Abstract
Saccharomyces cerevisiae cell division ends with destruction of a septum deposited during cytokinesis; this must occur only after the structure's construction is complete. Genes involved in septum destruction are induced by the transcription factor Ace2, which is activated by the kinase Cbk1, an Ndr/LATS-related protein that functions in a system related to metazoan hippo pathways. Phosphorylation of a conserved hydrophobic motif (HM) site regulates Cbk1; at peak levels in late mitosis we found that approximately 3% of Cbk1 carries this modification. HM site phosphorylation prior to mitotic exit occurs in response to activation of the FEAR (Cdc fourteen early anaphase release) pathway. However, HM site phosphorylation is not sufficient for Cbk1 to act on Ace2: the kinase is also negatively regulated prior to cytokinesis, likely by cyclin-dependent kinase (CDK) phosphorylation. Cbk1 cannot phosphorylate Ace2 until after mitotic exit network (MEN)-initiated release of the phosphatase Cdc14. Treatment of Cbk1 with Cdc14 in vitro does not increase its intrinsic enzymatic activity, but Cdc14 is required for Cbk1 function in vivo. Thus, we propose that Cdc14 coordinates cell separation with mitotic exit via FEAR-initiated phosphorylation of the Cbk1 HM site and MEN-activated reversal of mitotic CDK phosphorylations that block both Cbk1 and Ace2 function.
Cell cycle control systems ensure that the numerous processes of cell division occur with proper order and timing (22, 23, 34). The final events of cell division in Saccharomyces cerevisiae exemplify this integration and have provided an excellent model for mitotic progression. After chromosome segregation, cytokinesis in budding yeast is accompanied by deposition of a septum that forms a rigid wall between mother and daughter cells (5, 11, 49, 64). This septum is destroyed minutes after cytokinesis is completed to allow separation of the cells. It is critical that septum destruction does not begin until after cytokinesis is completed. The mechanism that coordinates septum destruction with the cell cycle is not understood.
Passage from mitosis to G1 requires a transition between states of high and low cyclin-dependent kinase (CDK) activity. In addition to inactivation of CDK itself, the sites on its target proteins are dephosphorylated. In budding yeast, the phosphatase Cdc14 plays a key role in mitotic exit by dephosphorylating proline-directed CDK sites (21, 26, 67). Cdc14 is sequestered in the nucleolus during most of the cell cycle and is released in two distinct phases during mitosis: an initial, relatively transient release of Cdc14 is controlled by the FEAR (Cdc fourteen early anaphase release) pathway, while a later more complete and sustained release of Cdc14 is regulated by the mitotic exit network (MEN).
The FEAR pathway consists of the polo-like kinase Cdc5, the separase Esp1, the kinetochore protein Slk19, and the nucleolar protein Spo12 (62), and it functions to promote the release of Cdc14 from the nucleolus into the nucleus during anaphase (4, 37, 54, 56, 57, 63, 68, 69). Once in the nucleus, Cdc14 dephosphorylates CDK targets to promote events required for anaphase progression (17, 33, 35). However, this release of Cdc14 by FEAR is transient and is not sufficient to drive cytokinesis and mitotic exit (62). The later-acting MEN sustains a prolonged release of Cdc14 to the cytoplasm and functions to ensure that nuclear division is properly completed prior to cytokinesis and septum formation (31). The MEN, initiated by activation of the GTPase Tem1, activates a signaling cascade of kinases, including Cdc15 and Dbf2/Mob1 (3, 6, 25, 39, 53, 66). Dbf2/Mob1 phosphorylates Cdc14 to inactivate its nuclear localization signal (NLS), promoting release of Cdc14 into the cytoplasm (46). Cytoplasmic Cdc14 dephosphorylates CDK targets to promote mitotic exit and the final events of cell division (7, 61, 67).
The final event of cell division is destruction of the septum between mother and daughter cells. Correct timing of this division event is partly achieved through control of transcription: the enzymes that degrade the septum are produced in a brief pulse of gene transcription as cells pass from mitosis to G1 phase. The activation of these genes is driven by the transcription factor Ace2, which localizes strongly to the daughter nucleus immediately before completion of actomyosin ring contraction (8, 16, 18, 19, 42, 50, 55). Ace2 is regulated by sequential inhibition and activation. First, prior to the metaphase/anaphase transition, CDK phosphorylation blocks Ace2's nuclear import (2, 50). Then, after downregulation of mitotic CDK, the Ndr/LATS kinase Cbk1 traps Ace2 in the daughter nucleus by phosphorylating sites that disable its nuclear export sequence (NES) (42). Determination of how the transition is made between Ace2's inhibition and activation is critical to understanding how this gene expression pattern is linked to late cell division.
Cbk1 is a component of a signaling system referred to as the RAM (regulation of ace2 and morphogenesis) network (8, 16, 48, 70), which is conserved in a broad range of eukaryotes. Cbk1, a member of the AGC family of kinases, is most closely related to Ndr family protein kinases, which includes mammalian Ndr1 and Ndr2, Drosophila melanogaster tricornered, Caenorhabditis elegans sax-1, cot1 from Neurospora crassa, and orb6 from Schizasaccharomyces pombe (for a review, see reference 24). Metazoan pathways related to the RAM network are involved in cell architecture control and neuron morphogenesis but are not well understood. Cbk1 is also similar to LATS/warts kinases, which are key components of the widely conserved hippo/MST signaling pathways.
Like other AGC family kinases, Cbk1 is controlled by two key phosphorylations: one in the kinase activation loop (T-loop), and another within a highly conserved C-terminal hydrophobic motif (the HM site, alternatively referred to as the CT motif) (29, 45, 52, 60, 71). Cbk1's T-loop modification, by intramolecular autophosphorylation, strongly activates the kinase but is only partially required for function. In contrast, HM site phosphorylation is essential for Cbk1 function and is dynamically regulated over the cell cycle, peaking at the time of cytokinesis (29). HM site phosphorylation requires all other components of the RAM network (29); however, the mechanisms that link phosphorylation of Cbk1's HM site to mitotic progression remain unknown.
To determine how the Ace2-driven transcriptional program is coordinated with cell cycle progression, we investigated the mitotic control of Cbk1. We show that HM site phosphorylation of Cbk1 requires activation of the FEAR network but not the MEN pathway. Using a gain-of-function CBK1 allele that eliminates the need for HM site phosphorylation, we demonstrate that this modification is not the sole mechanism for M-phase control of Cbk1's activation of Ace2. Although Cbk1 is modified at the HM site in late mitosis, we show that Cbk1 is unable to act on Ace2, likely by inhibitory CDK phosphorylation. Our findings indicate that MEN-triggered Cdc14 release plays a key regulatory role in this pathway, simultaneously reversing inhibition of both Cbk1 and Ace2. Thus, the RAM network is coordinately regulated by multiple inputs to ensure the proper ordering of late cell division events.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All strains generated and used in this study are listed in Table 1 and are derived from the W303 genetic background (ELY83). We generated deletion, hemagglutinin (HA)-tagged and green fluorescent protein (GFP)-tagged strains by standard gene replacement or tagging (38). To construct the CBK1-T743E (ELY1398) strain, we used two-fragment PCR to replace cbk1Δ::TRP1 (ELY870) with the mutated CBK1 allele and the genetic marker KanMX. We sequenced the endogenous locus to confirm proper integration. To generate the CBK1-T743E-GFP (ELY1516) strain, we transformed ELY1398 by gene replacement to replace the KanMX cassette with GFP-HIS3 and sequenced the endogenous locus to confirm proper integration and retention of the mutation. To generate cdc15-2 CBK1-T743E (ELY1447), we mated ELY1372 and ELY1399 (MATα CBK1-T743E::KanMX) and then sporulated and genotyped resulting haploids by marker and temperature sensitivity. We introduced plasmids by standard lithium acetate transformation; plasmids used are listed in Table 2.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| ELY83 | MATaleu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 (W303) | D. Drubin |
| ELY1372 | MATacdc15-2 | D. Morgan |
| ELY915 | MATacdc5-1 bar1Δ::HIS | D. Morgan |
| ELY1370 | MATacdc14-1 bar1Δ::HIS | D. Morgan |
| ELY1398 | MATaCBK1-T743E::KanMX | This study |
| ELY870 | MATacbk1Δ::TRP1 | This study |
| ELY1414 | MATaCBK1-T743E::KanMX [pELW754] | This study |
| ELY875 | MATaCBK1-GFP::KanMX | This study |
| ELY1516 | MATaCBK1-T743E-GFP::HIS3 | This study |
| ELY1565 | MATacdc15-2 ACE2-HA::KanMX | This study |
| ELY1566 | MATacdc14-1 ACE2-HA::KanMX | This study |
| ELY1340 | MATa ADE2::GAL-CDC20 cdc20Δ::LEU2 ACE2-HA::KanMX | This study |
| ELY1428 | MATacdc15-2 [pELW754] | This study |
| ELY1429 | MATacdc15-2 [pELW818] | This study |
| ELY1431 | MATacdc15-2 [pELW893] | This study |
| ELY1538 | MATacdc15-2 CBK1-T743E::KanMX [pELW754] | This study |
| ELY1539 | MATacdc15-2 CBK1-T743E::KanMX [pELW818] | This study |
| ELY1613 | MATacdc15-2 CBK1-T743E::KanMX [pELW893] | This study |
| ELY1541 | MATacdc15-2 cbk1Δ::TRP1 [pELW754] | This study |
| ELY1542 | MATacdc15-2 cbk1Δ::TRP1 [pELW818] | This study |
| ELY1614 | MATacdc15-2 cbk1Δ::TRP1 [pELW893] | This study |
| ELY1395 | MATaace2Δ::HIS3 | This study |
| ELY1275 | MATacbk1Δ:: TRP1 [pELW894] [pELW895] | This study |
| ELY1203 | MATacdc15-2 [pELW827] | This study |
| ELY1272 | MATacbk1Δ::TRP1[pELW767] | This study |
| ELY1615 | MATacbk1Δ::TRP1[pELW887] | This study |
| ELY1616 | MATacbk1Δ::TRP1[pELW889] | This study |
TABLE 2.
