Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that possesses a large arsenal of virulence factors enabling the pathogen to cause serious infections in immunocompromised patients, burn victims, and cystic fibrosis patients. CbrA is a sensor kinase that has previously been implied to play a role with its cognate response regulator CbrB in the metabolic regulation of carbon and nitrogen utilization in P. aeruginosa. Here it is demonstrated that CbrA and CbrB play an important role in various virulence and virulence-related processes of the bacteria, including swarming, biofilm formation, cytotoxicity, and antibiotic resistance. The cbrA deletion mutant was completely unable to swarm while exhibiting an increase in biofilm formation, supporting the inverse regulation of swarming and biofilm formation in P. aeruginosa. The cbrA mutant also exhibited increased cytotoxicity to human lung epithelial cells as early as 4 and 6 h postinfection. Furthermore, the cbrA mutant demonstrated increased resistance toward a variety of clinically important antibiotics, including polymyxin B, ciprofloxacin, and tobramycin. Microarray analysis revealed that under swarming conditions, CbrA regulated the expression of many genes, including phoPQ, pmrAB, arnBCADTEF, dnaK, and pvdQ, consistent with the antibiotic resistance and swarming impairment phenotypes of the cbrA mutant. Phenotypic and real-time quantitative PCR (RT-qPCR) analyses of a PA14 cbrB mutant suggested that CbrA may be modulating swarming, biofilm formation, and cytotoxicity via CbrB and that the CrcZ small RNA is likely downstream of this two-component regulator. However, as CbrB did not have a resistance phenotype, CbrA likely modulates antibiotic resistance in a manner independent of CbrB.
Pseudomonas aeruginosa is an important opportunistic human pathogen, causing serious diseases in patients with impaired immunity and mucosal defenses. This Gram-negative bacterium is the dominant pathogen in chronic cystic fibrosis pulmonary infections, persisting in the lungs and inducing serious inflammation that destroys healthy host tissue (1, 39, 42). P. aeruginosa infections are particularly difficult to treat due to the bacterium's intrinsic resistance to a broad spectrum of antimicrobial agents and its repertoire of virulence factors (12).
Motility is strongly associated with the virulence of P. aeruginosa. It enables the bacterium to colonize different environments, such as those of the lungs of cystic fibrosis patients, and contributes to the ability of the bacterium to attach to and to form biofilms on a variety of biotic and abiotic surfaces (33). P. aeruginosa is unusual in that it is capable of three major forms of motility depending on the medium viscosity. P. aeruginosa utilizes its single polar flagellum to swim in aqueous environments and at low agar concentrations (<0.3% [wt/vol]). The bacteria also possess type IV pili to enable twitching motility on solid surfaces or interfaces. Besides swimming and twitching, P. aeruginosa has recently been observed to swarm on semisolid (viscous) surfaces (0.5 to 0.7% [wt/vol] agar) in a coordinated manner. Swarmer cells are differentiated from vegetative cells in that swarmer cells are elongated and hyperflagellated with two polar flagella (19). This form of motility is induced in P. aeruginosa under nitrogen limitation and when certain amino acids, such as glutamate, aspartate, proline, or histidine, are provided as the sole nitrogen source (19). To date, swarming of P. aeruginosa has been identified to require the flagella and the type IV pili as well as the production of the biosurfactant rhamnolipids (5, 19, 36, 40, 48).
A recent screen of the P. aeruginosa strain PA14 transposon insertion mutant library identified 233 mutants that exhibited alterations in swarming phenotype compared to the wild type (55). The swarming-associated genes functioned not only in flagellum or type IV pilus biosynthesis but also in diverse processes such as transport, secretion, and metabolism. This, together with data demonstrating that hundreds of genes are dysregulated during swarming, including genes encoding most virulence factors, antibiotic resistance loci, and distinct metabolic processes, indicated that swarming is not just a form of motility but rather a complex adaptation and an alternative growth state. Among the mutants with genes that had altered swarming motility were 35 mutants with transposon insertions in genes encoding regulators. Only a few of these regulatory mutants showed significant defects in the production of type IV pili, flagella, or rhamnolipid, each of which is known to be involved in swarming, suggesting that the majority of these regulators control other factors important in swarming (55). Of the 35 transcriptional regulators that were altered in swarming behavior, one interesting mutant with a mutation in the cbrA gene was chosen to be investigated in great detail.
Metabolically versatile pseudomonads effectively utilize a broad range of organic compounds as carbon and/or nitrogen sources. Expression of the components of catabolic pathways involved in utilization of these compounds is subject to catabolite repression (4). In contrast to Escherichia coli and Bacillus subtilis, succinate and other tricarboxylic acid (TCA) cycle carboxylates are preferable to glucose as carbon sources for Pseudomonas spp. Moreover, studies have shown that pseudomonads utilize a different mechanism of catabolite regulation, which involves a Crc protein that binds mRNA and acts as a translational repressor (15, 27, 53). The CbrAB two-component system in P. aeruginosa was first identified to be involved in controlling the expression of a number of catabolic pathways involved in carbon and nitrogen utilization (22, 32). Mutations in the sensor kinase CbrA or the response regulator CbrB rendered the bacterium incapable of growing on a variety of organic compounds as the sole carbon source (32). Recently, Sonnleitner et al. discovered that the P. aeruginosa genome encodes a small RNA, CrcZ, which binds to and sequesters the Crc protein with high affinity and thus relieves catabolite repression of a variety of degradative genes, such as amiE (46). The same group also found that expression of CrcZ is controlled by the CbrAB system.
In addition to its role in metabolism, the CbrAB system has been demonstrated to be involved in the metabolic regulation of the type III secretion system (T3SS) and its effectors, exoenzymes S and T (41). A two-component system, LipQR, was discovered in Pseudomonas alcaligenes and demonstrated to be involved in the regulation of lipase expression (20). The LipQR system exhibits significant homology to the CbrAB system in P. aeruginosa (20). Analysis of the transcriptome profile of P. aeruginosa exposed to sublethal concentrations of tobramycin revealed downregulation of the cbrA gene (K. N. Kindrachuk, L. Fernandez, M. Bains, and R. E. W. Hancock, submitted for publication). Furthermore, a previous PA14 screen revealed that a cbrA mutant is swarming deficient and exhibits hyperbiofilm formation (55). These results led us to propose that CbrA may be involved in substantially more than just catabolite regulation in P. aeruginosa.
In this paper, we demonstrate that CbrA is involved in the regulation of not only carbon and nitrogen metabolism but also various virulence and virulence-related processes in P. aeruginosa. We constructed a cbrA deletion mutant and showed that this mutant displayed swarming deficiency while exhibiting increased biofilm formation and in vitro cytotoxicity toward human bronchial epithelial (HBE) cells. The cbrA mutant also demonstrated increased resistance to a variety of clinical antibiotics. Microarray analysis of the cbrA mutant provided insight into the basis for these observed phenotypes. Based on detailed phenotypic and genetic studies of the cbrB mutant, we propose that CbrA most likely modulates swarming, biofilm formation, and cytotoxicity via the response regulator CbrB while CbrA may cross talk with another regulatory system to modulate antibiotic resistance.
MATERIALS AND METHODS
Tissue culture, bacterial strains, and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Cultures were routinely grown in Luria-Bertani (LB) broth, tryptone broth (10 g/liter Bacto tryptone), BM2 minimal medium [62 mM potassium phosphate buffer, pH 7, 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose], or BM2-swarming medium comprising BM2 with 0.1% (wt/vol) Casamino Acids substituted for 7 mM (NH4)2SO4. Escherichia coli S17-1 λpir was used as the donor strain in bacterial conjugations (45). P. aeruginosa competent cells were prepared as previously described (3). For plasmid or transposon selection or maintenance, antibiotics were added to growth media at the indicated concentrations: E. coli, 10 μg/ml gentamicin and 100 μg/ml ampicillin; P. aeruginosa, 30 μg/ml gentamicin, 100 μg/ml tetracycline, and 500 μg/ml carbenicillin.
TABLE 1.
