Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a food-borne pathogen that can cause severe health complications and utilizes a much lower infectious dose than other E. coli pathotypes. Despite having an intact ure locus, ureDABCEFG, the majority of EHEC strains are phenotypically urease negative under tested conditions. Urease activity potentially assists with survival fitness by enhancing acid tolerance during passage through the stomach or by aiding with colonization in either human or animal reservoirs. Previously, in the EHEC O157:H7 Sakai strain, a point mutation in ureD, encoding a urease chaperone protein, was identified, resulting in a substitution of an amber stop codon for glutamine. This single nucleotide polymorphism (SNP) is observed in the majority of EHEC O157:H7 isolates and correlates with a negative urease phenotype in vitro. We demonstrate that the lack of urease activity in vitro is not solely due to the amber codon in ureD. Our analysis has identified two additional SNPs in ureD affecting amino acid positions 38 and 205, in both cases determining whether the encoded amino acid is leucine or proline. Phylogenetic analysis based on Ure protein sequences from a variety of urease-encoding bacteria demonstrates that the proline at position 38 is highly conserved among Gram-negative bacteria. Experiments reveal that the L38P substitution enhances urease enzyme activity; however, the L205P substitution does not. Multilocus sequence typing analysis for a variety of Shiga toxin-producing E. coli isolates combined with the ureD sequence reveals that except for a subset of the O157:H7 strains, neither the in vitro urease-positive phenotype nor the ureD sequence is phylogenetically restricted.
Enterohemorrhagic Escherichia coli (EHEC) is a highly infectious pathogen that colonizes the large intestine in humans and can cause severe clinical manifestations, such as hemorrhagic colitis and hemolytic-uremic syndrome (21, 26). E. coli O157:H7 has been responsible for the majority of EHEC outbreaks in the United States; however, other EHEC serogroups, such as O26, O111, and O145, are prevalent in other countries and are increasingly found to be associated with outbreaks in the United States (3, 14, 22). The bovine intestinal tract is the principle reservoir of EHEC, but other livestock animals, such as sheep and goats, have been found to transiently harbor this pathogen (15). Outbreaks of EHEC disease have been associated primarily with contaminated food, either insufficiently cooked beef products (5, 51) or leafy vegetables contaminated in the farm environment (19, 24, 31). With a calculated infectious dose of only 10 to 100 CFU (50, 51, 52), persistence of EHEC even at low levels in the bovine gastrointestinal tract can have a major public health impact. The sequenced EHEC genomes (16, 40) have been found to contain the ure locus, encoding genes for producing the enzyme urease. Urease, a cytoplasmic enzyme, catalyzes the hydrolysis of urea to form ammonia and carbon dioxide, providing both an increase in pH and a readily assimilated source of nitrogen (32). Recent studies have shown that bacterial ureases may encode other biological activities independently of their ureolytic activity, such as induction of platelet aggregation and activation of lipooxygenases (37). The EHEC ure gene cluster, ureDABCEFG, contains the seven genes necessary to form active urease. The inactive apoprotein, (UreABC)3, is activated by the addition of two Ni2+ ions to each of the three active sites in the enzyme. Incorporation of the nickel cofactor is accomplished using the accessory proteins encoded by ureD, ureE, ureG, and ureF (32). In order for proper assembly of the urease metallocenter, all four of the accessory proteins must be functional. Although urease has been demonstrated to be an important virulence factor in a number of bacteria, such as Helicobacter pylori (11), Yersinia enterocolitica (10), Proteus mirabilis (9), Brucella species (1, 45), and Klebsiella pneumoniae (27), the role of urease in EHEC pathogenesis is not defined. It is possible that, like some other bacteria, EHEC exploits the enzyme urease as an additional factor to aid with surviving the low pH conditions as it passes through the stomach. Alternatively, or in addition, expression of urease might enhance EHEC competitive survival for colonization in either the bovine or human intestinal tract or aid with survival in the farm environment.
The lack of information on the role of EHEC urease may be due to the fact that very few isolated EHEC strains demonstrate a urease-positive phenotype in vitro. Neither the EHEC O157:H7 EDL933 nor the EHEC O157:H7 Sakai strain, the first two EHEC genomes to be fully sequenced (16, 40), shows detectable urease activity in vitro. However, as reported in the first experimental work involving EHEC urease (18), urease activity was conferred on E. coli DH5α (amber suppressor strain) harboring a plasmid containing the entire EDL933 ure gene cluster. Therefore, it was determined that the EDL933 ure genes are capable of producing functional urease, given the correct conditions for expression. Nakano et al. compared the DNA sequence of the entire ure gene cluster from the Sakai strain, which has 100% DNA sequence identity with the EDL933 ure gene cluster (EDL933 encodes two separate but identical gene clusters), with that of a strain showing positive urease activity (36). The sequence analysis identified a single nucleotide substitution in ureD, C742T, resulting in a substitution of the amber stop codon for glutamine in the Sakai strain. This same C742T substitution in ureD is highly conserved among EHEC isolates with a urease-negative phenotype in vitro. The truncation of the UreD protein due to the presence of the amber codon is believed to be the cause of the urease-negative phenotype observed in these strains (36). Irrespective of their urease phenotype, EHEC isolates retain the entire ure gene cluster with a high degree of sequence conservation among strains for which the entire ure locus or specific ure genes have been sequenced (13, 38). This sequence conservation, combined with the continued metabolic burden imposed by expression of the ure genes, suggested that urease may play an important role in the pathogenesis or survival of this organism and that perhaps there exists a mechanism by which EHEC isolates carrying this mutation are able to produce active urease in vivo.
In comparing ureD sequences from isolates representing various EHEC serotypes, two additional single nucleotide polymorphisms (SNPs) have been identified. Phylogenetic analysis based on Ure protein sequences from a wide variety of urease-producing bacteria combined with experiments involving expression of ureD with the different SNP combinations elucidates the role of these SNPs in EHEC urease enzyme activity. Using a combination of multilocus sequence typing (MLST) and ureD gene sequencing, we demonstrate that the ureD SNPs are not restricted to a single branch of diarrheagenic E. coli. We demonstrate that the amber codon in ureD is not the defining genetic feature resulting in the lack of urease production and have identified additional SNPs that contribute to the urease-negative phenotype.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli cultures were grown in Luria-Bertani (LB) broth or on LB agar plates for construction of mutants and extraction of genomic DNA. For urease expression experiments, the E. coli strains were grown at 37°C in either M9 minimal medium or low-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10 μM NiCl2. The following antibiotics were used at the indicated concentrations, where appropriate: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; streptomycin (Sm), 50 μg/ml; and tetracycline (Tc), 2 or 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| S17-1λpir | λpir lysogen of S17-1, recA RP4 2-Tc::Mu-Km::Tn7 | 47 |
| TOP10 | F−mcrA recA1 araD139 Δ(ara leu) endA1 | Invitrogen |
| EHEC strains | ||
| Sakai Δstx2A | Sakai strain with stx2A deletion | This study |
| Sakai Δure | Urease gene cluster deletion mutant of Sakai Δstx2A | This study |
| Sakai ureD(Q) | Sakai Δstx2A with T742C mutation in ureD, resulting in replacement of amber codon with glutamine in UreD; aadA inserted at attTn7, conferring Smr | This study |
| Sakai ureD(P,Q) | Sakai ureD(Q) with T113C mutation in ureD, resulting in L38P mutation in UreD | This study |
| Sakai tet(Am) | Sakai Δstx2A carrying mutant (Q56amber) Tcr cassette at attTn7 | This study |
| Plasmids | ||
| pSRS1 | Derived from suicide plasmid CVD442 by addition of mcs; contains Apr and sacB | This study |
| pSRS10 | Suicide vector for deletion of stx2A | This study |
| pKD3 | Cmr cassette flanked by FRTa sites | 8 |
| pKD46 | Red recombinase Relper plasmid, RepA101(Ts), Apr | 8 |
| pSRS20 | Suicide vector for construction of Sakai ureD(Q) | This study |
| pGRG24 | Plasmid for transgene insertion of aadA at attTn7 site on chromosome | 29 |
| pACYC184 | Tcr cassette | New England Biolabs |
| pSRS50 | First suicide vector for construction of Sakai ureD(P,Q) | This study |
| pSRS60 | Second suicide vector for construction of Sakai ureD(P,Q) | This study |
| pGFIB-1 | Vector for cloning tRNA for constitutive expression in E. coli, Apr | National Institute of Genetics, Japan (28) |
| pSUPE | pGFIB-1 expressing the supE tRNA | This study |
| pGRG36 | Plasmid for insertion of cloned transgene at attTn7 site on chromosome | 29 |
| pBADHisA | Apr, araBAD promoter based expression vector | Invitrogen |
| pLLamber | pBADHisA expressing 6× His UreD(L,L,amber) | This study |
| pLLQ | pBADHisA expressing 6× His UreD(L,L,Q) | This study |
| pPLQ | pBADHisA expressing 6× His UreD(P,L,Q) | This study |
| pPPQ | pBADHisA expressing 6× His UreD(P,P,Q) | This study |
| pLPQ | pBADHisA expressing 6× His UreD(L,P,Q) | This study |
FRT, FLP recombination target.
