Abstract
To help uncover the mechanisms underlying the staggered expression of cartilage-specific genes in the growth plate, we dissected the transcriptional mechanisms driving expression of the matrilin-1 gene (Matn1). We show that a unique assembly of evolutionarily conserved cis-acting elements in the Matn1 proximal promoter restricts expression to the proliferative and prehypertrophic zones of the growth plate. These elements functionally interact with distal elements and likewise are capable of restricting the domain of activity of a pancartilaginous Col2a1 enhancer. The proximal elements include a Pe1 element binding the chondrogenic L-Sox5, Sox6, and Sox9 proteins, a SI element binding Nfi proteins, and an initiator Ine element binding the Sox trio and other factors. Sox9 binding to Pe1 is indispensable for functional interaction with the distal promoter. Binding of L-Sox5/Sox6 to Ine and Nfib to SI modulates Sox9 transactivation in a protein dose-dependent manner, possibly to enhance Sox9 activity in early stages of chondrogenesis and repress it at later stages. Hence, our data suggest a novel model whereby Sox and Nfi proteins bind to conserved Matn1 proximal elements and functionally interact with each other to finely tune gene expression in specific zones of the cartilage growth plate.
Sox proteins play critical roles in lineage specification during development (18, 21, 25). They have an Sry-related high-mobility-group (HMG) box domain, which binds the minor groove of DNA with low affinity. They may act as architectural proteins to organize transcriptional complexes (25). Three Sox proteins direct chondrocyte specification and differentiation, but it is still unclear how they orchestrate the sequential induction of cartilage-specific genes in developing endochondral bones.
Endochondral bones form through tightly intertwined morphogenetic and differentiation events (11, 20, 24, 37). First, mesenchymal cells condense, commit to the chondrocyte lineage, and undergo chondrocyte early differentiation to form cartilage primordia of future bones. They then sequentially differentiate into proliferating, prehypertrophic, hypertrophic, and terminal cells and ultimately die to allow replacement of cartilage by bone. Importantly, the multiple layers of cells that comprise cartilage primordia proceed through the multiple steps of differentiation in a staggered manner. They thereby establish growth plates (GP), i.e., a series of adjacent tissue zones comprising cells at progressively more advanced stages of maturation. The process is tightly regulated both spatially and temporally to allow GP to continue to grow in one end and to be progressively replaced by bone in the other end throughout fetal and postnatal growth (24). Bone growth is determined by the number of cells proliferating in the columnar zone and progressing toward hypertrophy. It involves complex functional interactions between fibroblast growth factor (FGF), Ihh, parathyroid hormone-related protein (PTHrP), and other factors and signaling pathways that allow chondrocytes to constantly modify their gene expression profile (11, 20, 37). Mutations in these factors and pathways cause severe forms of dwarfism and skeletal malformation diseases (20, 33). Elucidating the transcriptional mechanisms involved in specifying gene expression in specific GP zones has thus special importance to allow development of suitable therapies for such diseases.
The composition of the cartilage extracellular matrix (ECM) progressively changes from one GP zone to the next. This is largely due to staggered expression of the genes encoding the specific components of this matrix (8, 24, 39). Col2a1 (collagen-2 gene) is activated as soon as prechondrocytes differentiate, whereas Agc1 (aggrecan gene) and most other cartilage ECM genes are turned on in early chondroblasts (24). In contrast, Matn1 (matrilin-1 gene) exhibits a narrower spatiotemporal activity (30, 31, 39, 42). It has the unique feature of being expressed exclusively in the overtly differentiated chondroblasts of the columnar and prehypertrophic GP zones (4, 5, 19). Chondrocytes turn all these genes off as they undergo hypertrophy and then activate Col10a1 (collagen-10 gene).
Sox9, L-Sox5, and Sox6 form a trio of transcription factors that are both required and sufficient to induce chondrogenesis (2, 7, 14, 38). Their main functions are to bind and thereby directly induce activation of Col2a1, Agc1, and several other cartilage ECM genes (1, 13, 23-26). Sox9 features a family-specific HMG box DNA-binding domain and a homodimerization domain, which mediate its binding to pairs of inverted Sox motifs (13, 26). It also features a potent transactivation domain. L-Sox5 and Sox6 are highly related to each other but only distantly related to Sox9 through their HMG box domain. They feature a dimerization domain, distinct from that of Sox9, and lack a transactivation domain. They bind more-variable Sox motifs on cartilage-specific enhancers and cooperate with Sox9 in transactivation by increasing the efficiency of Sox9 binding to its own sites on DNA (13). It remains unknown, however, whether and how the activity of this Sox trio is controlled to confer markedly different expression patterns on the various cartilage ECM genes. We used here the Matn1 promoter as a model to reach deeper insight into gene regulation orchestrated by the Sox trio.
Matrilin-1 (also called cartilage matrix protein [CMP]) belongs to a family of multidomain adaptor proteins (10, 22, 44). It facilitates assembly of the cartilage ECM by forming collagen-dependent and -independent filaments and interacting with aggrecan. It also forms complexes with biglycan, and decorin, linking collagen-6 microfibrils to aggrecan and collagen-2 (45). We previously showed that the Matn1 promoter features several blocks of sequences highly conserved in amniotes (34). A 334-bp short promoter is insufficient to direct reporter gene activity in cartilage in transgenic mice (34) but can be activated at a low level in the Matn1-specific GP zones upon addition of an intronic enhancer (19). Stronger activity is obtained by using a 2-kb promoter with or without the intronic enhancer (19). These data suggest that the proximal promoter may contain the cis-acting elements driving Matn1 expression in the growth plate but requires distal and intronic enhancers to be activated. This short promoter features highly conserved promoter element 1 (Pe1), recognized by the Sox trio, and two silencer elements (SI and SII) binding Nfi proteins (34, 41).
Here we demonstrate that the short promoter has a central role in conferring on Matn1 its restricted spatiotemporal expression pattern. We show the respective roles of the Sox-binding sites in the Pe1 and the Ine elements and Nfi-binding sites in the SI element. We show that L-Sox5/Sox6 and Nfi differentially modulate promoter activation by Sox9, according to the relative levels of the proteins. Our data thereby provide new insights into the transcriptional mechanisms that underlie staggered gene expression in the cartilage GP.
MATERIALS AND METHODS
Cell culture.
Chicken embryo chondroblasts (CEC) and fibroblasts (CEF) and mesenchymes were prepared and cultured as described previously (41). Low-density mesenchyme (LDM) and high-density mesenchyme (HDM) cultures were made similarly, by plating 1 × 106 cells and 5 × 106 cells, respectively, in 35-mm plates containing F-12 medium-Dulbecco's modified Eagle medium (DMEM) (1:1; HyClone Laboratories) supplemented with 10% fetal bovine serum (FBS; Sigma and Gibco Laboratories). COS-7 cells were cultured under standard conditions. HDM cultures consisting of early proliferative (stage Ia) chondroblasts and CEC cultures rich in late proliferative (stage Ib) chondroblasts express Matn1 at low and high levels, respectively (31, 41, 42). LDM, CEF, and COS-7 cultures served as Matn1-nonexpressing controls.
Oligonucleotides and plasmid constructions.
Nucleotide sequences for wild-type and mutant versions of Pe1 and SI and consensus HMG and SOX9 competitors were described previously (34, 41). Sequences of oligonucleotides for wild-type Ine, Ine derivatives, and mutant versions of Ine are depicted in Fig. 3B.
All positions are given in bp from the first T of the chicken Matn1 TATA motif. The TR70 (−2011/+67) and NAD1 (−334/+67) Matn1-LacZ constructs were reported previously (19, 34). PS-NAD1 was produced by inserting the −2011/−948 Matn1 sequence upstream of NAD1. Eight tandem copies of a 48-bp Col2a1 enhancer element (ECol2a1) were inserted upstream of NAD1 to obtain 8×ECol2a1-NAD1. ΔIneM1-TR70 was made by replacing the NAD1 promoter with the −2011/+67 fragment of ΔIneM1-AC8Luc (see below).
Luciferase reporters FO15Luc and AC8Luc, driven by the short and long Matn1 promoters, respectively, as well as ΔPe1M1-FO15Luc and ΔPe1M4-FO15Luc, carrying point mutations in the Sox motif and spacer of Pe1, respectively, were described (34). To produce 8×ECol2a1-FO15Luc, eight copies of ECol2a1 were inserted upstream of FO15Luc. Mutations were introduced into Pe1, Ine, and SI elements of reporters by PCR-based QuikChange site-directed mutagenesis (Stratagene) using oligonucleotides carrying the desired mutations. All constructs were verified by restriction enzyme analysis and DNA sequencing.
Generation and histological analysis of transgenic mice.