Plasmids used in this study
| Name | Insert |
|---|---|
| pELW754 | ACE2-GFP::LEU2 |
| pELW818 | ace2-T575A/S701A/S714A-GFP::LEU2 (ace2-AAA) |
| pELW893 | ace2-T575A/S701A/S714A/S122E/S137D- GFP::LEU2 (ace2-AAA-NSM) |
| pELW827 | pGAL-GST-CDC14::LEU2 |
| pELW894 | pGAL-GST-CBK1::LEU2 |
| pELW895 | pGAL-13MYC-MOB2::HIS3 |
| pELW767 | pRS316-CBK1::URA3 |
| pELW887 | pRS316-cbk1-9A::URA3 |
| pELW889 | pRS316-cbk1-6E::URA3 |
We cultured cells in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal selection medium (0.67% yeast nitrogen base without amino acids, 2% glucose, and 0.2% amino acid drop-in; US Biological). For mitotic arrest, we added nocodazole (Sigma, St. Louis, MO) to YPD medium at a 15-μg/ml final concentration and allowed growth for 1.5 to 2 h until >90% of cells were large budded. To release from nocodazole, we filtered and washed cells into fresh prewarmed medium at the indicated temperatures. For the experiments described below in Fig. 1, we added nocodazole to 15 μg/ml to cells released from synchrony after 70 min to arrest cells in the next mitosis. We harvested cells at the indicated times for protein analysis, fixing a portion with formaldehyde for analysis of budding index. For arrest of cells carrying temperature-sensitive alleles, we grew cultures to mid-log phase and transferred them to 37°C for 2 to 2.5 h until >90% of cells were large budded; for release, we divided the culture into equal volumes and placed them at 24°C, removing samples at the indicated times for protein or RNA analysis.
FIG. 1.
Cbk1 is phosphorylated at the HM site upon release from nocodazole arrest. (A) HM site phosphorylation is detected after release from nocodazole arrest. Wild-type (CBK1) cells were arrested for 2 h in nocodazole and released into fresh medium at 30°C. After 70 min, fresh nocodazole was added to arrest cells in the following mitosis. Cells were harvested at the indicated times, and immunoprecipitated Cbk1 was probed with an anti-pT743 antibody (top panel) or anti-Cbk1 antibody (bottom panel). The percentage of Cbk1 phosphorylated at T743 is shown below each lane. (B) HM site phosphorylation occurs at the M/G1 transition just prior to bud emergence. The budding index upon release from nocodazole arrest is shown over time. (C) Ace2 target gene expression correlates with HM site phosphorylation. RNA was harvested from cells collected at the indicated times, and transcript levels were determined by QPCR. The increases in CTS1 and DSE1 transcripts relative to nocodazole arrest (time zero) are indicated. Standard errors of the means (SEM) for two experiments are shown. (D) Quantification of Cbk1 by immunoblotting. A standard curve was generated by plotting pixel intensity (Odyssey) to picomoles of Cbk1. Error bars represent errors of three independent immunoblot assays. A representative blot is shown. (E) Quantification of pT743 by immunoblotting. A standard curve was generated by plotting pixel intensity (Odyssey) to picomoles of phosphorylated Cbk1. Error bars represent results from three independent immunoblot assays. A representative blot is shown for the phosphorylated peptide and a nonphosphorylated control.
We constructed a centromeric Ace2-GFP plasmid (pELW754) by first cloning the Ace2-GFP coding sequence and sequences 500 bp upstream of the start codon and 300 bp downstream of the stop codon into pCR2.1 (Invitrogen). We then inserted the resulting fragment into the YCplac111 (CEN LEU2) vector between HindIII and XbaI sites. To generate Ace2 alleles, we introduced the following substitutions by site-directed mutagenesis using Pfu Turbo (Stratagene): ace2-AAA, T575A, S701A, and S714A (pELW818); ace2-AAA-NSM, S122E, S137D, T575A, S701A, and S714A (pELW893). We generated Cbk1-9A with T93A, T109A, S143A, S164A, S251A, S409A, T574A, T615A, and S711A (pELW887) and Cbk1-6E with S164E, S251E, S409E, T574E, T615E, and S711E (pELW889) alleles by site-directed mutagenesis using Pfu Turbo (Stratagene) on pRS316-CBK1::URA3 (pELW889). Primers used in this study are listed in Table 3. We amplified coding sequences of CDC14, CBK1, and MOB2 with Ex Taq polymerase (Takara) and cloned by the drag and drop method (28) into pGREG545 (pGAL-GST-CDC14; pELW827), pGREG545 (pGAL-GST-CBK1; pELW894), and pGREG523 (pGAL-13MYC-MOB2; pELW895) and verified all constructs by sequencing. To induce overexpression of CDC14, we grew cdc15-2 cells containing pELW827 to mid-log phase in selection medium and transferred cells to 37°C for 2.5 h to arrest. The culture was split in half and resuspended in prewarmed selection medium containing glucose or galactose for 3 h at 37°C.
TABLE 3.
Primers used in this study
| Name | Sequence |
|---|---|
| Ace2_T575A_Fwd | GAAATAATACCTAGGACGGCGCCAATGAAA |
| Ace2_T575A_Rev | CTTGGTTATTTTCATTGGCGCCGTCCTAGG |
| Ace2_S701A,714A Fwd | AAACTTACGGCGCCCAAAAAAAGCCTGCTTGACAGCCCGCATGACACAGCTCCCGTAAAA |
| Ace2_S701A,714A Rev | TTTTACGGGAGCTGTGTCATGCGGGCTGTCAAGCAGGCTTTTTTTGGGCGCCGTAAGTTT |
| Cbk1_T93A_Fwd | CTGAATTATCCAGCTGCACCACCACC |
| Cbk1_T93A_Rev | TATTATGTGGTGGTGGTGCAGCTGG |
| Cbk1_T109A_Fwd | AAATCAGATGATTAATGCCCCACCG |
| Cbk1_T109A_Rev | TCGACGGCGGTGGGGCATTAATCATCTG |
| Cbk1_S143A_Fwd | GCACAATTACCCCAATTGGCACCGGG |
| Cbk1_S143A_Rev | TATTGGCCCGGTGCCAATTGGGG |
| Cbk1_S164A_Fwd | TTTAAATGGCAGTTCCTCGAGTGCTCCG |
| Cbk1_S164A_Rev | GGTTGGTGGAACGGAGCACTCGAGGAACTGCC |
| Cbk1_S251A_Fwd | CAGCAGCAATCACAAGCGCCCGTTCAG |
| Cbk1_S251A_Rev | CGCTCTGAACGGGCGCTTGTGATTG |
| Cbk1_S409A_Fwd | TGGCTGGAAGTGATGCTCCTTGGGTG |
| Cbk1_S409A_Rev | GCGAAACCACCCAAGGAGCATCACTTC |
| Cbk1_T574A_Fwd | TTCTACCGTAGGAGCTCCAGATTATATTG |
| Cbk1_T574A_Rev | AGGAGCAATATAATCTGGAGCTCCTACGG |
| Cbk1_T615A_Fwd | TCTGTTCCGAGGCGCCACAGGAAACG |
| Cbk1_T615A_Rev | TGTACGTTTCCTGTGGCGCCTCGG |
| Cbk1_S711A_Fwd | GGAGAATGTTCCAGATGCGCCGGCTATG |
| Cbk1_S711A_Rev | AGCTTGTGCCATAGCCGGCGCATCTG |
| Cbk1_S251E_Fwd | GCAATCACAAGAGCCAGTGCAGAGCG |
| Cbk1_S251E_Rev | TTAAAGCCGCTCTGCACTGGCTCTTG |
| Cbk1_S409E_Fwd | GGCTGGAAGTGATGAGCCTTGGGTGG |
| Cbk1_S409E_Rev | CGAAACCACCCAAGGCTCATCACTTC |
| Cbk1_T574E_Fwd | TACCGTAGGTGAACCGGATTATATTG |
| Cbk1_T574E_Rev | GGAGCAATATAATCCGGTTCACCTAC |
| Cbk1_T615E_Fwd | TCTGTTCCGAAGAGCCTCAGGAAACG |
| Cbk1_T615E_Rev | CTGTACGTTTCCTGAGGCTCTTCGG |
| Cbk1_S711E_Fwd | GAATGTTCCAGATGAGCCAGCCATGG |
| Cbk1_S711E_Rev | CTTGTGCCATGGCTGGCTCATCTGG |
| Cbk1_S164E_Fwd | CTTCTAGTGAACCGTTTCACCAACCG |
| Cbk1_S164E_Fwd | GTTTGCGGTTGGTGAAACGGTTCACTAG |
| Cbk1 fragment F | TCGACGCGCCAGTCAAGGAAGATTTACCTTTTATTGGCTACACTTACGCCAGATTTGACTATTTGGCAAGAAAAAATGCGTTGGC |
| Cbk1 fragment R | GGCCGCCAACGCATTTTTTCTTGCCAAATAGTCAAATCTGGCGTAAGTGTAGCCAATTAAAGGTAAATCTTCCTTGACTGGCGCG |
Immunoprecipitation, kinase, and phosphatase assays.
For total cell lysate, we collected equivalent numbers of cells (measured by the optical density at 600 nm) for each strain and prepared proteins as previously described (36). Equal volumes were loaded and separated on 8% SDS-PAGE gels. For immunoprecipitations, we prepared cell lysate by bead beating as previously described (29), measured protein concentrations for normalization by the Bradford assay (Bio-Rad), and added anti-Cbk1 antibody at a 1:50 dilution along with 80 μl of a 1:1 recombinant protein G-Sepharose (Invitrogen) slurry. For analysis of Ace2 phosphorylation at S122, we arrested temperature-sensitive strains at 37°C. As a positive control for Ace2 S122 phosphorylation, we depleted Cdc20 in a PrGal-CDC20 strain by growth in 2% glucose for 2 h, followed by release in 2% galactose medium for 35 min, a point of maximal phosphorylation of Ace2 by Cbk1 (43). We immunoprecipitated Ace2-HA from cells as described above with the following changes: anti-HA antibody (12CA5) was used at 1:50 with 50 μl of a 1:1 slurry of recombinant protein G-Sepharose (Invitrogen) in yeast lysis buffer (see below) that also contained 20 μM cantharidin and 1 mM EDTA. For all immunoprecipitations we rotated bead-lysate mixtures at 4°C for 2 h followed by two washes in ice-cold yeast lysis buffer containing protease and phosphatase inhibitors (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 120 mM β-glycerolphosphate, 2 mM sodium orthovanadate, 20 mM sodium molybdate, 3 mM benzamidine, 1 mM phenylmehtylsulfonyl fluoride, 1 μg/ml pepstatin, 0.5 mM leupeptin, and 2 μg/ml chymostatin). We then washed the beads twice in ice-cold lysis buffer lacking inhibitors, resuspended them in lambda phosphatase buffer, and split the samples in half, treating one portion with 300 units lambda phosphatase (New England BioLabs) for 30 min at 37°C. We then resuspended the beads in 2× SDS-PAGE sample buffer and resolved the samples on 8% SDS-PAGE gels.