Construction of urease mutants.
Previous to this work, an in-frame deletion in the stx2A gene was constructed in the wild-type EHEC Sakai strain, resulting in the strain Sakai Δstx2A (S. R. Steyert, unpublished work). For simplicity, this strain is referred to as Sakai in this experimental work and constructed strains are derivatives of this Δstx2A strain. A mutant strain, Sakai ureD(Q), was constructed in which a ureD point mutation, T742C, results in substitution of glutamine for the amber codon in the UreD protein. A three-step process was used for construction of the strain with this point mutation. First, bacteriophage lambda Red-mediated recombination (8, 33) was used to simultaneously delete approximately 100 bp of DNA surrounding the nucleotide to be mutated and insert a Cmr cassette. Primers UREDP1 and UREDP2 (see Table S1 in the supplemental material), each including a 40-bp flanking region, were used on the pKD3 template for this procedure. Plasmid pKD46 was used for expression of the Red recombinase system (8, 33). The second step in mutant construction involved homologous recombination using the suicide plasmid pSRS20 to delete the Cmr cassette and insert the ureD sequence carrying the point mutation (T742C in ureD). The suicide plasmid was constructed by ligating the DNA sequence containing the 100-bp deleted region along with flanking sequences ∼500 bp upstream and downstream into pSRS1. The inserts were prepared from the PCR product using a primer containing the point mutation. The two primer pairs UDM1F/UDM1R and UDM2F/UDM2R (see Table S1 in the supplemental material) were used for the construction. After confirmation of the point mutation on the chromosome by sequencing, the third step, insertion of the aadA gene conferring Smr into the attTn7 site, was accomplished using plasmid pGRG24 and the transposition method developed by McKenzie and Craig (29). This strain, Sakai ureD(Q), was then used to create strain Sakai ureD(P,Q) which has an additional point mutation, T113C, resulting in the L38P substitution in UreD. This strain was constructed in two steps, each using a suicide plasmid. For the first step, approximately 150 bp surrounding the intended T113C mutation site was deleted and a Tcr cassette was inserted. Flanking regions each consisting of ∼500 bp were PCR amplified from Sakai genomic DNA with primer pairs SV1F1F/SV1F1R and SV1F2F/SV1F2R (see Table S1 in the supplemental material), and the Tcr cassette was amplified from pACYC184 using primer pair TetF/TetR (see Table S1 in the supplemental material). These three PCR products were digested as appropriate, such that upon ligation into digested pSRS1, the Tcr cassette is positioned between the flanking regions, creating plasmid pSRS50. After conjugation with the aid of E. coli S17-1λpir and homologous recombination, followed by counterselection, the suicide plasmid used in the second step, pSRS60, was used to remove the Tcr cassette and insert the ureD sequence carrying the point mutation. pSRS60 was constructed by ligating the DNA sequence containing the 150-bp deleted region along with flanking sequences into pSRS1. The two primer pairs SV2F1F/SV2F1R and SV2F2F/SV2F2R (see Table S1 in the supplemental material), one primer of which incorporated the mutation, were used for the construction. The point mutation was confirmed in the resulting mutant strain, Sakai ureD(P,Q), by sequencing.
Urease screen and assay.
Our initial experiments were with EHEC strains, E. coli strains that produce Shiga toxin and contain the locus of enterocyte effacement (LEE) pathogenicity island. Subsequently, we began including strains of E. coli that lack the LEE island but that still produce Shiga toxin, known collectively as Shiga toxin-producing E. coli (STEC). The STEC isolates were screened for positive urease activity in 96-well plates using medium containing 4.0 g/liter yeast extract and 29.0 g/liter urea agar base (BD). One colony from each isolate grown overnight on LB plates was added to 200 μl medium, and the plate was covered with film to limit medium evaporation and incubated overnight at 37°C. Urease activity was quantified for the strains found to be urease positive using a previously described colorimetric assay based on reaction of phenol-hypochlorite in which ammonia is released during urea hydrolysis (53). Overnight cultures were concentrated by centrifugation, washed twice in 50 mM HEPES buffer, pH 7.5, resuspended in the same buffer, and lysed using a French press. The total protein concentration in the lysates was determined using the Bio-Rad DC protein assay, according to the manufacturer's specifications and with bovine serum albumin as the standard. A lysate volume containing 50 μg protein was combined with the phenol-hypochlorite assay buffer, consisting of 50 mM HEPES, pH 7.5, and 25 mM urea, and the mixture was compared to standards prepared from NH4Cl. E. coli MG1655 was used as a negative control, and EHEC O157:H7 strain IN1 and/or Mo28 was used as a positive control. Urease activities reported are from three or more independent measurements, with each measurement consisting of the average of two time points during urea hydrolysis.
Suppression of amber codon in ureD.
The glnX tRNA gene was PCR amplified from E. coli DH5α using primers GlnXF and GlnXR (see Table S1 in the supplemental material) and cloned into pGFIB-1. This was followed by site-directed mutagenesis using primers Sems and Sema (see Table S1 in the supplemental material), resulting in a G35A mutation in glnX to obtain plasmid pSUPE carrying the stop codon suppressor tRNA supE in place of glnX. A reporter strain was constructed on the basis of mutation of the first glutamine to an amber codon in the Tcr gene of the plasmid pACYC184. After the site-directed mutagenesis using primers Tcms and Tcma (see Table S1 in the supplemental material), the mutant Tcr gene was PCR amplified from pACYC184 using primers TetF and TetR (see Table S1 in the supplemental material), ligated into the SmaI site of pGRG36, and transposed into the attTn7 site of the chromosome of the Sakai Δstx2A strain using the method developed by McKenzie and Craig (29). Plasmids pGFIB-1 and pSUPE were each electroporated into this reporter strain, Sakai tet(Am), now carrying a Tcr cassette with an internal amber codon.
Reverse transcription-PCR (RT-PCR).
RNAprotect (Qiagen) was added to overnight cultures grown in M9 minimal medium supplemented with 1 μM NiCl2 and concentrated by centrifugation at 3,700 × g for 10 min. RNA was extracted using a Qiagen RNA extraction kit and treated twice with DNase I. The SuperScript III first-strand synthesis system (Invitrogen) was used to convert RNA into first-strand cDNA. The cDNA was then used to amplify a 304-bp product from the 5′ region of ureD using primer pair RTureDF/RTureDR (see Table S1 in the supplemental material) and a 200-bp product from the rpoB gene using primer pair RTrpoBF/RTrpoBR (see Table S1 in the supplemental material) as a loading control. The primer pairs used for the other six ure genes are listed in Table S1 in the supplemental material.
Expression of cloned ureD.