P. aeruginosa strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference and/or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PA14 (WT) | Wild-type P. aeruginosa PA14 | 23 |
| cbrA mutant | PA14 transposon insertion mutant, ID33836 | 23 |
| ΔcbrB mutant | PA14 transposon insertion mutant, ID44074 | 23 |
| Δcrc mutant | PA14 transposon insertion mutant, ID44185 | 23 |
| ΔcbrA mutant | ΔcbrA chromosomal deletion mutant of PA14; Gmr | This study |
| ΔcrcZ mutant | ΔcrcZ chromosomal deletion mutant of PA14 | This study |
| cbrA/B+ strain | ΔcbrA mutant with pUCP18::cbrAB+; Cbr | This study |
| cbrB+ strain | ΔcbrB mutant with pUCP19::cbrB+; Cbr | This study |
| crc+ strain | Δcrc mutant with pUCP18::crc+; Cbr | This study |
| crcZ+ strain | ΔcrcZ mutant with crcZ+::mini-Tn7 chromosomal integration; Gmr | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG φ80lacZΔM15ΔlacX74 | Invitrogen |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 λ−thi-1 gyrA96 relA | Invitrogen; 46 |
| S17-1 | galU galK rpsL(Strr) endA1 nupG thi pro hsdR hsdM+recA (RP4-2Tc::Mu Km::Tn7) λpir | 45, 46 |
| HB101 | thi-1 hsdS20(rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | 46 |
| SM10/λpir | thi-1 thr-1 leu-6 tonA21 lacY1 supE44 recA chromosomal RP4-2 [Tcr::Mu Kmr::Tn7]λpir | 46 |
| Plasmids | ||
| pUCP18, pUCP19 | E. coli-Pseudomonas shuttle vector, Apr Cbr | 50 |
| pUCP18::cbrAB+ | pUCP18 with cbrAB fragment | This study |
| pUCP19::cbrB+ | pUCP19 with cbrB fragment | This study |
| pUCP18::crc+ | pUCP18 with crc fragment | This study |
| pEX18Ap | Suicide plasmid carrying sacBR, Apr | 16 |
| pME3087 | Suicide vector, ColE1 replicon, Mob; Tcr | 46 |
| pME9673 | pME3087 with a 160-bp deletion in crcZ | 46 |
| pRK2013 | Helper plasmid, ColE1 replicon, Tra; Kmr | 46 |
| pME3280a | Chromosomal integration vector, mini-Tn7; Gmr Apr | 46 |
| pME9818 | pME3280a with crcZ in mini-Tn7 | Laetitia Abdou |
| pUXBF-13 | Helper plasmid containing Tn7 transposition functions, R6K replicon; Apr | 46 |
Ap, ampicillin; Cb, carbenicillin; Gm, gentamicin; Km, kanamycin; Tc, tetracycline.
The simian virus 40 (SV40)-transformed, immortalized human bronchial epithelial (HBE) cell line 16HBE14o- was a gift from D. Gruenert (University of California, San Francisco, CA). 16HBE14o- cells were cultured in minimum essential medium (MEM) with Earle's salts (Life Technologies Invitrogen), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Life Technologies Invitrogen) and 2 mM l-glutamine (Life Technologies Invitrogen). 16HBE14o- cells were routinely cultured to 85 to 90% confluence in 100% humidity and 5% CO2 at 37°C and were used between passages 9 and 15.
General DNA manipulations.
Routine genetic manipulations were carried out using standard procedures (43). Primers were synthesized by AlphaDNA Inc. (Montreal, QC, Canada), and their sequences are available from us on request. Plasmid DNA was isolated using QIAprep spin miniprep kits (Qiagen Inc., Mississauga, ON, Canada), and agarose gel fragments were purified using a QIAquick gel extraction kit (Qiagen). Restriction endonucleases were from New England BioLabs (Mississauga, ON, Canada). DNA sequencing was carried out by the UBC NAPS unit.
Recombinant DNA manipulations.
For construction of a cbrA deletion mutant, an in-frame deletion of cbrA was obtained via splicing by an overlap extension PCR strategy (17). Briefly, primers were designed to amplify the gentamicin cassette of pPS858. Primers were also designed to amplify approximately 1-kb fragments located upstream and downstream of cbrA from PA14 genomic DNA with additional short sequences of overlap with the gentamicin cassette. Next, the three DNA fragments were fused together and the final product was boosted by a third PCR. The resulting fragment was cloned into pEX18Ap carrying a sacB sucrose sensitivity gene (16). This plasmid was transformed into E. coli S17-1 λpir and conjugated into P. aeruginosa PA14 to generate an in-frame deletion of the cbrA gene in the PA14 strain by allelic exchange. Selection for double recombinants was performed on plates containing gentamicin and 5% (wt/vol) sucrose. The deletion was confirmed by PCR and sequencing.
The suicide vector pME9673, obtained from Dieter Haas's laboratory, contained the deleted crcZ promoter and the 5′ region of crcZ (46). To construct a crcZ deletion mutant, a crcZ deletion was introduced from plasmid pME9673 into the chromosomal crcZ locus of P. aeruginosa strain PA14 by gene replacement as described previously (46). Briefly, the plasmid pME9673 was mobilized from E. coli DH5α into P. aeruginosa PA14 with the help of E. coli HB101/pRK2013. PA14 transconjugates carrying a chromosomally integrated copy of pME9673 were selected on tetracycline. Excision of the vector by a second crossover (i.e., tetracycline-sensitive derivatives) was subsequently obtained by enrichment with carbenicillin. The chromosomal crcZ deletion was confirmed by PCR and sequencing.
The PA14 cbrA deletion mutant and cbrB and crc transposon mutants were complemented by amplifying cbrAB, cbrB, or crc, including an upstream region of 400 base pairs, from PA14 genomic DNA by PCR and cloning each fragment into the broad-host-range vector pUCP18 or pUCP19 (50). The resulting hybrid plasmids, pUCP18::cbrAB+, pUCP19::cbrB+, and pUCP18::crc+, were transferred into the cbrA, cbrB, and crc mutants, respectively, by electroporation (3).
The crcZ deletion mutant was complemented by chromosomal insertion of a mini-Tn7 carrying the functional crcZ+ gene into the Tn7 attachment site of the mutant. The suicide plasmid, pME9818, which contained the cloned crcZ+ gene including the promoter region, was kindly provided by the laboratory of Dieter Haas (L. Abdou and D. Haas, unpublished data). Briefly, pME9818, carried by host E. coli S17-1, was transferred into the PA14 crcZ mutant by conjugation. Transposition of the mini-Tn7 carrying the crcZ+ gene into the chromosome of the crcZ mutant was facilitated by the E. coli SM10λpir helper carrying pUXBF-13. The P. aeruginosa strain carrying a crcZ+ insertion was selected with gentamicin and chloramphenicol and confirmed by PCR.
Motility experiments.
Swimming and twitching of P. aeruginosa PA14 wild type (WT) and mutants were examined on LB plates with 0.3% (wt/vol) agar and 1% (wt/vol) agar, respectively. Swarming for PA14 strains was examined on BM2-swarming plates containing BM2 with 0.1% (wt/vol) Casamino Acids substituted for 7 mM (NH4)2SO4 and 0.5% (wt/vol) agar. To test the effects of the carbon source on the swarming motility of the cbrA and cbrB mutants, glucose was replaced with 0.4% (wt/vol) glycerol, 0.4% (wt/vol) mannitol, or 20 mM succinate. For iron complementation studies, Fe(II) sulfate was added to the swarming medium to a final concentration of 100 μM. Swimming and swarming motility were assayed by spotting 1 μl of mid-logarithmic-growth-phase cultures grown in LB broth or BM2-swarming medium onto the respective motility plates. Twitching motility was assessed by stab-inoculating a single colony grown overnight on LB broth at 37°C. The resultant diameters of the swim or twitch zones were measured after incubation for 20 h and 24 h at 37°C. The resultant swarming colonies were analyzed by measuring the surface coverage on agar plates after 20 h at 37°C. For each form of motility, 3 independent experiments were performed with 3 replicates for each mutant.
Biofilm assays and rapid attachment.
Biofilm formation was analyzed using an abiotic solid-surface assay as described elsewhere (9, 34). Overnight cultures were diluted in BM2-biofilm medium [62 mM potassium phosphate buffer, pH 7, 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose, 0.5% (wt/vol) Casamino Acids] in polystyrene microtiter plates (Falcon) and incubated at 8 h or 20 h at 37°C to study the initiation and mature biofilm formation, respectively. To test the effects of the carbon source on biofilm formation of the cbrA and cbrB mutants, glucose was replaced with 0.4% (wt/vol) glycerol, 0.4% (wt/vol) mannitol, or 20 mM succinate. For iron complementation studies, Fe(II) sulfate was added to the biofilm medium to a final concentration of 100 μM. Biofilms were stained with crystal violet, and absorbance was measured at 600 nm using a microtiter plate reader (Bio-Tek Instruments Inc.).
Rapid attachment was assayed as described previously with modifications (34). Overnight cultures were first diluted 1/100 into fresh BM2-biofilm medium and grown to an optical density at 600 nm (OD600) of 0.5, and 100 μl was added to each well of a 96-well polystyrene microtiter plate. Cells were allowed to attach for 30 min at room temperature prior to staining with crystal violet as described above.
CR assay.
Congo red (CR) binding assays were performed as previously described (9). Briefly, tryptone-grown overnight cultures were diluted to OD600s of 0.025 and 1, 5, and 10 μl were spotted onto CR plates (10 g/liter tryptone broth with 10 g/liter agar, 40 μg/ml Congo red, and 20 μg/ml Coomassie brilliant blue). The plates were incubated for 24 h at 37°C, followed by 48 h at room temperature to assess colony morphology.
MIC determination and killing experiments.
MICs were measured using standard broth microdilution procedures (26) in BM2-swarming medium. Growth was scored following 24 h of incubation at 37°C. For measuring MICs against polymyxin B, a modified assay was used to prevent artificially high MICs due to aggregation of the antibiotic and binding to polystyrene (52).
To perform killing experiments, cells of P. aeruginosa were grown to an OD600 of 0.5 in BM2-swarming liquid medium or on BM2-swarming agar plates for 18 h (37). These cultures were diluted into 1× BM2-salts containing 1 μg/ml polymyxin B sulfate (Sigma). Samples were shaken at 37°C, and aliquots were taken at specified times, plated for survivors on LB agar, and grown overnight at 37°C. All experiments were repeated at least 3 times.
