Abstract
The plant-pathogenic bacterium Dickeya dadantii (formerly Erwinia chrysanthemi) produces a large array of plant cell wall-degrading enzymes. Using an in situ detection test, we showed that it produces two feruloyl esterases, FaeD and FaeT. These enzymes cleave the ester link between ferulate and the pectic or xylan chains. FaeD and FaeT belong to the carbohydrate esterase family CE10, and they are the first two feruloyl esterases to be identified in this family. Cleavage of synthetic substrates revealed strong activation of FaeD and FaeT by ferulic acid. The gene faeT appeared to be weakly expressed, and its product, FaeT, is a cytoplasmic protein. In contrast, the gene faeD is strongly induced in the presence of ferulic acid, and FaeD is an extracellular protein secreted by the Out system, responsible for pectinase secretion. The product of the adjacent gene faeR is involved in the positive control of faeD in response to ferulic acid. Moreover, ferulic acid acts in synergy with polygalacturonate to induce pectate lyases, the main virulence determinant of soft rot disease. Feruloyl esterases dissociate internal cross-links in the polysaccharide network of the plant cell wall, suppress the polysaccharide esterifications, and liberate ferulic acid, which contributes to the induction of pectate lyases. Together, these effects of feruloyl esterases could facilitate soft rot disease caused by pectinolytic bacteria.
Pectinolytic enterobacteria cause diseases in a wide range of plants, including many crops of economic importance such as vegetables and ornamentals (37). The soft rot disease produced by Dickeya dadantii (formerly Erwinia chrysanthemi) results from the general disorganization of the plant cell wall (13). This symptom is generated by a set of bacterial enzymes able to degrade the plant cell wall polysaccharides. Pectate lyases, which depolymerize pectin, play an essential role in soft rot disease. Pectin, the major polysaccharide of the primary cell wall and the middle lamella, is necessary for the cohesion of the plant tissue. Dickeya dadantii produces different types of pectinases, including pectin methylesterases, pectin acetylesterases, exo- and endopectate lyases, exopolygalacturonases, and a rhamnogalacturonate lyase (13). Most of the plant cell wall-degrading enzymes are secreted by the bacteria, and it is a type II secretion system, named Out, that specifically exports most pectinases to the external medium (18). The transcription of the pel genes, encoding pectate lyases, is tightly regulated by environmental and metabolic stimuli (13). Pectin degradation products are the major signals allowing Dickeya dadantii to trigger pectate lyase induction. In addition, uncharacterized compounds present in plant extracts are able to increase the induction of pectate lyases by acting in synergy with pectin catabolic products (3, 25). Previous studies have led to the identification of several regulators controlling pel transcription (KdgR, PecS, PecT, Pir, CRP, Gac, etc.) that often affect other virulence factors (13, 21, 25, 28, 29, 34).
The structural complexity of the plant cell wall matrix requires the concerted action of several enzymes for its complete breakdown. In addition to pectinases, D. dadantii produces the cellulase CelZ, the xylanase XynA, and the endogalactanase GanA, which contribute to the degradation of plant cell wall polysaccharides (8, 18). The action of depolymerases is often limited by the presence of esterifications, which need to be removed prior to depolymerization. For example, the production of pectin methyl and acetylesterases greatly facilitates pectate lyase action in D. dadantii (30). Thus, esterases are considered important accessory enzymes. Feruloyl esters are a type of modification commonly found in pectins and xylans (16). Considering its large spectrum of plant cell wall-degrading enzymes, it seemed possible that D. dadantii might also produce feruloyl esterases (4-hydroxy-3-methoxycinnamoyl-sugar hydrolase) (EC 3.1.1.73) able to cleave this type of esterification.
Ferulic acid is the major phenolic acid esterified to carbohydrates in the plant cell wall (16). It is found mainly in arabinan and galactan chains of pectin hairy regions (5). The ferulic acid esters can form dehydrodimers, which permit covalent cross-linking of the polysaccharides that they esterify. Such diferulic acid bridges contribute to the formation of the pectic network, which is based on different types of cross-linkings (5). Enzymes responsible for cleaving the ester link between ferulate and the polysaccharidic chain have been identified in plants and associated microorganisms. Feruloyl esterases are of great interest in biotechnology for many industrial and medicinal applications (2). To date, feruloyl esterases have been characterized mainly in fungi (2). Fewer studies have been performed on bacterial feruloyl esterases, with a few enzymes described in diverse genera, such as Bacillus (9) or Butyrivibrio (7).
The potential role of feruloyl esterases in the modification of plant cell wall integrity led us to investigate whether D. dadantii produces such esterases. Using a functional test to detect feruloyl esterase activity, we identified two D. dadantii genes, named faeD and faeT, encoding such enzymes. The enzymatic activity of the proteins FaeD and FaeT was analyzed, and their cellular localization was determined. Analysis of gene expression demonstrated that ferulic acid is involved not only in the induction of faeD but also, in synergy with pectin catabolite products, in an increased induction of the pel genes encoding the major virulence factors.
MATERIALS AND METHODS
Bacterial strains and genetic techniques.