For Cdc14 treatment, we immunoprecipitated Cbk1 from a strain expressing glutathione S-transferase (GST)-Cbk1 and 13Myc-Mob2 under the control of the galactose promoter. Reactions were completed as previously described (32). Briefly, beads were washed three times into phosphatase buffer (25 mM HEPES [pH 7.4], 0.1 mg/ml bovine serum albumin [BSA], 150 mM NaCl, 2 mM MnCl2). Bacterially purified GST-Cdc14 was added to 0.36 μg/μl and allowed to incubate for 35 min at 30°C. Beads were washed three times in ice-cold yeast lysis buffer and resuspended in 2× SDS-PAGE sample buffer (see Fig. 7). For the experiment shown below in Fig. 8, Cbk1 was immunoprecipitated from cdc15-2 arrested cells and treated as above. Samples were then washed three times in kinase base buffer containing phosphatase inhibitors (10 mM Tris [pH 8.0], 150 mM NaCl, 120 mM β-glycerolphosphate, 2 mM sodium orthovanadate, 20 mM sodium molybdate) (see Fig. 8).
We performed kinase assays as previously described (29) with 2 μg GST-Ace242-242(S113A/S137A) for 1 h (see Fig. 3). In the experiment shown below in Fig. 8, we used 2 μg GST-Ace2102-306-His6 as substrate, and samples were removed from a single reaction mixture at 5, 10, and 15 min after Ace2 addition and quenched with 5× sample buffer and incubation at 85°C for 20 min (see Fig. 8). For the experiments shown in Fig. 8A and B, phosphorimager counts (ImageQuant v5.2; Molecular Dynamics) were used to plot the kinetics of phosphorylation. In the experiment shown in Fig. 8C, normalized phosphorylation was expressed as phosphorimager counts divided by the Cbk1 protein level as determined by Western blotting and quantification with the Odyssey software (v2.0; Li-Cor, Inc.).
Immunoblotting.
For all Western blot assays, proteins were transferred to BioTraceW polyvinylidene difluoride (PVDF; Pall) membrane. Membranes were probed as previously described (29) with the following changes. A Cbk1-specific polyclonal rabbit antibody (anti-Cbk1) was raised against a GST-tagged N-terminal fragment (residues 1 to 333) of Cbk1 expressed in Escherichia coli, and the crude serum was used at 1:1,000 (service provided by Open Biosystems). We used affinity-purified phospho-specific rabbit antibody (anti-pT743 [29]) at 1:1,000, Ace2 anti-pS122 antibody (43) at 1:200, anti-GST antibody (Sigma) at 1:1,000, and phosphoglycerate kinase (PGK) monoclonal antibody (Invitrogen) at 1:1,000. We performed all blotting assays in 1% BSA (Sigma) in Tris-buffered saline with 0.1% Tween (TBST). Primary antibodies were incubated with blots for 1 h at room temperature, followed by incubation with Alexa 680-conjugated goat anti-rabbit secondary (Invitrogen) or IRDye 800-conjugated goat anti-mouse secondary (Rockland, Inc.) at 1:3,000 in TBST for 30 min. Blots were washed three times for 3 min with TBST between antibody additions and prior to imaging. Blots were imaged and quantified with the Odyssey software (v2.0; Li-Cor, Inc.), and images of blots were processed using Photoshop (Adobe).
Quantification of phosphorylation at T743.
We cloned a Cbk1(731-756; S745A, T751A) fragment by using synthesized oligonucleotides (IDT DNA) into a modified pET vector containing N-terminal GST and C-terminal hexahistidine (His6) tags and expressed this fusion protein in E. coli Rosetta(DE3)pLysS. We purified this fusion protein by chromatography on Ni-nitrilotriacetic acid resin (Qiagen) followed by glutathione-Sepharose 4B (GE Healthcare) using the manufacturer's protocols. The protein was dialyzed into 50 mM Tris, 100 mM NaCl, 1 mM dithiothreitol (DTT), pH 8.0 (for in vitro kinase assays), flash-frozen in liquid nitrogen, and stored at −80°C.
We constructed a plasmid for expression of amino acids 1 to 333 of Cbk1 fused C-terminally to maltose binding protein by cloning an appropriate fragment generated by PCR from a plasmid containing wild-type CBK1 into pMAL-c2X (New England BioLabs). We expressed the MBP-Cbk1(1-333) fragment in E. coli BL21(DE3)RIL and immobilized it on amylose resin (New England BioLabs). We eluted MBP-Cbk1(1-333) with 6 M Urea in 1× phosphate-buffered saline (PBS), measured the optical density at 280 nm by using a NanoDrop spectrophotometer, and determined the concentration using the protein's extinction coefficient, calculated based on its amino acid composition. We then flash-froze aliquots of the protein in liquid nitrogen and stored them at −80°C.
To obtain a standard curve for the Thr743 phospho-specific antibody (anti-pT743), we first in vitro phosphorylated GST-Cbk1(731-756; S745A, T751A)-His6 fragment with bacterially purified Kic1 (J. Hsu, unpublished data). For these reactions, we combined 20 pmol of purified Kic1 kinase with 100 pmol of purified Cbk1 fragment as substrate in a 30-μl reaction mixture containing 50 mM Tris (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 50 μM ATP, and 10 μCi [γ-32P]ATP. We incubated reaction mixtures at room temperature for 30 min and quenched the reactions by addition of 5× SDS-PAGE sample buffer with immediate mixing followed by 10 min of incubation at 85°C. We performed 2-fold serial dilutions of this material, starting with 12.5 pmol of substrate, resolved proteins on a 12% SDS-PAGE gel, and transferred them to a PVDF membrane (Pall). We evaluated the extent of Cbk1 labeling by using a Storm 860 Imager (Molecular Dynamics) to quantify the amount of 32P incorporated into the Cbk1 fragment. We measured pixel intensities by using ImageQuant software (Molecular Dynamics) and compared volume over gel backgrounds in regions of interest (ROIs) by encompassing the Cbk1 fragment bands with ROIs encompassing simultaneously acquired spots of known 32P quantities. We then detected blotted phosphorylated Cbk1 with anti-pT743, evaluating signal strength with a LiCor Odyssey system as described above. We obtained a standard curve by plotting anti-pT743 pixel intensity as a function of pmol of phosphorylated Cbk1 fragment, using data from three independent dilution experiments.
To obtain a standard curve for the Cbk1 antibody (anti-Cbk1), we performed 2-fold serial dilutions of MBP-Cbk1(1-333), starting with 0.25 pmol of material, and resolved proteins on 8% SDS-PAGE gels followed by transfer to a PVDF membrane (Pall). We analyzed total Cbk1 amount with anti-Cbk1 using a LiCor Odyssey system as described above. We obtained a standard curve by plotting the anti-Cbk1 pixel intensity as a function of pmol of blotted Cbk1, again assessing the results of three independent experiments.
QPCR.
We prepared RNA from log-phase or arrested cultures by hot acid phenol extraction as previously described (15). We treated 2 μg of RNA with 10 units of RNase-free DNase I (Roche) and converted it to cDNA with Moloney murine leukemia virus reverse transcriptase (RT; Promega). We performed quantitative RT-PCRs (QPCRs) by using CTS1, DSE1, and ACT1 primers (42), generating standard curves from serial dilutions of yeast genomic DNA using linear regression analysis of cycle threshold (CT) values. We quantified cDNA template concentrations using the standard curve for each primer set. We internally normalized samples to the concentration of ACT1 and then normalized values to control samples as indicated.
Microscopy and image analysis.
We grew cells to mid-log phase, or arrested them for 2.5 h at 37°C in minimal medium with selection as necessary, and performed fluorescence and differential interference contrast microscopy using an Axiovert 200 M microscope (Carl Zeiss MicroImaging, Inc.) with a Cascade II-512B camera (PhotoMetrics, Inc.). Images were taken with a 100×/1.45-numerical aperture oil immersion objective. We acquired images using the Openlab software (v5.5.0; Improvision) and measured fluorescence intensity using ImageJ. For mother-daughter analysis, we incubated cells with rhodamine-labeled concanavalin A (Sigma) at 1:1,000 for 5 min, washed cells three times, and resuspended them in fresh medium (29). For Ace2 nuclear-to-cytoplasmic quantification, cdc15-2 cells expressing Ace2-GFP or Ace2-AAA-GFP were arrested for 2.5 h at 37°C and immediately imaged. Images were then acquired every 5 min at ambient room temperature. The intensities of GFP fluorescence in the cytoplasm and nucleus were measured in cells at the arrest and then when Ace2 exhibited peak nuclear accumulation. This varied for individual cells but was between 30 and 45 min. For cell separation quantification, cells were sonicated prior to microscopy, and >100 cell clumps were counted per strain as previously described (42).
RESULTS
M/G1 phosphorylation of Cbk1's hydrophobic motif site requires FEAR but not MEN.
Previous analysis using G1-synchronized cells showed that phosphorylation of Cbk1's HM site increases late in cell division (29). To better understand the mitotic timing of this critical regulatory phosphorylation, we arrested wild-type cells at the metaphase/anaphase transition with the microtubule-depolymerizing agent nocodazole and examined HM site phosphorylation after release from this arrest. To ensure that this synchrony examined a single coherent cell cycle, we added additional nocodazole to the culture 70 min after release, resulting in a second M-phase arrest. We did not observe HM site phosphorylation when we used an anti-pT743 antibody (29) in nocodazole-arrested cells, and we found that phosphorylation of Cbk1's HM site was detectable 50 min after release (Fig. 1A). Phosphorylation of the HM site occurred during the M-to-G1 transition, prior to bud emergence (Fig. 1B). Quantitative RT-PCR analysis of the transcript levels of the Ace2-regulated genes CTS1 and DSE1 showed that Cbk1-mediated activation of this transcription factor occurred approximately coincident with HM site phosphorylation, peaking between 60 and 70 min after release from nocodazole (Fig. 1C). Phosphorylation of the HM site persisted throughout G1 and during bud emergence and then diminished as cells entered the next mitosis, becoming undetectable in cells arrested with a second addition of nocodazole (Fig. 1A). This persistence of HM site phosphorylation into late G1 was expected, as this modification is critical for Cbk1's role in bud morphogenesis and polarized growth (29).