All animal experiments were conducted according to the ethical standards of the Animal Health Care and Control Institute, Csongrád County, Hungary. C57BL/6, CBA, CD-1, and FVB mice were obtained form Charles River Laboratories, Hungary. Transgenic mice were generated essentially as described previously (19). On embryonic day 15.5 (E15.5), foster mothers were sacrificed by cervical dislocation and the transgenes were detected by PCR in founder (G0) embryos. These embryos were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and photographed as whole mounts with a Leica MZFLIII stereomicroscope equipped with a DC300F camera. Cryosections were counterstained with 0.5% eosin and analyzed using a Nikon Eclipse E600 microscope equipped with Spot RT Slider camera as described previously (19). Figures were made with Adobe Photoshop 8.0 and CorelDraw X4 software.
EMSA and supershift experiments.
Full-length cDNAs for L-Sox5 and Sox6 were inserted in frame into the pGEX expression vector. Glutathione S-transferase (GST)-tagged L-Sox5, Sox6, and SOX9 were expressed and purified, and crude cell extracts were made as described previously (34). Twenty to 30 fmol end-labeled DNA probes was incubated with 1 to 2.5 μg GST-SOX9 or GST-L-Sox5 or 3 μg crude CEC or CEF cell extracts in the presence of 100 to 500 ng poly(dG-dC)·(dG-dC) and separated on prerun 5% or 6.6% PAGE gel. In competition electrophoretic mobility shift assays (EMSA), 50- and 500-fold molar excesses of cold competitors were added. Supershift experiments were performed as described previously (34) using Sox9, L-Sox5, and Sox6 antisera (26).
In vivo footprinting.
CEC and CEF cells and HDM cultures were treated with dimethylsulfate (DMS) or irradiated with UV light and subjected to genomic footprinting as described previously (34). Briefly, 30 μg of in vivo- and in vitro-treated DNA samples cleaved with piperidine were amplified by ligation-mediated PCR (LM-PCR) (32) between −227 and +140 using linker primers LP11 and LP25 and gene-specific nested primers PU1 and PU2 (upper strand), PL1 and PL2 (lower strand), and PU3 or PL3 (hybridization probe) (34) (see Fig. 4A).
Transient expression assay.
CEC, CEF, and COS-7 cells were transfected with the Ca-phosphate coprecipitation method 4 to 6 h after plating (34, 41). HDM and LDM cultures were transfected similarly, but 24 h after plating. Briefly, 2 μg (CEC and CEF) or 5 μg (HDM, LDM, and COS-7) reporters was added with 0.5 μg pRL-TK vector (Promega) as an internal control to correct for transfection efficiency. Control plates were transfected with FO15Luc. Firefly and Renilla luciferase activities were measured using Luminoscan Ascent (ThermoLabsystem 2.6) and luciferase assay systems (Promega) 72 h (HDM and LDM) or 48 h (other cells) posttransfection.
Unless indicated otherwise, cotransfections were performed with 2 μg or 5 μg AC8Luc and increasing amounts (50 to 250 ng) of effector plasmids pcDNA5′UT-FLAG-L-Sox5 (pFSox5) and pcDNA5′UT-FLAG-Sox6 (pFSox6) (26) without or with 250 ng pCDMA-SOX9 (pSOX9) (26). In a typical experiment, 250 ng pSOX9 and 125 ng each of pFSox5 and pFSox6 effector plasmids were used. Other experiments were performed with 0 to 300 ng effector plasmids expressing human CTF-1 (pCTF-1) (36) or mouse Nfia, Nfib, Nfic, and Nfix (pNfia, pNfib, pNfic, and pNfix), homologous to chicken Nfia1.1, Nfib2, Nfic2, and human NFIX2, respectively (9). Transfection mixtures were adjusted with empty vectors to the same amount of total DNA. Luciferase activities were expressed as fold values relative to that for FO15Luc, taken as 1, unless noted otherwise. Transfections were performed in duplicate or triplicate and repeated 3 to 10 times with at least two different DNA preparations. Results are presented as means ± standard errors of the means (SEM).
Combined forced expression and Western analysis.
To estimate the relative expression levels of Sox and Nfi proteins, we used pcDNA5′UT-FLAG-SOX9 (pFSOX9) (26) and we made pFNfib by inserting fragments of Nfi expression plasmids (9) into pcDNA5′UT-FLAG. COS-7 cells were cotransfected as described above with AC8Luc, 1 μg pFSOX9, and increasing amounts of effector plasmids pFSox5 and pFSox6 or pFNfib. Transfected cells were lysed in 100 μl buffer containing 14 mM HEPES (pH 7.9), 1.5 mM MgCl2, 6 mM KCl, 0.44 mM NaCl, 0.08 mM EDTA, 2.3 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, and a protease inhibitor cocktail (Sigma-Aldrich; P2714). Supernatants were used to measure luciferase activities and in Western blots with rabbit anti-FLAG (Sigma) antisera.
Statistical analysis was carried out using one-way analysis of variance (ANOVA) with KyPlot version 2.0 beta 15.
QRT-PCR.
Total RNA was isolated from cultured cells using an RNA isolation kit (Macherey-Nagel). Quantitative real-time PCR (QRT-PCR) was performed on a RotorGene 3000 instrument (Corbett Research) with gene-specific primers and the SYBR green protocol (16). Briefly, 2 μg of DNase-treated RNA was reverse transcribed using the High-Capacity cDNA Archive kit (Applied Biosystems). Reactions were done with FastStart SYBR green Master mix (Roche Applied Science) at a primer concentration of 250 nM as follows: 15 s at 95°C and 45 cycles of 95°C for 15 s, 60°C for 25 s, and 72°C for 25 s. The quality of the reaction was checked by melting temperature analysis. Individual threshold cycle (CT) values were normalized to the average CT values of three internal control genes (glyceraldehyde 3-phosphate dehydrogenase [GAPDH], 18S rRNA, and 28S rRNA genes). The final relative gene expression ratios were calculated as ΔΔCT values (comparison of the normalized ratios). Gene-specific primer sequences are shown in Table S1 at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf.
Genome sequence analysis.
The MAF format 44-way vertebrate multiple alignment file of human chromosome 1 was downloaded from the University of California, Santa Cruz (UCSC), website (http://hgdownload.cse.ucsc.edu/downloads.html). The corresponding Matn1 promoter region was extracted from this file using the MAF2FASTA program of the MULTIZ package (http://www.bx.psu.edu/miller_lab/) using the corresponding human positions. The Ine and Pe1 region was extracted and further refined manually. The 75% majority rule consensus and the sequence logo were generated with the Geneious Pro program.
RESULTS
Proximal elements restrict spatiotemporal activation of the Matn1 promoter by homologous and heterologous enhancers.
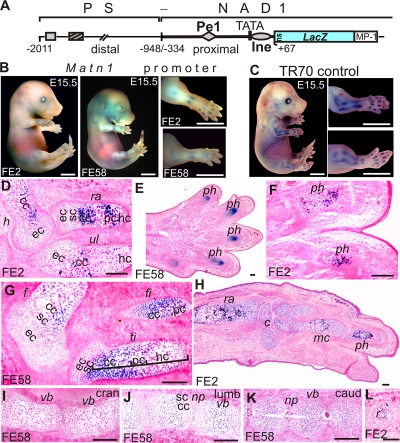
We previously reported that a LacZ transgene driven by a Matn1 short promoter (−334/+67; NAD1) exhibited low activity in mouse embryos (34). The addition of the Matn1 −2011/−948 sequence (PS) markedly increased transgene activity (Fig. 1 A and B), but this activity remained lower than that of a transgene harboring the full-length (−2011/+67; TR70) promoter (19) (Fig. 1C). Histological analysis revealed that the transgene activity also increased proximodistally in the limbs and craniocaudally in the vertebral bodies, as with TR70 (Fig. 1D to L). This activity was restricted to columnar and prehypertrophic chondrocytes. The −2011/−1134 region increased the short promoter activity less markedly than the −2011/−948 region but showed similar zonal and proximodistal specificity (see Fig. S1 at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf). We therefore concluded that elements in the −2011 to −334 region enhance the activity but not the tissue specificity of the Matn1 promoter.
FIG. 1.
Upstream elements increase the activity of the Matn1 proximal promoter in vivo. (A) Schematic of PS-NAD1 depicting conserved DNA blocks (rectangles, diamond, and oval) in the distal and proximal Matn1 promoter regions. (B and C) Expression of PS-NAD1 (B) and TR70 (C; 2-kb Matn1 promoter) transgenes in founder embryos (FE) stained with X-Gal at E15.5. (D to L) Histological analysis of PS-NAD1 embryo cryosections. In the developing limbs, X-Gal staining increases proximodistally from humerus (h), ulna (ul), and femur (f) to radius (ra), tibia (ti), and fibula (fi) and further from carpals (c) and metacarpals (mc) to phalanges (ph) (D to H). Staining is absent in cranial (cran) vertebral bodies (vb) but increases from lumbar (lumb) to caudal (caud) regions (I to K). LacZ activity is restricted to the GP zones of columnar chondroblasts (cc) and prehypertrophic chondrocytes (pc), while it is low or absent in the zones of epiphyseal (ec) and source chondroblasts (sc) and hypertrophic chondrocytes (hc) (D, G, and J to L). np, nucleus pulposus; r, rib. Bars, 2 mm (B and C) and 200 μm (D to L).