The bacterial strains of D. dadantii or Escherichia coli and the plasmids used in this study are listed in Table 1. The phi-EC2 generalized transducing phage was used for transduction (27).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/phenotype | Reference/origin |
|---|---|---|
| D. dadantii strains | ||
| 3937 | Wild type | Laboratory collection |
| A350a | rafR ganB | 8, 12, 15 |
| A1077 | A350 kdgR::Cm | Laboratory collection |
| A1798 | A350 pelD::uidA-Km | 14 |
| A2507 | A350 crp::Cm | 28 |
| A3784 | A350 faeT::uidA-Km | This work |
| A3810 | pir::Cm | S. Reverchon |
| A3916 | A350 faeD::uidA-Km | This work |
| A3994 | A350 faeR::Cm | This work |
| A3959 | A350 faeD::uidA-Km faeT::Cm | This work |
| A3953 | pecS::Cm | 29 |
| A4415 | pelD::uidA-Km | 14 |
| A4116 | pecT::Cm | 34 |
| A4239 | gacA::Cm | 21 |
| A5099 | faeD::uidA-Km | This work |
| A5100 | faeT::uidA-Km | This work |
| A5101 | faeR::Cm | This work |
| A5123 | faeR::Cm faeD::uidA-Km | This work |
| A5124 | faeR::Cm faeT::uidA-Km | This work |
| A5126 | faeR::Cm pelD::uidA-Km | This work |
| A5216 | faeD::uidA-Km faeT::Cm | This work |
| E. coli strains | ||
| NM522 | Δ(lac-proAB) Δ(mcrB-hsdSM)5 supE thi [F′ proAB lacIqlacZΔM15] | Laboratory collection |
| BL21(DE3) | E. coli B, F−dcm ompT hsdS gal λ(DE3), T7 polymerase gene under the lacUV5 promoter | 33 |
| Plasmids | ||
| pUC18 | Apr | Laboratory collection |
| FE1 | pUC18 derivative with a 12.3-kb insert, faeD+ | This work |
| FE2 | pUC18 derivative with a 16.7-kb insert, faeD+ | This work |
| FE3 | pUC18 derivative with a 17-kb insert, faeT+ | This work |
| pT7-5 | T7 phi10, Apr | 35 |
| pT7-6 | T7 phi10, Apr | 35 |
| pNA30 | pT7-6 derivative with a 1.6-kb BamHI HpaI fragment, faeD+ | This work |
| pNA55 | pT7-5 derivative with a 2.3-kb PstI SmaI fragment, faeT+ | This work |
Media and growth conditions.
Bacteria were grown in LB or in M63 medium (24). When required, the media were solidified with agar (15 g liter−1). D. dadantii cells were usually incubated at 30°C, and E. coli cells were usually incubated at 37°C. Carbon sources were added at 2 g liter−1. When required, antibiotics were usually added at the following concentrations: kanamycin (Km), 20 μg ml−1; ampicillin (Ap), 50 μg ml−1; and chloramphenicol (Cm), 20 μg ml−1.
Plant extract was prepared by autoclaving 10 g of chrysanthemum leaves in 100 ml of M63 (10% extract). The extract was diluted by 10-fold in the growth medium.
Feruloyl esterase screening test.
A test for screening feruloyl esterase-producing colonies was adapted from the work of Donaghy et al. (9). After 24 h of growth on M63 solid medium, colonies were covered with 6 ml of molten soft agar (4 g liter−1), containing 0.05 g liter−1 of ethyl ferulate (ethyl-4-hydroxy-3-methoxy-cinnamate) to ensure a cloudy overlay throughout the plate. Plates were incubated at 30°C for 2 to 4 h, until a clear zone around the colonies indicated feruloyl esterase production.
Enzyme assays.
Pectate lyase activity was determined by monitoring spectrophotometrically the formation of unsaturated products from polygalacturonate at 235 nm (36). Specific activity is expressed as micromoles of unsaturated products liberated per minute per milligram of bacterial dry weight (μmol min−1 mg−1).
β-Glucuronidase activity was measured by following the degradation of p-nitrophenyl-β-d-glucuronide into p-nitrophenol at 405 nm (1). Specific activity is expressed as nanomoles of product liberated per minute per milligram of bacterial dry weight (nmol min−1 mg−1).
Esterase activity was measured by following the degradation of p-nitrophenyl-acetate (PNPA) into p-nitrophenol at 405 nm. The appearance of the product was monitored for 20 min at 37°C. The standard assay mixture consisted of 50 mM phosphate buffer at pH 6.5, 2 mM PNPA, and the extract in a total volume of 1 ml. Specific activity is expressed as nanomoles of product liberated per minute per milligram of protein (nmol min−1 mg−1). The Km and Vmax values for FaeD and FaeT were determined under the standard conditions using substrate concentrations between 0.1 and 2 mM. Different assay conditions were also tested on FaeD and FaeT activity. The optimum pH was determined using 50 mM phosphate buffer from pH 6 to 7.5. Various buffer concentrations, 50 to 400 mM, were tested. The influence of various divalent cations was assessed in the presence of Ca2+, Cu2+, Fe2+, Mg2+, or Mn2+ at 0.1 and 1 mM concentrations relative to the corresponding chloride salt. To complex the cations, EDTA was added at a final concentration of 10 mM. The effect of ferulic acid on the esterase activity was tested at concentrations ranging from 0.1 to 0.5 mM. Under each condition, the spontaneous cleavage of the substrate was tested in parallel by omitting the extract addition.
Recombinant DNA techniques.
Preparation of plasmid or chromosomal DNA, restriction digestions, ligations, DNA electrophoresis, and transformations were carried out as previously described (11). For the construction of the D. dadantii genomic library, the chromosomal DNA was partially digested with Sau3A, and 10- to 20-kb fragments were ligated into plasmid pUC18 previously digested with BamHI and treated with alkaline phosphatase (32).
For nucleotide sequence determination, deletions were generated with restriction nucleases, and sequence completion was performed by Eurofins MWG Operon (Grenoble, France). The nucleotide sequences of 3-kb HindIII-SalI and of 3.3-kb BglII-SmaI fragments encompassing faeD and faeT, respectively, were determined.
Genetic fusions were constructed on the cloned genes by insertion of uidA-Km cassettes (1) into the SacII site of faeD or into the Eco47III site of faeT. The faeR gene was inactivated by insertion into the internal BamHI site of the CKC15 Cm cassette (21). Plasmids were introduced into D. dadantii cells by electroporation. The insertions were integrated into the D. dadantii chromosome by marker exchange recombination after successive cultures in low phosphate medium in the presence of the appropriate antibiotic. Verification of the correct recombination of the insertions was performed by PCR.
Overproduction of the proteins FaeD and FaeT.