To estimate the extent of HM site phosphorylation of Cbk1 during the M-to-G1 transition, we generated standard curves for both the anti-pT743 and the anti-Cbk1 antibodies by using a known amount of a T743 phosphorylated fragment of Cbk1 and an N-terminal fragment of Cbk1, respectively (Fig. 1D and E). We found that at the peak of Cbk1 phosphorylation (60 min after release from nocodazole), roughly 3% of immunoprecipitated Cbk1 was phosphorylated at T743. This indicates that only a small pool of Cbk1 is likely phosphorylated at the end of mitosis and that this is sufficient to activate Ace2.
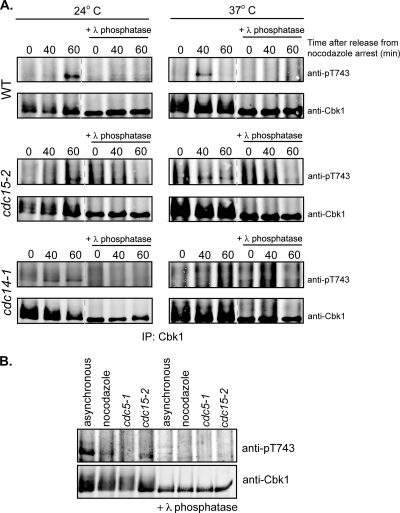
The timing of Cbk1 HM site phosphorylation after release from nocodazole arrest is consistent with regulation by the FEAR or MEN pathway, which act sequentially to coordinate the events of late mitosis. We therefore used conditional alleles of components of these pathways to assess their requirement for phosphorylation of the Cbk1 HM site, exploiting the temperature-sensitive cdc14-1 and cdc15-2 alleles to arrest cells just prior to the M/G1 transition. While these conditional alleles cause cytologically similar telophase arrest at restrictive temperature, they are distinct from one another: cdc14-1 cells arrest without initiation of the FEAR pathway, while cdc15-2 cells arrest after FEAR activation but prior to the MEN-mediated release of Cdc14 from the nucleus.
We first blocked wild-type, cdc15-2, and cdc14-1 cells at the metaphase/anaphase transition with nocodazole, and then released cells from this arrest at permissive or restrictive temperatures (24°C or 37°C, respectively) (Fig. 2A). At the permissive temperature, we detected phosphorylation of the Cbk1 HM site 60 min after release in wild-type, cdc14-1, and cdc15-2 cells, consistent with the results shown in Fig. 1A. In wild-type and cdc15-2 cells released from nocodazole arrest at 37°C, we observed HM site phosphorylation after 40 min. However, in the cdc14-1 cells released at 37°C, we observed no detectable increase in phosphorylation of Cbk1's HM site. HM site phosphorylation was also undetectable in arrested cdc5-1 cells, which arrest prior to FEAR function (Fig. 2B). Thus, phosphorylation of the Cbk1 HM site requires FEAR activation but does not require activation of the MEN.
FIG. 2.
HM site phosphorylation requires FEAR but not MEN activation. (A) Phosphorylation of the HM site was examined in FEAR- or MEN-arrested cells. The indicated strains were first synchronized with nocodazole and then released into prewarmed medium at a nonpermissive (37°C) temperature to arrest without activating the FEAR (cdc14-1) or the MEN (cdc15-2) pathways. Cells released at permissive temperature (24°C) and wild-type cells were used as controls for HM site phosphorylation. Cells were harvested at the indicated times after release from nocodazole arrest, and immunoprecipitated Cbk1 was probed with an anti-pT743 antibody or anti-Cbk1 antibody. Lambda phosphatase-treated immunoprecipitates are marked. (B) HM site phosphorylation requires activity of the FEAR kinase Cdc5. Cbk1 immunoprecipitated from wild-type asynchronous or nocodazole-arrested cells or cdc5-1 or cdc15-2 cells was probed with an anti-pT743 antibody or anti-Cbk1 antibody. Lambda phosphatase treated immunoprecipitates are shown in the lanes on the right side.
Phospho-mimetic substitution at Cbk1's HM site hyperactivates the kinase but does not affect Ace2 asymmetry or function.
HM site phosphorylation is necessary for Cbk1's control of Ace2's nuclear localization and function. Therefore, we hypothesized that both the onset and duration of Ace2-driven transcription are due to the temporally controlled phosphorylation of the HM site. To determine if the constitutive presence of Cbk1 activated by HM site phosphorylation either hyperactivated Ace2 or disrupted its function, we constructed a Cbk1 allele in which the HM site phospho-acceptor threonine at position 743 is replaced by glutamic acid (CBK1-T743E). While not a perfect mimic, in many cases a negatively charged side chain can simulate the effects of constitutive phosphorylation. Cells in which the endogenous CBK1 open reading frame is replaced by CBK1-T743E were viable, and expression of this allele was similar to the wild-type protein (Fig. 3A). We examined the in vitro kinase activity of Cbk1-T743E immunoprecipitated from asynchronous cells, using a bacterially expressed fragment of Ace2 containing Cbk1 phosphorylation sites as a substrate. The mutant protein not only exhibited increased autophosphorylation but also showed increased kinase activity toward this Ace2 fragment compared with wild-type Cbk1 (Fig. 3B).
FIG. 3.
Phenotypic analysis of the CBK1-T743E allele indicated that the kinase has increased activity in vitro but did not hyperactivate Ace2. (A) Cbk1-T743E was expressed at levels comparable to wild-type Cbk1. Total cell lysates prepared from wild-type, CBK1-T743E, or cbk1Δ strains were probed with an anti-Cbk1 antibody. PGK1 was used as a loading control (bottom panel). (B) Cbk1-T743E exhibited increased kinase activity in vitro. Cbk1 was immunoprecipitated from wild-type (CBK1), CBK1-T743E, and cbk1Δ strains and used in an in vitro kinase assay with a fragment of Ace2. The top panel is an immunoblot of total Cbk1. The bottom panels are autoradiographs of Cbk1 autophosphorylation (middle panel) and phosphorylation of the Ace2 fragment (bottom panel). (C) The CBK1-T743E strain has a wild-type cell separation phenotype. Wild-type (CBK1), cbk1Δ, and CBK1-T743E strains were grown to mid-log phase and imaged by differential interference contrast (DIC). The number of connected cells in a group was determined for wild-type and CBK1-T743E strains. Standard deviations are shown for three independent trials. (D) The CBK1-T743E strain does not exhibit hyperactivation of Ace2 target transcription. QPCR was performed on RNA isolated from the indicated strains. Transcription of CTS1 relative to cbk1Δ is shown. Error bars indicate standard errors of the means of five experiments. There was no significant difference between CTS1 transcript levels in CBK1 and CBK1-T743E strains (P = 0.43).
We next characterized the CBK1-T743E allele's in vivo function by assessing its ability to promote cell separation and Ace2-driven transcription. We found that CBK1-T743E cells grown in liquid culture were mainly present in groups of one to five associated cell bodies; this distribution was indistinguishable from that of wild-type cells (Fig. 3C). Consistent with this, the transcript levels of Ace2 target genes in the CBK1-T743E strain were similar to those in wild-type cells (Fig. 3D). Importantly, we found no evidence for elevated levels of Ace2-driven transcripts, which would be expected from ectopic or prolonged activation of the transcription factor by constitutively active Cbk1.
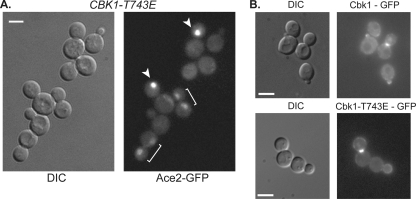
Cbk1 is functionally restricted to the daughter cell cytoplasm (70). One possible mechanism for this partitioning is dephosphorylation of Cbk1's HM site in late mitosis specifically in the mother cell, effectively inactivating any kinase that diffuses from the daughter to the mother. If so, CBK1-T743E, which carries a constitutive negative charge at this position, should disrupt the segregation of Ace2-GFP. However, we found that Ace2 distribution was normal in CBK1-T743E cells, with strong localization in the daughter cell nucleus (Fig. 4A, white arrowheads) preceded by faint localization in both mother and daughter cell nuclei (Fig. 4A, white brackets). Consistent with this, Cbk1-T743E-GFP localizes asymmetrically to the daughter cell cortex, bud neck, and daughter nucleus, similar to the wild-type protein (Fig. 4B). Thus, Ace2's asymmetric localization does not require dynamic or spatially restricted phosphorylation of Cbk1's HM site.
FIG. 4.
The CBK1-T743E strain exhibits wild-type localization of Ace2 and Cbk1. (A) Ace2 localization was not disrupted in the CBK1-T743E strain. CBK1-T743E expressing Ace2-GFP was imaged for Ace2 localization. Arrows indicate daughter cell localization, and brackets indicated mother-daughter pairs. (B) Cbk1-T743E-GFP exhibited wild-type localization to the daughter cell cortex, nucleus, and bud neck. A representative image is shown for localization of Cbk1-T743E-GFP and wild-type Cbk1-GFP in cells grown to mid-log phase at 30°C.
As this work was in preparation, Panozzo et al. reported that the CBK1-T743E allele does not negatively affect Ace2 function, and our findings confirm this (51). However, the other group did not examine the protein levels or enzymatic activity of this allele or address its effects on transcript levels of Ace2-driven genes. Our additional analysis indicates that while CBK1-T743E is a gain-of-function allele for kinase activity, it does not promote hyperactivation of Ace2-driven transcription.
Hydrophobic motif phosphorylation is not sufficient for Cbk1 to activate Ace2 in mitosis.
Phosphorylation of Cbk1's HM site is necessary for Ace2's localization to the daughter cell nucleus and concomitant induction of the transcription factor's target genes (29). However, it is not clear if HM site phosphorylation is sufficient for Cbk1 to act on Ace2 or if additional regulatory events have to occur as cells pass from mitosis to G1 for this interaction to take place. Confounding matters, Ace2 is controlled by two opposing mechanisms: mitotic phosphorylation by the cyclin-dependent kinase Cdk1/Cdc28 keeps the transcription factor out of the nucleus (2, 50), and subsequent Cbk1 phosphorylation of Ace2's NES drives its nuclear retention (42). Therefore, to assess Cbk1's regulation of Ace2 more directly, we eliminated mitotic CDK control of Ace2's nuclear entry by changing phospho-acceptor residues in three key CDK sites to alanine. This allele, referred to as ace2-AAA, eliminates mitotic CDK inhibition of Ace2 nuclear import (50, 55). With these inhibitory CDK sites removed, nuclear accumulation and transcriptional activity of Ace2 are regulated only by Cbk1's inactivation of the Ace2 NES (42).
