Abstract
Comparative genome analysis of Enterococcus faecium recently revealed that a genomic island containing the esp gene, referred to as the esp-containing pathogenicity island (esp PAI), can be transferred by conjugation and contains a partial Tn916-like element and an integrase gene, intA. Here, we characterize the role of intA in the excision of the esp PAI. An intA insertion-deletion mutant in E. faecium E1162 (E1162ΔintA) was constructed and in trans complemented with wild-type intA (E1162ΔintA::pEF30). Circular intermediates (CI) of excised esp PAI were determined using inverse PCR analysis on purified chromosomal DNA from strains E1162, E1162Δesp, E1162ΔintA, and E1162ΔintA::pEF30. In E1162 and E1162Δesp, CI of the esp PAI were detected. No CI were detected in E1162ΔintA, while in the complemented strain E1162ΔintA::pEF30 CI formation was restored, indicating that intA is essential for excision and subsequent mobilization of the esp-containing genomic island in E. faecium. Based on the fact that this island can be mobilized and is self-transmissible, we propose to change the name of the esp PAI to ICEEfm1.
For a long time, the Gram-positive species Enterococcus faecium was considered a harmless commensal of the mammalian gastrointestinal (GI) tract. However, in the last 2 decades E. faecium has emerged as one of the leading nosocomial pathogens (16, 18, 20). Molecular epidemiological studies using multilocus sequence typing (MLST) identified host-specific genogroups, including a hospital-associated E. faecium subpopulation (18-20). Recently, whole-genome sequencing revealed the total sequence of a previously partially sequenced genomic island containing the enterococcal surface protein gene, esp, designated the esp pathogenicity island (esp PAI) in strain E1162 (GenBank accession number ABQJ00000000) (6, 17). In this strain, the esp PAI (contig 156; GenBank accession number ABQJ01000139) (Fig. 1A) is flanked by direct repeats (DR) of 54 bp, is integrated at the 3′ end of the rpsI gene, and contains a partial, conjugation module-containing, Tn916-like element. Conjugation experiments using E1162Δesp as a donor revealed that the E1162 esp PAI is self-transmissible and integrates in a site-specific manner (17). This indicates that the esp PAI in E1162 is an integrative conjugative element (ICE) rather than a genome or pathogenicity island, which is a chromosomal region acquired through horizontal gene transfer that is no longer or never was self-transmissible (2).
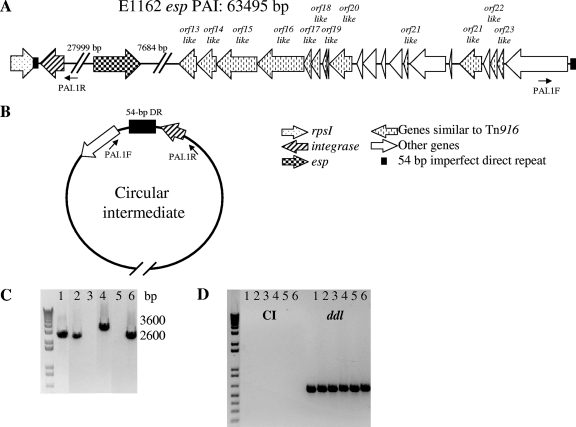
FIG. 1.
(A) Structure of the E. faecium E1162 esp PAI. Arrows indicate coding sequences (CDS) and direction of transcription; only CDS that are relevant for this study are depicted. Indicated are binding sites for the outward-facing primers (PAI.1R and PAI.1F) used to detect circular intermediates. (B) A schematic representation of circular intermediate (CI) formation. Primers (PAI.1R and PAI.1F) for the detection of circular intermediates are indicated. (C) Detection of circular intermediates using inversed PCR analysis on purified chromosomal DNA with outward-facing primers at the borders of the esp PAI. (D) Detection of circular intermediates using inversed PCR analysis on purified chromosomal DNA after S1 nuclease treatment; ddl PCR analysis was performed as a positive control. Lanes: 1, conjugation mixture of E. faecium strain E1162Δesp with BM4105RF; 2, E1162; 3, E1162ΔintA; 4, E1162ΔintA::pEF30; 5, E1162ΔintA::pEF25; and 6, the transconjugant (TC).
The term ICE has been introduced for all elements that excise by site-specific recombination into a circular form, self-transfer by conjugation, and subsequently integrate into the host genome, very often at the 3′ end of tRNAs, whatever the specificity and mechanism of integration and conjugation (1, 2). ICEs are characterized by the presence of an integrase/excisionase gene belonging to the tyrosine or serine family of recombinases, a conjugation module, and a modular structure (9). The intA gene (locus tag EfmE1162_2326) is the only integrase encoded on the esp PAI. The IntA protein is homologous to a family of phage and phage-related integrases. BLAST searches with the amino acid sequence of the IntA protein revealed that this protein belongs to the tyrosine family of recombinases that is commonly located on the ICE, like Tn916 (3, 4, 10, 13, 14). Remarkably, the closest homolog of IntA was found with an integrase of Enterococcus faecalis E1Sol (83% amino acid identity). This integrase was also found directly downstream of the rpsI gene of E. faecalis and is probably also part of a mobile element because of the presence of a partial Tn916 element, which includes the tetracycline resistance gene tetR, downstream of the integrase. We therefore hypothesized that intA is involved in the excision and formation of circular intermediates (CI) of the esp PAI.