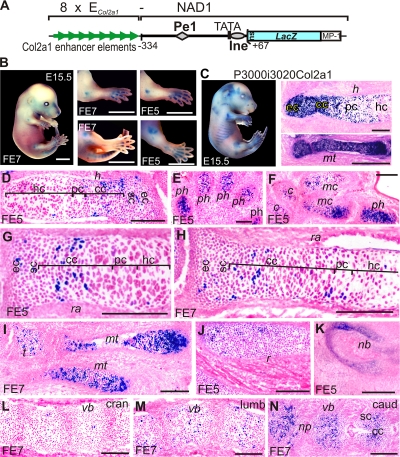
Next we tested activation of the Matn1 short promoter by 8 tandem copies of the pancartilaginous Col2a1 minimal enhancer (8×ECol2a1-NAD1). Interestingly, 8×ECol2a1-NAD1 dramatically differed in its expression pattern from p3000i3020Col2a1, which contains the Col2a1 promoter and enhancer (47) (Fig. 2 B and C). It was expressed exclusively in distal structures, like digits, caudal vertebral bodies, and nasal cartilage and only in columnar and prehypertrophic GP zones (Fig. 2D to N). Its sporadic or weak activity in epiphyseal and source chondroblasts and lack of activity in condensed mesenchymal cells, perichondrium cells, and prechondrocytes differed markedly from the high activity of the Col2a1 transgene in these cells (47, 48). Thus, the Matn1 short promoter inhibited the Col2a1 enhancer in proximal structures and at early chondrocyte differentiation stages. Even 16 copies of ECol2a1 neither increased the transgene activity nor altered its restricted spatiotemporal expression (data not shown).
FIG. 2.
The short promoter restricts the Col2a1 enhancer activity in vivo. Schematic (A) and expression (B) of the 8×ECol2a1-NAD1 transgene in comparison with the pattern of P3000i3020Col2a1 (C) driven by the Col2a1 promoter and enhancer (47). (D to N) Histological analysis of 8×ECol2a1-NAD1 cryosections. In the developing limbs (D to I), X-Gal staining is relatively weak in the humerus and radius but sharply increases toward the distal phalanges. LacZ activity is completely repressed or limited to a few cells in the cranial and lumbar vertebral bodies but is high in the caudal ones and in the distal part of nasal bone (nb) (K to N). Expression is highest, limited to groups of cells, in the columnar and prehypertrophic zones, but it is strongly reduced in the source and epiphyseal chondroblasts, except for sporadic staining in the distal epiphysis of the humerus (D to N). t, tarsal; mt, metatarsal. Other abbreviations are as defined for Fig. 1. Bars, 2 mm (B) and 200 μm (D to N).
To sum up, the Matn1 short promoter plays a critical role in restricting cartilage-specific expression, and its activity is enhanced by distal elements. It is even capable of restricting the activity of a powerful Sox-driven panchondrocytic Col2a1 heterologous enhancer to distal structures and specific GP zones.
The Sox trio binds to the initiator element in vitro.
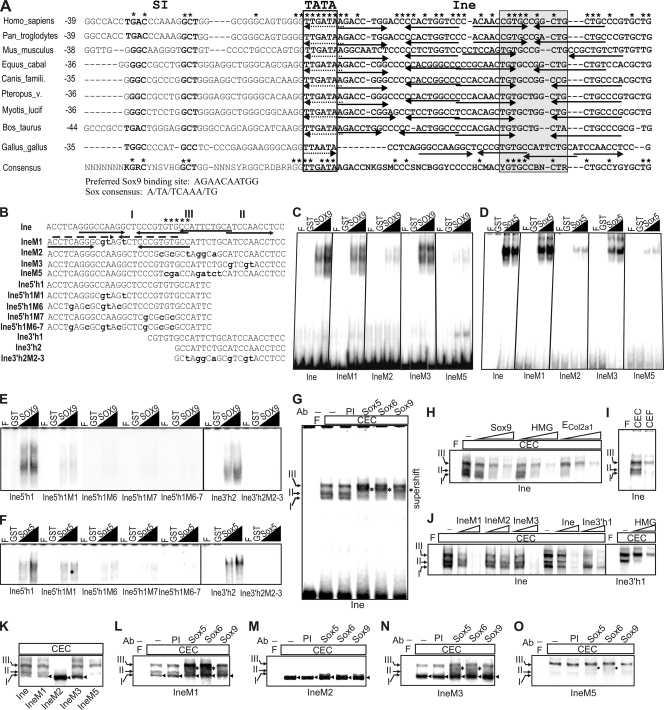
To uncover the powerful mechanism employed by the Matn1 short promoter, we dissected its elements. We reasoned that Pe1, which is the most conserved element in amniotes, could be involved in Sox-mediated functions, because it bears a palindrome resembling the preferred Sox9-binding site (29) (see Fig. S2B at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf) and because it is recognized by the Sox trio in vitro and protected in genomic footprinting (34). In addition, we found two conserved pairs of inverted Sox motifs in the Ine element of mammals (Fig. 3A; see Fig. S2A at the URL listed above). The sequence is poorly conserved in the chicken, but the chicken promoter also features two pairs of Sox motifs and a conserved GTGCC motif in the Ine element and an Nfi site upstream of TATA (Fig. 3A). These motifs could thus also play conserved regulatory roles.
FIG. 3.
Binding of Sox proteins to Ine in vitro. (A) Ine sequences of selected amniotes and the 75% majority rule consensus for amniotes (see the whole alignment in Fig. S2A at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf). TATA and the most conserved motif are boxed; NFI motifs are in boldface. Positions are given from TATA. Nucleotides fully conserved in mammals or in amniotes are marked by asterisks at the top and above the consensus sequence, respectively. Arrows and dotted arrows depict motifs similar to the preferred Sox9-binding site (29) and Sox consensus (21), respectively. Equus_cabal, Equus caballus; Canis_famili., Canis familiaris; Pteropus_v., Pteropus vampyrus; Myotis_lucif, Myotis lucifugus. (B) Sequences of Ine and its shorter or mutant derivatives. The conserved GTGCC motif, mutant nucleotides, the 5′ (I) and 3′ (II) paired Sox sites and unrelated factor-binding site (III) are denoted. (C to F) EMSA of nucleoprotein complexes formed with purified GST-fused Sox proteins on Ine and its derivatives. (G to O) Binding of CEC nuclear proteins to Ine and Ine mutants. (G) Supershift analysis with Sox antibodies (Ab). (H) Competition EMSA with 50-, 100-, and 500-fold molar excesses of the indicated cold competitors. (I) Comparison of CEC and CEF nucleoprotein complexes. (J) Competition EMSA on Ine and Ine3′h1 with the cold probes indicated. (K to O) EMSA (K) and supershifts (L to O) of the wild-type and mutant Ine. The supershifts (asterisks) and new complexes of Ine mutants (arrowheads) are marked. F, free probe; PI, preimmune serum.
We delineated the Sox-binding sites of Ine by EMSA using purified Sox proteins (Fig. 3B to F). GST-fused SOX9, L-Sox5, and Sox6 efficiently bound Ine in vitro (Fig. 3C and D and data not shown). Mutations M2 and M5, which disrupted both inverted Sox motifs, abolished complex formation, whereas M1 and M3, which disrupted only one Sox motif, had partial effect. Probes carrying the 5′ (Ine5′h1) or 3′ (Ine3′h2) Sox motif showed weaker binding than Ine (Fig. 3E and F). Mutation M1 less severely reduced complex formation with L-Sox5 than with SOX9. This was possibly due to the presence of an upstream Sox-like motif, as mutations in this motif (M6), in the 5′ half site (M7), or in both sites (M6-7) had a more drastic consequence than M1 (Fig. 3E and F). Mutation M2-3 abolished Sox binding to the 3′ site. We concluded that both Sox sites might be functional and might act cooperatively.
CEC nuclear proteins formed three major complexes with Ine. Complexes I and II were supershifted with Sox9, L-Sox5, and Sox6 antibodies (Fig. 3G) and competed with HMG, Sox9, or ECol2a1 probes (Fig. 3H). They thus likely contain the Sox trio. In contrast, complex III was not specific to chondrocytes and did not contain Sox factors (Fig. 3G to I).