The question we sought to answer was can Cbk1 phosphorylate Ace2 in arrested cdc15-2 cells, in which phosphorylation of the kinase's HM site occurs? Our expectation was that wild-type Ace2 would not localize to nuclei at this arrest due to mitotic CDK inhibition of the Ace2 NLS. However, if Cbk1 can phosphorylate Ace2's NES at this arrest, the Ace2-AAA protein should accumulate in nuclei and activate transcription of target genes. In contrast, if Cbk1 needs to undergo additional regulation, Ace2-AAA should exhibit faint nuclear localization that increases dramatically in daughter cells upon release from arrest.
Wild-type Ace2-GFP was cytoplasmically localized in 100% of arrested cdc15-2 cells (Fig. 5A). This was unsurprising because cdc15-2 cells arrest prior to large-scale release of Cdc14, which mediates dephosphorylation of the CDK sites in wild-type Ace2 (43, 55). By examining localization of Ace2-AAA-GFP, we found that cdc15-2 cells grown at permissive temperature exhibit the same Ace2-AAA-GFP nuclear localization pattern as wild-type cells (data not shown). In cdc15-2 cells arrested at restrictive temperature, we did not see strong daughter-specific nuclear localization of Ace2-AAA-GFP, and we found it weakly accumulated in both mother and daughter (16%) or daughter nuclei (21%) (Fig. 5A). This is consistent with Ace2's previously observed localization pattern, which goes from a cytoplasmic distribution to faint nuclear localization before exhibiting strong accumulation to daughter cell nuclei (10, 29, 42, 70). In addition, the fluorescence intensity of Ace2-AAA-GFP in the nucleus of the arrested cells was significantly lower than in the daughter nucleus of cells upon release from the arrest (Fig. 5B). This absence of strong daughter-specific localization demonstrates that the MEN is essential for Cbk1 to inactivate the transcription factor's NES.
FIG. 5.
Cbk1 cannot inactivate Ace2's NES at the MEN arrest. (A) Ace2 localization at the cdc15-2 arrest showed that Cbk1 does not inactivate Ace2's NES. cdc15-2 cells expressing Ace2-GFP or Ace2-AAA-GFP were stained to differentiate mothers from daughters (see Materials and Methods) and were grown at 24°C or arrested for 2.5 h at 37°C. Cells were counted as mother and daughter localized (M + D), daughter localized (D), or unlocalized. No cells exhibited mother-only localization. The numbers of counted cells are shown. (B) Ace2 does not exhibit strong daughter-specific localization in MEN-arrested cells. cdc15-2 cells expressing Ace2-GFP or Ace2-AAA-GFP were arrested at 37°C and imaged over time at 24°C. The nuclear-to-cytoplasmic fluorescence intensity ratio was determined at the arrest (gray bars) and upon release, when individual cells exhibited peak Ace2 nuclear accumulation (white bars). Quantification was only performed on Ace2-AAA-GFP cells exhibiting nuclear localization at the arrest. (C) Ace2's NES is not phosphorylated without activation of FEAR or MEN. The indicated strains were arrested, and Ace2-HA was immunoprecipitated. Protein was probed with an antibody for phosphorylation at Ace2 S122. Protein was harvested from cells released from a PrGal-Cdc20 block as a positive control for Ace2 phosphorylation. Lambda phosphatase-treated samples are shown on the right. (D and E) Ace2's target genes are not expressed in MEN-arrested cells. cdc15-2 cells with wild-type CBK1, CBK1-T743E, or cbk1Δ containing the indicated ACE2 allele were arrested at 37°C. QPCR was performed on isolated RNA, and CTS1 (D) and DSE1 (E) transcript levels are shown relative to a control strain lacking ACE2. Standard errors of the means of three independent trials are shown.
To confirm more directly that Cbk1 could not phosphorylate Ace2's NES in a late mitotic arrest, we used a recently generated an antibody that specifically recognizes phosphorylated serine 122 in Ace2's NES, which has been shown to be a key Cbk1 phosphorylation site (anti-pS122) (42, 43). At the cdc15-2 and cdc14-1 arrest, immunoprecipitated Ace2-HA was not phosphorylated at this residue (Fig. 5C), while Ace2-HA was phosphorylated when isolated from cells released from mitotic arrest induced by Gal-CDC20 shutoff at a time of maximal phosphorylation (43) (Fig. 5C). These data further indicate that Cbk1 does not phosphorylate Ace2's NES in cells arrested prior to mitotic exit.
We examined the transcription factor's activity to corroborate our analysis of Ace2 localization and NES phosphorylation, comparing transcript levels of the Ace2-driven genes CTS1 and DSE1 in arrested cdc15-2 cells carrying the ACE2 and ace2-AAA alleles. As a positive control, we expressed an ace2 gain-of-function allele in which phospho-acceptor residues in two Cbk1 sites in Ace2's NES were changed to acidic amino acids (ace2-AAA-NSM [NES site mutant]). This allele lacks negative regulation of nuclear import by CDK and mimics constant Cbk1 NES phosphorylation, resulting in Cbk1-independent nuclear accumulation of the mutant protein and transcription of Ace2-regulated genes (42). We found that both Ace2 and ace2-AAA were inactive in arrested cdc15-2 cells: CTS1 and DSE1 transcript levels were indistinguishable in arrested cdc15-2 cells expressing either ACE2 or ace2-AAA (Fig. 5D and E). Transcript levels are shown relative to those in asynchronous ace2Δ cells. This baseline level was indistinguishable in asynchronous cbk1Δ and arrested cdc15-2 cbk1Δ cells, demonstrating a lack of RAM-independent expression of Ace2-driven genes at the arrest (data not shown). We also observed similar results in arrested cdc14-1 cells (data not shown).
These findings, combined with localization of Ace2 and phosphorylation of its S122 NES site, indicate that Cbk1 regulation of Ace2 requires MEN activation. However, Cbk1's HM site phosphorylation is somewhat less robust in arrested cdc15-2 cells, and it is therefore possible that cells arrested by MEN abrogation do not achieve sufficient levels of activated Cbk1. We therefore assessed transcription of Ace2 target genes as described above in cells expressing the CBK1-T743E allele, which mimics constitutive phosphorylation of the HM site and drives Ace2 localization and activation normally (Fig. 3 and 4). We found that the CBK1-T743E allele did not activate expression of Ace2-driven genes at this arrest: transcript levels were comparable to those of the control strain even in the presence of the ace2-AAA allele, which lacks negative regulatory CDK phosphorylation sites (Fig. 5D and E).
In contrast, arrested cdc15-2 strains expressing ace2-AAA-NSM exhibited significant increases in Ace2-driven transcription over levels seen in strains expressing ACE2 (Fig. 5D and E). Even in cells lacking CBK1, the ace2-AAA-NSM allele was able to activate transcription at the arrest (Fig. 5D and E). Thus, combining inactivation of Ace2's NES with absence of its CDK-mediated nuclear import block is sufficient for the transcription factor to activate its target genes. Taken together, these data indicate that in the absence of FEAR or MEN activation, Cbk1 cannot inactivate Ace2's NES. Furthermore, the results show that HM site phosphorylation, while necessary for Cbk1 function, is not by itself sufficient for the kinase to activate Ace2.
Cdc14 may promote Cbk1's regulation of Ace2 following mitotic exit.
These results suggest that final activation of Cbk1 depends either directly on Cdc15 itself or on a process downstream of MEN activation. We previously demonstrated that Cbk1 is a hyperphosphorylated protein, and we proposed a model in which this phosphorylation inhibits Cbk1's function in vivo (29). One important function of the MEN is to reverse the phosphorylations performed by the mitotic cyclin-dependent kinase Cdc28/Cdk1 (39, 46, 67). Thus, at the cdc15-2 arrest, mitotic CDK phosphorylations may persist and might contribute to the inhibition of Cbk1. To determine how the phosphorylation state of Cbk1 might be regulated at the M/G1 transition, we examined Cbk1's electrophoretic mobility from arrested cdc15-2 cells. We found that Cbk1 exhibited increased electrophoretic shifting that was eliminated by treatment with lambda phosphatase, a mobility shift indicative of phosphorylation (Fig. 6A). Upon release from cdc15-2 arrest, Cbk1's electrophoretic shifting decreased between 30 and 45 min after release, indicating that the protein was less phosphorylated. In contrast, HM site modification persisted (Fig. 6A).
FIG. 6.
Cbk1 activation requires activation of MEN. (A) Cbk1 exhibits dephosphorylation upon release from MEN arrest. The cdc15-2 strain was released from arrest, and cells were harvested every 15 min. Immunoprecipitated Cbk1 was probed with anti-Cbk1 (top panel) or anti-pT743 (middle panel) antibody. Lambda phosphatase-treated samples are shown on the bottom. (B) Activation of Ace2 target gene expression correlates with Cbk1 dephosphorylation. The cdc15-2 strain was released from arrest, and cells were harvested every 15 min. QPCR was performed to determine the transcript levels CTS1 and DSE1 relative to the levels in arrested cells (0 min). Standard errors of the means from three independent experiments are shown. (C) Strong daughter-specific localization of Ace2 correlates with dephosphorylation of Cbk1. cdc15-2 cells expressing Ace2-GFP were released from arrest, and images were acquired every 2 min. Representative cells were followed for the full time course.
Consistent with the idea that elimination of inhibitory phosphorylation is necessary for Cbk1 to act on Ace2, the timing of Cbk1 dephosphorylation correlates with Ace2 daughter-specific localization and transcriptional activity. Between 20 and 30 min after release from cdc15-2 arrest, Ace2-GFP strongly localized to the daughter cell nucleus (Fig. 6C). In addition, levels of CTS1 and DSE1 transcripts increased upon release from the arrest and peaked around 45 min (Fig. 6B). Peak levels of these Ace2-driven transcripts lag behind maximal transcription factor localization. Their levels are also sustained for a period after Ace2 exits the nucleus, a latency that is likely an inherent function of the kinetics of the transcripts' accumulation and decay (43). Similarly, we found that these transcripts persist after loss of Cbk1 HM site phosphorylation. These data demonstrate that Cbk1's activation of Ace2 occurred only after MEN activation and support the model that Cbk1 requires both HM site phosphorylation and removal of inhibitory phosphorylations in order to activate Ace2.