The ureD gene was PCR amplified using genomic DNA extracted from the Sakai, Sakai ureD(Q), Mo28, and 88-0643 strains using primer pair UreDF/UreDR (see Table S1 in the supplemental material), and the product was cloned into the pBADHisA vector. The PCR product was cloned into the vector such that the ATG of the NcoI site rather than the native TTG is used at the start methionine and the 6× His tag was contained in the 5′ primer rather than on the vector. This cloning of the amplified product from each isolate results in plasmids pLLamber, pLLQ, pPLQ, and pPPQ, respectively. A fifth plasmid, pLPQ, was constructed by site-directed mutagenesis using a QuikChange II site-directed mutagenesis kit (Stratagene). Primers LPQF and LPQR (see Table S1 in the supplemental material), which produce the C113T mutation in ureD, were used with plasmid pPPQ for this procedure. This combination of SNPs was not observed in any isolate but was included to represent possible SNP variations. These plasmids were electroporated into the Sakai ureD(Q) strain. Cultures of these strains were grown overnight in LB medium supplemented with ampicillin, diluted 1:100 in low-glucose DMEM supplemented with ampicillin and NiCl2, grown to an optical density at 600 nm (OD600) of 1.0, induced with 0.002% arabinose, and grown for an additional 4 h. Aliquots were then removed and concentrated by centrifugation for the urease assay and for Western blot analyses.
Western analysis.
Bacterial pellets from induced cultures were suspended in volumes of H2O and Laemmli sample buffer (Bio-Rad), required to adjust for differences in OD600 at the end of inductions. N-terminal 6× His-tagged UreD protein (6× His UreD) was purified from lysates obtained from overexpression of UreD from pLLQ in E. coli TOP10 cells for 4 h after arabinose induction. Pelleted cells were suspended in 100 mM HEPES buffer, pH 7.5, containing 8 M urea, and the cells were disrupted by sonication. 6× His UreD was purified by mixing lysates with Ni-coated beads (Promega MagneHis protein purification system), washing three times with buffer (100 mM HEPES, pH 7.5, 10 mM imidazole, 8 M urea, 1 mM dithiothreitol), and then eluting using buffer as described for the washes but containing 500 mM imidazole. The eluant was dialyzed against 100 mM HEPES buffer, pH 7.5, in a 10,000-molecular-weight-cutoff Slide-A-Lyzer dialysis cassette (Pierce). A UreD protein preparation with a concentration of 300 μg/ml was obtained. Protein samples and MagicMark XP Western protein standard were resolved by SDS-PAGE using 12% polyacrylamide running gels and 3.9% stacking gels with Tris-glycine buffer. Proteins were transferred onto Immobilon-FL polyvinylidene fluoride membranes (Millipore) using a Trans-Blot SD semidry transfer cell (Bio-Rad) and blocked overnight at 4°C in a 1:1 mixture of 1× phosphate-buffered saline (PBS) and blocking buffer (Odyssey). His-tagged proteins were detected using a monoclonal tetra-His anti-mouse antibody (Qiagen), followed by an Alexa Fluor 680-conjugated goat anti-mouse secondary antibody. A polyclonal GroEL anti-rabbit antibody (Sigma) along with an Alexa Fluor 680-conjugated goat anti-rabbit secondary antibody was also used as a loading control. Antibodies were diluted, and blots were washed in 1× PBS-Tween 20. An Odyssey infrared imaging system was used to visualize the proteins.
Phylogenetic analysis of bacterial urease sequences.
DNA sequences of the urease gene cluster were obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank) for 51 bacterial species, including several EHEC O157:H7 isolates (see Table S2 in the supplemental material). Sequences were aligned using the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) with the neighbor-joining method for each of the seven ure genes, and these alignments were used to view phylogenetic trees using the Treeview program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Additionally, the DNA sequences were translated and the protein sequences were concatenated in the order UreDABCEFG for each isolate. The peptide order was standardized to take into account the variation in the gene order observed in a number of clusters. An unrooted cluster diagram was generated in Treeview using the ClustalW alignment generated from the concatenated protein sequences.
Multilocus sequence typing and sequencing of ure genes.
For each strain, internal regions of the seven housekeeping genes aspC, clpX, fadD, icdA, lysP, mdh, and uidA were amplified and sequenced using the primers listed at the STEC Reference Center (http://www.shigatox.net). Genomic DNA was extracted from each strain using an Easy-DNA kit (Invitrogen), the seven genes were PCR amplified using PfuUltra II Fusion HS DNA polymerase (Stratagene), and the PCR products were cleaned using a QIAquick 96 PCR purification kit (Qiagen) prior to sequencing. The ureD gene was also PCR amplified for each strain using primer pair Us1F/Us1R (see Table S1 in the supplemental material). Forward and reverse sequences were aligned to create a consensus sequence for each PCR product. Sequences of the MLST gene regions for strains MG1655, CL-37, TW08566, DEC3A, DEC4E, DEC5D, DEC5E, DEC8A, DEC8B, DEC8C, DEC8E, DEC10A, DEC10B, DEC10E, and DEC11C were obtained from the STEC Reference Center website, and the genome sequences for strains Sakai, EDL933, 86-24, and TW14359 were mined from GenBank. The sequences of the seven loci were concatenated and then aligned on the basis of the neighbor-joining method using ClustalW. An unrooted cluster diagram was generated using Treeview. Treeview was also used to construct a phylogenetic tree with E. coli MG1655 as an outgroup. For selected strains, the entire urease gene cluster was sequenced. This was accomplished by using six sets of primers with overlapping products (Us1F/Us1R through Us6F/Us6R, listed in Table S1 in the supplemental material). Primer pair UPF/UPR (see Table S1 in the supplemental material) was used to sequence the 670-bp region just upstream of the start codon for ureD for some strains.
Detection of stx1, stx2, and eae genes.
The E. coli strains used in the MLST analysis were also screened by PCR for the presence of the stx1, stx2, and eae genes. Primer sequences (30, 39, 46, 48), amplification conditions, and amplicon sizes are listed in Table S3 in the supplemental material. PCR products were electrophoresed on 1% agarose gels, and those reactions with bands of the appropriate size were scored as having the genes present. Reactions with amplicons that were not present or that were of different sizes were scored as not having the genes. EHEC Sakai was used as a positive control, and E. coli MG1655 was used as a negative control. Screening for the Shiga toxin genes was first performed with primer sets stx1F/stx1R and stx2F/stx2R (see Table S3 in the supplemental material). Primer pair SK1/SK2 (see Table S3 in the supplemental material) was used for detection of intimin since these primers are considered universal eae primers that target a conserved region of eae (57). Because of the number of STEC stx and eae variants, isolates demonstrating a negative result in this first PCR screen were subjected to a second screen with primer pairs stx1a/stx1b, stx2a/stx2b, and eaeAF/eaeAR (see Table S3 in the supplemental material). PCR was also performed on isolate H.I.8, known to harbor the stx2f variant, and other isolates negative for stx2 using primer pair 128-1/128-2 (see Table S3 in the supplemental material). Finally, a third primer pair, AE9/AE10 (see Table S3 in the supplemental material), was used for detection of eae in those isolates still showing a negative result after the PCRs employing the previous two sets of primers for eae detection.
RESULTS
Internal amber codon in ureD is not solely responsible for lack of urease activity in vitro in Sakai strain.
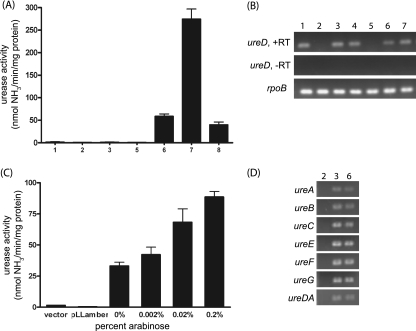
To investigate the importance of urease in EHEC pathogenesis and examine regulation of the urease genes, we constructed mutant strains of the Sakai strain since the genome sequence for this strain was available and it was demonstrated to carry only one ure locus. Since Nakano et al. reported that the internal amber codon in Sakai ureD is the reason for a lack of in vitro urease activity (36), we constructed a strain, Sakai ureD(Q), in which a point mutation, T742C, results in a codon encoding glutamine replacing the amber codon in ureD, correcting the premature truncation of the UreD protein. As a negative control, a complete ure gene cluster deletion mutant, Sakai Δure, was constructed. The Sakai strain (carrying the wild-type ure gene cluster) and mutant Sakai Δure and Sakai ureD(Q) strains were tested for in vitro urease activity after overnight growth in M9 minimal medium. As expected, neither the Sakai strain nor the Sakai Δure strain showed detectable urease activity. However, unexpectedly, the Sakai ureD(Q) strain also exhibited a urease-negative phenotype (Fig. 1A). This lack of urease activity could not be explained by a lack of transcription of ureD since several strains tested, including Sakai ureD(Q), produce the ureD transcript (Fig. 1B). Furthermore, RT-PCR analysis of the other six ure genes reveals that all seven of the ure genes are transcribed in Sakai ureD(Q) (Fig. 1D).
