Abstract
Colicin M (Cma) lyses Escherichia coli cells by inhibiting murein biosynthesis through hydrolysis of the phosphate ester between C55-polyisoprenol and N-acetylmuramyl (MurNAc)-pentapeptide-GlcNAc in the periplasm. To identify Cma functional domains, we isolated 54 point mutants and small deletion mutants and examined their cytotoxicity levels. Activity and uptake mutants were distinguished by osmotic shock, which transfers Cma into the periplasm independent of the specific FhuA receptor and the Ton system. Deletion of the hydrophobic helix α1, which extends from the compact Cma structure, abolished interference with the antibiotic albomycin, which is transported across the outer membrane by the same system as Cma, thereby identifying α1 as the Cma site that binds to FhuA. Deletion of the C-terminal Lys-Arg strongly reduced Cma translocation across the outer membrane after binding to FhuA. Conversion of Asp226 to Glu, Asn, or Ala inactivated Cma. Asp226 is exposed at the Cma surface and is surrounded by Asp225, Asp229, His235, Tyr228, and Arg236; replacement of each with alanine inactivated Cma. We propose that Asp226 directly participates in phosphate ester hydrolysis and that the surrounding residues contribute to the active site. These residues are strongly conserved in Cma-like proteins of other species. Replacement of other conserved residues with alanine inactivated Cma; these mutations probably altered the Cma structure, as particularly apparent for mutants in the unique open β-barrel of Cma, which were isolated in lower yields. Our results identify regions in Cma responsible for uptake and activity and support the concept of a three-domain arrangement of Cma.
Colicins are plasmid-encoded protein toxins released by Escherichia coli and taken up by sensitive E. coli cells (8, 35). Colicins are important traits of E. coli since they are produced by 40 to 50% of the natural isolates. Colicins are also the only proteins imported by E. coli. These enzymes or pore-forming proteins are equipped with receptor binding and translocation domains for their import into sensitive cells. They kill cells by degrading DNA or RNA in the cytoplasm, dissipating the membrane potential by forming pores in the cytoplasmic membrane, degrading murein, or inhibiting murein biosynthesis in the periplasm. Cma cleaves the phosphate bond between C55-polyisoprenol and N-acetylmuramyl (MurNAc)-pentapeptide-GlcNAc (10) in the periplasm near the outside of the cytoplasmic membrane. The released C55-polyisoprenol no longer translocates MurNAc-pentapeptide-GlcNAc across the cytoplasmic membrane. Normally, C55-polyisoprenol leaves the biosynthetic reaction as a pyrophosphate ester in the periplasm and enters the reaction cycle as a monophosphate ester at the inner side of the cytoplasmic membrane. The same lipid reaction cycle incorporates O antigen into lipopolysaccharide, which is also inhibited by colicin M (14).
Cma differs from the other colicins in various aspects. With 271 amino acid residues, it is the smallest of the colicins, most of which are composed of 500 to 700 residues (8). It is the only colicin that inhibits murein and O-antigen biosynthesis (13, 14, 25). It forms a compact structure (36), which makes it difficult to delineate the functional domains; most other colicins have extended conformations with well-separated domains. The fold of Cma is unique among colicins and even among all known proteins (36).
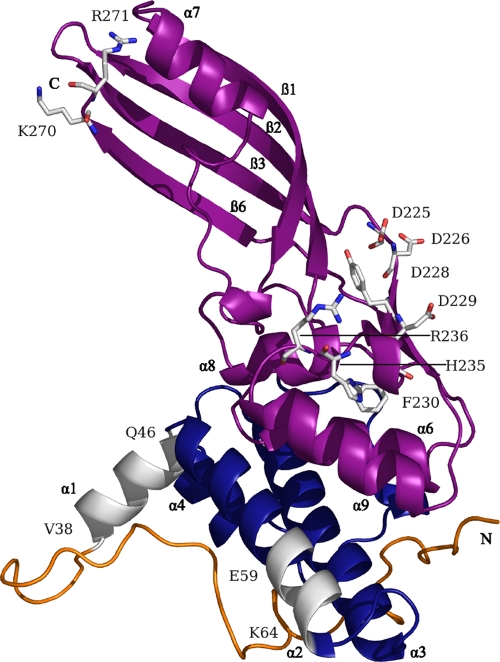
The boundaries of the Cma functional domains have been derived from the crystal structure (Fig. 1) (36) and the sequence identity (26 to 45%) of the C-terminal half (residues 124 to 271) to uncharacterized Cma-like proteins of Pectobacterium carotovorum, Pseudomonas syringae, Pseudomonas aeruginosa, various strains of Burkholderia, and a Pseudomonas phage (see Fig. S1 in the supplemental material). However, the predicted activity domain is not similar in sequence and conformation to known phosphatases (36). The N-terminal and central domains of these proteins have only a few identical residues (2, 3, 36), which reflects the import specificities of the various bacterial genera. Cma homologues of various Pseudomonas species, like Cma, release C55-polyisoprenol from lipid II (3).
FIG. 1.