The T7 promoter-T7 RNA polymerase system (35) was used to overproduce the esterases. The 1.6-kb BamHI-HpaI fragment overlapping faeD and the 2.3-kb PstI-SmaI fragment overlapping faeT were inserted into the pT7-6 and pT7-5 vectors, respectively. The nucleotide sequences of the resulting plasmids, pNA30 and pNA55, respectively, were verified. The plasmids pNA30 (faeD+) and pNA55 (faeT+) were introduced into the E. coli strain BL21(DE3), which contains a chromosomal copy of the T7 RNA polymerase gene under the control of the lacUV5 promoter (33). After E. coli transformation, the feruloyl esterase activity of the recombinant clones was verified by using the screening test in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). The BL21(DE3) cells containing the plasmids were grown at 30°C in LB medium with ampicillin (200 μg ml−1). When the optical density at 600 nm reached 0.4 to 0.6, the synthesis of T7 RNA polymerase was induced by the addition of IPTG at a final concentration of 2 mM, and cells were grown for an additional 2 h.
Cellular fractionation.
Different cellular fractions were obtained from E. coli BL21(DE3)-induced cells. The bacterial cells were recovered by centrifugation for 2 min at 8,000 rpm. The pellet was suspended in 0.7 ml of 80 mM Tris-HCl buffer at pH 8.0 and 0.1% Triton X-100, and the cells were broken by sonication. Centrifugation for 2 min at 10,000 × g led to the recovery of the supernatant containing the soluble proteins. The pellet corresponding to the insoluble proteins and cell debris was suspended in 0.7 ml of 80 mM Tris-HCl buffer, pH 8. The periplasmic fraction was recovered by osmotic shock (6).
Analytical procedures.
The protein concentrations in each fraction were determined by the Bradford method using a commercial protein assay kit (Bio-Rad) and bovine serum albumin (BSA) as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on slab gels (4% stacking gel and 12% separating gel) using the Mini-Protean II system (Bio-Rad). The proteins were revealed by staining with Coomassie blue G-250.
Pathogenicity tests.
Plant infections were inoculated as previously described (12, 38). Ten chicory leaves were infected for each strain using 106 bacteria per inoculation site. After incubation in a dew chamber for 24 h at 30°C, the length of rotted tissue was measured to estimate the disease severity. Similarly, 10 potato tubers were infected for each strain using 5 × 106 bacteria per inoculation site. Following incubation in a dew chamber for 48 h at 30°C, the weight of rotted tissue was measured to estimate the disease severity. After infections, the plant-macerated tissue was recovered and used to perform bacterial cell numeration by dilution plating and β-glucuronidase assay (38). The specific activity was calculated as nmol of product formed per 109 bacteria. The wild-type strain used as a negative control showed no detectable β-glucuronidase activity. A fusion in the pelD gene was used as a positive reference in each experiment (14).
RESULTS AND DISCUSSION
Isolation of two D. dadantii genes encoding feruloyl esterases.
A culture medium containing ethyl ferulate has been previously used to detect feruloyl esterase activity in Bacillus spp. (9). In order to detect colonies degrading ferulate esters, we adapted a similar plate assay for the enterobacterium D. dadantii. Since the presence of ethyl ferulate in the growth medium strongly inhibited the growth of both D. dadantii and E. coli, we overcame this inhibition by adding ethyl ferulate in an overlay poured onto the plates after 24 h of growth. The presence of this substrate resulted in an opaque appearance, and clear haloes appeared around the colonies degrading ethyl ferulate after a few hours of incubation (Fig. 1). D. dadantii 3937 gave a positive response to this test. In contrast, E. coli strains were totally negative. Consequently, this test was used to screen a D. dadantii genomic library introduced into E. coli NM522. Three clones giving a positive reaction, FE1, FE2, and FE3, were selected among about 800 transformants (10- to 20-kb inserts). FE1 and FE2 had 12.3- and 16.7-kb inserts, respectively, including a 5.5-kb common region. FE3 had a 17-kb insert distinct from those of FE1 and FE2. The esterase gene common to FE1 and FE2 was named faeD, and that found in FE3 was named faeT. These genes were localized more precisely by subcloning (Fig. 2). A second screening of the D. dadantii genomic library was performed on about 1,000 novel clones. Four novel positive clones were obtained, and their restriction analysis indicated that they carried the gene faeD. This test appeared to be specific for enzymes degrading ferulate esters, since a negative response was obtained with E. coli containing other known D. dadantii esterase genes, such as pemA, pemB, paeX, paeY, estC, and estV (data not shown).
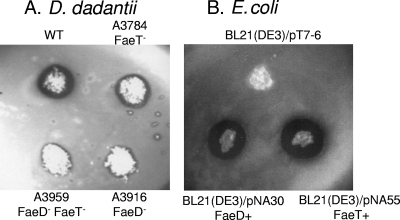
FIG. 1.
In situ detection of feruloyl esterases. The test was performed on D. dadantii colonies grown on M63 medium supplemented with glycerol (A) and on E. coli after growth on M63 medium supplemented with glucose, Casamino Acids, and IPTG (B). The wild-type D. dadantii 3937 strain (WT) gave a positive response, and E. coli strain BL21(DE3)/pT7-6 was used as a negative control.
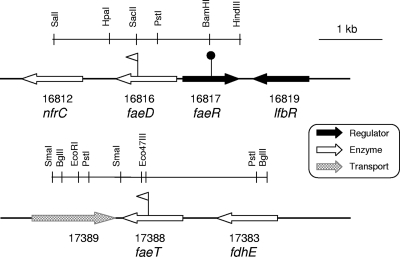
FIG. 2.
Genetic organization of the faeD and faeT regions in the D. dadantii 3937 genome. The flags indicate the positions of the uidA-Km insertions, resulting in gene fusions (triangles), and the Cm insertions in the regulatory gene (circles).
Characterization of the D. dadantii genes faeD and faeT.