Ine3′h1, in which the 5′ Sox site was mutated, formed two major CEC nucleoprotein complexes (Fig. 3J). These complexes migrated like complexes II and III, and formation of the former one was competed by the HMG probe. As complex I was neither efficiently disrupted nor formed with Ine3′h1, we concluded that it likely formed on the 5′ Sox site. IneM1, which was mutated in the 5′ Sox site, decreased the formation of complex I (Fig. 3K), as judged by reduced supershift formation, especially with Sox9 antibody (compare Fig. 3G and L). Interestingly, a new complex, not supershifted with Sox antibodies, migrated close to or slightly below complex I with probes IneM1, IneM2, and IneM3 (Fig. 3K to N, arrowheads). As IneM1 and IneM3 carried an intact Sox site, they efficiently displaced both the Sox-specific and unrelated complexes of Ine (Fig. 3J). IneM3, which carried mutations in the 3′ Sox site, did not produce the Sox-specific complex II, but supershifts with L-Sox5 and Sox6 antibodies indicated that it formed complex I (Fig. 3K and N). IneM2, however, which carried mutations in both Sox sites and in the conserved GTGCC motif, neither formed complexes I to III (Fig. 3K and M) nor competed for those (Fig. 3J). Complex III may contain the unidentified factor binding to the GTGCC motif. Supporting this hypothesis, only complex III efficiently formed with IneM5, which carried mutations in both Sox sites but had part of the GTGCC motif intact (Fig. 3K and O).
We concluded that Sox factors cooperatively bind the Ine 5′ and 3′ Sox sites. SOX9 binds efficiently only when both sites are intact. Apart from two CEC-specific Sox complexes, an unrelated complex forms on the conserved GTGCC motif in mesenchymal cells. This motif is not listed in the TRANSFAC database and likely interacts with a nonchondrocytic factor.
Cartilage-specific in vivo occupancy of Ine and SI.
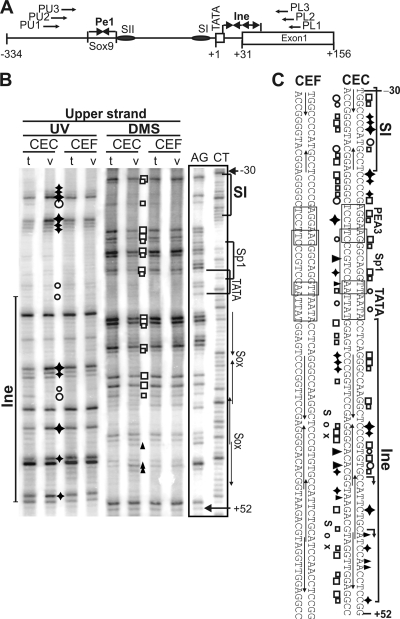
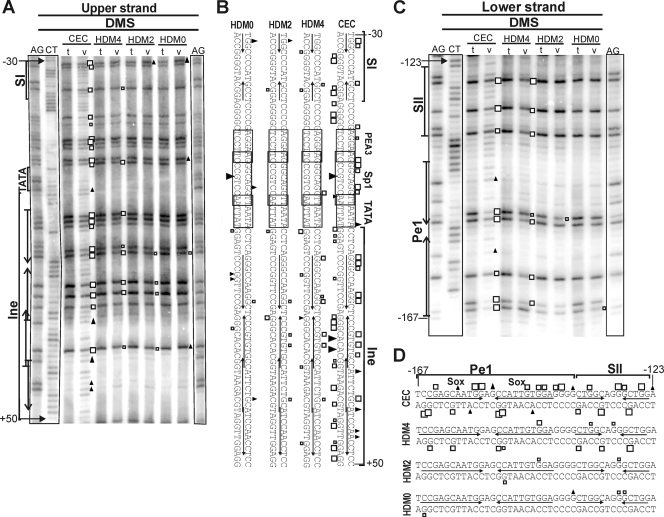
To determine occupancy of the Matn1 promoter in intact cells, we performed in vivo footprinting. We treated CEC and CEF genomic DNA with DMS or UV light to modify G residues at the N-7 position or produce 6-4 photoproducts at TC and CC dinucleotides, respectively (Fig. 4). Bound proteins blocking these modifications appeared as footprints on LM-PCR genomic sequencing ladders compared to LM-PCR of naked CEC and CEF DNA treated with the same reagents in vitro. Differences in the modification patterns between the in vivo- and in vitro-treated samples indicated in vivo DNA-protein contacts at specific nucleotides in the promoter area in CEC cultures (Fig. 4B and C). Protection, combined with hyperreactivity on the opposite strand, revealed protein binding to the Sox motifs of Ine and to the conserved TGTGCC motif at the start site. Both treatments revealed in vivo occupancy at TATA, at the Nfi contact points of the reported SI element (41), at putative PEA3, Sp1, and GC-rich motifs downstream of SI, and in the Nfi spacer region. In contrast, no footprints were detected in CEF.
FIG. 4.
Tissue-specific occupancy of Ine1 and SI in genomic footprinting. (A) Schematic depicting the primers used in footprinting and the short promoter elements. (B) Footprints on the upper DNA strand. AG and CT are Maxam-Gilbert ladders. DNA from CEC and CEF cultures treated in vivo (v) with DMS (open boxes or solid triangles) or UV light (open circles or solid diamonds) is compared with the in vitro (t) DNA samples treated with these reagents after isolation from CEC and CEF. Differences in the modification patterns between in vivo and in vitro treatments appear as hyperactivities (solid diamonds or triangles) or protections (open circles or boxes), revealing specific in vivo DNA-protein contacts. (C) Summary of in vivo footprinting on both strands.
In HDM cultures undergoing early chondrogenesis, none or weak occupancy was seen at TATA, Ine, and SI elements on days 0 and 2 (Fig. 5A and B). By day 4, however, the 5′ Sox site of Ine started to be occupied and stronger protection was also observed for the Sox motifs of Pe1 and Nfi contact points of SII (Fig. 5C and D).
FIG. 5.
Slow gradual occupancy of the short promoter elements during chondrogenesis. Shown is a comparison of in vivo footprints formed with DMS in the vicinity of the Ine (A) and Pe1 elements (C) in CEC and in day 0, 2, and 4 HDM cultures. (B and D) Summary of in vivo footprinting on both strands. Other symbols are as in Fig. 4.
We concluded that the CEC-specific in vivo footprints at the Ine Sox sites are likely due to stage-specific binding of Sox proteins. The TGTGCC motif, the Nfi site, and other potential ubiquitous factor-binding sites near TATA were also occupied. Gradual protection at the Pe1 and Ine Sox sites and at SII and SI Nfi sites in HDM culture suggests that these elements participate in Matn1 activation during chondrogenesis.
The Pe1 Sox site and SI Nfi site are indispensable for promoter activation in transiently transfected chondrocytes.
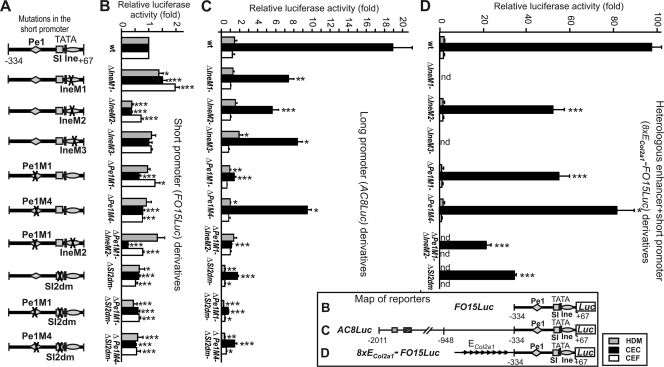
To study the contribution of short promoter elements to promoter activity, we introduced point mutations into Ine, Pe1, and SI and measured their effect on the activity of short (FO15Luc) and long (AC8Luc) promoter constructs. IneM2, which carried mutations in the Sox- and ubiquitous factor-binding sites of Ine, and a double Pe1M1/IneM2 mutation most effectively reduced the activity of the short promoter in CEC (Fig. 6 A and B). The −2011/−334 sequence enhanced the short promoter activity ∼19-fold in AC8Luc in CEC but hardly did so in low- or nonexpressing cultures (Fig. 6C). IneM1, IneM2, and IneM3 cut the long promoter activity by one half or more in CEC. The effect of Pe1M4, which carried a mutation in a factor-binding site in Pe1 (34), was similar, but Pe1M1, in which the Sox site of Pe1 was disrupted, dropped the long promoter activity 13-fold, abolishing CEC-specific enhancement from upstream elements. Pe1M1/IneM2 decreased the activity (P < 0.05) to a level even closer to that of FO15Luc. Thus, the Sox sites in Pe1 and Ine are needed to mediate promoter activation from upstream elements.
FIG. 6.
Effect of Ine, Pe1, and SI mutations on reporter activities in transfected cells. (A) Schematic of single or double mutations introduced into the short promoters of reporters FO15Luc, AC8Luc, and 8×ECol2a1-FO15Luc driven by the short or long Matn1 promoter or multiple copies of ECol2a1 fused to the short promoter, respectively, as seen on their full maps (bottom). (B to D) Luciferase activities of wild-type (wt) and mutant reporters in the low-, high-, and nonexpressing HDM, CEC, and CEF cultures, respectively, are presented as fold values relative to that for FO15Luc. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with wild-type reporter). nd, not determined.