FIG. 1.
Urease activity and RT-PCR results for the Sakai ureD mutants. (A, B, and D) Lanes: 1, Sakai; 2, Sakai Δure; 3, Sakai ureD(Q); 4, Sakai Δure attTn7::ure (chromosomal complement of Sakai Δure; unpublished work); 5, E. coli MG1655; 6, EHEC IN1; 7, EHEC Mo28; 8, Sakai ureD(P,Q). (A) Cultures were grown overnight in DMEM supplemented with NiCl2, and the urease activities of cell lysates were measured. EHEC strains IN1 and Mo28 were used as positive controls, and MG1655 was used as a negative control. (B) RNA from overnight cultures was extracted and used for ureD RT-PCR experiments. (C) Sakai ureD(Q) harboring empty plasmid, pLLamber, or pLLQ was grown overnight, diluted 1:100 in M9 minimal medium supplemented with NiCl2 and ampicillin, grown to an OD600 of 0.45, induced with arabinose, and then grown for an additional 4 h. The urease activities of the Sakai ureD(Q) cell lysates were measured. The urease activity in lysates of Sakai ureD(Q) harboring pLLamber (the Sakai wild-type ureD) remained negligible at all concentrations of arabinose; thus, only that for 0.2% is included. (D) RNA from overnight cultures was extracted and used for detecting the transcript by RT-PCR.
Since the urease-negative phenotype was unexpected in the Sakai ureD(Q) strain, corroboration of the result was sought by suppression of the internal amber codon in ureD by expression of the supE tRNA in the Sakai Δstx2A strain. The supE tRNA inserts glutamine at amber codons (55). Plasmid pGFIB-1, which was developed for the purpose of expressing tRNAs and which contains a cloning site that can be utilized to put the cloned gene under the control of a promoter that is constitutive in E. coli, was chosen for expression of supE. A reporter strain carrying a chromosomal copy of a Tcr cassette with an internal amber codon, Sakai tet(Am), was constructed to verify expression of supE tRNA. As expected, the reporter strain was found to be Tc sensitive when carrying the empty pGFIB-1 plasmid but Tc resistant when carrying the plasmid pSUPE expressing the supE tRNA. This Sakai tet(Am) strain remained urease negative even when expressing the supE tRNA, verifying the result obtained for the strain with the point mutation, Sakai ureD(Q). Thus, neither mutation of the amber codon to glutamine nor readthrough of the amber codon in ureD is sufficient to restore urease activity in vitro in the Sakai strain.
It was previously reported that the EHEC Sakai strain harboring a plasmid with ureD cloned from a phenotypically urease-positive strain exhibits urease activity (36). One way to explain this result in a manner consistent with our data thus far would be that the urease-positive phenotype is due to the higher copy number of the plasmid, whereas the wild-type Sakai strain has only one chromosomal copy of ureD. To test this possibility, ureD from the Sakai ureD(Q) strain was cloned into pBADHisA to put the genes under the control of the arabinose-inducible promoter, and this plasmid was electroporated into the Sakai ureD(Q) strain. A dose-dependent response was observed (Fig. 1C), as increasing the arabinose concentration in the medium results in increasing urease activity, indicating that the quantity of UreD is limiting in Sakai ureD(Q). Cloning the wild-type ureD from the Sakai strain, which contains the amber codon, and overexpressing this ureD under the same experimental conditions does not give a urease-positive phenotype, thus showing the necessity of the C-terminal portion of the UreD protein for enzyme activity.
Three SNPs were identified in EHEC ureD.
Two EHEC O157:H7 strains, Mo28 and IN1, had previously been identified to be phenotypically urease positive (18). The ureD gene from urease-positive strain Mo28 was cloned and expressed in the Sakai ureD(Q) strain. Interestingly, complementing with the Mo28 ureD conferred greater urease activity on the Sakai ureD(Q) strain than complementing with cloned Sakai ureD(Q) [222.9 and 76.1 nmol NH3/min/mg protein for Mo28 ureD and Sakai ureD(Q), respectively]. Comparison of the sequences of these two cloned ureD genes revealed the only difference to be a T-to-C base change, resulting in a leucine-to-proline substitution, L38P, in the Mo28 strain. After identifying the L38P substitution in UreD in the Mo28 strain, we sequenced ureD in the IN1 strain, revealing the same T-to-C base change. A collection of EHEC isolates was screened for urease activity, resulting in the identification of two more urease-positive isolates, S102-9 and 537/89. We sequenced the ureD genes of these isolates and found that, like IN1 and Mo28, these strains each encode glutamine rather than an amber codon in the C terminus of UreD and the proline substitution in the N terminus at position 38. In addition, these two strains, S102-9 and 537/89, have an additional SNP, a T-to-C base change, that alters amino acid 205 to be either leucine or proline. The three SNPs identified in ureD in these EHEC strains affect amino acid positions 38, 205, and 248.
Incorporation of proline at position 38 enhances urease activity, but proline at position 205 does not confer increased urease activity.
We constructed an additional point mutation that altered the leucine at position 38 to the more conserved proline found in Gram-negative urease-positive species. The ureD mutation T113C was created in Sakai ureD(Q) so that the resulting strain has proline in place of the leucine (L38P) and glutamine rather than the amber codon at position 248. This strain, Sakai ureD(P,Q), was grown overnight in DMEM, and the urease assay was performed. Although this strain exhibits modest urease activity (Fig. 1A) compared to the other strains, mutation of the leucine to proline led to consistent measurable activity in vitro.
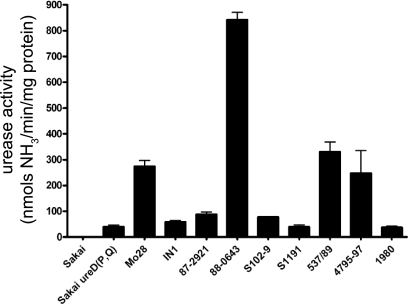
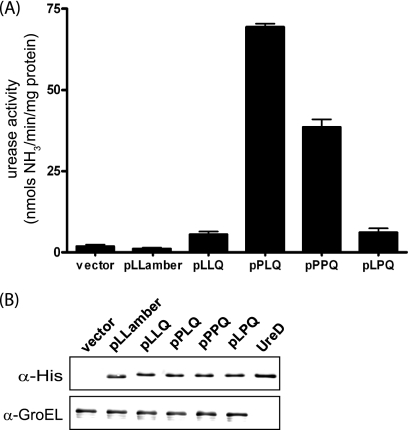
It was tempting to assume that a proline residue at position 205 in the UreD protein would further enhance urease enzyme activity since the two strains having a proline rather than a leucine in that position show greater urease activity after overnight growth (Fig. 2). However, there may be many factors contributing to overall urease activity in a particular strain. To assess the contributions of the various SNPs to the urease activity, ureD genes having the different combinations of residues at positions 38, 205, and 248 were cloned or constructed in a plasmid, putting the construct in the same genetic background and under the control of the arabinose-inducible promoter in which it is expressed with an N-terminal 6× His tag. These plasmids were electroporated into Sakai ureD(Q) and induced, followed by determination of the urease activities of the cell lysates carrying the plasmids with the different SNP combinations (Fig. 3A). Aliquots of the induced cultures were also subjected to analysis by Western blotting (Fig. 3B), demonstrating that the differences in measured urease activity were not due to protein expression differences. Enhanced urease activity is observed in the strains harboring the cloned UreD with proline at position 38. However, the proline at position 205 does not confer increased urease activity.
FIG. 2.
Urease activities of the urease-positive STEC isolates. Cultures of the urease-positive strains included in the MLST analysis were grown overnight in DMEM supplemented with NiCl2. The urease activities of cell lysates was measured by four or more independent assays.
