Abstract
We compared ileal Clostridium perfringens quantification results produced by real-time PCR and culture-based methods in broiler chickens in a challenge model of necrotic enteritis. Assessment of the relative standard deviations (RSDs) revealed that the real-time PCR assay generated a smaller standard deviation and thus was more precise than the culture-based method. Linear regression analysis indicated that the bacterial counts of these two methods were highly correlated (R2 = 0.845). We suggest that real-time PCR could be a replacement of the culture method for quantifying C. perfringens in the intestinal tracts of broiler chickens.
Clostridium perfringens, a Gram-positive spore-forming and anaerobic nonmotile rod bacterium, is an important pathogenic bacterium in humans and livestock. It can be found in animal and human intestinal tracts and feces and in environmental samples (such as soil and wastewater samples). C. perfringens types A, B, C, D, and E produce many different toxins that may be involved in pathogenesis (18). In poultry, this bacterium often causes necrotic enteritis (NE) which leads to losses of over $2 billion each year in the world's broiler industry (5). Bacteria in the gastrointestinal tracts of chickens play an important role in health (15). Therefore, quantification of the major bacterial communities in the chicken gut is essential for monitoring changes in microbial ecology in experiments involving C. perfringens challenge. Traditionally, analysis of gastrointestinal communities has depended on bacterial culture-based counting methods or microscopy. The methodologies involved are time-consuming and require researchers to possess substantial microbiological expertise. Furthermore, potential bias is present, since only those bacteria whose physiological and metabolic requirements are reproducible in vitro can be cultivated (25).
Molecular approaches have been applied to rapid characterization of bacteria. These approaches include denaturing gradient gel electrophoresis (DGGE) (10, 24), temperature gradient gel electrophoresis (TGGE) (29), conventional PCR (13), and terminal restriction fragment length polymorphism (T-RFLP) (23). These approaches are able to identify relevant bacterial groups, but the drawback of these techniques is that they are not fully quantitative and so cannot act as a stand-alone alternative method to in vitro culture and enumeration. In contrast, real-time PCR can be used to quantify bacteria, as the number of target gene copies can be determined in DNA extracted from samples. Hence, real-time PCR has recently been used to enumerate bacteria in environmental samples (3, 21) and in animal gastrointestinal tracts and feces (6, 7, 9, 26).
This study compared the precision of real-time PCR quantification of C. perfringens (PCR targeting the 16S rRNA genes of C. perfringens) with the culture-based colony counting method. The analysis of relative standard deviations (RSDs) indicated that real-time PCR quantification was more precise and reproducible than the traditional culture-based method. Therefore, this efficient and cost-effective molecular approach can replace the culture-based counting method for quantification of C. perfringens in the intestinal tracts of chickens in a challenge model of NE disease.
Sampling of ileal digesta of the birds.
The animal experiment was conducted as described recently (28). Briefly, 1,350 birds were raised for 5 weeks with the birds in each cage assigned to one of nine treatment groups with six birds per treatment (25 birds/cage). On day 9, the birds in groups that would be challenged were given per os three Eimeria species (Bioproperties Pty Ltd., Glenorie, New South Wales, Australia), and on days 14, 15 and 16, they were inoculated per os with approximately 108 CFU of a pathogenic strain of C. perfringens type A (CSIRO Livestock Industries, Geelong, Victoria, Australia). The experimental design and the treatment acronyms are shown in Table 1. On days 13 and 17, 2 birds were randomly chosen in each cage and sacrificed for sample collections. Approximately 1 g of the ileal digesta was collected for microbial culture, and a section of approximately 3 cm of ileum (including digesta) was taken at the midpoint between Meckel's diverticulum and cecal tonsils per bird for quantitative PCR analysis of C. perfringens.
TABLE 1.
