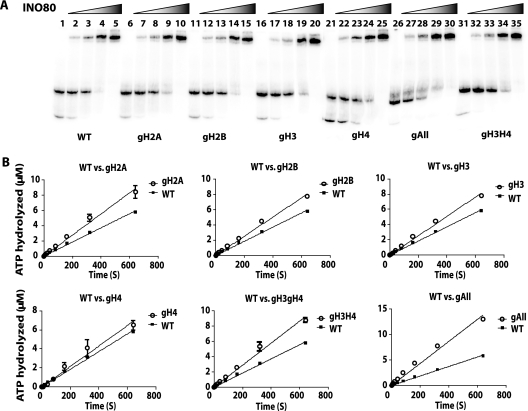
The role of histone tails in stimulating the ATPase activity of INO80 was explored by measuring the rate of ATP hydrolysis with different histone tails missing. The rate of ATP hydrolysis was modestly increased with the deletion of H2A, H2B, and H3 histone tails (Fig. 7B). The effect of all histone tails being removed stimulated the ATPase activity of INO80 ∼2-fold over nucleosomes with all tails present, and this was a stronger effect than with any individual tail being removed (Table 1). Surprisingly, several histone tails have a modestly negative impact on the ATPase activity of INO80 which, when combined, can be additive.
TABLE 1.
Rates of ATP hydrolysis and nucleosome movement of INO80 with nucleosomes having all histone tails or missing particular histone tails
| Activity measured | Rate (fold change)a for histone tail |
||||||
|---|---|---|---|---|---|---|---|
| WT | gH2A | gH2B | gH3 | gH4 | gH3H4 | gAII | |
| ATP hydrolysis (nM s−1) | 9.30 ± 0.13 | 13.87 ± 0.44 (1.49) | 12.62 ± 0.23 (1.36) | 12.72 ± 0.24 (1.37) | 10.77 ± 0.46 (1.16) | 14.47 ± 0.38 (1.55) | 21.63 ± 0.58 (2.32) |
| Remodeling (pM s−1) | 4.11 ± 0.59 | 16.93 ± 2.18 (4.04) | 3.63 ± 0.35 (0.88) | 4.88 ± 0.49 (1.16) | 6.48 ± 1.10 (1.56) | 3.09 ± 0.24 (0.72) | 12.05 ± 1.47 (2.88) |
The fold changes (globular versus WT) in activity rates of nucleosomes without versus with histone tails are indicated.
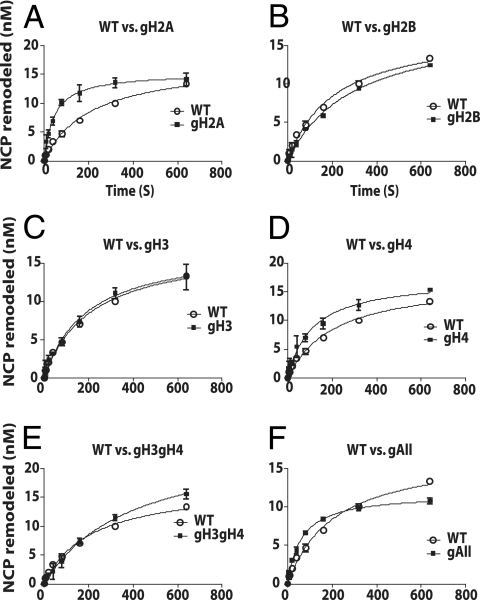
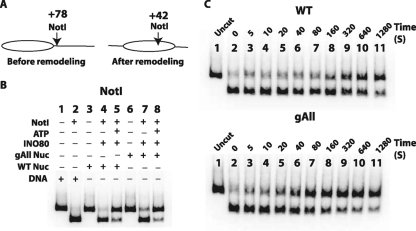
Next, the rate of INO80 remodeling was measured under complete INO80 binding conditions. Unlike ISWI, INO80 did not require intact N-terminal histone tails for its remodeling activity, as seen by gel shift analysis (Fig. 8; see also Fig. S3 in the supplemental material); however, the rate of remodeling was increased 4-fold when the histone H2A tail was deleted (Fig. 8 and Table 1). It may be that the H2A tail interacts with or blocks a region required for INO80 remodeling and therefore has a negative regulatory role. Deletion of all histone tails changed the electrophoretic mobility of the nucleosome to such an extent that made it difficult to determine if INO80 is able to mobilize these nucleosomes. Instead, a restriction enzyme accessibility assay using the NotI site that is exposed prior to remodeling (Fig. 9A and B, lanes 4 and 7) and becomes inaccessible after INO80 remodeling (Fig. 9A and B, lanes 5 and 8) was used to measure nucleosome movement. Time course experiments with restriction endonuclease accessibility assays found that INO80 remodels nucleosomes without any histone tails almost 3-fold faster than nucleosomes with histones tails present (Fig. 9C and Table 1). INO80 differs from ISWI family members in that it does not require the H4 histone tail.