The sequence of a 3-kb DNA fragment including faeD and that of a 3.3-kb fragment overlapping faeT were determined. The faeD and faeT sequences were identical to those reported for ID16816 and ID17388, respectively, in the 3937 genome sequence (GenBank accession no. CP002038.1). Insertion of a uidA-Km cassette into the SacII site interrupting faeD suppressed the positive response to the esterase screening test, confirming that this gene encodes the esterase activity. Similarly, insertion of a uidA-Km cassette in the Eco47III site of faeT confirmed the correct identification of this esterase gene.
The gene faeD is divergently transcribed from a gene encoding a regulator of the LysR family, which we named faeR (ID16817) (Fig. 2). The genes faeD and faeR are separated by 174 nucleotides (nt) and could share the same regulatory region. The gene faeD is followed by a sequence typical of a rho-independent termination site, and it is separated by a 581-nt intergenic space from the gene nrfC (ID16812) encoding an iron-sulfur protein which is part of the formate-dependent nitrite reductase complex. The gene faeR is adjacent to lfbR, encoding a previously analyzed LacI family regulator of unknown function (38). The gene faeT is separated by 661 nt from the upstream gene, fdhE (ID17383), encoding a chaperone of formate dehydrogenase, and it is separated by 99 nt from the downstream divergent gene encoding a potential transporter. Thus, considering their genetic organization (Fig. 2), both faeD and faeT probably constitute independent transcriptional units.
The genes faeD and faeT encode predicted proteins of 324 and 326 amino acids, respectively. The products of both ID16816 and ID17388 were annotated as a putative lipase/esterase (http://asap.ahabs.wisc.edu/asap/ASAP1.htm). FaeD and FaeT shared 44% identity between them and about 27% identity with the D. dadantii pectin acetylesterase PaeX (Fig. 3). Comparison of their sequences with those of the classified carbohydrate esterases (http://www.cazy.org/) indicated that they belong to the large family CE10. The primary sequence of these proteins includes the motif GXSXG, characteristic of the active site of serine hydrolases (4) (Fig. 3). The main differences between FaeD and FaeT are observed in their N-terminal regions. In contrast to FaeT, the N-terminal sequence of FaeD shows the characteristics of a classical signal sequence of exported proteins, with a putative cleavage site between the two alanine residues at positions 27 and 28.
FIG. 3.
Alignment of FaeD, FaeT, and PaeX. The N-terminal signal sequence and the G-S-G motif of serine hydrolases are underlined. The stars indicate the conserved residues that are putative constituents of the catalytic triad of a serine hydrolase, S, D, and H.
The locus faeD-faeR is conserved in the genomes of the Dickeya and Pectobacterium species D. dadantii Ech586 and Ech703, D. zeae Ech1591, P. carotovorum PC1 and Wpp14, P. wasabiae WWPP163, and P. atrosepticum SCRI1043 (67 to 72% identity). The gene faeT is conserved in D. dadantii Ech586 and Ech703 and in D. zeae Ech1591 (79% identity). In contrast, there is no faeT ortholog in the Pectobacterium species.
Characterization of the esterases FaeD and FaeT.
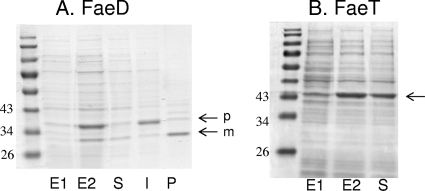
The faeD and faeT genes were inserted under the control of the T7 promoter in the pT7-6 and pT7-5 vectors, respectively. The resulting plasmids, pNA30 (faeD+) and pNA55 (faeT+), were used to overproduce the FaeD and FaeT proteins by IPTG induction in E. coli BL21(DE3). The feruloyl esterase plate assay showed that colonies of BL21(DE3) containing either pNA30 or pNA55 are surrounded by large haloes, while a negative response was obtained with the empty pT7-6 vector (Fig. 1). Different cellular fractions were prepared and analyzed by SDS-PAGE (Fig. 4). In the case of FaeD, two bands of overproduced proteins were observed, with apparent sizes of 31 and 35 kDa. This result is in agreement with the data deduced from the faeD nucleotide sequence, predicting a precursor form of 35,045 Da and a mature form obtained after cleavage of the predicted signal sequence of 32,270 Da. The 35-kDa band was located mainly in the fraction corresponding to insoluble or membrane proteins, and the 31-kDa band was recovered mainly in the periplasmic fraction (Fig. 4). A previous analysis of the 3937 secretome demonstrated that FaeD is an extracellular protein secreted by the Out system involved in the secretion of most D. dadantii pectinases (18). The first step of this type II secretion is the exportation of the secreted protein through the inner membrane by the general Sec system. The conservation of this step in E. coli confirmed the functionality of the FaeD signal sequence.
FIG. 4.
Overproduction and cellular localization of the FaeD and FaeT proteins in E. coli. The proteins were separated by SDS-PAGE, and the gels were stained with Coomassie blue. The positions of the molecular weight standards are indicated. The arrows indicate the positions of the overproduced proteins (p, precursor form; m, mature form). (A) Overproduction of FaeD. Cell lysates of BL21(DE3)/pNA30 before induction (E1) and after induction (E2) with IPTG. Cell fractionation obtained after IPTG induction: soluble (S), insoluble (I), and periplasmic (P) fractions. (B) Overproduction of FaeT. Cell lysates of BL21(DE3)/pNA55 before (E1) and after (E2) induction with IPTG and the soluble protein fraction obtained after induction.
In the case of FaeT, a single band of overproduced protein was observed. Its size of 35 kDa corresponded to that of the protein predicted from the faeT nucleotide sequence (34,686 Da). More than 80% of the 35-kDa protein was found in the fraction containing the soluble proteins, suggesting that FaeT is a cytoplasmic protein.