Considering that the SI element was protected in genomic footprinting in CEC culture (Fig. 4) and bound Nfi proteins in vitro (41), we also mutated its Nfi contact points. Mutation SI2dm, either alone or in combination with Pe1M1 or Pe1M4, markedly reduced the short promoter activity in mesenchymal cells (Fig. 6A and B). This mutation also dropped the long promoter activity by 10-fold in CEC and similarly in other cultures, indicating non-tissue-specific inhibition (Fig. 6C). Double mutation Pe1M1/SI2dm further diminished the activity (P < 0.001) to the basal promoter level in mesenchymal cells, suggesting an additive or synergistic effect. Thus, disruption of the Nfi site of the SI silencer element abolished both the tissue- and stage-specific promoter activity.
We concluded that, although Ine recognition by Sox factors may be involved, Sox factor binding to Pe1 seems to be more crucial for promoter activation in CEC culture rich in late proliferative chondroblasts. In addition, binding of the ubiquitous Nfi to SI near the TATA box may be similarly crucial. The position-specific conservation of motifs similar to the NFI consensus (35) near TATA in amniotes (see Fig. S2A at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf) further supports the importance of SI in the regulation of the gene. The significant, but less dramatic, effect of other mutations suggests that the binding of factors to the Pe1 spacer and to the conserved motif of Ine may also be needed for full promoter activity.
Sox and Nfi sites of the short promoter are important for enhancement by ECol2a1.
Next we tested the activation of the short promoter by a heterologous cartilage enhancer. Eight copies of ECol2a1 robustly increased the short promoter activity in CEC but had no effect in CEF or in HDM cultures consisting of early proliferative chondroblasts (Fig. 6D). Lining up with transgenic mouse data, these results indicate that the Matn1 short promoter also restricted the broad cartilage-specific enhancement by ECol2a1 to late proliferative chondroblasts in tissue culture.
Mutations Pe1M1, IneM2, SI2dm, and Pe1M1/IneM2 decreased the relative activity of 8×ECol2a1-FO15Luc by 43.6%, 46.6%, 64.9%, and 78%, respectively, in CEC culture (Fig. 6D). Thus, our data show that, whereas Sox factor binding to Pe1 is crucial for the interaction between the homologous distal and proximal promoter elements, Sox9 binding to Pe1 and Ine is less essential for mediating enhancement from ECol2a1. Disrupting all three Sox sites of Pe1 and Ine or the Nfi site of SI, however, highly diminished the enhancement, supporting the hypothesis that the short promoter elements may also interact with the heterologous enhancer via the bound Sox and Nfi factors.
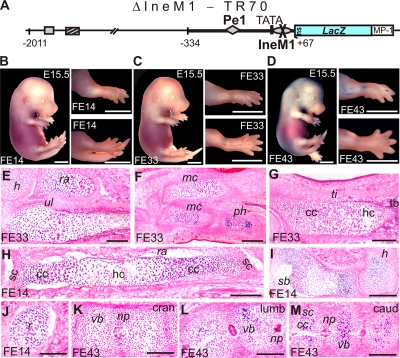
Dramatic decrease of transgene activity by mutation of the 5′ Sox site in Ine.
A transgene carrying the IneM1 mutation displayed very low activity in founder embryos, but this activity remained restricted to the columnar and prehypertophic GP zones, as with TR70 (Fig. 7). Thus, consistent with the reduced Sox-specific complex formation (Fig. 3L), the IneM1 mutation hampered promoter activation in vivo but did not alter the zone- and distal structure-dependent expression pattern of the promoter. The 5′ Sox site of Ine is thus needed for optimal promoter activation in vivo.
FIG. 7.
Low zonal activity of the ΔIneM1-TR70 transgene. (A to D) Schematic (A) and low activity of the transgene (B to D). (E to M) Histological analysis of cryosections. Weak X-Gal staining in the developing shoulder blade (sb) and limbs slightly increases toward phalanges (E to I). The increase is more pronounced from cranial to caudal vertebral bodies (K to M). Staining is seen in the columnar and prehypertrophic zones (G, H, and M). For other abbreviations, see the Fig. 1 legend. Bars, 2 mm (B to D) and 200 μm (E to M).
Accumulation of Nfi and Sox mRNAs during in vitro chondrogenesis.
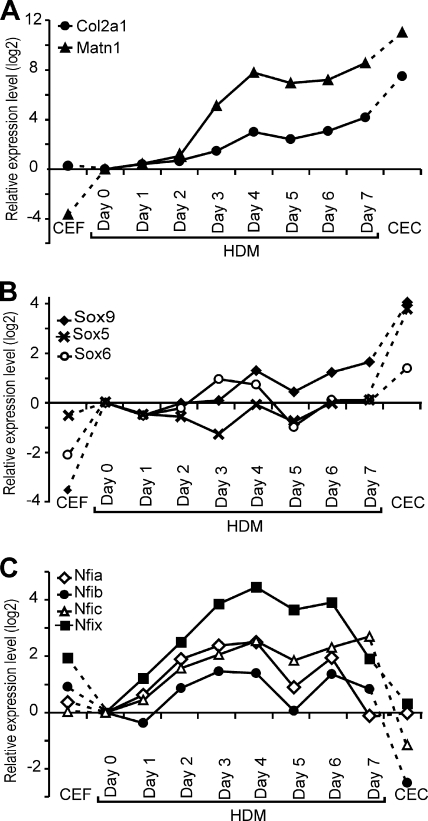
We compared the kinetics of expression of Matn1 and other genes in chondrogenic cultures by QRT-PCR. In CEFs, the steady-state mRNA levels of Matn1 and the Sox trio were very low, while those for Nfi, but not Nfic, were elevated relative to those in the committed mesenchyme (HDM, day 0) (Fig. 8 A to C). In HDM culture, the Col2a1 mRNA level slowly but continuously accumulated during differentiation, whereas the Matn1 mRNA level dramatically increased between days 2 and 4 (Fig. 8A). Upon differentiation of CEC, the Col2a1 and Matn1 relative mRNA levels rose to values of 181-fold and 2,057-fold, respectively. Sox9 and Col2a1 mRNAs accumulated with similar kinetics, but the low levels of L-Sox5 and Sox6 mRNAs increased sharply only in CEC culture, except for a small, transient boost of Sox6 mRNA at days 3 and 4 in HDM culture, just preceding the first peak in the Matn1 mRNA level (Fig. 8B). The relative Nfi mRNA levels also increased transiently by 2.6- to 22-fold, with two peaks at day 4 and days 6 and 7 in HDM culture, followed by a sharp decline in CEC culture to close to 1 (Nfia and Nfix) or below 1 (Nfib and Nfic) (Fig. 8C).
FIG. 8.
QRT-PCR analysis of marker gene expression in chondrogenic cultures. (A to C) Marker mRNA levels were determined during chondrogenesis in HDM culture at time points indicated relative to the day 0 values and compared to mRNA levels of high-expressing CEC and nonexpressing CEF cultures. CT values were normalized and relative gene expression ratios were calculated according to Materials and Methods. Relative expression levels (ΔΔCT) are plotted as log2 ratios.
Thus, CEC culture, rich in late proliferative chondroblasts, is characterized by high Matn1 and Sox trio levels but low Nfi mRNA levels. However, day 4 HDM culture, consisting of early proliferative chondroblasts, exhibits high Nfi mRNA levels but lower Matn1, Sox9, and Sox6 mRNA levels and very low L-Sox5 mRNA expression. Sox6 and Nfi mRNA levels peaked in HDM culture at the time of Matn1 activation, suggesting a function in Matn1 regulation.
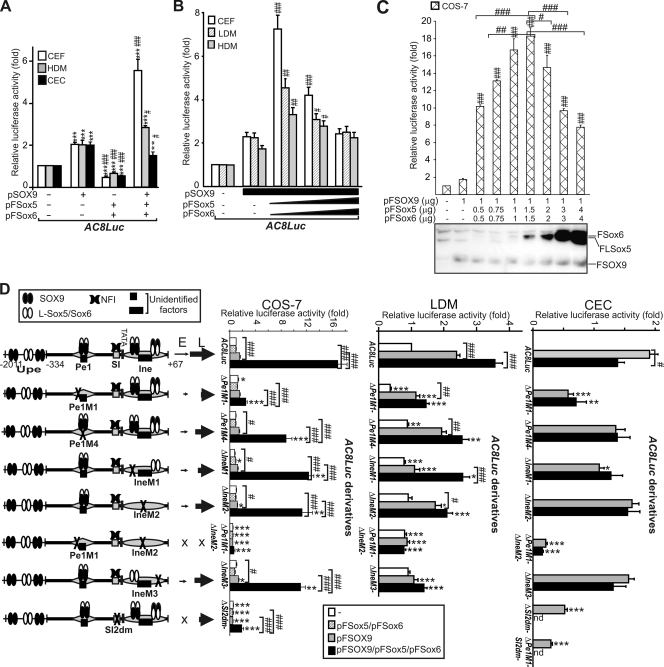
Dose-dependent synergy of L-Sox5/Sox6 with SOX9.