An intA insertion-deletion mutant was constructed in a clinical E. faecium strain (E1162), using the temperature-sensitive vector (pTEX5500ts) as previously described (5, 8). The intA mutant was named E1162ΔintA. The other bacterial strains and plasmids used in this study are listed in Table 1. Culture conditions and genetics methods were described previously (5).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. faecium strains | ||
| E1162 | Clinical blood isolate; Ampr Vans Chls Gens Spcs; esp PAI+; CI+ | 5, 19 |
| E1162ΔintA | int insertion-deletion mutant of E1162; Chlr Gens; esp PAI+; CI− | This study |
| E1162ΔintA::pEF25 | int insertion-deletion mutant of E1162, complemented with “empty” vector; Spcr Chlr Gens; PAI+; CI− | This study |
| E1162ΔintA::pEF30 | int complementation strain of E1162ΔintA; Spcr Chlr Gens; esp PAI+; CI+ | This study |
| BM4105RF::PAI | Transconjugant; Efm PAI+ | 17 |
| Plasmids | ||
| pTEX5500ts | Shuttle plasmid; temperature sensitive in Gram-positive hosts; Chlr Genr | 8 |
| pEF25 | Shuttle plasmid pAT18 with spectinomycin resistance cassette cloned in the EcoRI site; Spcr Eryr | This study |
| pEF30 | pEF25::int; Spcr Eryr | This study |
Amp, ampicillin; Van, vancomycin; Chl, chloramphenicol; Gen, gentamicin; Spc, spectinomycin; esp PAI, E. faecium esp-containing pathogenicity island; CI, circular intermediates; +, presence; −, absence.
IntA is involved in recipient-independent formation of circular intermediates.
To determine whether conjugative transfer of the esp PAI occurs via CI, PCRs were performed on purified DNA from the conjugation mixture of E1162Δesp and BM4105RF using outward-facing primers designed on the borders of the PAI (Fig. 1A and B). This PCR yielded a single amplification product with the expected size of 2.6 kb that can occur only when CI forms are present (Fig. 1C). Subsequent sequencing confirmed the specificity of the PCR product. To investigate whether CI formation is dependent on the presence of a recipient strain and on the presence of a functional intA gene, in total three different donor strains, comprising E1162, the intA insertion-deletion mutant E1162ΔintA (Table 1), and the previously described transconjugant (TC) obtained from the mating of E1162Δesp with BM4105RF (17), were grown on filters in a manner similar to filter-mating experiments but in the absence of a recipient strain. CI PCR analysis revealed single bands for all donor strains except for E1162ΔintA (Fig. 1C). This indicates that CI formation is not dependent on the presence of the recipient strain in the conjugation mixture and that intA is involved in excision of the esp PAI (Fig. 1C). Sequencing of the CI PCR products of E1162 and TC confirmed that the PCR products were specific for the junction site.
In trans complementation of intA.
Complementation studies were performed to determine whether expression of intA in trans, from a plasmid (pEF30) (Table 1), was able to restore CI formation in the intA mutant strain. As a control the intA mutant strain was also complemented with plasmid pEF25 (Table 1), which does not contain wild-type intA. Both plasmids were transferred to E1162ΔintA by electroporation, resulting in E1162ΔintA::pEF25 and E1162ΔintA::pEF30 (Table 1). Restored CI formation was observed in E1162ΔintA::pEF30, but not in the intA mutant strain complemented with pEF25 (Fig. 1C), corroborating the finding that intA is involved in esp PAI excision. The size of the CI PCR product in E1162ΔintA::pEF30 is larger than that in the other donor strains due to the insertion of the cat resistance marker in the inactivated copy of intA in the esp PAI of E1162ΔintA.
Sequence analysis of CI PCR products.