FIG. 3.
Correlation of urease enzyme activity with UreD sequence. His-UreD was expressed from cloned ureD with different combinations of the three SNPs. Plasmid pLLamber carries cloned Sakai wild-type ureD. Cultures of Sakai ureD(Q) carrying these plasmids were grown overnight, diluted into DMEM supplemented with NiCl2, and induced with arabinose for 4 h, and samples were processed for the urease assay and Western blotting. (A) Urease activities of cell lysates were measured. The difference between the vector and pLLQ was statistically significant (P = 0.011), while no significant difference between the vector and pLLamber was found (P = 0.32). (B) Western blot using the anti-His antibody and anti-GroEL as a control shows that different levels of urease activity are a result of differences in UreD sequence rather than different levels of UreD protein expression. Sakai ureD(Q) carrying the empty cloning vector is shown in the first lane, and purified UreD is shown in the last lane.
Proline at amino acid position 38 is highly conserved among bacterial species.
Genes encoding proteins necessary for active urease enzyme have been discovered in both Gram-positive and Gram-negative bacteria, including alpha-, beta-, gamma-, and epsilonproteobacteria. Our results suggested that a proline residue at position 38 rather than the leucine found in the Sakai strain was significant for the function of the urease enzyme. A determination of how conserved this residue is in the UreD protein of ure locus-containing bacteria was undertaken. Upon examination of the protein sequence alignment, it was found that the proline at position 38 (using the EHEC UreD sequence as a reference for the numbering) is highly conserved (32 of 51 total isolates; 68% of listed species). It is found in all urease gene clusters containing gamma-, beta-, and epsilonproteobacteria included in the analysis, with the exception of the EHEC isolates. Conversely, the proline at position 205 is not as highly conserved (14 of 51 isolates; 30% of listed species) as that at position 38, but is found in many gammaproteobacteria, including the Klebsiella, Enterobacter, and Citrobacter isolates included in the analysis.
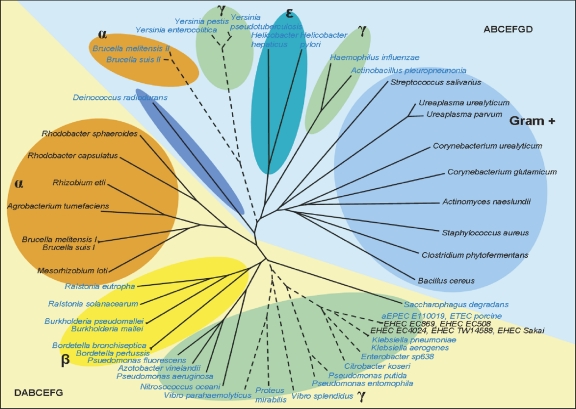
We also sought to determine how the EHEC ure sequences clustered phylogenetically with those of a variety of bacterial species. The ure locus can be found with two different gene orders on the basis of whether ureD is found on the 5′ or the 3′ end of the gene cluster, with the two possible gene orders identified being ureDABCEFG and ureABCEFGD. Additionally, within the ure locus of some bacterial species, even those that are known to produce active urease, insertion of nonurease genes is identified; thus, only bacterial species containing all seven ure genes in a single unit were included for further analysis (see Table S2 in the supplemental material). A total of 51 bacterial urease gene cluster sequences were included, 5 of which are from different EHEC isolates. For each of the seven genes, the EHEC sequence is most similar to the sequences of Klebsiella, Enterobacter, and Citrobacter species. The unrooted cluster diagram based on concatenated protein sequences is divided into two sections following the gene order within the bacterial species (Fig. 4, yellow versus blue backgrounds). Within this larger classification of the cluster diagram, smaller groupings are related to bacterial species classification.
FIG. 4.
Phylogenetic cluster diagram based on urease protein sequence alignment. Ure protein sequences for 51 bacterial species were concatenated in the order UreDABCEFG and aligned. The species having a proline at amino acid position 38 in UreD (numbering according to EHEC UreD) are shown in blue font, and those with a proline at position 205 in UreD are shown with a broken line. The gene orders within the ure gene clusters are designated in the lower left and upper right corners and are distinguished by the yellow and blue backgrounds.
STEC urease is not evolutionarily restricted.
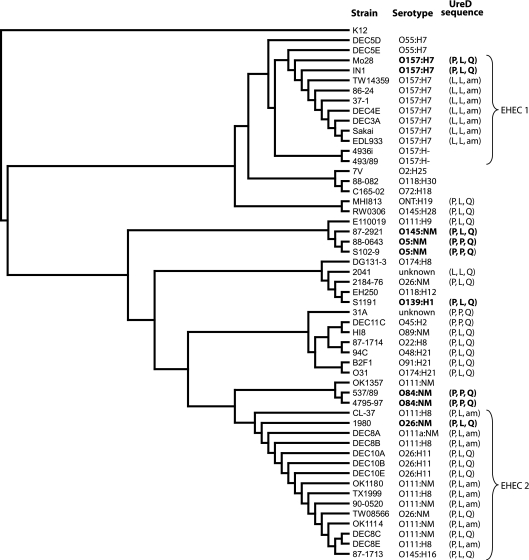
Since the SNPs identified in ureD affect the activity of the urease enzyme, we sought to determine the correlation between these SNPs, the in vitro urease-positive phenotype, and the results of a phylogenetic analysis based on the housekeeping genes. The majority of STEC strains isolated display a urease-negative phenotype in vitro (Table 2). Those with a urease-positive phenotype were included in the phylogenetic analysis with representative strains from the EHEC 1 and EHEC 2 clonal groups (42), as well as E. coli strains with various other serotypes. A total of 53 E. coli strains were included in the MLST analysis (Table 2). These strains were further characterized by PCR screening for the presence of stx1, stx2, and eae (Table 2). Because of the increasing number of stx1, stx2, and eae variants identified in STEC strains over the past several years, several sets of divergent primers were included in the screening. Weak or inconsistent PCR results have been reported as negative, but the possibility that some of these isolates contain a variant of eae that is not amplified with the existing primers cannot be ruled out. Figure 5 depicts the results of the MLST phylogenetic analysis, with the urease phenotype and UreD amino acids 38, 205, and 248 being indicated next to each isolate. As expected, the isolates with serotype O157:H7 cluster together (EHEC 1), and the isolates with either serotype O26 or O111 form a second tight cluster (EHEC 2), with the less common serotypes being distributed in the analysis (42). The DEC5 strains are thought to be the precursors to the EHEC 1 clonal group (25), and our results indicate that they have not yet acquired the ure gene cluster. This is in agreement with the findings described in a previous report stating the absence of the ure genes in nine E. coli O55:H7 strains and hypothesizing the evolutionary divergence of EHEC O157:H7 prior to the acquisition of the O island carrying the ure genes (12). The fact that the O157:H− strains do not carry the ure gene cluster suggests that this subgroup diverged before acquisition of the urease-containing O island. The EHEC 2 clonal group in particular formed a tight cluster with 100% DNA identity between some strains. Neither urease activity nor the assemblage of ureD mutations is consistently represented in a single evolutionary lineage; thus, we conclude that the urease-positive genotype is not evolutionarily restricted.
TABLE 2.