Location of the mutations in an improved crystal structure of Cma (PDB 2XMX). This structure showed an excellent defined structural homogeneity for all main chain and side chain atoms, whereas the former structure (PDB 3DA4) showed partial disorder, in particular in the flexible N-terminal domain. Note that the short β1- and β2-strands in the previous structure are no longer listed as ß-strands since they are too short. This resulted in a renumbering of the ß-strands, ß1 to ß6 instead of ß1 to ß8. The predicted structural and functional domains are indicated as follows: yellow, N-terminal translocation domain, which was extended from the previously proposed extension from residues 1 to 35 (36) to 1 to 37 according to the results of this paper; blue, central receptor binding domain; magenta, C-terminal activity domain extending from residues 124 to 271 (2) and helix α1 (V38-Q46), which when mutated resulted in inactive Cma derivatives; N, N-terminal end; C, C-terminal end; white, carbon atoms; red, oxygen atoms; blue, nitrogen atoms. For the sake of clarity, not all sites whose predicted functions are discussed in the text are indicated.
Although 90% of the colicin M synthesized remains inside cells and is not released into the culture medium (32), it does not kill the producer cells. Colicin M kills cells only after it has been taken up across the outer membrane into the periplasm (15). Uptake of Cma specifically involves binding to the FhuA outer membrane receptor protein and the electrochemical potential of the cytoplasmic membrane for translocation into the periplasm. TonB, ExbB, and ExbD serve to transfer energy from the cytoplasmic membrane into the outer membrane (4, 6, 23).
The finding that killing of cells by Cma strictly requires the periplasmic prolyl cis-trans isomerase/chaperone FlpA (17), the determination of the Cma crystal structure (36), the identification of the phosphatase activity (10), and the prediction of Cma-like proteins in species other than E. coli (2, 3, 36) prompted our interest in the functional regions of Cma proteins. To study the functional domains of Cma and to correlate them with structural domains, we isolated and characterized mutants in the various predicted domains, with special emphasis on the activity domain.
By constructing and analyzing Cma mutants we identified sites that affect binding to FhuA, translocation across the outer membrane, and phosphatase activity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli AB2847 aroB thi tsx malT and its fkpA40 deletion mutant, E. coli Mo3 (17), were used for the activity assays. E. coli BL21(DE3) fhuA F− ompT gal dcm hsdB(rB− mB−) λ(DE3) lon lacI lacUV5-phage T7 gene 1 (17) served to produce Cma and mutant Cma proteins (30). This strain encodes the T7 RNA polymerase under lac control, which, upon induction with isopropyl-β-d-thiogalactopyranoside (IPTG), transcribes cma cloned downstream of the T7 gene 10 promoter on plasmid pMLD237 (generously provided by D. Mengin-Lecreulx, Université Paris-Sud, Orsay, France). The fhuA mutation prevents killing of the producer by Cma.
Point mutants and small deletions of Cma were generated by site-directed mutagenesis of cma with appropriate primers, which are available upon request, using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland). The cma gene on plasmid pMLD237 encodes Cma with an N-terminal (His)6 tag. Mutagenized cma genes were introduced into E. coli DH5α [λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] (12) by transformation, and the mutations were verified by sequencing the cma genes isolated from the transformants. If the yields of Cma were low, we again sequenced the cma genes to exclude T7 gene 10 promoter mutations. To determine Cma activity, we isolated wild-type and mutant Cma proteins from transformants of the E. coli BL21(DE3) fhuA mutant carrying the wild-type or mutated cma genes on pMLD237.
Cma sensitivity tests.
Sensitivity of E. coli cells to Cma was tested with Cma and mutant derivatives purified by Ni-nitrilotriacetic acid (NTA) agarose chromatography or with crude extracts of cells that overexpressed Cma or mutant derivatives. Cma had to be isolated from disrupted cells since the cma transformants released less than 10% of the Cma produced. They lack a Cma lysis gene, which is also absent on natural pColBM plasmids (32). Cells were grown in LB medium (19) at 37°C to an optical density at 578 nm (OD578) of 0.5, IPTG was added to a final concentration of 1 mM, and the cells were further cultured for 3 h. Cells from an 8-ml culture were harvested by centrifugation and suspended in 0.5 ml of 0.1 M Tris-HCl, pH 7.5. An aliquot of 0.25 ml was disrupted by sonication, and cell debris was removed by centrifugation (16,000 × g at 4°C). Tenfold dilutions of Cma in water were spotted onto 20-ml LB agar plates seeded with 109 cells of the strains to be tested in 3 ml of soft agar. The results are given as percentage of wild-type Cma activity (100%).
Larger amounts of Cma were isolated from 3.2-liter cultures, and Cma in the crude cell extract was purified by Ni-NTA agarose chromatography as previously described (17).
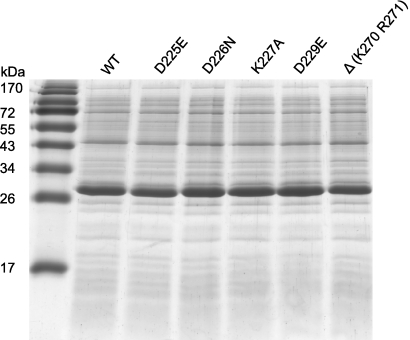
Synthesis of all mutant proteins was tested by SDS-PAGE of crude cell extracts. Since the strongest protein band was Cma (Fig. 2), crude extracts were tested, except for the 13 Cma mutant proteins listed in the footnote to Table 1, which were purified by Ni-NTA agarose chromatography. The purified proteins included mainly those which were assigned to the active center of Cma. From the purified proteins the same amounts of Cma were used in the assays. From the crude extracts similar amounts were used which, in the case of the active derivatives, resulted in the same activity levels in the spot test.
FIG. 2.
SDS-PAGE of crude cell extracts of the indicated wild-type (WT) and mutant strains harvested at the same OD578. The major band of 29 kDa is Cma.