FIG. 8.
The N-terminal histone H2A tail has a negative effect on INO80 movement of nucleosomes. The rates of nucleosome movement by INO80 with nucleosomes missing one or more histone tails were measured by gel shift analysis as described in Materials and Methods. INO80 (40 nM) was prebound to nucleosomes (33 nM) for 15 min at 30°C, followed by addition of 800 μM ATP. After the desired incubation time at 30°C, reactions were stopped by the addition of γ-S-ATP and sonicated salmon sperm DNA to final concentrations of 1.5 mM and 0.5 mg/ml, respectively. Samples were analyzed on a 5% native PAGE gel, and the percentage of nucleosomes with an altered mobility relative to the original was determined. Nucleosomes were missing the N-terminal tails of H2A (A), H2B (B), H3 (C), H4 (D), H3 and H4 (E), or all histones (F). In the case of the experiment shown in panel F, the extent of nucleosome movement could only be determined using the restriction enzyme accessibility assay.
FIG. 9.
INO80 does not require any of the histone tails for efficiently mobilizing nucleosomes. (A) The location of the NotI restriction endonuclease site is shown in terms of the number of bp from the dyad axis of the 601 nucleosome before and after INO80 remodeling. End-positioned nucleosomes with 70 bp of extranucleosomal DNA (0N70) were used for the experiment. The locations of mobilized nucleosomes were previously determined by site-directed mapping. (B) Nucleosomes (33 nM) with (WT) or without (gAll) all the histone tails were remodeled with INO80 (40 nM) and ATP (800 μM). Samples were incubated with NotI and deproteinized, and DNA was analyzed on a 6% native PAGE gel. (C) The nucleosomes were mobilized by INO80 for different lengths of time ranging from 5 s to 21 min and analyzed as described for panel B.
The increase in the rate of nucleosome movement observed with loss of the H2A histone tail does not correspond to an enhanced ability to hydrolyze ATP. Deletion of the H2A tail only increased the rate of ATP hydrolysis 1.5 times above that for nucleosomes with all histone tails and was 2.7 times less of a change than observed for the rate of remodeling (4.04 versus 1.49). Somehow, the H2A negatively regulates INO80 mobilization of nucleosomes without perturbing the ability of INO80 to hydrolyze ATP in a nucleosome-stimulated manner. In comparison, the increase in INO80 remodeling observed with nucleosomes missing all histone tails (2.88 times) was comparable to that observed in terms of rates of ATP hydrolysis (2.32 times). The increase in remodeling observed with all histones tails removed is therefore likely due to an increased ability to hydrolyze ATP.
The presence of the H2A-H2B dimer is required for INO80 remodeling.
Yeast SWI/SNF has been shown to mobilize an array of H3-H4 tetrasomes, although they are poorer remodeling substrates than nucleosomal arrays (5). The presence of H2A-H2B dimer within the nucleosome was found to be required for INO80 remodeling, but not for binding using tetrasomes assembled on the 601 positioning sequence with 70 bp of extranucleosomal DNA. INO80 bound to tetrasomes readily, as seen by gel shift analysis, at a level comparable to nucleosome binding (see Fig. S4A in the supplemental material). Tetrasome mobilization by INO80 was assayed by gel shift assay and restriction site accessibility. The gel shift assay showed no significant change in tetrasome electrophoretic mobility after remodeling with INO80 (see Fig. S4B). The restriction enzyme accessibility assays were carried out with NotI and PmlI, and there were no significant changes in the cleavage pattern upon INO80 remodeling (see Fig. S4C). These data indicate that INO80 cannot remodel tetrasomes and that the H2A-H2B dimer is likely recognized by INO80 and is critical for its activity. In comparison, SWI/SNF was able to mobilize tetrasomes under similar conditions to those used for INO80 (S. Kassabov, unpublished data).