Serotypes and virulence profiles of E. coli isolates included in the MLST phylogenetic analysis
| Strain | Serotype | Virulence profile |
|||
|---|---|---|---|---|---|
| Urease activity | stx1 | stx2 | eae | ||
| MG1655 | − | − | − | − | |
| Sakai | O157:H7 | − | + | + | + |
| EDL933 | O157:H7 | − | + | + | + |
| DEC3A | O157:H7 | − | + | + | + |
| IN1 | O157:H7 | + | + | + | + |
| 86-24 | O157:H7 | − | − | + | + |
| TW14359 | O157:H7 | − | − | + | + |
| Mo28 | O157:H7 | + | − | + | + |
| 37-1 | O157:H7 | − | + | − | + |
| DEC4E | O157:H7 | − | + | − | + |
| 4936i | O157:H− | − | − | + | + |
| 493/89 | O157:H− | − | − | + | + |
| DEC5D | O55:H7 | − | − | − | + |
| DEC5E | O55:H7 | − | − | − | + |
| DEC8A | O111a:NMa | − | + | − | + |
| DEC8B | O111:H8 | − | + | + | + |
| DEC8C | O111:NM | − | + | − | + |
| DEC8E | O111:H8 | − | + | − | + |
| CL-37 | O111:H8 | − | + | − | + |
| OK1114 | O111:NM | − | + | + | + |
| OK1180 | O111:NM | − | + | + | + |
| OK1357 | O111:NM | − | − | − | − |
| TX1999 | O111:H8 | − | + | + | + |
| 90-0520 | O111:NM | − | + | − | + |
| E110019 | O111:H9 | − | − | − | + |
| DEC10A | O26:H11 | − | + | − | + |
| DEC10B | O26:H11 | − | + | − | + |
| DEC10E | O26:H11 | − | + | − | + |
| 2184-76 | O26:NM | − | − | − | + |
| 1980 | O26:NM | + | − | − | + |
| TW08566 | O26:NM | − | − | + | + |
| 7V | O2:H25 | − | − | + | − |
| 88-0643 | O5:NM | + | + | − | + |
| S102-9 | O5:NM | + | + | − | + |
| MHI813 | ONT:H19 | − | + | + | − |
| 87-1714 | O22:H8 | − | + | + | − |
| DEC11C | O45:H2 | − | + | − | + |
| 94C | O48:H21 | − | + | + | − |
| C165-02 | O72:H18 | − | − | + | − |
| 537/89 | O84:NM | + | + | + | + |
| 4795-97 | O84:H4 | + | + | − | + |
| H.I.8 | O89:NM | − | − | + | + |
| B2F1 | O91:H21 | − | − | + | − |
| EH250 | O118:H12 | − | − | + | − |
| 88-082 | O118:H30 | − | − | + | + |
| S1191 | O139:H1 | + | − | + | − |
| 87-1713 | O145:H16 | − | + | − | + |
| RW0306 | O145:H28 | − | − | + | + |
| 87-2921 | O145:NM | + | + | + | + |
| DG131/3 | O174:H8 | − | + | + | − |
| O31 | O174:H21 | − | − | + | − |
| 2041 | Unknown | − | − | − | − |
| 31A | Unknown | − | + | − | + |
NM, nonmotile.
FIG. 5.
Correlation between phylogenetic analysis based on seven-gene MLST and SNPs in ureD. Sequences for the genes aspC, clpX, fadD, icdA, lysP, mdh, and uidA were concatenated and aligned. A phylogenetic tree based on the MLST analysis was constructed and rooted using E. coli MG1655. The ureD gene was sequenced, and the amino acids at positions 38, 205, and 248 are shown. The ureD gene could not be PCR amplified from the strains for which no UreD sequence is shown. The strains having a urease-positive phenotype in vitro are shown in boldface type.
Differences in measured urease activity cannot be explained by differences in ure gene sequences alone.
It was noted above that some isolates show greater urease activity than others even when they are grown under the same conditions and share the same ure genotype. The results of experiments depicted in Fig. 3 clearly show that the proline at position 205 does not lead to increased urease enzyme activity. However, the 88-0643 strain was found to exhibit higher urease activity than the Mo28 strain, despite having the proline at position 205. We measured the urease activities of the nine urease-positive isolates along with the urease activity of the Sakai ureD(P,Q) mutant (Fig. 2). The results indicate that there is significant strain-to-strain variation in urease activity that cannot be accounted for by the ureD sequence alone. This raises the possibility that there are other SNPs within the ure gene cluster that have an effect on overall urease enzyme activity. With this in mind, we sequenced the ure gene cluster for several of the urease-positive isolates. The resulting SNPs are shown in Table 3. Despite the genetic distance between some of these strains on the basis of the MLST-based phylogeny, the ureA, ureB, ureC, and ureE genes were found to have 100% DNA sequence identity and ureF and ureG each have only one nonsynonymous substitution. Thus, ureD is the most variable gene in the urease gene cluster. The results above discuss this variation in ureD; however, in order to investigate the possibility that the strain-to-strain variation in measured urease activity was due to differences in the promoter sequences, we sequenced the 670-bp region located 5′ of the start codon for ureD. We did not identify any mutations in the promoter region in any of the isolates listed in Table 3 or several other isolates, suggesting that the variation in urease production is rooted elsewhere in the genome.
TABLE 3.
ure gene cluster sequence comparisona
| Gene, amino acid | Sequence |
|||||
|---|---|---|---|---|---|---|
| Sakai | IN1 | Mo28 | 88-0643 | S102-9 | OK1114 | |
| ureD | ||||||
| 38 | Leu (CTG) | Pro (CCG) | Pro (CCG) | Pro (CCG) | Pro (CCG) | Pro (CCG) |
| 205 | Leu (CTG) | Leu (CTG) | Leu (CTG) | Pro (CCG) | Pro (CCG) | Leu (CTG) |
| 248 | Amber (TAG) | Gln (CAG) | Gln (CAG) | Gln (CAG) | Gln (CAG) | Amber (TAG) |
| ureF, 65 | Ala (GCC) | Ala (GCC) | Ala (GCC) | Ala (GCC) | Ala (GCC) | Val (GTC) |
| ureG | ||||||
| 34 | Thr (ACC) | Thr (ACT) | Thr (ACT) | Thr (ACC) | Thr (ACC) | Thr (ACC) |
| 99 | Glu (GAG) | Asp (GAT) | Asp (GAT) | Asp (GAT) | Asp (GAT) | Asp (GAT) |
ureA, ureB, ureC, and ureE all have 100% DNA sequence identity.
DISCUSSION
The work in this study was motivated by the goal of characterizing the regulation and the role in virulence of the ure genes in STEC. EHEC O157:H7 was recognized as an emerging pathogen in 1982 (43), and although EHEC O157:H7 has received the majority of attention, many STEC isolates contain the ure locus in their genomes (13, 35, 38). However, the majority of STEC isolates exhibit a urease-negative phenotype in vitro; thus, screening for urease activity is not a valuable method for clinical identification of STEC. However, because of the presence of the ure gene cluster in STEC but not in other diarrheagenic E. coli pathotypes, there has been interest in using ureC as a molecular biomarker for some of the commonly known virulent strains (13, 35). The urease-negative phenotype has been attributed to a truncation of UreD caused by an SNP in ureD resulting in an internal amber codon (36). The lack of a phenotype precluded studies on STEC urease gene regulation and what role urease may play in virulence. Although it could be advantageous for STEC to express higher levels of urease under certain environmental circumstances, constitutive high-level expression could be damaging in some situations.
The conservation of urease gene sequences in STEC has been noted in previous studies (13, 38). Phenotypically urease-negative STEC isolates, even those possessing the amber codon in ureD, appear to retain the complete ure gene cluster, with few strains demonstrating the accumulation of mutations that would be expected of nonfunctional coding regions. This suggests that a functional urease is expressed and is a selective advantage under one or more environmental conditions. Thus, there is a selective advantage or positive gene pressure for maintenance, and the ureD mutations are induced with in vitro growth or the ure locus is a recent acquisition. Construction and characterization of the Sakai ureD(Q) mutant strain in this work led to the unexpected result that the urease-negative phenotype in the Sakai strain is not solely due to the internal stop codon in ureD. It has been previously noted, however, that several isolates of the O26:H11 serotype encode a glutamine rather than the amber codon in ureD but still do not demonstrate urease activity in vitro (13). Sequence analysis of a limited number of urease-positive EHEC isolates identified two additional ureD SNPs that alter the UreD sequence at amino acid positions 38 and 205. Both SNPs result in a leucine-to-proline change; however, it was not known how important these particular amino acids were to UreD function. To answer this question, we took two approaches, a bioinformatic approach in which we examined other urease-positive gene clusters for conservation of the amino acid encoded at these positions in UreD and an experimental approach measuring the changes in urease enzyme activity depending on the amino acid encoded at these three SNP locations.