Cytotoxicity assays.
For the interaction assay, 16HBE14o- cells were seeded in 96-well plates (Corning Life Science, Corning, NY) at a density of 2 × 104 cells/well and grown at 37°C with 5% CO2 until 100% confluent (∼48 h). Bacteria were grown in LB broth until mid-logarithmic phase, washed with phosphate-buffered saline, and resuspended and diluted in MEM containing 1% FBS and 2 mM l-glutamine. The interaction assay was performed at a multiplicity of infection (MOI) of 2 bacteria/cell in MEM containing 1% FBS and 2 mM l-glutamine, and the assay mixture was incubated at 37°C with 5% CO2. At postinfection time points, medium was removed from the wells, placed in microtiter plates, and spun for 10 min at 3,000 rpm to pellet the bacteria and host cell debris. The level of lactate dehydrogenase (LDH) in the supernatant was then assayed in triplicate using a colorimetric cytotoxicity detection kit (Roche, Mannheim, Germany). As a positive control for maximum LDH release, cells were treated with 1% Triton X-100 (Sigma, Oakville, Canada), resulting in complete cell lysis, while untreated cells were used to assess background LDH release.
Growth curves.
P. aeruginosa mutants and wild type were grown overnight in LB medium, BM2-swarming medium, or BM2-biofilm medium. If necessary, cultures were diluted to obtain equal optical densities. Five-microliter portions of these cultures were added to 195 μl of fresh medium in 96-well microtiter plates. The growth of these cultures at 37°C under shaking conditions was monitored with a Tecan Spectrofluor Plus by determining the absorbance at 620 nm every 20 min for 24 h. Two independent experiments were performed with 3 replicates for each mutant.
Real-time quantitative PCR (RT-qPCR).
Total RNAs from the cbrA, cbrB, crcZ, and crc mutants were harvested under various conditions as follows: (i) for the swarming condition, cells were obtained from the leading edge of the dendritic swarm colonies of the PA14 wild type and the entire nonswarming colonies of the PA14 cbrA transposon mutant (ID33836), the cbrA deletion mutant, the cbrB transposon mutant, and the crcZ deletion mutant; (ii) for the polysaccharide synthesis-inducing condition, cells for RNA isolation were obtained by spotting 10 μl of diluted cultures grown in tryptone broth onto CR plates without Congo red or Coomassie brilliant blue; (iii) for the HBE cell infection condition, 16HBE14o- cells were seeded in tissue culture-treated petri dishes (Corning Life Science, Corning, NY) at a density of 2 × 104 cells/well and grown at 37°C with 5% CO2 until 100% confluent (∼48 h). Bacteria were grown in LB medium until mid-logarithmic phase, washed with phosphate-buffered saline, and resuspended and diluted in MEM containing 1% FBS and 2 mM l-glutamine. The interaction assay was performed at an MOI of 100 bacteria/cell in MEM containing 1% FBS and 2 mM l-glutamine, and the assay mixture was incubated at 37°C with 5% CO2. At 4 h postinfection, medium was removed from the dishes, placed in sterile Falcon tubes, and spun for 10 min at 3,000 rpm to pellet the bacteria. Subsequently, RNAs were isolated using RNeasy minicolumns (Qiagen) treated with DNase I (Invitrogen) to remove contaminating genomic DNA. Three micrograms of total RNA was combined with 0.5 μM deoxynucleoside triphosphates (dNTPs), 500 U SuperaseIN/ml (Ambion), and 10 μM dithiothreitol (DTT) in 1× reaction buffer and reverse transcribed with Superscript II reverse transcriptase (Invitrogen). The resultant cDNA was used as a template for qPCR. Analysis was carried out in the ABI Prism 7000 sequence detection system (Applied Biosystems) using the two-step RT-qPCR kit with SYBR green detection (Invitrogen). Fold change was determined using the comparative threshold cycle (CT) method by comparison to the PA1544 housekeeping gene.
DNA microarray experiment.
Microarray experiments were performed on three independent cultures. The cbrA deletion mutant and wild-type PA14 were grown on a BM2-swarming plate containing 0.5% (wt/vol) agar for 18 h at 37°C. RNA was harvested from the leading edge of the dendritic swarm colonies of the PA14 wild type and of the entire nonswarming colonies of the cbrA mutant. As described previously (37), cells were resuspended in BM2-swarming medium supplemented with RNAprotect reagent (Qiagen, Germany). Harvesting of cells, RNA isolation, cDNA synthesis, hybridization to P. aeruginosa PAO1 DNA microarray slides (aminosilane coated) from the Institute for Genomic Research (TIGR) Pathogenic Functional Genomics Resource Center, analysis of microarray slides using ArrayPipe version 1.7, and RT-qPCR were performed as described previously.
Pyoverdine assay.
Bacterial strains were grown in CAA medium (5 g/liter low-iron Bacto Casamino Acids [Difco], 1.54 g/liter K2HPO4·3H2O, 0.25 g/liter MgSO4·7H2O) at 37°C for 48 h. The supernatants were diluted 1/75 in 10 mM Tris-HCl, pH 7.5, and excited at 400 nm with a spectrofluorimeter (11).
Microarray accession number.
The ArrayExpress accession number is E-FPMI-22.
RESULTS
Construction of a PA14 cbrA deletion mutant.
Previous studies indicated that the PA14 cbrA transposon mutant (ID33836), obtained from Harvard University, was swarming deficient (55). As cbrA is located directly upstream of its cognate response regulator gene cbrB, it is possible that the swarming defect observed in the cbrA transposon mutant was simply due to a polar effect. Therefore, we investigated whether or not the transposon insertion in the PA14 cbrA gene affected the expression of cbrB. RNA was harvested from the PA14 wild type and the PA14 cbrA transposon mutant and then reverse transcribed into cDNA. Changes in the expression levels of cbrA and cbrB were determined via RT-qPCR. In the PA14 cbrA transposon mutant, cbrB expression was slightly downregulated (fold change of −1.9 ± 0.1 relative to the wild type), suggesting that the transposon insertion in cbrA affected not only the expression of CbrA but also that of CbrB.
Since the expression of CbrB was affected in the cbrA transposon mutant, it was necessary to generate a nonpolar, in-frame deletion of cbrA in the PA14 wild type in order to study the role of CbrA in P. aeruginosa. To generate a P. aeruginosa cbrA deletion mutant, we utilized the sacB-based method (16) that involved amplifying three overlapping DNA fragments, splicing these fragments together by overlap extension PCR, and cloning the resultant fragment into a suicide vector, pEX18Ap. The plasmid-borne deletion was then transferred to the P. aeruginosa strain PA14 chromosome by homologous recombination and selected on medium containing gentamicin and 5% sucrose. The resultant cbrA deletion mutant was verified by sequencing and PCR. RT-qPCR was performed to ensure that the expression of CbrB was not affected in the cbrA deletion mutant (data not shown).
The cbrA deletion mutant was impaired in swarming motility.
The abilities of the cbrA deletion mutant to swim, twitch, and swarm on 0.3%, 1%, and 0.5% agar, respectively, were examined. The cbrA deletion mutant exhibited minor defects in flagellum-mediated swimming and type IV pilus-mediated twitching motilities (data not shown). In contrast, this mutant's ability to swarm was completely abolished. The swarming-defective phenotype of this mutant could be restored to the wild-type level by introducing the wild-type cbrAB genes into the mutant (Fig. 1A). We also investigated whether the inability of the mutant to swarm was influenced by differences in production of rhamnolipids. Using the rhamnolipid agar plate method (6, 7), no difference was observed between the diameters of the halos formed due to rhamnolipid produced from the cbrA mutant and the PA14 wild type (data not shown). To investigate whether the inability of the cbrA mutant to swarm was due to its poor ability to utilize glucose as the carbon source, we replaced glucose, in the swarming medium, with succinate, a carbon source that had been demonstrated, and confirmed in our growth studies, to sustain the wild-type growth of the P. aeruginosa cbrA mutant. Furthermore, swarming of the cbrA mutant was tested on other carbon sources, including glycerol and mannitol. Replacement of glucose with other carbon sources did not restore the ability of the cbrA mutant to swarm (data not shown).
FIG. 1.
Swarming motility of the PA14 cbrA and cbrB mutants. Defective swarming motilities of the cbrA deletion mutant (ΔcbrA) and the cbrB transposon mutant (ΔcbrB) were restored to levels similar to that of the wild type, P. aeruginosa PA14 (WT), by transforming these mutants with plasmids containing the wild-type cbrAB genes or the wild-type cbrB gene.
The cbrA mutant exhibited enhanced biofilm formation.
P. aeruginosa forms biofilms on a number of surfaces, including tissues of the human host. Consequently, biofilm infections are virtually impossible to eradicate due to the biofilm's inherent resistance to conventional antibiotic therapies (28). Therefore, we investigated here the ability of the cbrA deletion mutant to form simple biofilms using static microtiter biofilm methods. These experiments demonstrated that the cbrA mutant showed a significant (P < 0.01 by Student's t test) but moderate (∼40 to 60%) enhancement in biofilm formation as early as 8 h (Fig. 2 A and B). This biofilm phenotype could be successfully complemented by introducing the wild-type cbrAB operon into the mutant. To determine whether the biofilm formation phenotype occurred during initial attachment stage or later during biofilm development, a rapid (30-min) attachment assay was performed. No difference in early attachment was observed between the PA14 wild type and the cbrA mutant (data not shown).