TABLE 1.
Activity of Cma mutantsa
| Cma protein | Cell lysis (%) | % Surviving colonies treated with Cma and osmotic shockb | Locationc | Affected function |
|---|---|---|---|---|
| Wild type | 100 | 0 | ||
| V30D | 100 | ND | N terminus | |
| G32K A33K | 10 | ND | N terminus | |
| L36D | 100 | ND | N terminus | |
| L37D | 10 | ND | N terminus | |
| L36K L37K | 0 | 1 | N terminus | Uptake |
| V40D | 10 | ND | α1 | |
| V40K V41K | 1 | ND | α1 | |
| V41P | 10 | ND | α1 | |
| F44D | 100 | ND | α1 | |
| Δ38-46 | 0.01 | 0.3 | α1 | Uptake |
| E59A | 10 | 0.5 | α2 | Uptake |
| D60A | 100 | ND | α2 | |
| Y61A | 100 | ND | α2 | |
| K63A | 100 | ND | α2 | |
| K64A | 100 | ND | α2 | |
| K63A K64A | 100 | ND | α2 | |
| H65A | 100 | ND | α2 | |
| T174Ad | 10 | ND | β1 | |
| S178Pd | 0.1 | ND | β1 | |
| S178Ad | 10 | ND | β1 | |
| F181Ad | 0.01 | ND | β1 | |
| G205Ad | 0.01 | ND | β2 | |
| T206Ad | 1 | ND | β2 | |
| L207Ad | 0.01 | ND | β3 | |
| W215Ad | 0.01 | ND | β3 | |
| W215Rd | 0 | 100 | β3 | |
| Y217Ad | 0 | ND | β3 | |
| G219Ad | 0.1 | ND | β3 | |
| R222Ad | 1 | ND | β3 | |
| S223A | 100 | ND | β3 | |
| S223T | 1 | ND | β3 | |
| Y224A | 0 | 45 | β3-β4 | Activity |
| D225A | 0 | 3 | β3-β4 | Activity |
| D225E | 0 | 65 | β3-β4 | Activity |
| D225N | 1 | ND | β3-β4 | |
| D226A | 0 | 28 | β3-β4 | Activity |
| D226E | 0 | ND | β3-β4 | |
| D226N | 0 | 98 | β3-β4 | Activity |
| K227A | 10 | 0.3 | β4 | Uptake |
| Y228A | 0 | 100 | β4 | Activity |
| D229A | 0.1 | 67 | β4 | Activity |
| D229E | 1 | ND | β4 | |
| D229N | 10 | ND | β4 | |
| F230A | 0 | 5 | β4 | Activity |
| N231A | 10 | ND | β4-α9 | |
| S233A | 100 | 0.3 | β4-α9 | |
| T234A | 100 | ND | β4-α9 | |
| H235A | 0 | 3 | β4-α9 | Activity |
| R236A | 0 | 5 | β4-α9 | Activity |
| Y255F | 100 | ND | ß5 | |
| K266A | 100 | ND | β6 | |
| S268A | 100 | ND | β6 | |
| G269Ad | 10 | ND | β6 | |
| Δ(K270 R271) | 1 | 0.3 | β6 | Uptake |
Cell lysis was tested by spotting a 10-fold dilution series of Cma and its derivatives on plates seeded with E. coli AB2847. The highest dilution of wild-type Cma that gave a clear zone of growth inhibition was taken as reference for the activity of the Cma mutant proteins. For example, a mutant was assigned an activity of 1 when it gave a lysis zone at a 103-fold dilution compared to wild-type Cma, which gave a lysis zone at a 105 dilution (listed as 100%). The values after Cma treatment by osmotic shock are related to the number of colonies in cultures treated with sucrose without Cma (see Materials and Methods). Crude extracts were tested except in the cases of wild-type Cma and the Cma derivatives E59A, Y224A, D225E, D226A, D226N, K227A, Y228A, D229E, F230A, S233A, H235A, R236A, and Δ(K270 R271), which were purified on Ni-NTA agarose. The affected function is not predicted for mutants whose activity was not tested by osmotic shock but is discussed in the text.
Values below 0.1% are listed as 0. ND, not determined.
Helix and/or β-strand(s) is indicated. N terminus, no secondary structure.
Cma derivative that was obtained in lower yields, which reduced toxicity.
Osmotic shock.