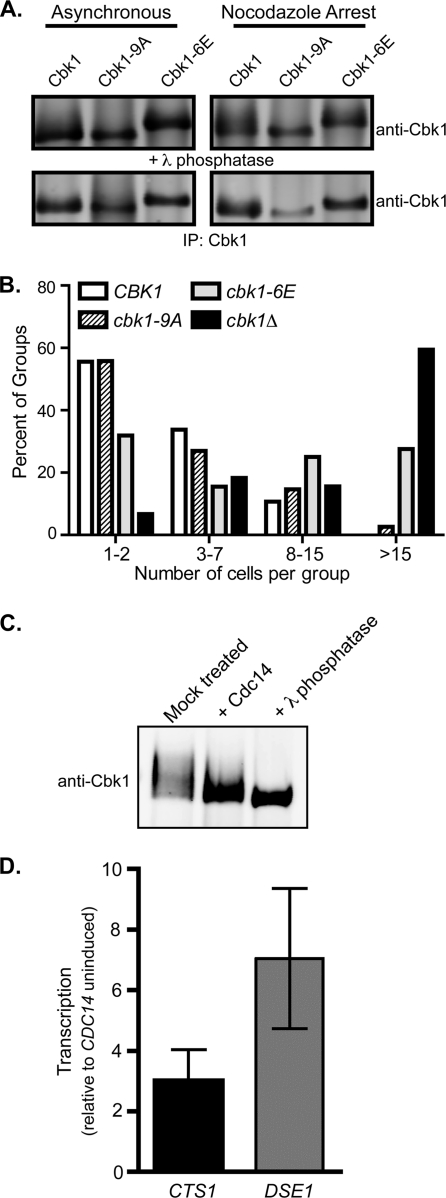
Cyclin-dependent kinases phosphorylate targets with the minimal motif (S/T)-P (13, 27). To determine if Cdc28/Cdk1 consensus motifs in Cbk1 are phosphorylated in vivo and exert regulatory effects, we changed sets of them to either alanine, which is nonphosphorylatable, or glutamic acid, which is phospho-mimetic in some cases. We changed putative phospho-acceptor residues in all nine (S/T)-P sites in Cbk1 to alanine (the cbk1-9A allele) and a subset of these sites to glutamic acid (the cbk1-6E allele). The Cbk1-9A and Cbk1-6E proteins were expressed at levels similar to wild-type Cbk1 (Fig. 7A). Cbk1-9A promoted normal cell separation (Fig. 7B), while Cbk1-6E exhibited a cell separation defect that was pronounced but not as severe as loss of cell separation seen in cbk1Δ cells (Fig. 7B). Cells expressing single glutamic acid substitutions at putative CDK sites did not show separation defects, suggesting that reduction of Cbk1 function requires a combination of acidic moieties at these sites (data not shown). In both asynchronous and nocodazole-arrested cells (in which hyperphosphorylation of Cbk1 is evident), Cbk1-9A ran as a single, unshifted band (Fig. 7A), while Cbk1-6E exhibited an electrophoretic mobility shift that was not reversible by phosphatase treatment. Overall, these findings indicate that hyperphosphorylation of Cbk1 in late mitosis may be mediated by Cdc28/Cdk1 and that multiple such phosphorylations can block Cbk1's function in vivo.
FIG. 7.
Removal of CDK phosphorylation on Cbk1 by Cdc14 may activate Cbk1 independently of the MEN. (A) Mutation of potential CDK phosphorylation sites on Cbk1. Nine potential CDK1 phosphorylation sites on Cbk1 were mutated to alanine (cbk1-9A), or six of these sites in the C terminus of Cbk1 were mutated to glutamic acid (cbk1-6E). Cbk1 was immunoprecipitated from asynchronous cells (left) or nocodazole-arrested cells (right). Both alleles were expressed at levels similar to the wild-type protein. The Cbk1-9A protein exhibited less electrophoretic mobility shifting compared to the wild-type protein. Lambda phosphatase-treated protein is shown below. (B) Glutamic acid substitution at CDK sites in Cbk1 is inhibitory. The number of connected cells in a group was determined for the indicated strains. The cbk1-9A allele promoted wild-type separation, while the cbk1-6E allele had a defect in cell separation. (C) Cdc14 can dephosphorylate Cbk1 in vitro. Immunoprecipitated Cbk1 was treated in vitro with bacterially purified Cdc14 or treated with lambda phosphatase as a control for complete dephosphorylation. (D) Overexpression of Cdc14 can activate Cbk1 and Ace2 independently of MEN. cdc15-2 cells containing a plasmid for PrGAL-GST-CDC14 were arrested at 37°C. Cells were maintained at the arrest, and CDC14 was induced (galactose) or uninduced (glucose). Cells were harvested, and QPCR was performed to determine transcript levels of CTS1 and DSE1 relative to uninduced cells. Standard errors of the means from three independent trials are shown.
Activation of the MEN triggers sustained release of the phosphatase Cdc14 into the cytoplasm. Once released, Cdc14 specifically dephosphorylates serine or threonine residues that are followed by a proline, targets of mitotic cyclin-dependent kinase (21, 26, 67). To determine whether Cdc14 can act directly on Cbk1, we immunoprecipitated Cbk1 and incubated it with purified bacterially expressed Cdc14. This treatment greatly reduced Cbk1's electrophoretic mobility, indicating that the kinase was phosphorylated at (S/T)-P sites. Treatment with the nonspecific lambda phosphatase further reduced Cbk1's electrophoretic mobility (Fig. 7C), indicating that Cbk1 is modified at additional sites (1, 9, 29, 58). Therefore, Cdc14 can dephosphorylate Cbk1 in vitro.
To determine if Cdc14 was sufficient to activate Cbk1 in the absence of MEN function, we overexpressed Cdc14 in cdc15-2 arrested cells by using an inducible PrGAL-CDC14 construct. We found that CTS1 and DSE1 levels were elevated when Cdc14 was induced in MEN-arrested cells relative to arrested cells in which the phosphatase is not induced, indicative of Cbk1 activation of Ace2 (Fig. 7D). Therefore, Cdc15 is not directly required for Cbk1 activation, and Cdc14 overexpression can bypass the effect of cdc15-2 arrest on Cbk1 activation of Ace2. Furthermore, these results suggest that activation of Cbk1 occurs downstream of mitotic exit, through dephosphorylation of CDK sites by Cdc14.
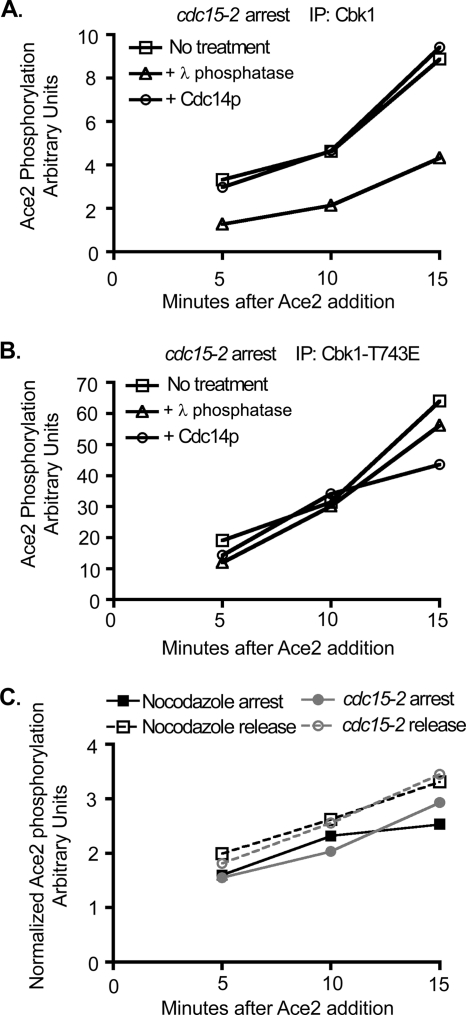
To determine if dephosphorylation of CDK sites modulates Cbk1's enzymatic activity, we examined the effect of Cdc14 treatment on the in vitro kinase activity of Cbk1 isolated from arrested cdc15-2 cells. We divided the immunoprecipitated Cbk1 into three equal fractions: one treated with Cdc14, one with lambda phosphatase, and one mock treated. After washing out the phosphatase, we examined phosphorylation of bacterially expressed Ace2 in the presence of phosphatase inhibitors. Untreated Cbk1 and Cdc14-treated Cbk1 phosphorylated Ace2 to the same extent (Fig. 8A), indicating that phosphorylation of putative CDK sites does not directly inhibit the enzyme's activity.
FIG. 8.
The intrinsic enzymatic kinase activity of Cbk1 is not increased upon dephosphorylation. (A and B) Dephosphorylation of Cbk1 by Cdc14 in vitro does not increase the intrinsic enzymatic activity of Cbk1. Cbk1 (A) or Cbk1-T743E (B) was immunoprecipitated from cdc15-2 arrested cells. Immunoprecipitated protein was split equally, and samples were incubated with phosphatase buffer alone, lambda phosphatase, or bacterially purified Cdc14 in vitro. Samples were washed into kinase buffer containing phosphatase inhibitors, and a kinase assay was performed with a fragment of Ace2. An aliquot of the assay mixture was removed at 5, 10, and 15 min after the addition of the substrate. Ace2 phosphorylation (arbitrary units) over time is shown. (C) The intrinsic enzymatic kinase activity of Cbk1 does not increase upon release from mitotic arrest. Cbk1 was immunoprecipitated from a wild-type strain arrested in nocodazole or at 60 min postrelease. Additionally, Cbk1 was immunoprecipitated from cdc15-2 arrested cells or at 45 min postrelease. A kinase assay was performed with a fragment of Ace2, and an aliquot of the assay mixture was removed at 5, 10, and 15 min after the addition of the substrate. Samples were normalized by protein concentration as determined by Western blotting for total Cbk1.