DISCUSSION
INO80 has been shown here for the first time to have nucleosome remodeling properties that suggest it may be a nucleosome spacing factor. The first of several properties consistent with INO80 spacing nucleosomes is the ability to move mononucleosomes toward the center of DNA no matter the length of DNA. This same characteristic is a hallmark of known nucleosome spacing factors ISW2 and ISW1a in yeast (20, 27, 51). There are, however, some key differences between INO80 and ISW2 in this regard. Although ISW2 centers nucleosomes reasonably well on DNA with nucleosomes having 70 bp of extranucleosomal DNA, when the length of extranucleosomal DNA is 142 bp ISW2 tends to randomize nucleosome positions (S. Hota, unpublished data). INO80, in contrast, is able to move nucleosomes 84 ± ∼10 bp with nucleosomes that have 168 bp of extranucleosomal DNA and thus to more precisely move nucleosomes to the center of DNA than ISW2, especially with longer extranucleosomal DNA. Even site-directed mapping of nucleosomes with 67 to 70 bp of extranucleosomal DNA has shown that nucleosomes have fewer translational positions after remodeling with INO80 than with ISW2 (56). Thus, although it has similar behavior in positioning mononucleosomes to the center of DNA, INO80 more tightly regulates its positioning of nucleosomes than does ISW2. The tight positioning of nucleosomes to the center of DNA implies that the mechanism used is unlikely to be due to a dimer of INO80 pulling in opposing directions, like that reported for ACF1 (37). The size of INO80 would also make it difficult for two INO80 complexes to be simultaneously bound to one nucleosome, and the Hill coefficient from the nucleosome binding data is ∼1, suggesting that only a monomer is bound.
Another characteristic that INO80 shares with ISW2 and ISW1a that is closely related to their directional preference in moving nucleosomes is the dependence of INO80 on the length of extranucleosomal DNA. Like ISW2, INO80 is unable to mobilize nucleosomes if the extranucleosomal DNA length is too short, even though the complex can still bind to nucleosomes (56). As shown in Fig. 2I and J, the efficiency of nucleosome movement increased markedly from 43 to 53 bp of extranucleosomal DNA, and then from 70 bp and longer there were no further enhancements. A key difference of INO80 versus ISW2 and ISW1a is that the length of extranucleosomal DNA required by INO80 for efficient nucleosome movement is 13 to 40 bp longer than that required by ISW2 and ISW1a. The minimal lengths of extranucleosomal DNA required for optimal nucleosome movement by ISW2 and ISW1a are 26 to 30 bp and 20 to 23 bp for minimal movement (56). The 33 bp of extranucleosomal DNA required for optimal nucleosome movement by ISW1a was also the same distance that ISW1a spaced nucleosomes. The same correlation, however, does not exist for INO80, as it spaces nucleosomes ∼30 bp apart but requires 70 bp of extranucleosomal DNA for efficient nucleosome movement.
Other differences that demarcate INO80 nucleosome spacing activity as distinct from ISW2 and ISW1a are its histone tail requirement and the activity with nucleosomes placed in the middle of DNA. ISW2 and ISW1a both require the histone H4 tail for efficient nucleosome mobilization, but INO80 does not. It may be significant that ISW1a and ISW2 are involved with the H4 tail, since the H4 tail is required for proper formation of higher-order chromatin structure (42). It is likely important that the main histone tail that affects INO80 mobilization of nucleosomes is the H2A tail and that it does so in a negative rather than positive way (Fig. 8 and Table 1). This raises the question whether posttranslational modifications such as acetylation of the histone H2A tail might counteract the generally negative effects of the H2A tail on INO80 remodeling. In this light it is interesting that acetylation by NuA4 of histones H4 and H2A has been found to be important for the recruitment of INO80 and SWR1 to DNA double-strand breaks (11). The other difference is that ISW1a is unable to move nucleosomes or to stimulate its ATPase when nucleosomes are positioned in the center of DNA, and thus the inability to mobilize nucleosomes is likely due to an inability to hydrolyze ATP. The ATPase activity of INO80 is, however, equally stimulated by nucleosomes in the center or on the end of DNA, so that the inability to be mobilized is not due to a decrease in ATPase activity. So, although INO80 spaces nucleosomes, it is unique in several ways from the ISW2 and ISW1a nucleosome spacing factors.