Osmotic shock not only releases periplasmic proteins into the shock medium (20) but also transfers added proteins into the periplasm (2, 5, 9, 33). The Cma sensitivity of cells after osmotic shock was tested with purified Cma samples. E. coli AB2847 in 10 ml of LB medium was grown to an OD578 of 0.5; cells were harvested by centrifugation and then suspended in 1 ml of 0.01 M Tris-HCl-0.03 M NaCl, pH 7 (buffer 1). Cells were pelleted by centrifugation, washed once with buffer 1, and centrifuged again. The washed cells were suspended in 0.5 ml of 33 mM Tris-HCl, pH 7 (buffer 2), and 0.1-ml aliquots were added to four tubes. Sucrose (40% in buffer 2; 0.1 ml) was added to two tubes, and buffer 2 (0.1 ml) was added to the other two tubes. Cells were suspended by vortexing and by further shaking for 10 min at 23°C and then harvested by centrifugation at 4°C. Cma (10 μg in 10 μl) was added to an Eppendorf tube containing a cell suspension treated with sucrose and a tube containing an untreated cell suspension. Then 0.3 ml of ice-cold 0.5 mM MgCl2 and 30 μl of 1 mM CaCl2 were added to each tube; the in vivo activity of Cma depends on Ca2+ (24). The contents of the tubes were immediately mixed by vortexing. Two additional samples received no Cma but were otherwise treated the same way. The four samples were kept on ice for 15 min and then serially diluted 10-fold in buffer 2 at 4°C. Aliquots of 0.1 ml were suspended in 3 ml of LB soft agar and spread on 20-ml LB agar plates. After overnight incubation at 37°C, colonies after appropriate dilution were counted. The values listed in Table 1 are related to the number of CFU of cells treated with sucrose without Cma (shock alone), normalized to 6%, the average number of surviving colonies in the sucrose experiments, as illustrated by the following example: for partially active Cma with a deletion of the C-terminal K270 R271 sequence [CmaΔ(K270 R271)] in the spot test, 110 × 106 (100%) colonies were counted in the untreated culture, 2.40 × 106 (2.18%) were counted in the culture treated with Cma, 4.5 × 106 (4.1%) were counted in the culture treated with sucrose, and 2 ×104 (0.02%) were counted in the culture treated with Cma and sucrose, of which 0.3% survivors were calculated.
Competition of Cma with albomycin.
Cells of E. coli Mo3 fkpA were shaken at 160 rpm in nutrient broth at 37°C to an OD578 of 0.3. Dodecyl maltoside (0.1% final concentration) was added to prevent precipitation of Cma. Cma (final concentration, 100 μg ml−1) was added 10 min before albomycin (1 μg ml−1) was added. At various time points the optical density of the cultures was measured at 578 nm.
Cleavage of Cma by proteinase K.
Twenty micrograms of purified Cma and Cma derivatives was incubated with 0.01 μg of proteinase K for 13 min on ice and then analyzed by SDS-PAGE.
CD.
Circular dichroism (CD) spectra of electrophoretically homogeneous wild-type and mutant Cma proteins were recorded in a Jasco spectral polarimeter, model J-810, at 190 to 240 nm and 20°C. Temperature-dependent denaturation was measured at 220 nm between 20 and 80°C.
Denaturation of Cma and Cma mutants by urea.
Proteins (1 mg ml−1) dissolved in 20 mM potassium phosphate, 0.5 mM MgCl2, 2 mM mercaptoethanol, and 150 mM NaCl or in the same buffer containing 1 to 8 M urea were incubated for 15 min at 20°C. The samples were centrifuged, and the fluorescence emission spectrum was measured between 300 and 500 nm (1 nm bandwidth), with excitation at 280 nm (3 nm bandwidth) in a Jasco spectrofluorometer FP-6500. The values of the peak maxima given are the average of five determinations.
Bioinformatics.
Sequences homologous to Cma were searched with PSI-BLAST (1) using database nr, the nonredundant weekly updated sequence database at the NCBI, at standard parameters. The 10 sequences found were aligned with CLUSTAL W (31). The graphic was drawn with ESPript, version 2.2 (http://escript.ibcp.fr) (11), using the improved Cma crystal structure (Protein Data Bank [PDB] code 2XMX) and for similarity calculations the parameter equivalent, global score 1.0, was used.
RESULTS
Helix α1 is involved in Cma uptake.
Helix α1, VQVVYSFFQ, from residue 38 to 46 (Fig. 1; see also Fig. S1 in the supplemental material), is hydrophobic and extends from the otherwise compact Cma structure (36). We previously speculated that the helix might fix Cma to the cytoplasmic membrane, where the substrate resides (36). To test this hypothesis, we mutagenized cma on plasmid pMLD237 by PCR with appropriate primers and isolated the Cma mutant proteins from transformants of the E. coli BL21(DE3) fhuA mutant carrying the plasmid-encoded cma genes. The fhuA mutation prevents lysis of the producer strain by released Cma since sensitivity of cells to Cma requires import across the outer membrane via FhuA. Cma was by far the most dominant protein in crude cell extracts examined by SDS-PAGE (Fig. 2). The identity of the strongest band with Cma was examined with anti-Cma antiserum by Western blotting (data not shown).
We purified Cma and some of the Cma mutant derivatives by affinity chromatography on Ni-NTA agarose, taking advantage of the N-terminal (His)6 tag carried on all Cma mutant proteins. We then assayed the activity of the Cma mutant proteins in a lysis assay in which formation of clear zones of growth inhibition was examined on plates seeded with the Cma-sensitive E. coli AB2847. The same amounts of Cma and Cma derivatives were used for the assays. Hydrophobic residues were replaced by charged residues to examine the role of the strong hydrophobicity of the helix, and proline was introduced to distort the helix. Cma mutants V40D and V41P, in the middle of helix α1, had 10% of the wild-type Cma activity, whereas F44D near the end of helix α1 was fully active (Table 1). Helix α1 is preceded by two leucine residues, L36 and L37; L36D did not reduce Cma activity, and L37D reduced activity to 10% of that of wild-type Cma. The double mutant L36K L37K [Cma(L36K L37K)] was inactive. To extend the analysis closer to the N terminus, we constructed the double mutant G32K A33K, which displayed 10% of the wild-type activity. When we deleted helix α1 (residues 38 to 46 [Cma(Δ38-46)]), the deletion did not affect the yield of the protein, but 105-fold more Cma(Δ38-46) was required to inhibit the growth of E. coli AB2847 than wild-type Cma (Table 1).