Experimental design of the treatments with fish meal feeding, Eimeria infection, and C. perfringens challengea
| Treatment groupb | Fish meal | Eimeria | C. perfringens |
|---|---|---|---|
| 1 (NFM−) | None | None | None |
| 2 (LFM−) | Low | None | None |
| 3 (HFM−) | High | None | None |
| 4 (NFM/Cp) | None | None | Yes |
| 5 (LFM/Cp) | Low | None | Yes |
| 6 (HFM/Cp) | High | None | Yes |
| 7 (NFM+) | None | Yes | Yes |
| 8 (LFM+) | Low | Yes | Yes |
| 9 (HFM+) | High | Yes | Yes |
The fish meal levels were 0, 250, and 500 g per kg of starter diet in the none, low, and high fish meal groups, respectively. On day 9, the birds in treatment groups 7, 8, and 9 were given per os a suspension of 5,000 sporulated oocysts of three Eimeria species (Eimeria acervulina, Eimeria maxima, and Eimeria tenella) (Bioproperties Pty., Glenorie, New South Wales, Australia) in 1 ml of sterile phosphate-buffered saline (PBS), and the birds in the other treatment groups were given 1 ml of sterile PBS instead of the Eimeria suspension. On days 14, 15, and 16, the birds in treatment groups 4 to 9 were inoculated per os with 1 ml of C. perfringens suspension at a concentration of 108 to 109 CFU/ml, and the birds in treatment groups 1, 2, and 3 received 1 ml of sterile thioglycolate broth in place of C. perfringens.
The treatment group abbreviations shown in parentheses are as follows: NFM−, not fed fish meal and negative for C. perfringens and Eimeria; LFM−, fed low level of fish meal and negative for C. perfringens and Eimeria; HFM−, fed high level of fish meal and negative for C. perfringens and Eimeria; NFM/Cp, not fed fish meal and challenged with C. perfringens only; LFM/Cp, fed low level of fish meal and challenged with C. perfringens only; HFM/Cp, fed high level of fish meal and challenged with C. perfringens only; NFM+, not fed fish meal and challenged by C. perfringens and Eimeria; LFM+, fed low level of fish meal and challenged by C. perfringens and Eimeria; HFM+, fed high level of fish meal and challenged by C. perfringens and Eimeria.
Quantification of ileal C. perfringens.
The quantification of ileal C. perfringens using the culture-based method followed the protocol described earlier (28). The C. perfringens bacteria were cultured and counted on Perfringens tryptose-sulfite-cycloserine and Shahidi-Ferguson Perfringens agar base mixed with egg yolk emulsion and Perfringens selective supplement (Oxoid). C. perfringens plates were incubated anaerobically for 48 h at 39°C prior to counting. Bacterial numbers were expressed as log10 CFU/gram of digesta.
Extraction of DNA from ileal content was conducted using a QIAamp DNA stool kit (Qiagen, Hilden, Germany) following the instructions of the manufacturer with slight modifications. First, 180 to 220 mg frozen digesta was taken from stored samples, and glass beads (300 mg) (0.1 mm; Biospec Products, Bartlesville, OK) were used to disrupt the cells in 400 μl of ASL lysis buffer by shaking the sample on a miniBeadBeater (Biospec Products, Bartlesville, OK) for 30 s. The cells were then lysed after adding 1 ml of ASL lysis buffer, stool particles were removed, and PCR inhibitors in the supernatant were absorbed by the InhibitEX tablet. DNA was precipitated by adding 200 μl of ethanol, captured on the QIAamp spin column, washed by 500 μl of washing buffers AW1 and AW2, and eluted in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]).
The quantitative real-time PCR assay was conducted by the method of Wise and Siragusa (27). TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA) was used. A pair of primers (CPerf165F [5′-CGCATAACGTTGAAAGATGG-3′] and CPerf269R [5′-CCTTGGTAGGCCGTTACCC-3′]; Invitrogen, Mulgrave, Victoria, Australia) and a dual labeled TaqMan probe (CPerf187F [5′-FAM-TCATCATTCAACCAAAGGAGCAATCC-TAMRA-3′ where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine]) (Applied Biosystems, Foster City, CA) that targets C. perfringens 16S rRNA genes were also used. The DNA extracted from ileal digesta was diluted five times in TE buffer, and then 2 μl of diluted sample was used in PCR amplification in a total volume of 20 μl. PCR was performed in a Rotorgene 6500 real-time PCR machine (Corbett, Sydney, Australia) with three replicates for each sample. A threshold cycle (CT) average from the replicate samples was used for data analysis. Serial dilutions of a C. perfringens DNA with known concentration and in a background of ileal DNA that contained no C. perfringens were used to construct a standard curve. The corresponding number of cells was calculated by taking the genome size of C. perfringens into consideration (6). Bacterial numbers were expressed as log10 genomic DNA copy number per gram of digesta (wet weight).
All data were analyzed using the statistical package Minitab for Windows 12.1 (Minitab Inc., State College, PA). To assess the correlation between the real-time PCR method and the culture method, simple linear regression of the treatment means produced by these two methods was performed. The precision of different measurements can be evaluated by the comparison of their relative standard deviations. It has been proposed that a higher RSD demonstrates poorer precision of the measurement or vice versa (11, 19, 22). To assess the precision of the real-time PCR method compared to the culture-based method, RSDs were calculated according to the following formula: percent RSD = (σ/x) × 100 where x is the mean value and σ is the standard deviation. The RSDs of both methods were compared for each treatment.