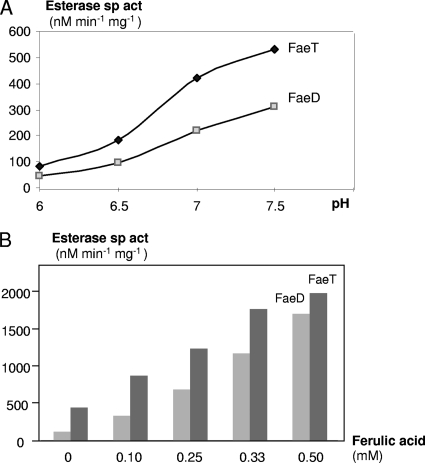
We used p-nitrophenyl-acetate (PNPA) as a substrate to perform esterase assays. The highest esterase activity was found in the periplasmic fraction for BL21(DE3)/pNA30 cells (FaeD+) and in the soluble fraction for BL21(DE3)/pNA55 (FaeT+). These fractions were retained for a biochemical analysis of FaeD and FaeT. An esterase assay at different pHs showed that the activity of FaeD and FaeT increased with the rise in pH (Fig. 5A), but the substrate became unstable at an alkaline pH. Thus, the standard assay was performed at pH 6.5 in 50 mM phosphate buffer. The initial velocities of the enzymes were determined at different PNPA concentrations under standard conditions. Apparent Km and Vmax values for FaeD were 1.7 mM and 625 nmol min−1 mg−1, respectively. For FaeT, apparent Km and Vmax values were 2.2 mM and 1,140 nmol min−1 mg−1, respectively.
FIG. 5.
Esterase activity of FaeD and FaeT. (A) Effect of pH. The esterase specific activity (sp act) was assayed with 2 mM PNPA as the substrate in 50 mM phosphate buffer at pH 6.0 to 7.5. (B) Activation by ferulic acid. The esterase assay was performed with 2 mM PNPA in 50 mM phosphate buffer at pH 6.5 in the presence of increasing concentrations of ferulic acid.
We used various compounds to test their potential effect on the esterase activity. No difference in enzymatic activity was observed after the addition of cations (Ca2+, Cu2+, Fe2+, Mg2+, or Mn2+) at a final concentration of 0.1 or 1 mM or of EDTA at a final concentration of 10 mM. In contrast, we observed a strong activation of both enzymes by ferulic acid (Fig. 5B). Addition of 0.5 mM ferulic acid gave a 15- and 4-fold increase of the FaeD and FaeT activities, respectively. This activation could result from a conformational change of the proteins FaeD and FaeT, giving rise to more active forms.
Phenotype and virulence of the faeD and faeT mutants.
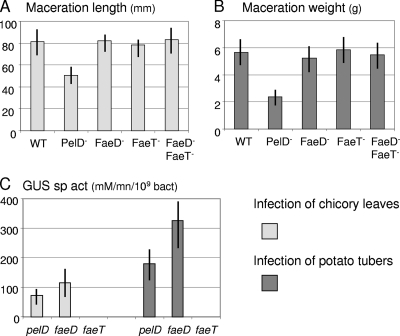
The plate assay was used to compare the feruloyl esterase activity levels of the feruloyl esterase single and double mutants (FaeD−, FaeT−, and FaeD− FaeT−) with that of the wild-type strain (Fig. 1). When faeT was inactivated, the size of the esterase halo clearly decreased. When faeD was inactivated, only very low residual activity was detected, as observed in the faeD faeT double mutant. Such low residual activity could be due to nonspecific esterases. Thus, FaeD appears to be responsible for most feruloyl esterase activity in D. dadantii.
The extent of soft rot caused by different mutants was tested on chicory leaves and potato tubers and compared with the symptoms caused by the wild-type strain 3937 (Fig. 6). We observed no significant difference in the degree of maceration caused by the faeD and faeT mutants. Moreover, these mutations did not affect bacterial growth in the plant tissues (data not shown). Similarly, the virulence of the faeR mutant and the faeD faeT double mutant on potato tubers or chicory leaves was not affected (Fig. 6 and data not shown). Transcriptional fusions with the reporter gene uidA were used to estimate the in planta expression of faeD and faeT. After infection, the expression of the faeD and faeT fusions in the macerated tissue was compared to that of the highly inducible pectate lyase gene pelD (14). The faeD fusion was transcribed in the macerated tissue at levels similar to those of pelD (Fig. 6C). In contrast, no expression of the faeT fusion could be detected. Similar data were observed for potato tuber infection. These results indicate that feruloyl esterase genes are not essential for the development of soft rot disease but that the faeD gene is expressed during plant infection.
FIG. 6.
Infection of chicory leaves and potato tubers with the Fae− mutants. (A) After a 24-hour infection with each mutant and the wild-type strain (WT), the length of macerated tissue was measured to estimate the disease severity. (B) Similarly, the weight of macerated tissue was measured after a 48-hour infection of potato tubers. (C) After infection, the β-glucuronidase (GUS) assay and a bacterial numeration were performed on the macerated tissue in order to estimate the expression of the uidA fusions in pelD, faeD, and faeT. The mean values and the standard deviations reported correspond to 10 infections with each strain.
Expression of the genes faeD and faeT under different conditions.
Different phenolic compounds, polysaccharides, and carbon sources were added to the growth medium to reveal any potential effects on faeD or faeT transcription. The expression of the faeD fusion was stimulated by about 3-fold in the presence of plant extracts or ethyl ferulate and 11-fold by ferulic acid (Table 2). It was not significantly affected by the addition of acetosyringone, benzoic acid, cinnamic acid, p-coumaric acid, vanillic acid, arabinogalactan, arabinose, galactose, galacturonate, mannose, pectin, polygalacturonate, rhamnose, salicine, xylose, or xylan. The faeT fusion was weakly expressed under all the conditions, and none of the compounds tested affected its expression (Table 2). This low expression level of faeT was confirmed by transcriptome analysis (G. Condemine, personal communication).
TABLE 2.