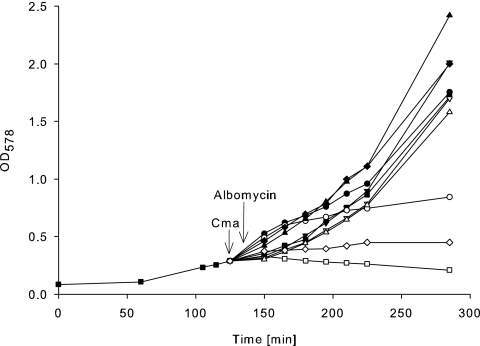
To distinguish between lack of uptake and lack of activity, we bypassed the specific requirement for uptake for the outer membrane FhuA receptor protein and the TonB, ExbB, and ExbD proteins by using osmotic shock (5). This method was originally developed and widely used to release periplasmic proteins into the medium (20) and can be used to transfer colicins unspecifically into cells (2, 5, 9, 22, 33). The method is not quantitative but clearly distinguishes between active and inactive Cma and between defects in uptake and enzymatic activity. Upon osmotic shock in the presence of Cma(Δ38-46), the number of surviving cells was reduced to 0.3% of the cells treated with only osmotic shock (Table 1). The very low activity of Cma(Δ38-46) against viable cells and its high activity after unspecific transfer into the periplasm indicate that the defect is in its uptake and not in its activity. We verified this conclusion using a competition assay between Cma(Δ38-46) and the antibiotic albomycin, which is taken up across the outer membrane by the same proteins as Cma. E. coli Mo3 (AB2847 fkpA40) was used, which is not lysed by Cma but is sensitive to albomycin. Wild-type Cma inhibited the killing of cells by albomycin, whereas Cma(Δ38-46) did not (Fig. 3). The mutant protein did not interfere with albomycin because it did not compete with albomycin for binding to FhuA. We concluded that helix α1 forms a hydrophobic interface through which Cma binds to FhuA. The hydrophobic helix α1 is not required to fix Cma to the cytoplasmic membrane as its deletion did not prevent killing of osmotically shocked cells. Inactive Cma(L36K L37K) was active when shocked into cells (Table 1) and inhibited albomycin (data not shown). Although the point mutations are adjacent to helix α1, these residues are involved not in binding to FhuA but in translocation across the outer membrane.
FIG. 3.
Binding of wild-type and mutant Cma proteins to the FhuA receptor in a competition assay with albomycin. Exponentially growing E. coli Mo3 fkpA40 in LB medium (▪) and in LB medium containing 1 μg ml−1 albomycin (□), 0.1 mg ml−1 wild-type Cma (▾), wild-type Cma and albomycin (▿), Cma(E59A) (•), Cma(E59A) and albomycin (○), CmaΔ(K270 R271) (▴), CmaΔ(K270 R271) and albomycin (▵), Cma(Δ38-46) (⧫), and Cma(Δ38-46) and albomycin (⋄). Growth was monitored spectrophotometrically. The arrows indicate the addition of the Cma samples and albomycin. The figure shows an example of reproducibly obtained data.
Analysis of region 59-EDYIKKH-65.
We functionally analyzed the Cma region 59-EDYIKKH-65 because it is conspicuously polar. Cma(E59A) displayed 10% of the wild-type activity, was active when transferred into the periplasm by osmotic shock (Table 1), and showed some inhibition of cell killing by albomycin (Fig. 3), which suggests that the residual activity was related to weak FhuA binding. Mutants D60A, Y61A, K63A, K64A, K63A K64A, and H65A were fully active (Table 1). We then tested whether the mutants were impaired in their interaction with the colicin M immunity protein Cmi. Cma is a basic protein (pI 8.9), and Cmi is an acidic protein (pI 4.9). The sequence KKH could participate in binding to Cmi, which contains acidic sequences such as DVEDE. However, cells expressing Cmi encoded on plasmid pPG105 were as resistant to the single Cma mutants as to wild-type Cma (data not shown), which renders it unlikely that this region interacts with Cmi.
Deletion of the C-terminal K270 R271 sequence renders Cma uptake deficient.
Previously, removal of the KR sequence at the C terminus of Cma with carboxypeptidase B inactivated Cma (9). Here, we constructed a genetic deletion, which resulted in CmaΔ(K270 R271) with an activity of 1% of that of wild-type Cma (Table 1). The mutant protein was active when osmotically shocked into cells. The protein reproducibly inhibited albomycin (Fig. 3), which showed that it binds to FhuA. The deletion affects a step in Cma uptake after binding to FhuA.
Mapping the active site of Cma.
The sequence 222-RSYDDKYDFNASTHR-236 contains an accumulation of conserved amino acids (see Fig. S1 in the supplemental material) and has been predicted to belong to the active site (3, 9, 22, 36). We replaced each of these amino acids individually with alanine. To exclude the possibility that inactivity was caused by the lack of uptake into the periplasm, inactive mutant Cma proteins were transferred into the periplasm by osmotic shock.
Nearly all Cma replacement mutants displayed strongly reduced activities when spotted onto indicator plates seeded with the Cma-sensitive E. coli AB2847 strain. The Cma derivatives that were transferred into the periplasm by osmotic shock remained inactive (Table 1), which indicated that the mutations affected the phosphatase activity and not the uptake of Cma. The mutations may directly affect residues that catalyze phosphate ester hydrolysis or the binding of the substrate, or they may change the geometry of the active site such that substrate binding and/or hydrolysis is impaired. Since the carboxyl groups in the side chain of the aspartate residues may be directly involved in phosphate ester hydrolysis, D225, D226, and D229 were replaced by glutamate, thereby keeping the carboxyl group on a side chain one CH2 group longer than in aspartate. Cma(D226E) remained completely inactive, which suggested that precise positioning of the carboxyl group was required for hydrolysis. Cma(D225E) after osmotic shock showed even lower activity than Cma(D225A), and Cma(D229E) was 10-fold more active than Cma(D229A).