Many animal or human pathogens carry the ure gene locus, yet despite the demonstrated importance of urease in either the survival or pathogenicity of a variety of bacterial species, reported phylogenetic analyses of urease sequences are minimal. A combination phylogenetic analysis based on single protein sequences and derived model protein structures has been used by one group in an attempt to study the evolutionary conservation of key residues or protein structures for the UreE, UreF, and UreG proteins (34, 44, 56). The protein sequence of UreC, the structural subunit containing the active site of urease and the largest of the urease proteins, has been used for phylogenetic characterization of ammonia-oxidizing bacteria (23) and in a study including 22 diverse bacterial species for phylogenetic characterization of the two ure gene clusters found in Brucella suis (7). The most comprehensive urease-based phylogenetic analysis included construction of a phylogenetic tree based on concatenated nucleotide sequence data of urease genes from 18 bacterial species, not including STEC, and demonstrated commonly shared ancestry between the urease genes of urease-positive thermophilic Campylobacter and Helicobacter (20). The current study encompasses a wide variety of bacterial species and includes all seven proteins encoded in the ure gene cluster. The finding that the EHEC ure gene cluster is most closely associated with that of Klebsiella was not surprising since it was already known that protein sequences based on translated nucleotide sequences of the EHEC ure genes are most similar to the Klebsiella aerogenes homologues, with predicted similarities of between 76 and 96% for the seven urease proteins (18). Identification that the proline at position 38 in UreD is highly conserved among Gram-negative bacteria correlates with our observation of a proline at that position in the EHEC strains displaying a urease-positive phenotype. The proline at position 205 noted in some but not all of our urease-positive strains is not as highly conserved in the UreD proteins of other urease-producing enteric bacteria. The protein sequence alignment also suggests that a highly conserved region of 26 amino acids in the UreD C terminus is important for UreD function. This result is in agreement with the experimental observation that urease activity cannot be complemented, even with overexpression, from UreD containing the internal stop at amino acid position 248.
The urease enzyme has long been studied and is known to have a trimeric structure composed of three copies each of the three structural protein subunits, UreA, UreB, and UreC (32, 41). The inactive urease apoprotein, (UreABC)3, is activated by addition of two Ni2+ ions to each of the three active sites in the enzyme. Incorporation of the nickel cofactor is accomplished using the accessory proteins encoded by ureD, ureE, ureG, and ureF. In order for proper assembly of the urease metallocenter, all four of the accessory proteins must be present and functional. The proposed pathway for activation of the enzyme involves the binding of UreD to the apoprotein, followed by binding of UreF and then UreG. UreE is a nickel-binding protein that delivers Ni2+ to the activation complex (41). After activation of the urease enzyme, the accessory proteins dissociate. The accessory protein UreD has been shown by chemical cross-linking to bind both UreC and UreB, whereas UreF binds only UreB (6). In addition, a yeast two-hybrid screen determined that UreD directly interacts in vivo with UreF (17). Binding of the chaperone proteins UreD and UreF changes the conformation of the apoprotein to expose the active site of urease, thereby allowing incorporation of Ni2+, as determined by a combination of chemical cross-linking, mutagenesis of UreB, and calculated flexibility analysis (41). Due to the insolubility of UreD when it is overexpressed alone, there is no reported crystal structure of UreD and it is the least characterized of the urease accessory proteins. However, recently, investigators have found a maltose binding protein-UreD fusion to be soluble and useful in characterization studies (4). Consistent with previous reports, these investigators found UreD to bind both UreF and UreG in vivo. The authors demonstrate that UreD also has Ni2+-binding capability and propose, supported by their additional work on UreG (2), that there is a sequential Ni2+ ion transfer from UreE to UreG to UreD to the urease apoenzyme active site. To our knowledge, it is not known which residues in UreD are involved in metal ion binding or in binding to UreF or UreG. Our experiments have demonstrated that position 38 is conserved in the majority of urease-positive species and that UreD (L38P) is clearly functional (Fig. 1A). Additionally, the data in Fig. 3A demonstrate that UreD can be functional with a leucine at position 38 with overexpression but has significantly reduced activity. Interestingly, the proline at position 205 appears to reduce the ability of UreD to activate the urease enzyme in conjunction with the other six Ure proteins from the Sakai strain, despite the fact that this proline appears to be conserved in other enteric bacterial ureases (Fig. 4).
We wondered whether the apparent discrepancy in urease activity of positive strains, such as 537/89 and 88-0643, which carry a proline at position 205, and the results of the UreD overexpression experiments reported in Fig. 3A might be due to compensating differences in any of the other Ure protein sequences compared to the Sakai strain Ure sequence. In order to investigate this further, the urease activities for all nine urease-positive isolates were measured, and the entire ure gene cluster was sequenced for four of these strains. Our results show that this discrepancy is not due to differences in other Ure protein sequences. Both strain pair IN1 and Mo28 and strain pair 88-0643 and S102-9 have identical ure gene cluster sequences yet exhibit different measured urease activities (Fig. 2). In addition, except for the amino acid at position 205 in ureD, isolates Mo28 and 88-0643 have identical ure gene cluster sequences; thus, the greater urease activity seen from 88-0643 cannot be explained by other SNPs within the ure gene cluster. In addition, the DNA sequences (670 bp) upstream of ureD were identical for these strains. Combining these results, it is clear that although the UreD protein sequence affects urease enzyme activity, there is at least one other trans-acting factor that affects the overall urease enzyme activity for a given isolate. Also of note, our results reiterate the finding of others that the ure gene cluster is carried by STEC strains with significant sequence conservation even among phylogenetically diverse isolates.
The MLST analysis was performed to determine whether the amber codon and the other base pair changes in ureD were occurring randomly among STEC strains or followed an evolutionary pattern. Figure 5 clearly demonstrates that neither an in vitro urease-positive phenotype nor the proline at position 205 is phylogenetically restricted. Although the amber codon is found in the majority of O157:H7 isolates and many O111 isolates, there are many STEC isolates carrying the ure gene cluster that do not have the amber codon in ureD (Fig. 5). Furthermore, with the exception of one isolate of unknown serotype, the leucine at amino acid position 38 is unique to one branch of the O157:H7 serotype cluster. The first two EHEC genomes to be sequenced belong to this cluster, contributing to the impression that this ureD sequence was typical of EHEC. Since the EHEC 1 and EHEC 2 clonal groups have evolved separately, our results suggest that separate events lead to the fixation of the amber codon in an ancestor within each subset of each of these clonal groups that contain the ure cluster. The results of this study, based on MLST phylogenetic analysis, demonstrate that even though urease-positive STEC isolates are not found in a single cluster, they all do have a proline at position 38 in ureD. Although this proline appears to be necessary for in vitro urease activity, it is not a reliable predictor of an in vitro urease-positive phenotype.
Detailed histories of the E. coli strains used for urease screening and phylogenetic analysis in this work were lacking in many cases. Additionally, we were unable to PCR amplify ureD from a small number of the isolates known to be STEC, and we were aware of the presence of the ure gene cluster in at least one atypical EPEC strain, suggesting that there is additional variability associated with this locus that is not captured in this study. Therefore, we sought to characterize the 53 strains used in the MLST analysis further by determining whether they carried genes encoding Shiga toxin or intimin. By comparing the data in Table 2 and Fig. 5, it can be observed that there are some strains that are stx positive but that do not appear to encode the ure gene cluster; thus, not all STEC strains carry the ure gene cluster. Previous investigators have reported a positive correlation between the presence of ureC and eae, with the ureC gene being found more frequently among STEC isolates harboring eae than those lacking eae (13, 38). The results from this study demonstrate only one strain, 88-082, that is eae positive but that does not appear to encode urease. In this study, we did identify seven isolates negative for eae that do encode urease in their genomes. However, these strains possess uncommon serotypes and variants of stx genes, and it is possible that they also encode unusual variants of eae that were not detected. One such strain, S1191, was originally isolated from a pig and encodes the Shiga toxin variant stx2e (54). Our result that this strain encodes urease is in contrast to the findings presented in a previous report which showed that all 13 human and 13 porcine isolates encoding stx2e were negative when they were screened for ureC (49). This underscores the caution that must be used when attempting to make correlations between genetic virulence markers due to the dynamic nature of the STEC genome.