Expression of the faeD and faeT transcriptional fusions
| Fusion/mutation | Potential inducera | β-Glucuronidase sp actb |
|---|---|---|
| faeD::uidA | None | 860 |
| Plant extract | 2,672 | |
| Ethyl ferulate | 2,176 | |
| Ferulic acid | 9,691 | |
| Polygalacturonate | 911 | |
| faeD::uidA faeR | None | 565 |
| Ferulic acid | 557 | |
| faeT::uidA | None | 12 |
| Plant extract | 15 | |
| Ferulic acid | 14 | |
| Polygalacturonate | 11 | |
| faeT::uidA faeR | None | 15 |
| Ferulic acid | 13 |
Strains were grown to late exponential growth phase in minimal medium containing glycerol. The potential inducers were added at the following concentrations: plant extract, 1%; ethyl ferulate, 0.1 mM; ferulic acid, 0.1 mM; polygalacturonate, 2 g liter−1.
The values reported (in nmol min−1 mg−1) are the averages of results from at least 3 independent experiments, with standard deviations corresponding to less than 20%. There is no significant difference between the faeT data. In comparison to the noninduced value, the expression of faeD was significantly different (P < 0.1) in the presence of plant extract, ethyl ferulate, or ferulic acid.
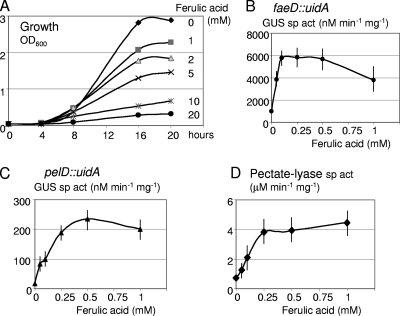
The effect of ferulic acid concentrations ranging from 0.05 to 1 mM was tested on faeD induction (Fig. 7 B). Maximal induction was reached at a concentration of 0.1 mM. Since phenolic acids are known to inhibit bacterial growth (26), concentrations ranging from 0.1 to 20 mM were tested on D. dadantii growth (Fig. 7A). While no inhibition at concentrations allowing faeD induction was observed, a clear inhibition occurred at concentrations higher than 1 mM, with both decreased growth rate and yield.
FIG. 7.
Effect of ferulic acid on growth, faeD transcription, and pectate lyase production. (A) Growth of the wild-type D. dadantii strain was monitored in M63 medium supplemented with glycerol and different concentrations of ferulic acid. OD600, optical density at 600 nm. (B to D) The β-glucuronidase (GUS) specific activity, reflecting the transcription of the faeD::uidA (B) or of the pelD::uidA (C) fusion, were determined at different concentrations of ferulic acid, as was the total pectate lyase specific activity (D).
When one of the regulatory genes kdgR, pecS, pecT, pir, or gacA was inactivated, the expression of the faeD fusion remained induced in the presence of ferulic acid, with induction factors of about 12, 10, 8, 11 or 7, respectively. Thus, none of these regulators seems directly involved in the induction by ferulic acid. In the presence of the crp mutation, faeD expression decreased by about 7-fold but remained weakly induced by ferulic acid. The gene faeR, which is divergent from faeD and encodes a LysR family regulator, was inactivated. The faeD fusion was introduced in the faeR::Cm mutant. When faeR was inactivated, the expression of the faeD fusion decreased, and it was no longer inducible in the presence of ferulic acid (Table 2). The expression of the faeT fusion remained low; it seemed to be unaffected by faeR inactivation (Table 2). Thus, FaeR is an activator of faeD expression, and it is responsible for faeD induction in the presence of ferulic acid.
Effect of ferulic acid on induction of the pectate lyase genes.
During analysis of faeD expression, we also tested the effect of the potential inducers on pectate lyase production. We observed that ferulic acid weakly induced pectate lyase activity (Table 3). In the presence of polygalacturonate, the effect of ferulic acid was even clearer, and induction reached a 5-fold factor (Table 3). Similar effects on the expression of a transcriptional fusion with pelD, which encodes one of the major pectate lyases, were observed (Table 3). Thus, induction by ferulic acid takes place at the transcriptional level, and ferulic acid acts in synergy with polygalacturonate to increase the induction of pectate lyase genes. Plant extracts were previously shown to provoke an increased pectate lyase induction in the presence of polygalacturonate, and this phenomenon was called hyperinduction, but the inducers were not identified (3, 14, 25). The presence of ferulic acid results in the same type of hyperinduction. However, the effects of ferulic acid and plant extract remained additive (Table 3), suggesting that ferulic acid is not the sole component of the plant extract with pectate lyase inducing activity. The effect of ferulic acid concentrations, ranging from 0.05 to 1 mM, was tested on the induction of the pelD fusion and on pectate lyase production (Fig. 7). The induction was maximal at ferulic acid concentrations of 0.25 to 0.5 mM. Thus, pectate lyase induction required a 5-fold-higher concentration of ferulic acid than faeD induction (Fig. 7).
TABLE 3.
Effect of ferulic acid on the production of pectate lyases
| Potential inducer(s)a | Sp actb |
|
|---|---|---|
| Pectate lyase | β-Glucuronidase (fusion pelD::uidA) | |
| None | 0.04 | 4 |
| Ferulic acid | 0.09 | 10 |
| Polygalacturonate | 1.04 | 29 |
| Polygalacturonate + ferulic acid | 5.01 | 235 |
| Plant extract | 0.24 | |
| Polygalacturonate + plant extract | 4.62 | |
| Plant extract + ferulic acid | 1.38 | |
| Polygalacturonate + ferulic acid + plant extract | 11.31 | |
The potential inducers were added at the following concentrations: plant extract, 1%; polygalacturonate, 2 g liter−1; ferulic acid, 0.5 mM.
The pectate lyase specific activity is expressed in μmol min−1 mg−1. The β-glucuronidase specific activity, reflecting the expression of the pelD transcriptional fusion, is expressed in nmol min−1 mg−1. These results are the average values from at least 3 independent experiments, with standard deviations corresponding to less than 20%.