To further examine the function of the carboxyl groups, we constructed the mutatnts Cma(D225N), Cma(D226N), and Cma(D229N). Cma(D226N) was inactive, whereas Cma(D225N) and Cma(D229N) displayed 1 and 10% of the wild-type activity, respectively. It is conceivable that D226 directly participates in phosphate ester hydrolysis, whereas D225 and D229 are not directly involved in catalysis but may bind the substrate or orient the catalytic side chains in an active conformation. The same reasoning applies for the inactive Cma mutant proteins Y224A, Y228A, H235A, and R236A, whose side chains may participate in binding or hydrolysis of the phosphate ester bond, or both. Since F230 does not contain a catalytic side chain, the aromatic side chain must be important for the correct conformation of the active site or for the binding of the substrate, or both. Cma(N231A) was partially active, and Cma(S233A) and Cma(T234A) were fully active. Serine residues in phosphatases frequently contribute to the cleavage of phosphate groups. The S223L replacement was previously found to inactivate Cma (22), whereas S233A, previously found to confer inactivity (22), did not inactivate Cma in the present study. Cma(S233A) also killed cells after osmotic shock. K227 is not essential for catalysis as Cma(K227A) displayed 10% of the Cma wild-type activity in the lysis test and was active after transfer into the periplasm by osmotic shock.
Stability of the mutant proteins.
In crude cell extracts, the yields of the proteins with mutations assigned to the active site were similar to the yield of wild-type Cma, which showed that the loss of activity was not caused by the loss of protein. The altered amino acids of the inactive Cma mutants could be located outside the active site but change the Cma structure such that the active site is no longer catalytic. To examine possible structural changes, we compared the circular dichroism (CD) spectra of selected purified Cma derivatives with the spectrum of purified wild-type Cma. In addition, we determined the denaturation of the proteins by heat and urea and their degradation by proteinase K. The CD spectra of the Cma derivatives with mutations in the putative active site did not differ from the wild-type Cma CD spectrum (see Fig. S2 in the supplemental material). The temperature-dependent melting curves of the proteins were virtually identical (see Fig. S3) and showed the same melting temperature (Tm) of 54°C. Urea denatured the wild-type and mutant Cma proteins between 5 and 6 M, except for CmaΔ(K270 R271), which was denatured at a slightly lower concentration (see Fig. S4). We set up the degradation with proteinase K such that a fraction of the protein was left undegraded to better reveal differences in the peptide pattern. Under these conditions, a major degradation product of 24 kDa (complete protein, 29 kDa) was previously identified that lacks the N-terminal portion (9). The degradation products of the proteins did not differ (see Fig. S5). These results suggested that the inactivity of the Cma derivatives with a mutation in the putative active site was not caused by structural changes beyond the replaced side chains.
Additional amino acids conserved among the Cma-like proteins of the strains shown in Fig. S1 in the supplemental material were mutated to alanine (Table 1). Among them were F181A, G205A, L207A, W215A, W215R, G219A, and Y217A, which were inactive. These amino acids are contained in the β1-, β2-, or β6-strands (Table 1), which are part of the Cma-typical open β-barrel formed by four β-strands (36). The strongly reduced activity, however, could mainly be attributed to the reduced yield of these proteins in soluble form. Cma S178P, F181A, W215A, and W215R gave very weak bands after SDS-PAGE, and the amounts of G205A, T206A, and L207A were approximately 20% of wild-type Cma (data not shown). Most Cma proteins with mutations in the open β-barrel were unstable.
DISCUSSION
Comparisons of colicin sequences have revealed that functional domains have been swapped between colicins, resulting in new colicins (summarized in references 6 and 8). Although the three-domain arrangement in the compact crystal structure of Cma (36) is less obvious than in other colicins, it was derived from previous mutational studies. The TonB box was identified by site-specific mutations (22). A C-terminal sequence contains the receptor binding and activity domains but lacks the translocation domain, as shown by a C-terminal proteolytic fragment that protects cells from being killed by wild-type Cma and by phage T5, which binds to FhuA, but is inactive against cells that are killed when the fragment is transferred into the periplasm by osmotic shock (9). The activity domain was recently confined to a genetically engineered fragment of residues 124 to 271, which is inactive against cells that are killed when the fragment enters the periplasm by osmotic shock (2). Randomly generated mutants were accumulated in a region tentatively assigned to the active site (22).