Next, we assessed activation of the Matn1 long promoter by cotransfected Sox proteins. While SOX9 doubled it, L-Sox5/Sox6 decreased the promoter activity by about one-half in mesenchymal cells (Fig. 9A). Coexpression of L-Sox5/Sox6 with SOX9 greatly or moderately increased the ability of SOX9 to activate the promoter in CEF and HDM cultures, respectively, but decreased it in CEC culture. This suggests synergy between Sox proteins at early differentiation stages. When we introduced a constant amount of pSOX9 and increasing amounts of pSox5 and pSox6 expression plasmids into CEF, LDM, and HDM cultures, synergistic activation peaked at a low ratio of pL-Sox5 and pSox6 versus pSOX9 and declined at an elevated ratio (Fig. 9B). Highest activation was seen in CEF (3.5-fold), followed by that in LDM and HDM cultures in inverse correlation with the endogenous Sox5 and Sox6 expression levels of these cultures (Fig. 8B), raising the possibility that L-Sox5/Sox6 may modulate the activation by SOX9 in a dose-dependent manner.
FIG. 9.
Functional importance of Sox-binding sites in cotransfection assays. (A to C) AC8Luc was cotransfected with Sox expression plasmids in various cultures as indicated. Western analysis with anti-FLAG antibody (C) shows the relative expressions of L-Sox5/Sox6 and SOX9 in the transfected COS-7 samples. (D) Effect of point mutations on the synergistic activation of the long promoter by L-Sox5/Sox6 and SOX9 coexpressed at optimal (2.7:1) molar ratio in COS-7 cells and at a higher ratio in LDM and CEC cultures. The schematic indicates factor binding to the short promoter elements and to the upstream elements (Upe) (not drawn to scale). Thin and thick arrows depict the transcription efficiencies at early (E) and at late (L) stages of chondrogenesis. Luciferase activities are given as fold values relative to that for AC8Luc. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the reporter cotransfected with vectors [A to C] or between the cotransfected mutants and the similarly cotransfected wild-type AC8Luc [D]); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 (compared with the SOX9-cotransfected reporters).
This hypothesis was confirmed by forced expression of the FLAG-tagged Sox trio in nonchondrocytic COS-7 cells and monitoring of protein expression in Western blots (Fig. 9C). Despite the small effect of SOX9 alone, L-Sox5/Sox6 synergized with SOX9 to activate the long promoter up to ∼18- to 20-fold at low molar excess. The activation was high from a 1:1 to 4:1 molar ratio of L-Sox5/Sox6 to SOX9 in repeated experiments, but the synergy dropped above a 5:1 molar ratio (Fig. 9C). When tested individually, L-Sox5 and Sox6 had similar effects (see Fig. S3 at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf).
We concluded that L-Sox5/Sox6 may finely tune the activity of the Matn1 promoter by increasing transactivation by SOX9 at a low molar ratio relative to SOX9 (early stages of chondrogenesis) and by decreasing transactivation by SOX9 at a high ratio (late stage).
Pe1 mutation hampers transactivation by SOX9, and Ine mutation decreases the synergy with L-Sox5/Sox6.
Next we studied the effect of Pe1, Ine, and SI mutations on the activity of the Sox trio. In COS-7 cells forced to express L-Sox5/Sox6 in optimal ratio relative to SOX9, the Pe1M1/IneM2 mutation decreased the synergistic activation of the long promoter by 96.1%, followed by the SI2dm and Pe1M1 mutations (85.5 to 89%) (Fig. 9D). The former mutations also repressed SOX9-mediated activation by ∼70%. Similar effects were obtained when L-Sox5/Sox6 was expressed in high excess relative to SOX9 in LDM and CEC cultures (Fig. 9D). Thus, disruption of the short promoter Sox sites abolished transactivation by the Sox trio even when upstream sites were intact. The effect of the Pe1M1 mutation was milder, suggesting that SOX9 binding to Pe1 is critical for transactivation by SOX9 in early and late stages of chondrogenesis.
Ine mutations diminished the synergistic activation of SOX9 with an optimal ratio of L-Sox5/Sox6 in COS-7 cells (Fig. 9D). In LDM culture, IneM1 and IneM3 abolished AC8Luc activation by SOX9, while IneM2 and IneM3 affected the synergistic activation by the Sox trio more drastically than IneM1. In keeping with the effect of Ine mutations in EMSA, this result indicates that the 3′ Sox site in Ine equally interacts with SOX9 and L-Sox5/Sox6, whereas the 5′ site preferably binds SOX9 in early chondroblasts. IneM1 also hampers activation by SOX9 in CEC culture (Fig. 9D). Notably, mutation of the SI Nfi site highly decreased SOX9- and Sox trio-mediated promoter activation in the cultures tested. The variable effect of Pe1M4 and the small effect of SOX9 in COS-7 cells suggest that ubiquitous and/or Sox partner factors may also bind the promoter elements.
We concluded that SOX9 binding to Pe1 likely plays a key role in mediating enhancement from distal elements. Based on the data, we suggest a model (see Fig. 10D). L-Sox5/Sox6 expressed at a low level and bound to Ine may synergistically increase activation by Pe1-bound SOX9 in early chondrogenesis. Later on, when produced in excess to SOX9, L-Sox5/Sox6 may decrease activation by SOX9, possibly by competing for binding to the same sites. In addition, Nfi binding to SI and binding of other factors to Pe1 and Ine may also be needed for efficient activation.
FIG. 10.
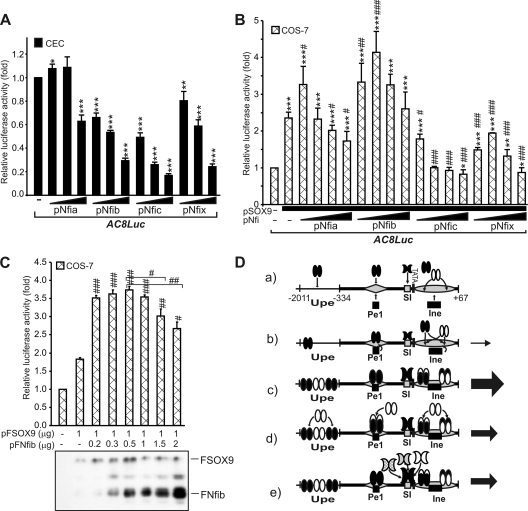
Modulation of the Matn1 promoter activity by cotransfected Nfi proteins. (A to C) AC8Luc was cotransfected with increasing amounts of Nfi expression plasmids without or with a constant amount of SOX9 expression plasmids in the cultures indicated. (C) Western analysis was made with anti-FLAG antibody to determine the relative ratio of Nfib and SOX9 expressed by force at optimal promoter activation. (D) Model for fine-tuning of the promoter activity by the Sox trio and Nfi. Shown are schematics of factor binding to DNA elements during Matn1 activation at the onset of chondrogenesis (a), in early (b) and late proliferative chondroblasts (c) at low and optimal occupancy of sites, respectively, and in the late stage at high occupancy of the Sox (d) or Nfi sites (e). See the text for a detailed description. Symbols are as defined for Fig. 9.
Nfi proteins modulate promoter activity.
Next we studied the effect of Nfi on AC8Luc activity in cotransfection assays. In CEC culture, all Nfi proteins, except Nfia at low concentration, robustly inhibited long promoter activity (Fig. 10A). When NFI and SOX9 were expressed at an optimal ratio, Nfib and Nfic decreased significantly transactivation by SOX9, but all Nfi proteins exerted 74% to 90% repression at higher ratio (see Fig. S4A at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf). Notably, CTF-1, an isoform of NFIC, only slightly inhibited activation by SOX9 (see Fig. S4B at the URL listed above), suggesting that the various Nfi splice variants may have different effects. In COS-7 cells, Nfia and Nfib in optimal amounts indeed cooperated with SOX9 and potentiated its transactivation of the long promoter (Fig. 10B). The activation, however, declined at higher levels of Nfia and Nfib. Forced expression of FLAG-tagged proteins in COS-7 cells revealed that the activation increased up to an ∼2:1 molar ratio of Nfib to SOX9 but significantly decreased above an ∼4:1 molar ratio (Fig. 10C).
These data suggest that Nfi proteins may increase or decrease SOX9-mediated transactivation of Matn1 depending on their abundance relative to SOX9 (Fig. 10D). The conservation of Nfi sites near TATA and Pe1 (see Fig. S2 at the URL mentioned in the previous paragraph) underlines the importance of Nfi proteins in the restricted cartilage-specific expression of Matn1 in amniotes.