FIG. 2.
Biofilm formation of the PA14 cbrA and cbrB mutants. Cells were incubated in 96-well microtiter plates containing BM2-biofilm medium for 8 h (A) and 20 h (B) at 37°C for the cbrA mutant and 20 h at 37°C for the cbrB mutant (C). Surface-associated biofilm formation was analyzed by crystal violet staining of the adherent biofilm followed by ethanol solubilization of the crystal violet and quantification (A600) of the stained well. Results shown are means with standard deviations for three biological experiments, each with eight technical repeats. **, statistically significant difference (P < 0.01) between the mutants and the wild type as determined by Student's t test.
The excess-biofilm-forming phenotype of the cbrA mutant led us to investigate further whether this mutant was altered for other biofilm-related functions. Chemical analyses of P. aeruginosa biofilms have suggested that the matrix is comprised of exopolysaccharides (EPS), DNA, RNA, proteins, and ions (47, 51). In P. aeruginosa, the psl and pel loci have been suggested to be involved in the production of the polysaccharide component of the matrix (9, 10). While alginate is a component of the extracellular matrix, studies have suggested that alginate is not a significant component of the extracellular polysaccharide present in the matrix of biofilms formed by P. aeruginosa under commonly used laboratory growth conditions (14, 54). Also, while P. aeruginosa strain PAO1 has both pel and psl loci, only the pel locus has been identified in the PA14 strain (9). To investigate whether the increased-biofilm-forming phenotype of this mutant was due to increased production of the pel-encoded extracellular matrix, we performed Congo red assays, as Congo red has been shown to bind the pel-encoded polysaccharide of P. aeruginosa PA14. The cbrA mutant showed substantially increased binding to Congo red compared to the PA14 wild type (Fig. 3).
FIG. 3.
Congo red binding. Strains were spotted on Congo red and Coomassie brilliant blue plates and incubated for 24 h at 37°C, followed by incubation for 48 h at room temperature. Representative images of the colony morphology of P. aeruginosa PA14 (WT), the cbrA mutant (ΔcbrA), the cbrB mutant (ΔcbrB), the complemented cbrA/B+ strain, and the complemented cbrB+ strain are shown.
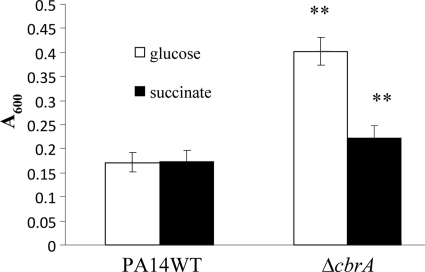
Furthermore, biofilm production of the cbrA mutant was assessed in different carbon sources, including succinate, mannitol, and glycerol. Interestingly, when glucose was replaced with succinate as the major carbon source, the cbrA mutant produced significantly less biofilm than in glucose at 20 h, while no such difference was observed for the wild type (Fig. 4). When mannitol or glycerol was provided as the carbon source, the cbrA mutant still produced significantly more biofilm than did the wild type (data not shown).
FIG. 4.
Influence of carbon source on biofilm formation of the PA14 cbrA deletion mutant. Cells were incubated at 37°C for 20 h in 96-well microtiter plates containing BM2-biofilm medium supplemented with 0.4% (wt/vol) glucose (open bars) or 20 mM succinate (filled bars). Surface-associated biofilm formation was analyzed by crystal violet staining of the adherent biofilm followed by ethanol solubilization of the crystal violet and quantification (A600) of the stained well. Results shown are means with standard deviations for three biological experiments, each with eight technical repeats. **, statistically significant difference (P < 0.01) between the mutants and the wild type as determined by Student's t test.
The cbrA mutant displayed enhanced cytotoxicity toward human bronchial epithelial cells.
P. aeruginosa possesses a large arsenal of virulence factors. One of the major virulence mechanisms employed by P. aeruginosa to intoxicate eukaryotic cells is the type III secretion system (T3SS). The T3SS, triggered by cell contact, injects toxins directly into the cytoplasm of the target cell. Moreover, the CbrAB system was previously demonstrated to be involved in the metabolic regulation of the T3SS and its effectors (41). Therefore, we examined the ability of the cbrA mutant to infect and destroy a monolayer of cultured 16HBE14o- epithelial cells. To measure the cytotoxic effects of the PA14 wild type and cbrA mutant on the epithelial cells, the amount of lactate dehydrogenase (LDH) released from the 16HBE14o- cells was quantified using an enzyme assay. The cbrA mutant displayed 3.5- and 2.2-fold-greater cytotoxicity than did the wild type at 4 h and 6 h postinfection, respectively (Fig. 5A). Introducing the wild-type cbrAB operon into the cbrA mutant restored cytotoxicity to wild-type levels at both time points. Furthermore, to investigate whether the cbrA mutant was able to infect the epithelial cells better than the wild type was, due to an improved ability to adhere to the epithelial cells, adhesion assays were performed. No significant differences in adherence to 16HBE14o- cells at 1 h were observed between the wild type and the mutant (data not shown).
FIG. 5.
In vitro cytotoxicity toward HBE cells. The abilities of the PA14 wild type (WT), the mutants (cbrA mutant [ΔcbrA] and cbrB mutant [ΔcbrB]), and the complemented strains (cbrA/B+ and cbrB+) to induce cell damage were determined by monitoring the release of intracellular lactate dehydrogenase (LDH) into the supernatant from HBE cells. Bacteria were cocultured with the cells, and LDH release was monitored at the time points indicated. Each result represents the mean of three independent biological repeats, each assayed in triplicate.
The cbrA mutant demonstrated increased resistance to polymyxins, aminoglycosides, and fluoroquinolones.
Recently, a microarray analysis indicated that among the many transcriptional changes, the cbrA gene was 2-fold downregulated (P < 0.05) in response to sublethal concentrations of tobramycin (Kindrachuk et al., submitted). Therefore, we were interested in investigating the role of CbrA in modulating resistance to a variety of clinical antibiotics, including aminoglycosides, cationic peptides, fluoroquinolones, cephalosporins, and carbapenems (Table 2). Intrinsic resistance of the mutant to these antibiotics was assessed by MIC assay in cells growing in BM2-swarming medium containing high (2 mM) Mg2+ to suppress the possibility of induction of genes by limiting Mg2+. Compared to the PA14 wild type, the cbrA mutant reproducibly exhibited a 2-fold-increased resistance to polymyxin B and colistin and 4-fold-increased resistance to ciprofloxacin and tobramycin. No difference was observed between the MIC values of the PA14 wild type and the mutant to piperacillin, tetracycline, cefepime, ceftazidime, and imipenem. Although the 2-fold change in MIC observed for the cbrA mutant to polymyxin B is often considered within the acceptable range of error of these assays, we confirmed the increased resistance of the cbrA mutant to polymyxin B by performing kill curve assays with cells taken from liquid swarm medium and from swarm plates (Fig. 6). The antibiotic susceptibility phenotype could be complemented to wild-type levels by introducing the wild-type cbrAB genes into the mutant.
TABLE 2.
MICs (μg/ml) of antibiotics toward P. aeruginosa grown in swarming mediuma
| Antibiotic | MIC (μg/ml) |
||
|---|---|---|---|
| PA14 (WT) | cbrA mutant | cbrB mutant | |
| Polymyxin B | 1 | 2 | 1 |
| Colistin | 1 | 2 | 1 |
| Tobramycin | 2 | 8 | 2 |
| Piperacillin | 4 | 4 | 4 |
| Tetracycline | 64 | 64 | 64 |
| Ciprofloxacin | 0.2 | 0.8 | 0.2 |
| Cefepime | 2 | 2 | 1 |
| Ceftazidime | 4 | 4 | 4 |
| Imipenem | 2 | 2 | 2 |
Results are shown as the mode of 4 to 6 independent experiments.
FIG. 6.
Polymyxin B resistance in the PA14 cbrA deletion mutant. Sensitivity to polymyxin B at 1 μg/ml was analyzed using cells from mid-log phase in swarm medium (A) or using cells directly off swarm plates and then plating diluted aliquots for survivors (B). For each condition, one representative experiment of four independent experiments that produced identical results is shown.
The moderate growth deficiency exhibited by the cbrA mutant was insufficient to explain its swarming, biofilm, or cytotoxicity phenotypes.