In the present study, we constructed inactive mutants in all three functional and structural domains. Deletion of helix α1 rendered Cma(Δ38-46) inactive against cells, but Cma(Δ38-46) retained activity after transfer by osmotic shock into the periplasm. Uptake of Cma(Δ38-46) was impaired, as shown by the lack of inhibition of albomycin uptake, which takes the same route across the outer membrane as Cma. The lack of interference with albomycin is caused by the lack of binding to the common receptor, FhuA. Helix α1 is best suited for binding to FhuA as it extends from the otherwise compact structure of Cma (Fig. 1). However, the involvement of a hydrophobic sequence in Cma binding to FhuA is unexpected because the surface loops of FhuA are predominantly hydrophilic. As seen in the crystal structure of the receptor protein BtuB bound to colicin E3, the 29 residues of BtuB that interact with the 27 residues of the receptor binding domain of colicin E3 are predominantly hydrophilic (18). The interacting residues of colicin E3 are near the tip of the coiled-coil region and are thus exposed, as are the residues of helix α1 of Cma. The crystal structure of the receptor binding domain of colicin Ia bound to the receptor protein Cir shows mainly a patch of four Asp and Glu residues at the tip of the receptor binding domain (34) that interact with two arginine residues of Cir, forming 18 hydrogen bonds (7). Since the receptor binding domain of Cma is not folded in a coiled coil, the highly exposed helix α1 fulfills a function similar to the coiled-coil tip of colicins Ia (7), E2 (27), and E3 (18, 29). It is likely that additional residues contribute to Cma binding to FhuA. Initial binding may cause structural changes in FhuA that expose hydrophobic regions of FhuA that interact with helix α1. Such large conformational changes occur upon binding of the colicin Ia receptor domain to the colicin Ia receptor Cir. Loops 7 and 8 of Cir move as a rigid body to open outwards by 37° compared with the same loops in the uncomplexed Cir structure (7).
The FhuA binding site does not extend beyond helix α1 toward the N terminus as inactive Cma(L36K L37K) inhibited albomycin uptake. Since Cma(L36K L37K) killed cells when it was transferred into the periplasm by osmotic shock, it is impaired in uptake. The residues involved in translocation across the outer membrane are immediately adjacent to the region that binds to FhuA. Binding of Cma and subsequent translocation involve hydrophobic interactions with FhuA. In contrast, CmaΔ(K270 R271) that bound to FhuA and killed cells when transferred into the periplasm by osmotic shock is located far from helix α1 (Fig. 1). The deletion must therefore specifically affect uptake after binding to FhuA by causing subtle conformational changes that affect translocation through long-range structural changes in Cma. The higher sensitivity to urea supports this notion. Of the same category is the previously isolated Cma(R115C) mutant which does not kill cells unless it is taken up by osmotic shock. It competes with wild-type Cma for binding to FhuA (22). This mutant and the other two translocation mutants, Cma(L36K L37K) and CmaΔ(K270 R271), show that mutations located outside the predicted translocation domain, residues 1 to 37 (Fig. 1), may strongly affect Cma uptake after binding to FhuA.
Our previous random mutagenesis of the cma gene with hydroxylamine (22) resulted in inactive Cma mutants that centered at residue 197 and included D226. Of the 10 inactive mutants isolated, three mutations were identified in G197 (G197S, G197D, and G197V). We hypothesized that these sites are part of the active site of Cma (22). G197 and D226 are strictly conserved in all Cma-like proteins (see Fig. S1 in the supplemental material). The proximity of conserved residues D226, D229, and Y255 in the crystal structure (36) also suggested that D226 as well as D229 and Y255 forms part of the active site, and this hypothesis was confirmed by analyzing the mutants.
Inactive Cma TonB box mutants were previously isolated by site-specific mutagenesis (22). One of the mutants, V6R, is active against strains mutated in TonB, TonB(Q160L) and TonB(Q160K). These TonB mutants also suppress TonB box mutations in FhuA (26) and BtuB (16), which indicates that there are sites of interaction between TonB and the TonB boxes of the receptors. Indeed, these interactions have been recently shown in crystal structures (21, 28). The TonB box of Cma assumes a conformation similar to the conformation of the FhuA TonB box and BtuB TonB box in complex with the TonB β-strand of the region around residue 160 (36). Since the TonB box is required for Cma uptake after binding to FhuA, it defines the translocation domain. This domain is contained in the flexible N-terminal region.
Very recently, Barreteau et al. (2) carried out an alanine-scanning mutagenesis of Cma with the aim of deciphering the catalytic domain. Of the 19 mutants in the mutagenized conserved sequence H135-E254, 13 are also within the 54 newly isolated mutants listed in Table 1. Barreteau et al. determined the cytotoxicity on plates, used osmotic shock, and determined phosphatase activities with 14C-labeled C55-polyisoprenyl-P-P-MurNAc-pentapeptide-GlcNAc. In all mutants studied, the cytotoxicity and the number of survivors after osmotic shock correlated with phosphatase activity. In particular, D226, Y228, D229, H235, and R236 displayed very low or no toxicity and phosphatase activity, which agrees with our assignment of the mutations to the activity domain. These residues are exposed to the surface of Cma in one region where they gain access to the substrate (Fig. 1).
We believe that D226 is directly involved in hydrolysis since our conversion of the aspartate residue to glutamate or asparagine abolished activity. Not only the carboxylate moiety but also its exact positioning is essential. D225 is less important than D226, even though Cma(D225E) and Cma(D225A) are inactive, because Cma(D225N) still exhibits 10% of the wild-type cytotoxicity. Accordingly, D225 occurs in only one other Cma-like protein, whereas D226 is strictly conserved in all Cma-like proteins (see Fig. S1 in the supplemental material). Although D229 is strictly conserved in all Cma-like proteins, it is less important than D226; Cma(D229A) was inactive and Cma(D229E) and Cma(D229N) showed 1 and 10% of the wild-type cytotoxicity, respectively. Y228 may play a structural rather than a catalytic role since in some Cma-like proteins it is replaced by phenylalanine, which does not contain a catalytic side chain. The same conclusion applies to F181 and Y217, which are essential for Cma activity. The aromatic residues are oriented to the inside of the Cma molecule and may play an important structural role.