DISCUSSION
By dissecting the control mechanism that directs Matn1 expression to specific GP zones, this study sheds new light on a distinctive regulatory network orchestrated by the chondrogenic Sox trio. Focusing on the role of short promoter elements, the present work, in line with former reports (19, 34, 41), reveals the following unique features of Matn1 regulation. (i) Remarkable sequence and positional conservation of proximal (short) and distal promoter elements strongly suggests an evolutionarily conserved transcriptional mechanism in amniotes. (ii) Fundamentally, the proximal promoter is responsible for conferring spatiotemporal expression. It exerts such a dominant effect that it is even capable of restricting spatially and temporally the activity of the otherwise pancartilaginous Col2a1 enhancer. (iii) This effect is likely due to a unique set of conserved proximal elements. The Sox site in Pe1, located 95 to 195 bp upstream of TATA, preferably binds SOX9 and is most crucial for promoter activity, while Sox sites in Ine located at the transcription start sites preferably bind L-Sox5/Sox6 and are also important. An Nfi site in SI near TATA is also needed for promoter enhancement, and conservation of Nfi motifs in SII near Pe1 suggests an important function. (iv) The most highly conserved Pe1 element plays a key role in SOX9-mediated transactivation from distal DNA elements, and L-Sox5/Sox6 bound to Ine and Nfi proteins bound to SI may modulate transactivation by SOX9 in a dose-dependent manner and may thereby fine-tune stage-specific promoter activity.
Cartilage-specific control elements with functional Sox sites in other genes in various locations, e.g., intronic, far-upstream, 5′ untranslated, or proximal promoter regions, were described previously (13, 15, 26-28, 46), but none shows similarity to the Matn1 control region. While the Matn1 short promoter is sufficient to specify the expression pattern of the gene, the Col2a1 promoter has no activity on its own and relies on an intronic enhancer capable of directing its activity, as well as that of a heterologous β-globin promoter, to all chondrocytic cells and each GP zone in transgenic mice (47, 48).
Comparison of orthologous promoter regions (known as phylogenetic footprinting) can reveal conserved motifs with important regulatory functions (6). As shown, e.g., for the Sox2 locus, conservation of extragenic sequences in amniotes can more reliably reflect their functional importance in development than the higher degree of conservation between mammals (17). A conserved cartilage-specific element has been identified, however, only in the far-upstream enhancer of the mammalian orthologs of Agc1, but it is not conserved in amniotes (13). Such a high degree of sequence and positional conservation among chicken and mammalian orthologs (34) (see Fig. S2 at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf) has not been found for other cartilage ECM genes, strongly suggesting a distance-dependent important function for Pe1 and Ine in amniotes. Pe1 and Ine include one or two pairs of oppositely oriented motifs sharing 6/10 to 8/10 or 5/10 to 7/10 nucleotide identity, respectively, with the preferred Sox9-binding site (29), while Sox9 sites of cartilage enhancers share only 4 or 5 nucleotides with the Sox consensus T/AT/ACAAT/AG (13). In line with our former report (34), the present mutational and functional data confirm the key role of the highly conserved Pe1 in SOX9 binding and SOX9-mediated enhancement from distal elements. Ine is less conserved, but it is also needed for high transgene activity. The 3′ paired Sox site of Ine had been shifted to a head-to-head position in the chicken ortholog, and it seems to interact rather with L-Sox5/Sox6 in EMSA and forced-expression studies. Notably, the TATA box also showed similarity to the Sox consensus in most of the Matn1 orthologs, indicating that, besides the conserved strong Sox9-binding site of Pe1, weaker Sox sites, which seem to be more diverged, are clustered near TATA, while the regulatory module might have been under evolutionary pressure and thus remained more conserved. To our knowledge, this is the first report on Sox sites clustered around the transcription start sites, strongly suggesting their importance in the assembly of the preinitiation complex (PIC).
Matn1 is regulated differently by the Sox trio than other cartilage genes (13, 24, 26). Whereas Sox9 is sufficient for the activation of Col2a1, Agc1, and Crtl1, Sox5 and Sox6 are required to turn on Matn1, as Matn1 mRNA was not detected in Sox5−/−; Sox6−/− mice (38). Activation of Sox6 precedes that of Matn1 in culture, underlining the importance of Sox6 in turning on Matn1. As in cartilage enhancers or the COMP promoter (13, 26, 27), L-Sox5 and Sox6 also synergize with SOX9 in the activation of the Matn1 promoter, but only in the early stage or at low molar excess. Their role, however, turns to repression in the late stage or at elevated molar ratio. Thus, in large excess, L-Sox5 and Sox6 may compete with SOX9 for the same binding sites, as for oligodendrocyte-specific genes (40).
In agreement with the transient activation of Nfi genes during in vitro chondrogenesis, dominant negative mutation of Nfib interfered with chondrogenesis (43). Overexpression of Nfib increased Sox9 and Col2a1 expression, but Nfi sites mediating this regulation have not been identified. By extending this and our former studies (41), here we provide the first evidence that, in addition to the Sox trio, Nfi proteins binding near TATA may also play a critical role in determining the chondrocyte stage-specific activity of the Matn1 promoter.
According to our model (Fig. 10D), the special geometric arrangement of proximal elements may explain the unique regulation of Matn1, as it allows fine-tuning of the promoter activity by L-Sox5/Sox6 and Nfi, depending on their abundances relative to that of SOX9. At the onset of chondrogenesis, binding of Sox and Nfi proteins might be needed to open the chromatin structure around TATA (Fig. 10D, a and b). This hypothesis is based on our observations (34) that in vivo footprints were absent from the short promoter in the nonexpressing CEF and that they gradually appeared in differentiating HDM culture, strongly suggesting that activation of Matn1 involves regulation at the chromatin level. In fact, the Nfi sites of SI and SII were not occupied in CEF, although the Nfi genes are expressed in CEF and Nfi proteins can bind SI and SII from CEF extracts in EMSA and in vitro footprinting (41). Based on their interaction with histones (3, 12), Nfi proteins may help disrupt the nucleosome structure during Matn1 activation.
At the early stage of chondrogenesis, when occupancy of the sites is low and SOX9 is expressed at high molar excess relative to L-Sox5/Sox6, SOX9 preferably binds Pe1 and the Ine-bound L-Sox5/Sox6 synergizes with SOX9 by likely increasing its efficiency for binding Pe1 (Fig. 10D, b). L-Sox5/Sox6 similarly secures Sox9 binding to the Agc1 and Col2a1 enhancers (13). Binding of Sox factors in the vicinity of TATA may bend the DNA and facilitate the binding of TATA-binding protein (TBP) and polymerase II during the assembly of PIC (see also Fig. S5 at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf). Clustering of Sox motifs in Ine may increase the probability of L-Sox5/Sox6 binding and help recruit SOX9 to Pe1 and TBP to TATA. Bending the DNA may also promote the binding of unidentified factors to Pe1 and Ine. Based on preliminary analysis of mutations, these factors may affect proximodistal transgene activity (data not shown). Further, as NFI proteins can activate transcription through direct interaction with basal transcription factors (e.g., CTF-1 with TFIIB and TBP via its proline-rich transactivation domain) and various coactivators and corepressors (12), Nfi binding to SI (and also possibly to SII near Pe1) may also help the assembly of PIC and the enhanceosome, thus highly contributing to activated transcription (see Fig. S5 at the URL listed above). Pe1 likely plays central role in enhanceosome formation and in SOX9-mediated promoter activation from distal elements, but, due to the low abundance of transcription factors, the transcription activity is low in early proliferative chondroblasts (Fig. 10D, b). The promoter activity is highest in late proliferative chondroblasts, when in vivo occupancy is optimal, high at Pe1, and Ine, and moderate at SI (Fig. 10D, c). Nfib exerted activation at an early stage in this study and another study (43), but we cannot exclude the possibility that another Nfi isoform is active in CEC culture, considering the drop in the relative expression level of Nfib and other Nfi mRNAs. At this late stage, when the Sox trio mRNA level is elevated, forced expression of L-Sox5/Sox6 in large molar excess to SOX9 can decrease transactivation by SOX9, possibly by competing with SOX9 for binding Pe1 and other elements (Fig. 10D, d). High occupancy of the Sox sites of Ine may even physically interfere with the recruitment of PIC to TATA. Overproduction of Nfi may also decrease promoter activity due to competition between activator (e.g., NfIb) and repressor Nfi isoforms, which may even sterically block TBP binding to TATA (Fig. 10D, e).
The unique molecular mechanism described here can facilitate the construction of GP zone-specific vectors and the development of biotechnological therapies for skeletal diseases.
Acknowledgments
We are grateful to B. de Crombrugghe for providing plasmid p3000i3020Col2a1 and SOX9, L-Sox5, and Sox6 antisera, to P. Berta for the GST-SOX9 plasmid, to R. Gronostajski for Nfia, Nfib, Nfic, and Nfix expression plasmids, and to N. Mermod for the CTF1 expression plasmid. We thank P. Szabó for introducing O.R. to genomic footprinting, A. Simon, E. Horváth, I. Kravjár, and K. Kávai for excellent technical assistance, and M. Tóth for the artwork.
This work was supported by grant OTKA PD50006 to E.K., by grants OTKA T049608 from the Hungarian National Scientific Research Foundation, ETT 008/2006 from the Medical Research Council of Hungary, and GVOP-3.1.1.-2004-05-0290/3.0 from the Economic Competitiveness Operative Program of the National Development Plan to I.K., and by NIH/NIAMS grant AR60016 to V.L. Á. Zvara and E. Barta were supported by János Bolyai fellowships from the Hungarian Academy of Sciences (BO/00381/07 and BO/00383/08). This work was also partly supported by grant AVINOMID from the Ányos Jedlik Programme of the National Office for Research and Technology (NKTH) to L.G.P.