Cbk1 activity is significantly enhanced by intramolecular phosphorylation of a conserved activation loop site (S570) (29). Consistent with this, removal of all phosphates by treatment with lambda phosphatase rendered Cbk1 significantly less active (Fig. 8A). We also performed an identical experiment with Cbk-T743E immunoprecipitated from arrested cdc15-2 cells (Fig. 8B). As in Fig. 3, this gain-of-function allele had significantly elevated in vitro enzymatic activity, and treatment with Cdc14 did not affect its activity. Intriguingly, lambda phosphatase treatment did not greatly reduce activity of this allele, suggesting that the intramolecular phosphorylation of Cbk1-T743E's activation loop site may be exceptionally rapid.
Finally, we assessed the in vitro kinase activities of Cbk1 immunoprecipitated from cells during both nocodazole and cdc15-2 arrest, as well as following release from both arrests at times corresponding with maximal HM site phosphorylation and Ace2 target transcription (60 min post-nocodazole release and 45 min post-cdc15-2 release) (Fig. 8C). We did not observe any significant changes in Cbk1's enzymatic activity between immunoprecipitations from arrested and postrelease cells. Overall, these findings indicate that mitotic CDK phosphorylations do not block Cbk1's ability to activate Ace2 by directly inhibiting the kinase's enzymatic activity, but they may prevent the proteins from interacting productively in vivo.
DISCUSSION
The transition from mitosis to G1 involves a massive reorganization of cell architecture in which diverse processes are precisely coordinated in space and time. Our analysis of the final event of budding yeast cell division, in which Ace2 drives M/G1 production of enzymes that destroy the septum, addresses mechanisms by which late cell cycle events are linked to mitotic progress. Intriguingly, destruction of the septum occurs minutes after it is built. Thus, this degradative process must be precisely coordinated with cytokinesis, since premature action of these enzymes would make it impossible to construct a functional septum.
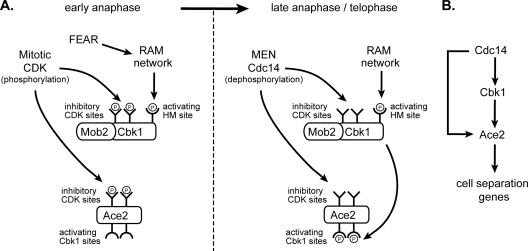
Our analysis provides a link between activation of the RAM network kinase Cbk1 and the cell cycle-regulated pathways FEAR and MEN, active in late mitosis. Our data support a model in which Cbk1's ability to regulate Ace2 is turned on during mitotic exit in two phases: at the initial phosphorylation of Cbk1's HM site and at subsequent activation or removal of inhibition. Jansen et al. previously demonstrated that HM site phosphorylation is essential for Cbk1 function and that it fluctuates over the course of cell division, with a peak in mitosis (29). We found that this occurs after the metaphase/anaphase transition and requires activation of the FEAR pathway but does not require the later-acting MEN. As diagrammed in Fig. 9, these data suggest that early and transient release of Cdc14 initiated by the FEAR pathway activates the RAM network, which promotes phosphorylation of Cbk1's HM site.
FIG. 9.
Regulation of Cbk1 and Ace2 in mitosis integrates signals from FEAR and MEN to ensure the proper timing of septum destruction. (A) In early anaphase, Mob2/Cbk1 and Ace2 are inhibited by mitotic CDK phosphorylation. Activation of the FEAR pathway triggers activation of the RAM network, leading to phosphorylation of Cbk1 at the HM site. Upon MEN activation in late anaphase/telophase, Cdc14 is released to the cytoplasm, where it dephosphorylates and activates Cbk1 and removes inhibitory phosphates on Ace2. Active Cbk1 phosphorylates the NES of Ace2, trapping Ace2 in the nucleus, where it drives transcription of the enzymes required for septum degradation. (B) Simultaneous activation of Cbk1 and Ace2 by Cdc14 constitutes a coherent feed-forward loop to activate the transcription of cell separation enzymes. Dephosphorylation of both Cbk1 and Ace2 by Cdc14 and activation of Ace2 by Cbk1 create a regulatory organization that may delay the output of Ace2 target gene activation.
How might the early anaphase release of Cdc14 promote phosphorylation of Cbk1's HM site? In addition to the Mob2 protein, which is constitutively bound to Cbk1, HM site phosphorylation requires all four upstream components of the RAM network: Tao3, Hym1, Sog2, and Kic1 (29). This system is related to the widely conserved furry/tricornered and hippo/wts/sav pathways of metazoans, in which hippo/MST-like kinases are primarily responsible for phosphorylating the HM site of an Ndr/LATS kinase (12, 20, 60, 65). We hypothesize that dephosphorylation by Cdc14 downstream of FEAR activates the upstream components of the RAM network, possibly by activating the kinase Kic1 (Fig. 9A). Intriguingly, this hippo-like kinase contains several potential CDK sites that are phosphorylated in vivo (1, 9, 58), which may provide an avenue for regulation by Cdc14. Our findings do not, however, exclude the possibility that a different Cdc14-regulated kinase phosphorylates Cbk1's HM site.
We and others have shown that the in vivo function of Cbk1 and closely related kinases requires phosphorylation at the HM site (29, 45, 71), consistent with the broad importance of this regulatory mechanism among AGC kinases. However, the role this phosphorylation plays in the cell is unclear. In prior work, our research group found that mutation of the HM phosphorylation site to alanine abolished Cbk1's function and led to constitutive hyperphosphorylation of other sites but did not appreciably decrease enzymatic activity of the immunoprecipitated protein. In contrast, mutations that greatly decreased the kinase's activity in vitro had relatively modest effects on its function (29). Other groups have similarly found that such mutations have distinct in vivo effects (60, 71). Thus, we previously suggested that HM site phosphorylation does not increase Cbk1's enzymatic activity, but rather promotes assembly of functional signaling complexes or recruits a phosphatase that reverses other inhibitory phosphorylations (29).
Our results here show that this view is partly incorrect. Cbk1-T743E, in which the HM site phospho-acceptor residue is changed to glutamic acid, was much more active in vitro than the wild-type enzyme, indicating that HM site phosphorylation increases Cbk1's kinase activity. We found that only about 3% of Cbk1 is phosphorylated at the HM site in vivo when this modification peaks in late mitosis. Thus, prior analysis of the enzymatic activity of immunoprecipitated total protein likely missed effects caused by the modification. In fact, it remains possible that true phosphorylation of the HM site would increase Cbk1's enzymatic activity more than glutamic acid substitution at the site. However, since cbk1 alleles that dramatically compromise the kinase's activity exhibit much milder effects than cbk1-T743A, it remains likely that modulation of enzymatic activity is not the sole function of HM site phosphorylation. We propose that a key function of HM site phosphorylation, in addition to elevating Cbk1 activity, may be to recruit the phosphatase Cdc14 and/or direct the formation of a functional complex with Ace2 and other substrates.
It is remarkable that a very small pool of Cbk1 is phosphorylated at the HM site. We propose that this reflects highly dynamic modification of the site in vivo, possibly indicating a mechanism by which the kinase is locally controlled. Upstream RAM components, including Kic1, are localized to the daughter cell cortex and bud neck (48) and thus may maintain HM site phosphorylation at these sites. The Cbk1 which diffuses away from these sites may be readily dephosphorylated by cytoplasmic phosphatases. Furthermore, Cbk1 HM site phosphorylation is dramatically reduced in cells lacking the downstream target Ace2 (29), suggesting that the kinase's interaction with its substrates protects HM site phosphorylation. Maintaining a localized pool of activated Cbk1 may restrict the regulation of targets locally and may prevent Cbk1 which diffuses from the cortex and bud neck from targeting substrates inappropriately.
Our results show that HM site phosphorylation is not sufficient for Cbk1 to activate Ace2 in mitosis. If the kinase's ability to regulate Ace2 were solely modulated by phosphorylation of its HM site, then Ace2-AAA should be robustly localized to the daughter nucleus and activate its transcriptional targets during a MEN arrest, in particular in cells expressing the CBK1-T743E gain-of-function allele. However, this was not the case: Ace2-AAA was only faintly nuclear at such an arrest, and Ace2 target genes were not expressed in cells arrested prior to MEN-driven Cdc14 release. Thus, we propose that additional control of Cbk1 occurs in response to mitotic exit, which allows the kinase to phosphorylate Ace2 and activate the gene expression program that cuts the connection between mother and daughter cells.
While the molecular nature of this final MEN control of Cbk1 remains unknown, we hypothesize that CDK phosphorylation keeps the Cbk1/Mob2 complex from acting on Ace2 in metaphase and early anaphase (Fig. 9A). Sustained release of Cdc14 triggered by the MEN might then remove this inhibition, unleashing the kinase to act on the transcription factor in vivo. In this view, Cdc14 made available by the FEAR pathway is insufficient to relieve this inhibition, while subsequent sustained release of the phosphatase by MEN activation permits the kinase to act on the transcription factor in vivo.
How might Cdc14 promote Cbk1's ability to regulate Ace2 as cells pass through mitotic exit? Cbk1 is highly phosphorylated in mitosis (29). We found that mutation of potential CDK phosphorylation sites eliminated the majority of this phosphorylation and that these sites may be inhibitory. Furthermore, some of this phosphorylation can be removed by in vitro treatment with Cdc14, indicating phosphorylation by proline-directed kinases. Recent results by Holt et al. have identified Cbk1 as a target of Cdc28/Cdk1 (27), and at least one (S/T)-P site on Cbk1 has been identified as a phosphorylation site in a high-throughput mass spectrometry study (1). Although we have been unable to prematurely activate Ace2 at the MEN arrest with the cbk1-9A allele (data not shown), the constitutively Cbk1-bound coactivator Mob2 also contains putative CDK consensus motifs, some of which are phosphorylated in vivo (9, 27). Therefore, it is possible that such CDK phosphorylation of the Cbk1/Mob2 complex blocks its assembly into a productive association with Ace2 in vivo, and only upon mitotic exit and Cdc14 activation is the productive complex formed.
In the overall logic of the system, Cdc14 releases both Cbk1 and Ace2 from inhibition by mitotic CDK. Simplification of the system to these three components, this pathway forms a coherent feed-forward loop (Fig. 9B). What function might this regulatory organization serve in the overall orchestration of mitotic events? Such network motifs, which are common in biological systems, can act as sign-sensitive delay elements (40, 41). This might be important for the appropriate timing of Ace2 target gene expression. Differences between the behavior of Ace2 and the closely related transcription factor Swi5 indicate that Ace2's regulatory system does in fact generate delayed output. In mitotically synchronized cells, Swi5 accumulates in nuclei significantly before Ace2 (55). Like Ace2, Swi5 is blocked from entering nuclei in mitosis by CDK phosphorylation (47), and Cdc14 reverses this (67). Unlike Ace2, Swi5 appears to require no additional regulatory input to robustly accumulate in nuclei and promote transcription of its target genes. Thus, this regulatory system may delay Ace2 activity until cytokinesis is well under way. Intriguingly, Cbk1 may participate in another feed-forward loop by inhibiting a protein that blocks translation of transcripts from Ace2-regulated genes (30), creating an interlocking regulatory system that controls expression of proteins that cut the connection between mother and daughter cells.