Previous studies identified CbrAB as an important regulatory element for the expression of several catabolic pathways and utilization of a variety of organic compounds as the sole carbon source (32). Li and Lu showed that a cbrAB mutant displayed weak growth when glucose was used as the sole carbon source, while growth on tricarboxylic acid (TCA) cycle intermediates was sustained (22). Therefore, we investigated whether any of the phenotypes observed for the cbrA mutant could be related to growth impairment when glucose was provided as the major carbon source in the swarming, MIC, and biofilm media. The growth of the cbrA mutant and the PA14 wild type was measured in the appropriate medium (BM2-swarm medium, LB medium, or BM2-biofilm medium) at 37°C using a Tecan Spectrofluor Plus to measure the absorbance at 620 nm every 20 min for 15 to 20 h under shaking conditions. The growth of the mutant and wild type was also determined by measuring the absorbance at 600 nm every 20 min during infection of 16HBE14o- cells. As shown in Fig. 7A, the cbrA mutant exhibited a very minor growth defect in LB medium. A moderate defect in growth under swarming conditions was observed for the mutant (Fig. 7B). The moderate growth defect of the mutant in this medium (∼50% change in growth rate), however, seemed insufficient to explain the complete abolition of swarming motility. The cbrA mutant also exhibited slight growth defects under biofilm-inducing (Fig. 7C) and HBE infection conditions, but these growth defects also seemed insufficient to explain the increased biofilm production and cytotoxicity of the mutant. Furthermore, we investigated whether changing the carbon source from glucose to succinate would restore the ability of the mutant to swarm and form biofilm to the wild-type level. The cbrA mutant indeed showed wild-type growth when glucose was replaced with succinate in the BM2-swarming and BM2-biofilm media, as suggested from previous studies (22), but exhibited the same swarming and biofilm defects as in glucose media.
FIG. 7.
Growth curves of P. aeruginosa PA14 wild type (WT) and cbrA deletion mutant. Cells were diluted and grown in LB broth (A), BM2-swarm medium (B), and BM2-biofilm medium (C). Growth was measured at 37°C using a Tecan Spectrofluor Plus.
Microarray analysis of the cbrA mutant.
To investigate how CbrA contributed to the various phenotypes observed, microarray studies were performed comparing the cbrA mutant to the PA14 wild type. For the microarray, RNAs from the PA14 wild type and the cbrA mutant were taken directly from BM2-swarm plates that had been incubated at 37°C for 20 h. The microarray revealed 236 genes that were differentially regulated by more than 2.0-fold (P ≤ 0.06) with 145 transcriptionally upregulated and 91 transcriptionally downregulated genes (see Table S1 in the supplemental material). A selection of these genes is presented in Table 3. Of note, PAO1 DNA microarray slides were used to analyze gene expression of PA14 cbrA mutant and PA14 wild type, as there are no PA14-specific microarray slides available. The PA14 genome (6.5 Mbp) is slightly larger than that of PAO1 (6.3 Mbp), but the PA14 and PAO1 genomes are very similar, with greater than 92% of all genes in PA14 also present in PAO1. The additional genes in the PA14 genome that are absent in PAO1 have been suggested to contribute to its enhanced pathogenicity, as PA14 is more virulent than PAO1 (13).
TABLE 3.
Selected genes significantly dysregulated in cbrA mutant as determined using microarray
| Category and gene IDa | Name | Fold changeb | P value | Function |
|---|---|---|---|---|
| Adaptation and protection | ||||
| PA1159 | −2.36 | 0.0353 | Probable cold shock protein | |
| PA2385 | pvdQ | −3.01* | 0.002 | 3-Oxo-C12-homoserine lactone acylase and pyoverdine biosynthesis |
| PA2386 | pvdA | −2.39* | 0.0011 | l-Ornithine N5-oxygenase |
| PA2397 | pvdE | −3.44 | 0.002 | Pyoverdine biosynthesis protein |
| PA2399 | pvdD | −2.24 | 0.0061 | Pyoverdine synthetase D |
| PA2920 | 2.14 | 0.0068 | Probable chemotaxis transducer | |
| PA3349 | 2.15 | 0.0013 | Probable chemotaxis protein | |
| PA4223 | pchH | −4.16* | 8.59E−05 | Probable ATP binding component of ABC transporter |
| PA4225 | pchF | −2.53 | 0.0005 | Pyochelin synthetase |
| PA4231 | pchA | −2.48 | 0.0009 | Salicylate biosynthesis isochorismate synthase |
| PA4356 | xenB | 2.27 | 0.0039 | Xenobiotic reductase |
| PA4468 | sodM | 3.55 | 8.86E−05 | Superoxide dismutase |
| PA4876 | osmE | −2.00 | 0.0161 | Osmotically inducible lipoprotein |
| Chemotaxis; cell wall/LPS/capsule | ||||
| PA1423 | 2.55 | 0.0007 | Probable chemotaxis transducer | |
| PA2920 | 2.14 | 0.0068 | Probable chemotaxis transducer | |
| PA3157 | wbpC | −2.3 | 0.0186 | Probable acetyltransferase |
| PA3349 | 2.15 | 0.0013 | Probable chemotaxis protein | |
| PA3545 | algG | 2.08 | 0.0068 | Alginate-C5-mannuronan-epimerase |
| Chaperones and heat shock proteins | ||||
| PA1596 | htpG | 2.21 | 0.0016 | Heat shock protein 90 |
| PA2830 | htpX | 3.27 | 0.0001 | Heat shock protein |
| PA3126 | ibpA | 3.26 | 6.23E−05 | Heat shock protein |
| PA4352 | 2.65 | 0.0003 | Putative universal stress protein | |
| PA4761 | dnaK | 3.57* | 5.33E−05 | Molecular chaperone |
| PA4762 | grpE | 2.95 | 0.0004 | Heat shock protein |
| PA5053 | hslV | 4.9 | 9.70E−05 | ATP-dependent protease peptidase subunit |
| PA5054 | hslU | 3.07 | 0.0054 | ATP-dependent protease ATP binding subunit |
| Antibiotic resistance and susceptibility | ||||
| PA1178 | oprH | 9.36* | 1.63E−06 | PhoP/Q and low-Mg2+-inducible outer membrane protein H1 precursor |
| PA1170 | phoP | 4.16* | 2.74E−05 | Two-component response regulator |
| PA1797 | 10.37* | 1.16E−06 | Putative function in adaptive polymyxin resistance | |
| PA2198 | 3.73 | 0.001 | Putative antibiotic biosynthesis monooxygenase | |
| PA3552 | arnB | 6.16* | 6.84E−06 | Hypothetical protein |
| PA3553 | arnC | 3.61 | 5.17E−05 | Probable glycosyl transferase |
| PA3554 | arnA | 2.44 | 0.0021 | Hypothetical protein |
| PA3555 | arnD | 4.49 | 1.57E−05 | Hypothetical protein |
| PA3556 | arnT | 3.13 | 0.0001 | Inner membrane l-Ara4N transferase |
| PA3557 | arnE | 4.51 | 2.97E−05 | Hypothetical protein |
| PA3558 | arnF | 2.45 | 0.0028 | Hypothetical protein |
ID, identification. Information is according to the P. aeruginosa genome website (http://www.pseudomonas.com).
Fold regulation of genes differentially expressed in cbrA mutant relative to PA14 wild type. A positive number indicates transcript upregulation and a negative number indicates transcript downregulation in the cbrA mutant. An asterisk indicates confirmation of gene regulation by RT-qPCR.
Analysis of the microarray data revealed dysregulation of genes involved in amino acid biosynthesis and metabolism, carbon compound catabolism, and central intermediary metabolism, consistent with the proposed role of CbrAB in the utilization of a variety of organic compounds as sole carbon source (22, 32). For example, hutU (urocanase), part of the hutUHIG operon involved in histidine catabolism, was moderately downregulated in the cbrA mutant (RT-qPCR revealed a fold change of −6.0 ± 0.6 relative to the wild type).
The cbrA microarray results were examined to identify genes that might influence antibiotic resistance of the mutant (since the microarray and MIC experiments utilized similar growth conditions). Moderate (3- to 10-fold) upregulation of the operons encompassing the two-component regulators oprH-phoPQ and pmrAB and of the downstream arnBCADTEF (lipopolysaccharide [LPS] modification) operon was observed in the microarray. To confirm the microarray results, RT-qPCRs were performed and revealed 9.3- ± 1.8-fold upregulation of oprH, 3.7- ± 0.5-fold upregulation of phoP, 3.1- ± 0.3-fold upregulation of phoQ, 10.2- ± 1.9-fold upregulation of arnB, and 4.0- ± 1.4-fold upregulation of pmrB. Activation of these operons is known to trigger bacterial resistance to cationic peptides and polymyxins in response to low-Mg2+ conditions by controlling the addition of aminoarabinose to lipid A, thereby reducing the net negative charge of LPS and limiting its interaction with polycationic peptides such as polymyxin B (29); however, their link to CbrAB had not been revealed previously. The involvement of the PhoPQ system in aminoglycoside resistance has also been defined (25, 38). Although the details of PhoPQ involvement in aminoglycoside resistance remain to be fully elucidated, PhoPQ appears to mediate resistance to aminoglycosides via a mechanism different from that involved with the polycationic peptides. The cbrA microarray also identified the upregulation of several heat shock protein genes, including htpG, ibpA, dnaK, grpE, hslV, and hslU. The upregulation of dnaK was confirmed by RT-qPCR, revealing 3.6- ± 0.2-fold upregulation in the mutant relative to the wild type. Recent studies have demonstrated that upregulation of heat shock genes prior to treatment with tobramycin led to increased resistance of P. aeruginosa to tobramycin (Kindrachuk et al., submitted). Although the cbrA microarray indicated upregulation of the mexX gene, known to be involved in involvement in aminoglycoside resistance (38), RT-qPCR failed to detect any significant changes in the transcriptional expression of this gene (fold change of 1.1 ± 0.3) in the cbrA mutant compared to the wild type.