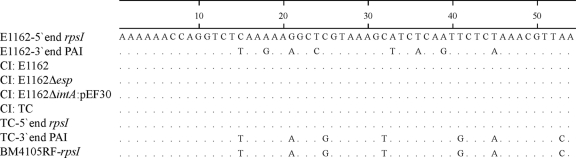
The whole-genome sequence project of E1162 enabled us to identify the borders of the esp-containing PAI (6, 17). In order to investigate whether CI formation occurs via recombination of the two imperfect 54-bp direct repeats leading to the presence of a single direct repeat at the junction site, as described for Tn916 elements (13), all CI PCR fragments were subjected to sequencing. Furthermore, the 5′- and 3′-end direct repeats of the esp PAI in the transconjugant and the integration site (the rpsI gene) of the recipient BM4105RF strain were sequenced. Sequence analysis revealed that all CI contained a single 54-bp direct repeat at the junction site. Interestingly, in all cases the sequences were identical to the 54-bp sequence at the 5′ end of the esp PAI of E1162 and different from the 54-bp sequence at the 3′ end of the esp PAI of E1162 and the rpsI integration site of BM4105RF (Fig. 2). The 3′-end sequence of the TC was identical to the BM4105RF rpsI integration site. This suggests that recombination always occurs via the 5′ end of the esp PAI.
FIG. 2.
Sequence alignment of the 54-bp direct repeat from the 5′-end rpsI and 3′-end PAI of strain E1162 and the transconjugant (TC), the rpsI in recipient strain BM4105RF, and the 54-bp direct repeat at the junction sites of the circular intermediates (CI) of E1162, E1162Δesp, E1162ΔintA::pEF30, and the transconjugant.
CI exist as ssDNA.
The identification of the 54-bp sequence of the 5′ end of the esp PAI suggests that transfer occurs via single-stranded DNA (ssDNA), as has been hypothesized previously (9, 12, 13). In order to investigate the presence of single-stranded CI, the purified chromosomal DNA obtained to determine CI was treated with S1 nuclease, which specifically degrades ssDNA and not double-stranded DNA (dsDNA). In the following CI PCR analysis, no PCR products were obtained, indicating degradation of CI, while the PCR result on the chromosomally encoded ddl gene remained positive (Fig. 1D). This indicates that the esp PAI forms a single-stranded CI.
Mechanistic differences in excision of the esp PAIs in E. faecium and E. faecalis.
Comparable to other ICEs, the esp PAI is flanked by two large direct (imperfect) repeats of 54 bp. Only the 54-bp DR at the 5′ end of the esp PAI is present at the junction site, suggesting that the esp PAI is similarly transferred as described for the ICE-like element Tn916. In contrast to the esp PAI, Tn916 does not integrate in a site-specific manner, but prefers A·T-rich regions by using nonspecific coupling sequences (2, 9, 11, 13, 17). Site-specific integration using longer direct repeats has been described for several ICEs (2, 9).
Also in E. faecalis, the esp gene is harbored on a large PAI of 150 kb in size (15). Except for the esp gene itself and a 10-kb completely conserved gene cluster, both esp PAIs are different (17). Recently, Manson et al. investigated the mechanism of transfer of the E. faecalis PAI (7), and a striking difference can be observed between the transfer of the E. faecium and E. faecalis esp PAIs. In contrast to that of E. faecium, transfer of the E. faecalis PAI is independent of the phage-related integrase and conjugation functions contained on the PAI. Instead, transfer occurred only from donor strains possessing a pheromone-responsive type of conjugative plasmid (7).
In summary, the E. faecium esp PAI is a self-transmissible element that requires the IntA integrase contained on this element for excision. Based on the fact that the esp PAI contains an integrase and a partial copy of Tn916, including the conjugation module, and the fact that this element is self-transmissible in a fashion comparable to that of ICEs, we propose to change the name of this esp PAI to ICEEfm1.
Acknowledgments
M. J. M. Bonten was supported by NWO-VICI 918.76.611, and W. van Schaik was supported by NWO-VENI 916.86.044
Footnotes
Published ahead of print on 10 December 2010.
REFERENCES
- 1.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 2.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 3.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 4.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 5.Heikens, E., M. J. Bonten, and R. J. Willems. 2007. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 189:8233-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leavis, H., et al. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manson, J. M., L. E. Hancock, and M. S. Gilmore. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269-12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nallapareddy, S. R., K. V. Singh, and B. E. Murray. 2006. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl. Environ. Microbiol. 72:334-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts, A. P., and P. Mullany. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251-258. [DOI] [PubMed] [Google Scholar]
- 10.Rocco, J. M., and G. Churchward. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 188:2207-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 13.Scott, J. R., and G. G. Churchward. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367-397. [DOI] [PubMed] [Google Scholar]
- 14.Senghas, E., J. M. Jones, M. Yamamoto, C. Gawron-Burke, and D. B. Clewell. 1988. Genetic organization of the bacterial conjugative transposon Tn916. J. Bacteriol. 170:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 16.Top, J., R. Willems, and M. Bonten. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297-308. [DOI] [PubMed] [Google Scholar]
- 17.van Schaik, W., et al. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Schaik, W., and R. J. Willems. 2010. Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16:527-532. [DOI] [PubMed] [Google Scholar]
- 19.Willems, R. J., et al. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willems, R. J., and W. van Schaik. 2009. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 4:1125-1135. [DOI] [PubMed] [Google Scholar]