How is Cbk1's ability to activate Ace2 restricted to the daughter cell? This remains unclear. Our findings indicate that Cbk1 must both be phosphorylated at the HM site and have inhibitory phosphorylations removed to act on Ace2 (Fig. 9). We propose that Cbk1 is phosphorylated at the HM site by upstream RAM components localized at the daughter cell cortex, specifically, the hippo/MST-like kinase Kic1. However, Ace2 localizes normally in Cbk1-T743E cells. Thus, this asymmetry must involve a separate mechanism that restricts this Cbk1 function to the daughter cell, perhaps acting in parallel to regulation of HM site phosphorylation during the M/G1 transition. We speculate that Cbk1 dephosphorylation by Cdc14 may be restricted to the daughter cell. While MEN-dependent release of Cdc14 to the cytoplasm is not daughter cell specific, it is not clear if the phosphatase's activity toward its substrates is spatially uniform. Furthermore, a small pool of Cdc14 is found at the bud neck (7). In a variant of a model proposed by Colman-Lerner et al. (16), this fraction of Cdc14 may act asymmetrically to release Cbk1 from mitotic inhibition.
Our results are notably consistent with recent work of Clemente-Blanco et al., who showed that Candida albicans Cdc14 is required for Ace2-driven transcription of genes involved in cell separation (14). As in budding yeast, the RAM network is required for Ace2 function in C. albicans (44, 59), and regulatory sites by which S. cerevisiae Cbk1 controls Ace2 are preserved in the C. albicans Ace2 ortholog (data not shown). While it is not clear if RAM network function is downstream of Cdc14 in C. albicans, the high degree of conservation of the system's components suggest that a similar regulatory organization might exist in this diverged yeast. Intriguingly, Cdc14 is not universally essential for exit from mitosis: while C. albicans Cdc14 clearly regulates Ace2, it is dispensable for mitotic exit. Thus, maintenance of this phosphatase's function in control of Ace2 and the RAM network may be under greater selective pressure than its role in initiation of cytokinesis and other mitotic exit phenomena.
Acknowledgments
Constructs for production of Cdc14 were a gift of David Morgan, who along with Angelika Amon generously provided important strains. We thank the members of the Weiss lab, especially Brian Yeh, for helpful discussions, and Jason Brickner and Rick Gaber for critical analysis of experiments and text.
This work was supported by NIH NRSA (5F32GM082011-03) to J.B. and NIH RO1 (5R01GM084223-02) to E.L.W. and by an ACS Research Scholar Grant to E.L.W. (GMC-111853).
Footnotes
Published ahead of print on 6 December 2010.
REFERENCES
- 1.Albuquerque, C. P., et al. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics 7:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambault, V., et al. 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14:699-711. [DOI] [PubMed] [Google Scholar]
- 3.Asakawa, K., S. Yoshida, F. Otake, and A. Toh-e. 2001. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics 157:1437-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzam, R., et al. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305:516-519. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian, M. K., et al. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 149:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardin, A. J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102:21-31. [DOI] [PubMed] [Google Scholar]
- 7.Bembenek, J., et al. 2005. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle 4:961-971. [DOI] [PubMed] [Google Scholar]
- 8.Bidlingmaier, S., E. L. Weiss, C. Seidel, D. G. Drubin, and M. Snyder. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodenmiller, B., et al. 2007. PhosphoPep: a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 3:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourens, M., et al. 2008. Mutations in a small region of the exportin Crm1p disrupt the daughter cell-specific nuclear localization of the transcription factor Ace2p in Saccharomyces cerevisiae. Biol. Cell 100:343-354. [DOI] [PubMed] [Google Scholar]
- 11.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 12.Chan, E. H., et al. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24:2076-2086. [DOI] [PubMed] [Google Scholar]
- 13.Chang, E. J., R. Begum, B. T. Chait, and T. Gaasterland. 2007. Prediction of cyclin-dependent kinase phosphorylation substrates. PLoS One 2:e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemente-Blanco, A., et al. 2006. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J. Cell Sci. 119:1130-1143. [DOI] [PubMed] [Google Scholar]
- 15.Collart, M. A., and S. Oliviero. 2001. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. Chapter 13:Unit13.12. [DOI] [PubMed] [Google Scholar]
- 16.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 17.D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117:455-469. [DOI] [PubMed] [Google Scholar]
- 18.Dohrmann, P. R., W. P. Voth, and D. J. Stillman. 1996. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol. Cell. Biol. 16:1746-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40:422-432. [DOI] [PubMed] [Google Scholar]
- 20.Emoto, K., J. Z. Parrish, L. Y. Jan, and Y. N. Jan. 2006. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443:210-213. [DOI] [PubMed] [Google Scholar]
- 21.Gray, C. H., V. M. Good, N. K. Tonks, and D. Barford. 2003. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 22:3524-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwell, L. H., J. Culotti, J. R. Pringle, and B. J. Reid. 1974. Genetic control of the cell division cycle in yeast. Science 183:46-51. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-634. [DOI] [PubMed] [Google Scholar]
- 24.Hergovich, A., M. R. Stegert, D. Schmitz, and B. A. Hemmings. 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7:253-264. [DOI] [PubMed] [Google Scholar]
- 25.Hofken, T., and E. Schiebel. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt, L. J., J. E. Hutti, L. C. Cantley, and D. O. Morgan. 2007. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell 25:689-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt, L. J., et al. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325:1682-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen, G., C. Wu, B. Schade, D. Y. Thomas, and M. Whiteway. 2005. Drag & Drop cloning in yeast. Gene 344:43-51. [DOI] [PubMed] [Google Scholar]
- 29.Jansen, J. M., M. F. Barry, C. K. Yoo, and E. L. Weiss. 2006. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J. Cell Biol. 175:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen, J. M., A. G. Wanless, C. W. Seidel, and E. L. Weiss. 2009. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr. Biol. 19:2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg, and D. O. Morgan. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9:2803-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaspersen, S. L., and D. O. Morgan. 2000. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10:615-618. [DOI] [PubMed] [Google Scholar]
- 33.Jin, F., D. Richmond, and Y. Wang. 2009. The multilayer regulation of the metaphase-to-anaphase transition. Cell Cycle 8:700-704. [DOI] [PubMed] [Google Scholar]
- 34.Johnston, G. C., J. R. Pringle, and L. H. Hartwell. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79-98. [DOI] [PubMed] [Google Scholar]
- 35.Konig, C., H. Maekawa, and E. Schiebel. 2010. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J. Cell Biol. 188:351-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 37.Liang, F., F. Jin, H. Liu, and Y. Wang. 2009. The molecular function of the yeast Polo-like kinase Cdc5 in Cdc14 release during early anaphase. Mol. Biol. Cell 20:3671-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Mah, A. S., J. Jang, and R. J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. U. S. A. 98:7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U. S. A. 100:11980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197-204. [DOI] [PubMed] [Google Scholar]
- 42.Mazanka, E., et al. 2008. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 6:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazanka, E., and E. L. Weiss. 2010. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol. Biol. Cell 21:2809-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNemar, M. D., and W. A. Fonzi. 2002. Conserved serine/threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 184:2058-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millward, T. A., D. Hess, and B. A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847-33850. [DOI] [PubMed] [Google Scholar]
- 46.Mohl, D. A., M. J. Huddleston, T. S. Collingwood, R. S. Annan, and R. J. Deshaies. 2009. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 184:527-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moll, T., G. Tebb, U. Surana, H. Robitsch, and K. Nasmyth. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66:743-758. [DOI] [PubMed] [Google Scholar]
- 48.Nelson, B., et al. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishihama, R., et al. 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185:995-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panozzo, C., M. Bourens, A. Nowacka, and C. J. Herbert. 2010. Mutations in the C-terminus of the conserved NDR kinase, Cbk1p of Saccharomyces cerevisiae, make the protein independent of upstream activators. Mol. Genet. Genomics 283:111-122. [DOI] [PubMed] [Google Scholar]
- 52.Pearce, L. R., D. Komander, and D. R. Alessi. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9-22. [DOI] [PubMed] [Google Scholar]
- 53.Pereira, G., T. Hofken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 6:1-10. [PubMed] [Google Scholar]
- 54.Queralt, E., C. Lehane, B. Novak, and F. Uhlmann. 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125:719-732. [DOI] [PubMed] [Google Scholar]
- 55.Sbia, M., et al. 2008. Regulation of the yeast Ace2 transcription factor during the cell cycle. J. Biol. Chem. 283:11135-11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shou, W., et al. 2002. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol. Biol. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shou, W., et al. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 58.Smolka, M. B., C. P. Albuquerque, S. H. Chen, and H. Zhou. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U. S. A. 104:10364-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song, Y., et al. 2008. Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol. Biol. Cell 19:5456-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stegert, M. R., A. Hergovich, R. Tamaskovic, S. J. Bichsel, and B. A. Hemmings. 2005. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 25:11019-11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203-232. [DOI] [PubMed] [Google Scholar]
- 62.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220. [DOI] [PubMed] [Google Scholar]
- 63.Traverso, E. E., et al. 2001. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 276:21924-21931. [DOI] [PubMed] [Google Scholar]
- 64.Vallen, E. A., J. Caviston, and E. Bi. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11:593-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vichalkovski, A., et al. 2008. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr. Biol. 18:1889-1895. [DOI] [PubMed] [Google Scholar]
- 66.Visintin, R., and A. Amon. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12:2961-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visintin, R., et al. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 68.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 69.Visintin, R., F. Stegmeier, and A. Amon. 2003. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell 14:4486-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss, E. L., et al. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziv, C., et al. 2009. Cell elongation and branching are regulated by differential phosphorylation states of the nuclear Dbf2-related kinase COT1 in Neurospora crassa. Mol. Microbiol. 74:974-989. [DOI] [PubMed] [Google Scholar]