Analysis of the microarray data also revealed moderate downregulation of several genes, PA0621, pvdD, PA3784, pchH, and pchF, which were possibly involved in the ciprofloxacin resistance phenotype observed in the cbrA mutant. A previous ciprofloxacin screen of the PA14 transposon mutant library by Breidenstein et al. showed that transposon mutants of PA0621, pvdD, PA3784, pchH, and pchF are more resistant than their wild-type parent strain toward ciprofloxacin (2). Flagella and type IV pili play important roles in biofilm and microcolony formation and are also required for swarming (19). Furthermore, studies have suggested that the flagellum secretion system plays a role in P. aeruginosa invasion of epithelial cells (8). However, the microarray did not reveal dysregulation in any flagellum- or type IV pilus-related genes. Furthermore, there was no dysregulation of the type III secretion apparatus or effector genes in the cbrA mutant.
This transcriptome analysis also highlighted several other interesting genes. These findings included moderate downregulation of the pyoverdine and pyochelin biosynthesis genes, including pvdA, pvdQ, and pchF. RT-qPCR confirmed downregulation of these genes in the mutant with values of −9.9 ± 1.3 for pvdA, −16.5 ± 2.4 for pvdQ, and −4.2 ± 0.6 for pchF. The downregulation of pvdQ in the cbrA microarray was of interest since a pvdQ mutant is swarming deficient (37) and has been suggested to play a role in biofilm formation and virulence of P. aeruginosa (18). Consistent with the qPCR data, we grew the cbrA mutant and the wild type in a low-iron medium to induce pyoverdine production and observed, using a spectrofluorimeter, that the mutant secreted less pyoverdine into the supernatant than did the wild type (Fig. 8). In a previous study, Jimenez et al. (18) showed that addition of iron in swarm plates restored the swarming of a swarming-impaired pvdQ mutant to the wild-type level. Therefore, we also tested whether addition of iron had any effect on swarming motility or the biofilm formation of the cbrA mutant. However, addition of iron did not restore swarming or biofilm formation of the cbrA mutant to the wild-type level (data not shown).
FIG. 8.
Pyoverdine production of the PA14 cbrA deletion mutant. Cells grown in Casamino Acids medium for 48 h at 37°C were diluted, and the resultant sample was excited at 400 nm with a spectrofluorimeter.
Transcriptional analysis, by RT-qPCR, of the cbrA mutant under various growth conditions.
The microarray experiment of the cbrA mutant versus PA14 wild type did not reveal transcriptional changes in the expression of genes known to be involved in virulence (e.g., the type III secretion system) or genes involved in the production of exopolysaccharides in P. aeruginosa. This was not completely unexpected, since the growth conditions used for the microarray experiment and the conditions used to assay exopolysaccharide production or in vitro cytotoxicity experiments were different and it is known that CbrAB influences the utilization of many different carbon and nitrogen sources. Therefore, we isolated RNA from bacterial cells during in vitro infection of HBE cells and cells growing on Congo red plates. Consistent with the increased binding phenotype observed on the Congo red plates (Fig. 3), RT-qPCR revealed 2.5- to 5-fold upregulation of the exopolysaccharide pelD and pelF genes in the cbrA mutant compared to the PA14 wild type (Table 4). Bacterial cells obtained during HBE cell infection revealed moderate upregulation of the type III secretion apparatus and effectors (exoT, exoY, exoU, pcrV, exsA, and popD) and the type I secretion apparatus, aprD (Table 4), consistent with the enhanced cytotoxicity of this mutant.
TABLE 4.
Dysregulated genes in the cbrA, cbrB, crcZ, and crc mutants during human bronchial epithelial (HBE) cell line infection and growth on Congo red plates as determined by RT-qPCR
| Expt and gene | Fold change relative to PA14 wild typea |
|||
|---|---|---|---|---|
| cbrA | cbrB | crcZ | crc | |
| HBE cell infection | ||||
| exoT | 2.9 ± 0.3 | 3.4 ± 1.0 | 3.8 ± 0.9 | −3.8 ± 1.2 |
| exoY | 2.7 ± 0.3 | 2.3 ± 0.6 | 2.1 ± 0.5 | −4.3 ± 0.3 |
| exoU | 2.5 ± 0.4 | 2.7 ± 0.3 | 2.0 ± 0.4 | −9.8 ± 1.0 |
| pcrV | 2.6 ± 0.3 | 2.2 ± 0.1 | 2.0 ± 0.3 | −5.8 ± 0.9 |
| exsA | 2.2 ± 0.2 | 2.7 ± 0.5 | 1.7 ± 0.6 | −2.7 ± 0.9 |
| pcrD | 1.4 ± 0.1 | 1.2 ± 0.5 | 1.2 ± 0.8 | −1.4 ± 0.5 |
| popD | 2.9 ± 0.5 | 2.3 ± 0.5 | 2.4 ± 0.4 | −6.6 ± 0.7 |
| pscF | 1.2 ± 0.1 | 1.4 ± 0.5 | 1.6 ± 0.9 | −2.4 ± 0.3 |
| pcrR | 1.3 ± 0.3 | 1.8 ± 1.1 | 1.2 ± 0.9 | −1.5 ± 0.5 |
| lasB | 1.2 ± 0.2 | 1.1 ± 0.6 | 1.5 ± 0.8 | −1.2 ± 0.5 |
| lipC | 1.2 ± 0.1 | −1.1 ± 0.3 | 1.1 ± 0.5 | −1.3 ± 0.3 |
| plcB | 1.6 ± 0.5 | 1.3 ± 0.3 | 1.7 ± 0.6 | −1.8 ± 0.6 |
| PA4528 | 1.5 ± 0.3 | 1.3 ± 0.1 | 1.1 ± 0.3 | −1.0 ± 0.5 |
| aprD | 2.5 ± 0.5 | 2.9 ± 0.4 | 2.4 ± 0.3 | −6.2 ± 0.8 |
| Congo red | ||||
| pelD | 2.6 ± 0.5 | 5.1 ± 0.5 | 2.7 ± 0.5 | −4.1 ± 1.0 |
| pelF | 2.5 ± 0.4 | 4.5 ± 0.4 | 2.8 ± 0.5 | −2.6 ± 0.8 |
Values are represented as averages of at least 3 biological samples.
CbrA mediated regulation of swarming, biofilm formation, and cytotoxicity in conjunction with CbrB.
The CbrB response regulator has been identified to play a role along with its cognate sensor kinase CbrA in the global metabolic regulation of carbon and nitrogen utilization in P. aeruginosa (22). Therefore, we examined whether CbrB also interacted with CbrA to modulate virulence and virulence-related processes in P. aeruginosa. Similarly to the cbrA mutant, the cbrB mutant displayed impairment in swarming and excessive biofilm formation (Fig. 1B and 2C). These phenotypes could also be complemented by introducing the wild-type cbrB allele into the cbrB mutant. Congo red assays revealed increased binding of Congo red to the cbrB mutant compared to the wild type (Fig. 3). Interestingly, on the Congo red plates, the cbrB mutant also showed wrinkled morphology while the cbrA mutant remained smooth like the wild type. Furthermore, the cbrB mutant exhibited 2-fold-enhanced in vitro cytotoxicity toward HBE cells (Fig. 5B). Similarly to the cbrA mutant, the cbrB mutant also exhibited moderate growth defects in swarming/MIC and biofilm media (data not shown). However, while the cbrA mutant showed increased resistance to a variety of antibiotics, the cbrB mutant displayed wild-type MIC values for all antibiotics tested (Table 2). Based on the microarray results of the cbrA mutant, we examined the expression level of a variety of genes in the cbrB mutant under swarming conditions by RT-qPCR. As expected, RT-qPCR revealed no dysregulation in the genes that were thought to explain the altered susceptibility of the cbrA mutant; these findings included a lack of dysregulation of the oprH-phoPQ operon, the pmrAB operon, and the LPS modification operon (data not shown). Similarly to the cbrA mutant, upregulation of energy metabolism genes, such as nirS and norC, was observed in the cbrB mutant. The expression of pvdQ was also downregulated in the cbrB mutant, while other genes involved in pyoverdine biosynthesis, such as pvdA, were not dysregulated in the cbrB mutant (data not shown). However, similarly to the cbrA mutant, pyochelin biosynthesis genes, such as pchG, were downregulated in the cbrB mutant. In addition, RNA was isolated from the cbrB mutant from Congo red plates, and during HBE cell infection, RT-qPCRs were performed. The cbrB mutant revealed upregulation of the pel operon under the Congo red condition, but to a greater extent than that with the cbrA mutant (Table 4). This result was consistent with the wrinkled morphology observed only in the cbrB mutant and the increased biofilm formation of the cbrB mutant compared to the cbrA mutant. RT-qPCR analysis of the cbrB mutant during HBE cell infection also revealed upregulation of the type III secretion system during in vitro infection (Table 4).
The CbrAB system regulated swarming, biofilm formation, and cytotoxicity through the posttranscriptional regulatory system Crc/CrcZ.