The list of differences between ISW2/1a and INO80 most likely reflects fundamental differences in the mechanisms used to space nucleosomes. For example, since the H4 tail does not stimulate either the ATPase or remodeling activities of INO80, it would seem that the H4 tail also does not promote binding of Ino80 near the SHL2 position of nucleosomal DNA, as it does for ISW2 or ISW1. The questions remain, then, as to where the ATPase domain might engage the nucleosome and how this affects the process of mobilizing nucleosomes. It will be important to determine how INO80 can so precisely center nucleosomes on DNA, whereas ISW2 and ISW1a are unable to do the same, and how this is reflected in terms of the mechanism of nucleosome spacing.
INO80 spacing of nucleosomes was directly shown by high-resolution mapping of nucleosome positions in short arrays of two or three nucleosomes. By using the 601 nucleosome positioning sequence to construct these arrays, it was possible to find that the minimal length of linker DNA required for nucleosome movement by INO80 is greater than 30 bp. INO80 changed nucleosome spacing from starting positions of 50 or 79 bp of linker DNA to a universal length of about 30 bp. In other words, regardless of their starting position, as long as there is enough linker DNA, INO80 uniformly spaces nucleosomes with a repeat length of ∼177 bp. It will be interesting to see how nucleosome spacing may be an important factor of INO80 in mediating activation of transcription or facilitating the stabilization of DNA replication forks.
It is not yet clear as to why more than one ATP-dependent chromatin remodeler is required for maximum gene activation if they essentially perform the same function in terms of chromatin structure. This dilemma is evident in the case of activation of the PHO5, PHO8, and PHO84 genes in yeast, where INO80 and SWI/SNF both activate transcription in independent manners (3, 53). Our study delineates the intrinsic differences between INO80 and SWI/SNF and points to how they may increase accessibility of chromatinized DNA in distinct ways. SWI/SNF has been shown recently to have nucleosome disassembly activity with di- and trinucleosome substrates that are similar to those used in this study (10). The disassembly activity of SWI/SNF is independent of any histone chaperone or DNA acceptor cofactor. Now, from these studies it is apparent that INO80 mobilizes nucleosomes without displacing them from DNA. Thus, the ability of INO80 to potentially open up promoter regions is likely to be a reflection of its ability to mobilize rather than disassemble nucleosomes as SWI/SNF does. However, in particular situations, INO80 could still be involved in vivo in disassembling nucleosomes and may require additional factors, such as histone chaperones, not present in our experiments.
How then can the spacing activity of INO80 and the disassembly activity of SWI/SNF work cooperatively to create or expand nucleosome-free regions? One model is that SWI/SNF tethered to a particular promoter site through its interaction with a gene-specific transcription factor such as Gal4 is able to disassemble nucleosomes within a short range of the Gal4 site. SWI/SNF alone is not able to disassemble the nucleosome beyond the reach of its protein tether and may be only able to disassemble two to three nucleosomes on its own. INO80 could potentially expand the size of this nucleosome-free region by moving nucleosomes to within the reach of SWI/SNF, so that they in turn can be disassembled from DNA. INO80 and SWI/SNF could work together in an assembly line fashion with INO80, providing nucleosomes to SWI/SNF for their subsequent release while maintaining the position of SWI/SNF in the genome. It will be important to test these and other models, given that we now better understand some of the fundamental nucleosome remodeling differences between INO80 and SWI/SNF.