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Akiyama, H. 2008. Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 18:213-219. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, H., M.-C. Chaboissier, J. F. Martin, A. Schedl, and B. de Crombrugghe. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16:2813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alevizopoulos, A., et al. 1995. A proline-rich TGF-β-responsive transcriptional activator interacts with histone H3. Genes Dev. 9:3051-3066. [DOI] [PubMed] [Google Scholar]
- 4.Aszódi, A., et al. 1996. Cloning, sequencing and expression analysis of mouse cartilage matrix protein cDNA. Eur. J. Biochem. 236:970-977. [DOI] [PubMed] [Google Scholar]
- 5.Aszódi, A., et al. 1994. The zonal expression of chicken cartilage matrix protein gene in the developing skeleton of transgenic mice. Matrix Biol. 14:181-190. [DOI] [PubMed] [Google Scholar]
- 6.Barta, E., et al. 2005. DoOP: databases of orthologous promoters, collections of clusters of orthologous upstream sequences from chordates and plants. Nucleic Acids Res. 33:D86-D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi, W., J. M. Deng, Z. Zhang, R. R. Behringer, and B. de Crombrugghe. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22:85-89. [DOI] [PubMed] [Google Scholar]
- 8.Cancedda, R., F. D. Cancedda, and P. Castagnola. 1995. Chondrocyte differentiation. Int. Rev. Cytol. 159:265-359. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhry, A. Z., A. D. Vitullo, and R. M. Gronostajski. 1998. Nuclear factor I (NFI) isoforms differentially activate simple versus complex NFI-responsive promoters. J. Biol. Chem. 273:18538-18546. [DOI] [PubMed] [Google Scholar]
- 10.Deák, F., R. Wagener, I. Kiss, and M. Paulsson. 1999. The matrilins: a novel family of oligomeric extracellular matrix proteins. Matrix Biol. 18:55-64. [DOI] [PubMed] [Google Scholar]
- 11.Goldring, M. B., K. Tsuchimochi, and K. Ijiri. 2006. The control of chondrogenesis. J. Cell. Biochem. 97:33-44. [DOI] [PubMed] [Google Scholar]
- 12.Gronostajski, R. M. 2000. Roles of the NFI/CTF gene family in transcription and development. Gene 249:31-45. [DOI] [PubMed] [Google Scholar]
- 13.Han, Y., and V. Lefebvre. 2008. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol. Cell. Biol. 28:4999-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, T., et al. 2004. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 50:3561-3573. [DOI] [PubMed] [Google Scholar]
- 15.Imamura, T., C. Imamura, Y. Iwamoto, and L. J. Sandell. 2005. Transcriptional co-activators CREB-binding protein/p300 increase chondrocyte Cd-rap gene expression by multiple mechanisms including sequestration of the repressor CCAAT/enhancer-binding protein. J. Biol. Chem. 280:16625-16634. [DOI] [PubMed] [Google Scholar]
- 16.Kálmán, J., et al. 2005. Gene expression profile analysis of lymphocytes from Alzheimer's patients. Psychiatr. Genet. 15:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Kamachi, Y., et al. 2009. Evolution of non-coding regulatory sequences involved in the developmental process: reflection of differential employment of paralogous genes as highlighted by Sox2 and group B1 Sox genes. Proc. Jpn. Acad. 85:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamachi, Y., M. Uchikawa, and H. Kondoh. 2000. Pairing Sox off with partners in the regulation of embryonic development. Trends Genet. 16:182-187. [DOI] [PubMed] [Google Scholar]
- 19.Karcagi, I., et al. 2004. Functional analysis of the regulatory regions of the matrilin-1 gene in transgenic mice reveals modular arrangement of tissue-specific control elements. Matrix Biol. 22:605-618. [DOI] [PubMed] [Google Scholar]
- 20.Karsenty, G., H. M. Kronenberg, and C. Settembre. 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25:629-648. [DOI] [PubMed] [Google Scholar]
- 21.Kiefer, J. C. 2007. Back to basics: Sox genes. Dev. Dyn. 236:2356-2366. [DOI] [PubMed] [Google Scholar]
- 22.Kiss, I., et al. 1989. Structure of the gene for cartilage matrix protein, a modular protein of the extracellular matrix. J. Biol. Chem. 264:8126-8134. [PubMed] [Google Scholar]
- 23.Kou, I., and S. Ikegawa. 2004. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J. Biol. Chem. 279:50942-50948. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre, V., and P. Smits. 2005. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res. C Embryo Today 75:200-212. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre, V., B. Dumitriu, A. Penzo-Méndez, Y. Han, and B. Pallavi. 2007. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 39:2195-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefebvre, V., P. Li, and B. de Crombrugghe. 1998. A long new form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17:5718-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, C., et al. 2007. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front. Biosci. 12:3899-3910. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., H. Li, K. Tanaka, N. Tsumaki, and Y. Yamada. 2000. Identification of an enhancer sequence within the first intron required for cartilage-specific transcription of the α2(XI) collagen gene. J. Biol. Chem. 275:12712-12718. [DOI] [PubMed] [Google Scholar]
- 29.Mertin, S., S. G. McDowall, and V. R. Harley. 1999. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 27:1359-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundlos, S., and B. Zabel. 1994. Developmental expression of human cartilage matrix protein. Dev. Dyn. 199:241-252. [DOI] [PubMed] [Google Scholar]
- 31.Muratoglu, S., et al. 1995. Expression of the cartilage matrix protein gene at different chondrocyte developmental stages. Eur. J. Cell Biol. 68:411-418. [PubMed] [Google Scholar]
- 32.Pfeifer, G. P., H. H. Chen, J. Komura, and A. D. Riggs. 1999. Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol. 304:548-571. [DOI] [PubMed] [Google Scholar]
- 33.Provot, S., and E. Schipani. 2005. Molecular mechanisms of endochondral bone development. Biochem. Biophys. Res. Commun. 328:658-665. [DOI] [PubMed] [Google Scholar]
- 34.Rentsendorj, O., et al. 2005. Highly conserved proximal promoter element harbouring paired Sox9-binding sites contributes to the tissue- and developmental stage-specific activity of the matrilin-1 gene. Biochem. J. 389:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roulet, E., et al. 2000. Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J. Mol. Biol. 297:833-848. [DOI] [PubMed] [Google Scholar]
- 36.Santoro, C., N. Mermod, P. C. Andrews, and R. Tjian. 1988. A family of human CAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature 334:218-224. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu, H., S. Yokoyama, and H. Asahara. 2007. Growth and differentiation of developing limb bud from perspective of chondrogenesis. Dev. Growth Differ. 49:449-454. [DOI] [PubMed] [Google Scholar]
- 38.Smits, P., et al. 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1:277-290. [DOI] [PubMed] [Google Scholar]
- 39.Stirpe, N. S., and P. F. Goetinck. 1989. Gene regulation during cartilage differentiation: temporal and spatial expression of link protein and cartilage matrix protein in developing limb. Development 107:23-33. [DOI] [PubMed] [Google Scholar]
- 40.Stolt, C. C., et al. 2006. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell 11:697-709. [DOI] [PubMed] [Google Scholar]
- 41.Szabó, P., et al. 1995. Identification of a nuclear factor-I family protein-binding site in the silencer region of the cartilage matrix protein gene. J. Biol. Chem. 270:10212-10221. [DOI] [PubMed] [Google Scholar]
- 42.Szüts, V., et al. 1998. Terminal differentiation of chondrocytes is arrested at distinct stages identified by their expression repertoire of marker genes. Matrix Biol. 17:435-448. [DOI] [PubMed] [Google Scholar]
- 43.Uchihashi, T., et al. 2007. Involvement of nuclear factor I transcription/replication factor in the early stage of chondrogenic differentiation. Bone 41:1025-1035. [DOI] [PubMed] [Google Scholar]
- 44.Wagener, R., et al. 2005. The matrilins—adaptor proteins in the extracellular matrix. FEBS Lett. 579:3323-3329. [DOI] [PubMed] [Google Scholar]
- 45.Wiberg, C., et al. 2003. Complexes of matrilin-1 and biglycan or decorin connect collagen VI mircofibrils to both collagen II and aggrecan. J. Biol. Chem. 278:37698-37704. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, P., S. A. Jimenez, and D. G. Stokes. 2003. Regulation of human COL9A1 gene expression. J. Biol. Chem. 278:117-123. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, G., et al. 1995. A 182 bp fragment of the mouse proα1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J. Cell Sci. 108:3677-3684. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, G., V. Lefebvre, Z. Zhang, H. Eberspaecher, and B. de Crombrugghe. 1998. Three high mobility group-like sequences within a 48-base pair enhancer of the Col2a1 gene are required for cartilage-specific expression in vivo. J. Biol. Chem. 273:14989-14997. [DOI] [PubMed] [Google Scholar]