Recently, Sonnleitner et al. found that the CbrAB/CrcZ/Crc system enables P. aeruginosa to utilize various carbon sources (46). In addition to its role in catabolite repression, O'Toole et al. showed that Crc is required for biofilm formation in P. aeruginosa (35). Furthermore, Linares et al. demonstrated that the Crc protein plays a role in the regulation of virulence in P. aeruginosa (24). They showed that a crc mutant is defective in type III secretion and is less virulent in a Dictyostelium discoideum model (24). Thus, these results led us to investigate whether the CbrAB system regulated swarming, biofilm formation, and cytotoxicity through the CrcZ/Crc system. First, we examined the ability of a PA14 crc transposon mutant to swarm, form biofilm, and infect HBE cells. As shown in Fig. 9A, the crc mutant was able to swarm as well as the wild type but consistently exhibited reduced branching of the swarming tendrils compared to the wild type. The crc mutant was also defective in biofilm formation and showed a reduced ability to bind Congo red (Fig. 9B and D). The crc mutant also displayed a reduced ability to infect and destroy a monolayer of cultured 16HBE14o- epithelial cells (Fig. 9C). As well, the crc mutant showed wild-type growth in swarm and biofilm media (data not shown). These phenotypes could also be complemented by introducing the wild-type crc allele into the crc mutant. These results suggest that the Crc protein had different effects than did the cbrA and cbrB mutants on swarming motility (modest effects on branching), while positively regulating biofilm formation and virulence of P. aeruginosa. RT-qPCR experiments on the crc mutant demonstrated downregulation of the type III secretion system during HBE cell infection and downregulation of the pel operon expression from Congo red plates (Table 4).
FIG. 9.
Swarming motility, biofilm formation, Congo red binding, and in vitro cytotoxicity of the PA14 Δcrc mutant. (A) Swarming motility was assessed by spotting 1 μl of mid-log cell cultures onto BM2-swarming plates with 0.5% (wt/vol) agar and incubating the plates at 37°C for 20 h. (B) Biofilm formation was assessed by diluting overnight cultures in BM2-biofilm medium and incubating the cultures at 37°C for 20 h. (C) Cytotoxicity toward HBE cells was determined by infecting a monolayer of HBE cells with mid-log bacterial cultures at an MOI of 2. (D) Congo red binding was assessed by spotting 1, 5, or 10 μl of diluted cultures onto Congo red plates and incubating the plates for 24 h at 37°C and for an additional 48 h at room temperature.
We also tested the ability of a crcZ deletion mutant to swarm, form biofilm, and infect HBE cells. In contrast to the crc mutant, the crcZ mutant exhibited phenotypes very similar to those of the cbrA and cbrB mutants, including substantial swarming deficiency, excessive biofilm formation, and enhanced cytotoxicity toward HBE cells (Fig. 10). These phenotypes could be complemented by introducing the wild-type crcZ allele into the crcZ mutant. In addition, growth studies revealed that the crcZ mutant exhibited moderate growth defects in swarming media (data not shown). RT-qPCR analysis of the RNA isolated from the crcZ mutant cells grown on Congo red plates revealed upregulation of the pel operon and upregulation of the type III secretion system during HBE cell infection (Table 4).
FIG. 10.
Swarming motility, biofilm formation, and in vitro cytotoxicity of the PA14 ΔcrcZ mutant. (A) Swarming motility of the PA14 wild type (WT) and crcZ mutant was determined by inoculating 1 μl of mid-log cell cultures onto BM2-swarming plates with 0.5% (wt/vol) agar and incubating plates at 37°C for 20 h. (B) Biofilm formation of the PA14 wild type (WT), PA14 crcZ mutant, and PA14 crcZ+ complemented strain was determined by diluting overnight cultures in BM2-biofilm medium and incubating the cultures at 37°C for 20 h. (C) Cytotoxicity toward HBE cells was determined by infecting a monolayer of HBE cells with mid-log bacterial cultures at an MOI of 2 for 6 h at 37°C with 5% CO2. **, statistically significant difference (P < 0.05) between the mutants and the wild type as determined by Student's t test. (D) Congo red binding was assessed by spotting 1, 5, or 10 μl of diluted cultures onto Congo red plates and incubating the plates for 24 h at 37°C and for an additional 48 h at room temperature.
We also tested the susceptibility/resistance of the crcZ and crc mutants toward the antibiotics tested for the cbrA mutant. However, no differences between the mutants and wild type were observed for any of the tested antibiotics (data not shown). These results indicate that the CbrAB system might be regulating swarming, biofilm formation, and cytotoxicity via CrcZ, with a partial inverse regulation by Crc.
DISCUSSION
In this study, we examined the role of the sensor kinase CbrA in motility, biofilm formation, cytotoxicity, and antibiotic resistance. A cbrA deletion mutant was swarming defective but exhibited enhanced biofilm formation and in vitro cytotoxicity toward human bronchial epithelial cells. Furthermore, the cbrA mutant exhibited increased resistance toward several common clinical antibiotics, including polymyxin B, tobramycin, and ciprofloxacin. The sensor kinase CbrA and adjacently encoded response regulator CbrB have been proposed to work together in regulating the utilization of a variety of organic compounds as sole carbon sources. Phenotypic and genetic analyses performed here indicated that CbrA regulated swarming, biofilm formation, and cytotoxicity via its cognate response regulator CbrB. In addition to CbrB, we provided evidence for the involvement of the small RNA CrcZ in the regulation cascade of these virulence and virulence-related phenotypes. In contrast, the antibiotic resistance phenotype observed in the cbrA mutant was absent in the cbrB and crcZ mutants, and we speculate that CbrA may cross talk with other response regulators to regulate antibiotic resistance. A proposed model for the regulation of swarming, biofilm formation, cytotoxicity, and antibiotic resistance by the CbrA/CbrB/CrcZ pathway is shown in Fig. 11. The situation is not as clear with Crc, which is known to be negatively regulated by crcZ RNA and to fully participate in regulation of carbon source utilization. If Crc were fully involved, we would expect there to be a reciprocal phenotype in the crc mutant. Instead, there was only a partial effect on swarming in our study, but a substantial decrease in biofilm formation and a complete absence of epithelial cell toxicity. Thus, downstream effects on crc may be part of the phenotype but do not appear to fully explain the phenotypes observed.
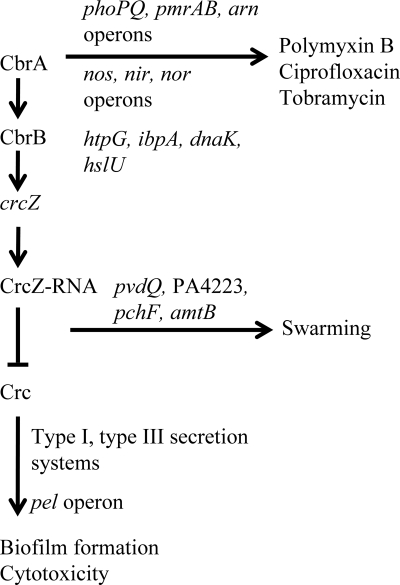
FIG. 11.
Proposed model for the involvement of the CbrA/CbrB/CrcZ regulatory cascade in the regulation of swarming, biofilm formation, cytotoxicity, and antibiotic resistance in P. aeruginosa. Under conditions that activate the CbrAB two-component system, the phosphorylated response regulator CbrB will activate the expression of CrcZ. In turn, CrcZ has a high affinity for the RNA binding protein Crc. The binding of CrcZ to Crc sequesters Crc, resulting in the inhibition of expression of mRNAs that encode factors to promote biofilm formation and virulence. In addition to sequestration of Crc, CrcZ is involved in the direct or indirect activation of the expression of genes to promote swarming motility. Antibiotic resistance, however, involves only the CbrA sensor kinase.
Complementation of the cbrA deletion mutant with the wild-type cbrAB operon was necessary since complementation with the PA14 wild-type cbrA allele alone only moderately but incompletely restored the phenotypes. We speculate that the reason that the cbrA mutant was unable to be fully complemented with cbrA alone is that in the cbrA complemented strain, there were multiple copies of the cbrA gene due to the presence of the multicopy vector. Hence, when CbrA was expressed, the disproportionate amount of CbrA relative to the amount of CbrB in the complemented strain, compared with the wild type (confirmed by RT-qPCR), might have changed the functional interaction of these components and even shifted the balance from phosphorylation to dephosphorylation (both functions of sensor kinases).
Several physical factors are currently known to be required for swarming, including flagella, type IV pili, and the production of the biosurfactant rhamnolipids. The ability of the cbrA mutant to swim and twitch at close to wild-type levels (55), and in microarrays to express relevant genes at the same level as that for the wild type, suggested that the cbrA mutant was not impaired in flagellum or type IV pilus biosynthesis. Furthermore, the cbrA mutant produced a wild-type level of rhamnolipids. These results indicate that CbrA normally promotes the expression of additional factors required for swarming in P. aeruginosa.