Analysis of pectate lyase production in mutants with inactivation of the regulator KdgR, PecS, PecT, Pir, Crp, or GacA indicated that none of them are necessary for the induction of pectate lyase genes by ferulic acid (data not shown). Similarly, analysis of the faeR mutant indicated that, in contrast to faeD regulation, FaeR is not involved in the induction of pectate lyases in the presence of ferulic acid (data not shown).
Conclusion.
The plant-pathogenic bacterium D. dadantii produces a large array of plant cell wall-degrading enzymes, including several pectinases. Using an in situ detection test, we showed that it also produces two feruloyl esterases, FaeD and FaeT. These enzymes have been previously analyzed in a variety of microorganisms, and they have potential biotechnological and medicinal applications (2). Carbohydrate esterases are classified in different families on the basis of their amino acid sequences (http://www.cazy.org/), and the previously characterized feruloyl esterases are members of the carbohydrate esterase family CE1. While recent selections were usually based on the search for CE1 homologues, we used a functional screening to isolate new enzymes. This led us to identify two esterases of the family CE10 (Fig. 3), which also includes the D. dadantii pectin acetylesterase PaeX (31) and several noncarbohydrate active enzymes. Thus, FaeD and FaeT are the first two members of a new family of feruloyl esterases. It will be interesting to study their properties in detail and to clarify their specificity on natural substrates for comparison with CE1 enzymes. We have already seen strong activation of the FaeD and FaeT esterases by ferulic acid (Fig. 5B), which was not observed for CE1 feruloyl esterases. Indeed, feruloyl esterases of the FaeD-FaeT family may have particular properties that could be of interest in extending the biotechnological applications of these enzymes.
Inactivation of the faeD and faeT genes indicated major activity of FaeD in D. dadantii (Fig. 1). In a previous analysis of the D. dadantii secretome, FaeD was identified among the extracellular proteins secreted by the Out system, which mediates the secretion of most pectinases (18). In the present report, we have shown that faeD expression is specifically induced by ferulic acid. Such induction is mediated by FaeR, a regulator of the LysR family encoded by the divergent faeD gene. The faeD-faeR locus is conserved in all the sequenced genomes of pectinolytic enterobacteria of the genera Dickeya and Pectobacterium. The extracellular location of FaeD allows it to act on feruloylated polysaccharides. This location explains why the small product, ferulic acid, rather than the large substrate acts as an inducer of the D. dadantii faeD gene. The basal level of faeD expression may lead to the release of ferulic acid from plant polysaccharides, which would then activate faeD transcription at low concentrations (Fig. 7B). Increased feruloyl esterase activity gives rise to higher ferulic acid concentrations, which in turn, induce the production of pectate lyases (Fig. 7D) that cause the maceration symptom.
The induction of feruloyl esterases by phenolic compounds has been previously observed. The gene cinB of Butyrivibrio fibrosolvens is specifically induced by feruloylated oligosaccharides which are also CinB substrates (7). In the fungus Aspergillus niger, feruloyl esterase production is stimulated by the presence of the product, free ferulic acid (10). In Aspergillus kawachii, ferulic acid increased the production of feruloyl esterases and also that of various plant cell wall-degrading enzymes (19).
In D. dadantii, ferulic acid induces pectate lyase production by an unknown mechanism, independent of FaeR. For this induction, ferulic acid acts in synergy with the pectate lyase substrate polygalacturonate (Table 3), which when cleaved, liberates strong inducers (13). Such synergy has been previously observed with crude plant extracts (3, 25), and it is probably due to the effect of several plant compounds, including ferulic acid. It was recently shown that some phenolic compounds affect the expression of the type III secretion system of D. dadantii. While cinnamic and o-coumaric acids induce this system (39), p-coumaric acid represses its expression (23). The GacAS system was thought to be responsible for this induction (39). In contrast, GacA is not involved in the induction of pectate lyases by ferulic acid. These data suggest the involvement of a number of phenolic signals and response regulators in D. dadantii. Plant phenolic compounds may play an important role as signals in the plant-D. dadantii interaction.
Phenolic acids constitute an important class of organic compounds produced by plants, and several are recognized as signaling molecules by plant-associated microorganisms. The role of acetosyringone in the induction of vir genes of agrobacteria has been widely documented, somewhat masking the involvement of other phenolic compounds. The Agrobacterium tumefaciens strain KU12, which has a large host range, is induced by the common phenolic compounds ferulic and p-coumaric acids (22). Some simple phenolics, including ferulic acid, were also identified as chemoattractants and inducers of the nod genes in certain rhizobial species (17). The variety of phenolic compounds, with differences in structures, amounts, and prevalence, may generate subtleties in the dialogues that take place during the plant-bacteria interactions.
Despite the fact that inactivation of the faeD or faeT gene did not reduce D. dadantii virulence, faeD is clearly expressed in the macerated tissue (Fig. 6C). Feruloyl esterases could have multiple influences on D. dadantii pathogenesis. The dissociation of internal cross-links in the polysaccharide network of the plant cell wall by feruloyl esterases could facilitate the access of the main-chain-degrading enzymes to the polysaccharide backbone. Suppression of the polysaccharide esterifications by feruloyl esterases could also favor the action of depolymerases. Finally, the liberation of ferulic acid by feruloyl esterases would contribute to the induction of pectate lyases, the main determinant of plant maceration. These different effects of feruloyl esterases facilitate soft rot disease caused by pectinolytic bacteria.
Acknowledgments
Appreciation is expressed to Guy Condemine, Vladimir Shevchik, and Valerie James for reading the manuscript. We thank the other members of the Lyon Erwinia group for helpful discussions and the members of the International Erwinia Consortium for the exchange of unpublished data concerning the D. dadantii 3937 genome.