Taken together, our results demonstrate that when a wide variety of STEC strains are included in an analysis, the presence of the amber codon in ureD is not as common an event as was previously believed. Furthermore, an in vitro urease-negative phenotype is not an indicator of the amber mutation in ureD. To the contrary, most non-O157:H7 isolates examined do not carry the mutation resulting in the amber codon in ureD (with the exception of a subset of O111 isolates [Fig. 5]). In addition, most STEC strains have a proline at position 38 in UreD, which we have demonstrated is highly conserved among Gram-negative bacterial ureases and which leads to greater urease enzyme activity. Overall, the results of this study indicate that many STEC strains do not carry the amber codon but do have the proline at position 38 in UreD; thus, under inducing conditions, possibly in vivo in the human or animal host, the urease will function appropriately and provide a significant advantage. Clearly, additional research is needed to elucidate the method by which the urease gene cluster is regulated in STEC.
Supplementary Material
Acknowledgments
We thank Mark Mammel for kindly providing the O157:H− strains.
This research was supported by NIH grants DK58957 and AI21657 to J.B.K. For part of the time this research was conducted, S.R.S. was a trainee under institutional training grant T32AI007540 from NIAID.
The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID or NIH.
Footnotes
Published ahead of print on 10 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bandara, A. B., et al. 2007. Brucella suis urease encoded by ure1 but not ure2 is necessary for intestinal infection of BALB/c mice. BMC Microbiol. 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boer, J. L., S. Quiroz-Valenzuela, K. L. Anderson, and R. P. Hausinger. 2010. Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry 49:5859-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderon, V. E., et al. 2009. Outbreak caused by cad-negative Shiga toxin-producing Escherichia coli O111, Oklahoma. Foodborne Pathog. Dis. 7:107-109. [DOI] [PubMed] [Google Scholar]
- 4.Carter, E. L., and R. P. Hausinger. 2010. Characterization of the Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J. Bacteriol. 192:2294-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1994. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992-1993. MMWR Morb. Mortal. Wkly. Rep. 42:258-263. [PubMed] [Google Scholar]
- 6.Chang, Z., J. Kuchar, and R. P. Hausinger. 2004. Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J. Biol. Chem. 279:15305-15313. [DOI] [PubMed] [Google Scholar]
- 7.Contreras-Rodriguez, A., et al. 2008. Enzymatic, immunological and phylogenetic characterization of Brucella suis urease. BMC Microbiol. 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dattelbaum, J. D., C. V. Lockatell, D. E. Johnson, and H. L. T. Mobley. 2003. UreR, the transcriptional activator of the Proteus mirabilis urease gene cluster, is required for urease activity and virulence in experimental urinary tract infections. Infect. Immun. 71:1026-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Koning-Ward, T. F., and R. M. Robins-Browne. 1995. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect. Immun. 63:3790-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, F. D., et al. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73:7252-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, A. W., et al. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 43:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, A. W., et al. 2006. Urease genes in non-O157 Shiga toxin-producing Escherichia coli: mostly silent but valuable markers for pathogenicity. Clin. Microbiol. Infect. 12:478-495. [DOI] [PubMed] [Google Scholar]
- 14.Gould, L. H., et al. 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recommend. Rep. 58(RR-12):1-14. [PubMed] [Google Scholar]
- 15.Grauke, L. J., et al. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, T., et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Heimer, S. R., and H. L. T. Mobley. 2001. Interaction of Proteus mirabilis urease apoenzyme and accessory proteins identified with yeast two-hybrid technology. J. Bacteriol. 183:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heimer, S. R., et al. 2002. Urease of enterohemorrhagic Escherichia coli: evidence for regulation by Fur and a trans-acting factor. Infect. Immun. 70:1027-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilborn, E. D., et al. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159:1758-1764. [DOI] [PubMed] [Google Scholar]
- 20.Kakinuma, Y., et al. 2007. Cloning, sequencing and characterization of a urease gene operon from urease-positive thermophilic Campylobacter (UPTC). J. Appl. Microbiol. 103:252-260. [DOI] [PubMed] [Google Scholar]
- 21.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 22.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 23.Koper, T. E., A. F. El-Sheikh, J. M. Norton, and M. G. Klotz. 2004. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 70:2342-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasekara, B. R., et al. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopold, S. R., et al. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713-8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning, S. D., et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maroncle, N., C. Rich, and C. Forestier. 2006. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res. Microbiol. 157:184-193. [DOI] [PubMed] [Google Scholar]
- 28.Masson, J. M., and J. H. Miller. 1986. Expression of synthetic suppressor tRNA genes under the control of a synthetic promoter. Gene 47:179-183. [DOI] [PubMed] [Google Scholar]
- 29.McKenzie, G. J., and N. Craig. 2006. Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercado, E. C., et al. 2004. Non-O157 Shiga toxin-producing Escherichia coli isolated from diarrhoeic calves in Argentina. J. Vet. Med. B 51:82-88. [DOI] [PubMed] [Google Scholar]
- 31.Michino, H., et al. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787-796. [DOI] [PubMed] [Google Scholar]
- 32.Mobley, H. L. T., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Microbiol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musiani, F., B. Zambelli, M. Stola, and S. Ciurli. 2004. Nickel trafficking: insights into the fold and function of UreE, a urease metallochaperone. J. Inorganic Biochem. 98:803-813. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, M., et al. 2001. Association of the urease gene with enterohemorrhagic Escherichia coli strains irrespective of their serogroups. J. Clin. Microbiol. 39:4541-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano, M., T. Iida, and T. Honda. 2004. Urease activity of enterohaemorrhagic Escherichia coli depends on a specific one-base substitution in ureD. Microbiology 150:3483-3489. [DOI] [PubMed] [Google Scholar]
- 37.Olivera-Severo, D., G. E. Wassermann, and C. R. Carlini. 2006. Ureases display biological effects independent of enzymatic activity. Is there a connection to diseases caused by urease-producing bacteria? Braz. J. Med. Biol. Res. 39:851-861. [DOI] [PubMed] [Google Scholar]
- 38.Orth, D., K. Grif, M. P. Dierich, and R. Wurzner. 2006. Prevalence, structure and expression of urease genes in Shiga toxin-producing Escherichia coli from humans and the environment. Int. J. Hyg. Environ. Health 209:513-520. [DOI] [PubMed] [Google Scholar]
- 39.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perna, N. T., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 41.Quiroz-Valenzuela, S., S. C. K. Sukuru, R. P. Hausinger, L. A. Kuhn, and W. T. Heller. 2008. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch. Biochem. Biophys. 480:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 43.Riley, L. W., et al. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 44.Salomone-Stagni, M., B. Zambelli, F. Musiani, and S. Ciurli. 2007. A model-based proposal for the role of UreF as a GTPase-activating protein in the urease active site biosynthesis. Proteins 68:749-761. [DOI] [PubMed] [Google Scholar]
- 45.Sangari, F. J., A. Seoane, M. C. Rodriquez, J. Aguero, and J. M. G. Lobo. 2007. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect. Immun. 75:774-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, H., et al. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 48.Slanec, T., A. Fruth, K. Creuzburg, and H. Schmidt. 2009. Molecular analysis of virulence profiles and Shiga toxin genes in food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 75:6187-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonntag, A.-K., et al. 2005. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profiles and interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 71:8855-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teunis, P. F. M., I. D. Ogden, and N. J. C. Strachan. 2008. Hierarchical dose response of E. coli O157:H7 from human outbreaks incorporating heterogeneity in exposure. Epidemiol. Infect. 136:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tilden, J., et al. 1996. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am. J. Public Health 86:1142-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuttle, J., et al. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]
- 54.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida, M., K. Kashiwagi, G. Kawai, A. Ishihama, and K. Igarashi. 2002. Polyamines enhance synthesis of the RNA polymerase σ38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 40:37139-37146. [DOI] [PubMed] [Google Scholar]
- 56.Zambelli, B., F. Musiani, M. Savini, P. Tucker, and S. Ciurli. 2007. Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry 46:3171-3182. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, W. L., et al. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.