Microarray analysis of the cbrA mutant, compared to the PA14 wild type under swarming conditions, revealed the downregulation of a number of genes that have been identified to be upregulated in swarmer cells (37), suggesting that these genes may be regulated through CbrA and involved in the swarming growth state. Among others, these genes included a probable ATP-binding component of the ABC transporter, PA4223; genes involved in pyochelin biosynthesis, pchG and pchF; genes involved in pyoverdine biosynthesis, pvdQ, pvdE, and pvdD; and a gene encoding an ammonium transporter, amtB. Of these genes, only the PA4223 and pvdQ mutants displayed swarming deficiencies, consistent with previous studies (37, 55). PvdQ is of particular interest, as this protein plays a dual role in pyoverdine biosynthesis, as well as quorum sensing, with an acylhomoserine lactone acylase activity. Recently, the role of PvdQ in swarming was further studied by Jimenez et al. (18). This group showed that under iron limitation, the pvdQ mutant was defective in swarming, and they indicated that swarming could be restored by exogenous addition of iron, suggesting that the role of PvdQ in swarming was closely linked to the pyoverdine/iron pathway (18). In our current study and the study by Overhage et al. (37), we found that, with the exception of pvdQ, the genes required for pyoverdine biosynthesis, such as pvdE and pvdD, exhibited wild-type swarming behavior. Moreover, addition of iron to the swarming medium did not change the swarming phenotype of the cbrA mutant. Therefore, we suggest that the role of PvdQ as a quorum signal quencher might be more influential in swarming. As suggested previously, PvdQ may play a role in maintaining the relative concentrations of the two homoserine lactone quorum sensing signals, required for swarming differentiation. Alternatively, the degradation product induced by PvdQ may act a signal during swarmer cell differentiation (37). We are currently investigating these possibilities.
The cbrA deletion mutant produced significantly more biofilm than did the wild-type strain in an abiotic biofilm assay (Fig. 2A and B). Congo red assays revealed that the hyperbiofilm phenotype of the cbrA mutant might be due to increased production of the pel-encoded exopolysaccharide (Fig. 3). CbrA did not appear to play a role during initial attachment stage, as a rapid attachment assay revealed no difference in the abilities of the mutant and the wild type to attach to the wells of the polystyrene plates. This result was expected, since the cbrA mutant had a functional flagellum and pilus. Overall, our results suggested that CbrA normally negatively regulates biofilm formation. In contrast, a recent screen of the PA14 transposon mutant library by Musken et al. revealed that a cbrA mutant exhibited a reduced-biofilm phenotype (31). The reason for the difference in biofilm phenotype may be due to the different media used to grow the biofilms. While we used minimal medium to cultivate formation of biofilm at the air-liquid interface, Musken's group used a rich medium (LB broth) to promote biofilm formation at the bottom of the microtiter plate. The intricate relationship between swarming motility and biofilm formation in P. aeruginosa is complex. Although biofilm formation is a surface-associated sessile behavior and swarming is a surface-associated motile behavior, both processes are suggested to involve similar components at certain stages and under specific conditions (49). For example, both swarming motility and the initiation of biofilm formation have been shown to require flagella (33). Moreover, there is evidence that swarming motility can contribute to the early stages of P. aeruginosa biofilm formation (44). There are a number of studies that suggest that these surface-associated behaviors are inversely regulated and mediated through the signaling molecule cyclic-di-GMP (c-di-GMP) (21, 30, 49). It has been demonstrated that the intracellular levels of this signaling molecule influence a number of bacterial behaviors, with the common theme being that the accumulation of c-di-GMP promotes sessile behaviors, such as biofilm formation, while the degradation of c-di-GMP favors motile behaviors, such as swarming. Recent studies have shown that BifA, a c-di-GMP phosphodiesterase, participates with SadC, a c-di-GMP diguanylate cyclase, to control the level of cellular c-di-GMP in regulating biofilm formation and swarming motility (21, 30). As the cbrA mutant also exhibits a severe swarming defect and a hyperbiofilm phenotype, it will be of interest to examine whether CbrA plays a role in regulating the level of cellular c-di-GMP. Regardless, we previously found numerous other mutations that either exhibited a reciprocal relationship between swarming and biofilm formation or showed similar effects on the two processes (36, 55), so it seems likely that control of these processes is multideterminant.
The antibiotic resistance phenotypes observed for the cbrA mutant were not observed for the cbrB mutant. Intriguingly, these results mirror what we have seen with other two-component regulators, where the sensor kinase and response regulator have different phenotypes (e.g., phoQ is constitutively resistant to polymyxin and aminoglycosides, while phoP is null) (25). Using RT-qPCR, we confirmed substantial changes in the cbrA mutant, but in contrast in the cbrB mutant, we observed no significant changes in the transcriptional expression of the oprH-phoPQ operon, the pmrAB operon, or the LPS modification operon (arn operon). Thus, the upregulation of these operons in the cbrA mutant can be concluded to play a major role in the resistance of this mutant to the majority of the antibiotics tested, including polymyxin B, colistin, and tobramycin. We plan to further investigate the mechanism of potential cross talk between CbrA and PhoP.
The involvement of CbrA and CbrB in a number of important adaptation-related processes in P. aeruginosa suggests that these regulators contribute in maintaining the overall physiological balance of the bacterium. By enabling the bacteria to utilize a variety of organic compounds as carbon sources and to undergo swarming motility, while sustaining an optimal level of biofilm production, cytotoxicity, and antibiotic resistance, CbrA optimizes the efficiency of the bacteria to adapt to various environments.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the Canadian Institutes for Health Research (CIHR) and the Canadian Cystic Fibrosis Foundation (CCFF). A.T.Y.Y. received studentships from the CCFF and the Natural Sciences and Engineering Research Council of Canada (NSERC). R.E.W.H. holds a Canada Research Chair.
We thank Laetitia Abdou and Dieter Haas for providing the necessary plasmids and primers for construction of the PA14 crcZ deletion mutant and complementation of the mutant. We thank Shaan Gellatly for her help with the in vitro human bronchial epithelial cell infection experiments. We also thank the Institute for Genomic Research (TIGR) for providing the microarray slides.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Boucher, R. C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146-158. [DOI] [PubMed] [Google Scholar]
- 2.Breidenstein, E. B., B. K. Khaira, I. Wiegand, J. Overhage, and R. E. W. Hancock. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 4.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in the pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 5.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 6.Deziel, E., G. Paquette, R. Villemur, F. Lepine, and J. Bisaillon. 1996. Biosurfactant production by a soil Pseudomonas strain growing on polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 62:1908-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deziel, E., et al. 1999. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1440:244-252. [DOI] [PubMed] [Google Scholar]
- 8.Fleiszig, S. M., S. K. Arora, R. Van, and R. Ramphal. 2001. FlhA, a component of the flagellum assembly apparatus of Pseudomonas aeruginosa, plays a role in internalization by corneal epithelial cells. Infect. Immun. 69:4931-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gooderham, W. J., et al. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155:699-711. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3:247-255. [DOI] [PubMed] [Google Scholar]
- 13.He, J., et al. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentzer, M., et al. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hester, K. L., et al. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 17.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez, P. N., et al. 2010. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology 156:49-59. [DOI] [PubMed] [Google Scholar]
- 19.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzeslak, J., G. Gerritse, R. van Merkerk, R. H. Cool, and W. J. Quax. 2008. Lipase expression in Pseudomonas alcaligenes is under the control of a two-component regulatory system. Appl. Environ. Microbiol. 74:1402-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchma, S. L., et al. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, W., and C. Lu. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189:5413-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberati, N. T., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linares, J. F., et al. 2010. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 12:3196-3212. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlane, E. L., A. Kwasnicka, and R. E. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor, C. H., J. A. Wolff, S. K. Arora, and P. V. Phibbs, Jr. 1991. Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J. Bacteriol. 173:7204-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mah, T. F., et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 29.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 30.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musken, M., S. Difiore, A. Dotsch, R. Fischer, and S. Haussler. 2010. Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiology 156:431-441. [DOI] [PubMed] [Google Scholar]
- 32.Nishijyo, T., D. Haas, and Y. Itoh. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 40:917-931. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overhage, J., S. Lewenza, A. K. Marr, and R. E. W. Hancock. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 Mini-Tn5-lux mutant library. J. Bacteriol. 189:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overhage, J., M. Bains, M. D. Brazas, and R. E. W. Hancock. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole, K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajan, S., and L. Saiman. 2002. Pulmonary infections in patients with cystic fibrosis. Semin. Respir. Infect. 17:47-56. [DOI] [PubMed] [Google Scholar]
- 40.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietsch, A., M. C. Wolfgang, and J. J. Mekalanos. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 72:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe, S. M., S. Miller, and E. J. Sorscher. 2005. Cystic fibrosis. N. Engl. J. Med. 352:1992-2001. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Shrout, J. D., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 45.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 46.Sonnleitner, E., L. Abdou, and D. Haas. 2009. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 106:21866-21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 48.Tremblay, J., A. P. Richardson, F. Lepine, and E. Deziel. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9:2622-2630. [DOI] [PubMed] [Google Scholar]
- 49.Verstraeten, N., et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496-506. [DOI] [PubMed] [Google Scholar]
- 50.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 51.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 52.Wiegand, I., K. Hilpert, and R. E. W. Hancock. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163-175. [DOI] [PubMed] [Google Scholar]
- 53.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and P. V. Phibbs, Jr. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 173:4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wozniak, D. J., et al. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung, A. T. Y., et al. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191:5592-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.