The results of previous studies (2, 3, 9, 22, 36) and of the current study provide consistent results about the three-domain structure of Cma. The N-terminal domain is responsible for translocation across the outer membrane, the central domain is responsible for binding to the FhuA receptor, and the C-terminal domain contains the phosphatase. Residues located in the catalytic center were identified in the surface-exposed link between β3 and β4 and within β3 and β4. The residues are extended over a relatively large area, which may reflect the size of the large substrate. Cma most probably recognizes mainly -O-P-P-MurNAc-pentapeptide-GlcNAc since the C55-isoprenoide is embedded in the lipid bilayer.
Supplementary Material
Acknowledgments
We thank Andrei Lupas for generous support, Kornelius Zeth for helpful discussions, Silke Patzer for advice, and Karen A. Brune for critically reading the manuscript.
This work was supported by the Max Planck Society and the German Science Foundation (BR330/25-1).
Footnotes
Published ahead of print on 10 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Altschul, S., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreteau, H., et al. 2010. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J. Biol. Chem. 285:12378-12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreteau, H., et al. 2009. Human and plant pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J. Bacteriol. 191:3657-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V. 1995. Energy-coupled transport and signal-transduction through the gram-negative outer-membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., J. Frenz, K. Hantke, and K. Schaller. 1980. Penetration of colicin M into cells of Escherichia coli. J. Bacteriol. 142:162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., et al. 2007. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 26:2594-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascales, E., et al. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreher, R., V. Braun, and B. Wittmann-Liebold. 1985. Functional domains of colicin M. Arch. Microbiol. 140:343-346. [DOI] [PubMed] [Google Scholar]
- 10.El Ghachi, M., et al. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281:22761-22772. [DOI] [PubMed] [Google Scholar]
- 11.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Harkness, R. E., and V. Braun. 1989. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J. Biol. Chem. 264:6177-6182. [PubMed] [Google Scholar]
- 14.Harkness, R. E., and V. Braun. 1989. Inhibition of lipopolysaccharide O-antigen synthesis by colicin M. J. Biol. Chem. 264:14716-14722. [PubMed] [Google Scholar]
- 15.Harkness, R. E., and V. Braun. 1990. Colicin M is only bactericidal when provided from outside the cell. Mol. Gen. Genet. 222:37-40. [DOI] [PubMed] [Google Scholar]
- 16.Heller, K. J., R. J. Kadner, and K. Günter. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 17.Hullmann, J., S. I. Patzer, C. Römer, K. Hantke, and V. Braun. 2008. Periplasmic chaperone FkpA is essential for imported colicin M toxicity. Mol. Microbiol. 69:926-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurisu, G., et al. 2003. The structure of BtuB with bound E3 R-domain implies a translocon. Nat. Struct. Biol. 10:948-954. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Nossal, N. G., and L. A. Heppel. 1966. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J. Biol. Chem. 241:3055-3062. [PubMed] [Google Scholar]
- 21.Pawelek, P. D., et al. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399-1402. [DOI] [PubMed] [Google Scholar]
- 22.Pilsl, H., et al. 1993. Domains of colicin M involved in uptake and activity. Mol. Gen. Genet. 240:103-112. [DOI] [PubMed] [Google Scholar]
- 23.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 24.Schaller, K., R. Dreher, and V. Braun. 1981. Structural and functional properties of colicin M. J. Bacteriol. 146:54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaller, K., J.-V. Höltje, and V. Braun. 1982. Colicin M is an inhibitor of murein biosynthesis. J. Bacteriol. 152:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schöffler, H., and V. Braun. 1989. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217:378-383. [DOI] [PubMed] [Google Scholar]
- 27.Sharma, O., et al. 2007. Structure of the complex of the colicin E2 R-domain and its BtuB receptor. The outer membrane colicin translocon. J. Biol. Chem. 282:23163-23170. [DOI] [PubMed] [Google Scholar]
- 28.Shultis, D. D., M. D. Purdy, C. N. Banchs, and M. C. Wiener. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396-1399. [DOI] [PubMed] [Google Scholar]
- 29.Soelaiman, S., K. Jakes, N. Wu, C. Li, and M. Shoham. 2001. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 8:1053-1062. [DOI] [PubMed] [Google Scholar]
- 30.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J., D. Higgiin, and T. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thumm, G., T. Ölschläger, and V. Braun. 1988. Plasmid pColBM-C1139 does not encode a colicin lysis protein but contains sequences highly homologous to the D protein (resolvase) and the oriV region of the mini-F plasmid. Plasmid 20:75-82. [DOI] [PubMed] [Google Scholar]
- 33.Tilby, M., I. Hindenach, and U. Henning. 1978. Bypass of receptor-mediated resistance to colicin E3 in Escherichia coli K-12. J. Bacteriol. 136:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiener, M., D. Freymann, P. Ghosh, and R. M. Stroud. 1997. Crystal structure of colicin Ia. Nature 385:461-464. [DOI] [PubMed] [Google Scholar]
- 35.Zakharov, S. D., and W. A. Cramer. 2004. On the mechanism and pathway of colicin import across the E. coli outer membrane. Front. Biosci. 9:1311-1317. [DOI] [PubMed] [Google Scholar]
- 36.Zeth, K., C. Römer, S. I. Patzer, and V. Braun. 2008. Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli. J. Biol. Chem. 283:25324-25331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.