This work was supported by a Ph.D. grant from the Syrian government to S.H. and by grants from the Centre National de la Recherche Scientifique and from the Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Bardonnet, N., and C. Blanco. 1992. 'uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 72:243-247. [DOI] [PubMed] [Google Scholar]
- 2.Benoit, I., E. G. Danchin, R. J. Bleichrodt, and R. P. de Vries. 2008. Biotechnological applications and potential of fungal feruloyl esterases based on prevalence, classification and biochemical diversity. Biotechnol. Lett. 30:387-396. [DOI] [PubMed] [Google Scholar]
- 3.Bourson, C., S. Favey, S. Reverchon, and J. Robert-Baudouy. 1993. Regulation of the expression of a pelA::uidA fusion in Erwinia chrysanthemi and demonstration of the synergistic action of plant extract with polygalacturonate on pectate lyase synthesis. J. Gen. Microbiol. 139:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528-530. [DOI] [PubMed] [Google Scholar]
- 5.Caffall, K. H., and D. Mohnen. 2009. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344:1879-1900. [DOI] [PubMed] [Google Scholar]
- 6.Copeland, B. R., R. J. Richter, and C. E. Furlong. 1982. Renaturation and identification of periplasmic proteins in two-dimensional gels of Escherichia coli. J. Biol. Chem. 257:15065-15071. [PubMed] [Google Scholar]
- 7.Dalrymple, B. P., and Y. Swadling. 1997. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR). Microbiology 143:1203-1210. [DOI] [PubMed] [Google Scholar]
- 8.Delangle, A., A.-F. Prouvost, V. Cogez, J.-P. Bohin, J.-M. Lacroix, and N. Hugouvieux Cotte-Pattat. 2007. Characterization of the Erwinia chrysanthemi gan locus involved in galactan catabolism. J. Bacteriol. 189:7053-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaghy, J., P. F. Kelly, and A. M. McKay. 1998. Detection of ferulic acid esterase production by Bacillus spp. and lactobacilli. Appl. Microbiol. Biotechnol. 50:257-260. [DOI] [PubMed] [Google Scholar]
- 10.Faulds, C. B., R. P. deVries, P. A. Kroon, J. Visser, and G. Williamson. 1997. Influence of ferulic acid on the production of feruloyl esterases by Aspergillus niger. FEMS Microbiol. Lett. 157:239-244. [DOI] [PubMed] [Google Scholar]
- 11.Hugouvieux-Cotte-Pattat, N. 2004. The RhaS activator controls the Erwinia chrysanthemi 3937 genes rhiN, rhiT and rhiE involved in rhamnogalacturonan catabolism. Mol. Microbiol. 51:1361-1374. [DOI] [PubMed] [Google Scholar]
- 12.Hugouvieux-Cotte-Pattat, N., and S. Charaoui-Boukerzaza. 2009. Catabolism of raffinose, sucrose, and melibiose in Erwinia chrysanthemi 3937. J. Bacteriol. 191:6960-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 14.Hugouvieux-Cotte-Pattat, N., H. Dominguez, and J. Robert-Baudouy. 1992. Environmental conditions affect the transcription of the pectinases gene of Erwinia chrysanthemi 3937. J. Bacteriol. 174:7807-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1989. Isolation of Erwinia chrysanthemi mutants altered in pectinolytic enzyme production. Mol. Microbiol. 3:1587-1597. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, T. 1997. Structure and functions of feruloylated polysaccharides. Plant Sci. 127:111-127. [Google Scholar]
- 17.Kape, R., M. Parniske, and D. Werner. 1991. Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl. Environ. Microbiol. 57:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazemi-Pour, N., G. Condemine, and N. Hugouvieux-Cotte-Pattat. 2004. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 10:3177-3186. [DOI] [PubMed] [Google Scholar]
- 19.Koseki, T., N. Mimasaka, K. Hashizume, Y. Shiono, and T. Murayama. 2007. Stimulatory effect of ferulic acid on the production of extracellular xylanolytic enzymes by Aspergillus kawachii. Biosci. Biotechnol. Biochem. 71:1785-1787. [DOI] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Lebeau, A., et al. 2008. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ. Microbiol. 10:545-559. [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. W., S. Jin, W. S. Sim, and E. W. Nester. 1995. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 92:12245-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., et al. 2009. The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microbiol. 75:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, New York, NY.
- 25.Nomura, K., W. Nasser, H. Kawagishi, and S. Tsuyumu. 1998. The pir gene of Erwinia chrysanthemi EC16 regulates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc. Natl. Acad. Sci. U. S. A. 95:14034-14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravirala, R. S., et al. 2007. Efflux pump gene expression in Erwinia chrysanthemi is induced by exposure to phenolic acids. Mol. Plant Microbe Interact. 20:313-320. [DOI] [PubMed] [Google Scholar]
- 27.Resibois, A., M. Colet, M. Faelen, T. Schoonejans, and A. Toussaint. 1984. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102-112. [DOI] [PubMed] [Google Scholar]
- 28.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS, a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127-1139. [DOI] [PubMed] [Google Scholar]
- 30.Shevchik, V. E., and N. Hugouvieux-Cotte-Pattat. 1997. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi 3937. Mol. Microbiol. 24:1285-1301. [DOI] [PubMed] [Google Scholar]
- 31.Shevchik, V. E., and N. Hugouvieux-Cotte-Pattat. 2003. PaeX, a second pectin acetylesterase of Erwinia chrysanthemi 3937. J. Bacteriol. 185:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchik, V. E., et al. 1999. Characterization of the exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J. Bacteriol. 181:1652-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 34.Surgey, N., J. Robert-Baudouy, and G. Condemine. 1996. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J. Bacteriol. 178:1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tardy, F., W. Nasser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toth, I. K., K. S. Bell, M. C. Holeva, and P. R. Birch. 2003. Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4:17-30. [DOI] [PubMed] [Google Scholar]
- 38.Van Gijsegem, F., A. Wlodarczyk, A. Cornu, S. Reverchon, and N. Hugouvieux-Cotte-Pattat. 2008. Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Mol. Plant Microbe Interact. 21:1471-1481. [DOI] [PubMed] [Google Scholar]
- 39.Yang, S., et al. 2008. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS One 3:e2973. [DOI] [PMC free article] [PubMed] [Google Scholar]