Supplementary Material
Footnotes
Published ahead of print on 6 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bakshi, R., et al. 2004. In silico characterization of the INO80 subfamily of SWI2/SNF2 chromatin remodeling proteins. Biochem. Biophys. Res. Commun. 320:197-204. [DOI] [PubMed] [Google Scholar]
- 2.Bao, Y., and X. Shen.2007. INO80 subfamily of chromatin remodeling complexes. Mutat. Res. 618:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaric, S., et al. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282:27610-27621. [DOI] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, L. A., et al. 2000. Roles of the histone H2A-H2B dimers and the (H3-H4)(2) tetramer in nucleosome remodeling by the SWI-SNF complex. J. Biol. Chem. 275:11545-11552. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R. 2004. Around the world of DNA damage INO80 days. Cell 119:733-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapier, C. R., et al. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clapier, C. R., K. P. Nightingale, and P. B. Becker. 2002. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang, W., M. N. Kagalwala, and B. Bartholomew. 2006. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol. Cell. Biol. 26:7388-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dechassa, M. L., et al. 2010. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38:590-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downs, J. A., et al. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 12.Ebbert, R., A. Birkmann, and H. J. Schuller. 1999. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32:741-751. [DOI] [PubMed] [Google Scholar]
- 13.Eberharter, A., R. Ferreira, and P. Becker. 2005. Dynamic chromatin: concerted nucleosome remodelling and acetylation. Biol. Chem. 386:745-751. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazzio, T. G., M. E. Gelbart, and T. Tsukiyama. 2005. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 25:9165-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165-173. [DOI] [PubMed] [Google Scholar]
- 17.Ford, J., O. Odeyale, and C. H. Shen. 2008. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem. Biophys. Res. Commun. 373:602-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford, J., et al. 2007. A SWI/SNF- and INO80-dependent nucleosome movement at the INO1 promoter. Biochem. Biophys. Res. Commun. 361:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangaraju, V. K., and B. Bartholomew. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 618:3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangaraju, V. K., and B. Bartholomew. 2007. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol. Cell. Biol. 27:3217-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgel, P. T., T. Tsukiyama, and C. Wu. 1997. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 16:4717-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyon, J. R., et al. 1999. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol. Cell. Biol. 19:2088-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamiche, A., et al. 2001. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. U. S. A. 98:14316-14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hishida, T., et al. 2004. Direct evidence that a conserved arginine in RuvB AAA+ ATPase acts as an allosteric effector for the ATPase activity of the adjacent subunit in a hexamer. Proc. Natl. Acad. Sci. U. S. A. 101:9573-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., et al. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson, Z. O., et al. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16:465-477. [DOI] [PubMed] [Google Scholar]
- 27.Kagalwala, M. N., et al. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23:2092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassabov, S. R., et al. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22:7524-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 30.Lorch, Y., J. W. LaPointe, and R. D. Kornberg. 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49:203-210. [DOI] [PubMed] [Google Scholar]
- 31.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 32.Mizuguchi, G., et al. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 33.Morrison, A. J., et al. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, A. J., and X. Shen. 2005. DNA repair in the context of chromatin. Cell Cycle 4:568-571. [PubMed] [Google Scholar]
- 35.Papamichos-Chronakis, M., and C. L. Peterson. 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 15:338-345. [DOI] [PubMed] [Google Scholar]
- 36.Parsons, C. A., et al. 1992. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl. Acad. Sci. U. S. A. 89:5452-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Racki, L. R., et al. 2009. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 462:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen, X., et al. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 39.Shen, X., et al. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12:147-155. [DOI] [PubMed] [Google Scholar]
- 40.Shen, X. 2004. Preparation and analysis of the INO80 complex. Methods Enzymol. 377:401-412. [DOI] [PubMed] [Google Scholar]
- 41.Shimada, K., et al. 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18:566-575. [DOI] [PubMed] [Google Scholar]
- 42.Shogren-Knaak, M., et al. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844-847. [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. L., and C. L. Peterson. 2005. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 25:5880-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinboeck, F., et al. 2007. The nuclear actin-related protein of Saccharomyces cerevisiae, Arp4, directly interacts with the histone acetyltransferase Esa1p. J. Biochem. 141:661-668. [DOI] [PubMed] [Google Scholar]
- 45.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 46.Szerlong, H., et al. 2008. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 15:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trujillo, K. M., and M. A. Osley. 2008. INO80 meets a fork in the road. Nat. Struct. Mol. Biol. 15:332-334. [DOI] [PubMed] [Google Scholar]
- 48.Tsukiyama, T., et al. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Attikum, H., et al. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 50.Varga-Weisz, P. D., et al. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 51.Vary, J. C., Jr., et al. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincent, J. A., T. J. Kwong, and T. Tsukiyama. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wippo, C. J., et al. 2009. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell. Biol. 29:2960-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, E. Y., et al. 2007. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol. Cell. Biol. 27:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zofall, M., et al. 2006. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13:339-346. [DOI] [PubMed] [Google Scholar]
- 56.Zofall, M., J. Persinger, and B. Bartholomew. 2004. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol. Cell. Biol. 24:10047-10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.