Efficiency of the quantitative PCR assay.
The sensitivity and amplification efficiency of the real-time PCR assay were tested by amplification of serial dilutions of C. perfringens DNA samples. When the threshold cycles were plotted against the log10 values of the initial number of C. perfringens DNA copies in the PCR to construct a standard curve for the assay, linearity was observed with the following regression curve: y = −3.434x + 34.850, where y is the CT values and x is the amount of bacteria (log10 value). A significant coefficient of correlation (R2 = 1.00) was attained, and a high amplification efficiency (96%) was achieved. Therefore, it is considered that the assay produced an accurate quantification of C. perfringens when the initial amount of C. perfringens DNA in the PCR fell in the range of 7.6 × 107 to 7.6 × 10−1 copies.
Comparison of C. perfringens values produced by real-time PCR and culture-based methods.
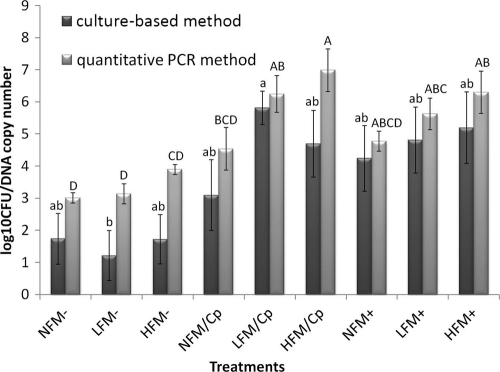
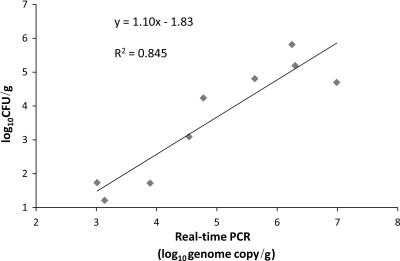
As shown in Fig. 1, the changes in the log10 values of CFU/g of C. perfringens among the treatments at day 17 briefly showed similar patterns between the real-time PCR and culture-based methods. The regression results showed that the real-time PCR method correlated highly with the results of the culture-based method (R2 = 0.845), and the regression curve had a slope of 1.10 (Fig. 2), indicating the validity of the real-time PCR method for the quantification of C. perfringens.
FIG. 1.
C. perfringens counts in response to experimental treatments on day 17 measured by culture and real-time PCR. The changes in the log10 values of CFU/g of C. perfringens in the treatment groups briefly showed similar patterns by real-time PCR and the culture-based method. Data are expressed as means ± standard errors (SE) (error bars) (n = 6). Treatment group abbreviations: NFM−, not fed fish meal and negative for C. perfringens and Eimeria; LFM−, fed low level of fish meal and negative for C. perfringens and Eimeria; HFM−, fed high level of fish meal and negative for C. perfringens and Eimeria; NFM/Cp, not fed fish meal and challenged with C. perfringens only; LFM/Cp, fed low level of fish meal and challenged with C. perfringens only; HFM/Cp, fed high level of fish meal and challenged with C. perfringens only; NFM+, not fed fish meal and challenged by C. perfringens and Eimeria; LFM+, fed low level of fish meal and challenged by C. perfringens and Eimeria; HFM+, fed high level of fish meal and challenged by C. perfringens and Eimeria. Bars with different letters above them correspond to values that are significantly different for culture (lowercase) and real-time PCR (uppercase) methods.
FIG. 2.
Simple linear regression of log10 DNA versus log10 CFU of C. perfringens in the birds subjected to different treatments. Data are means for six birds for each treatment group. The results for the real-time PCR method showed a high correlation with the results for the culture-based method (log10 CFU/g) (R2 = 0.845), and the regression curve had a slope of 1.10, indicating the validity of the real-time PCR method for the quantification of C. perfringens.
To assess the precision of the real-time PCR method compared to the culture-based method, the RSDs of both methods were calculated, and the values are shown in Table 2. For all treatments, the RSDs of real-time PCR analysis were smaller than those of the culture-based method, suggesting that the real-time PCR was more precise than the culture-based method. For example, the RSD of the culture-based method was as high as 10.8 times that of the real-time PCR method in the high-fish-meal-fed birds without C. perfringens and Eimeria infections (HFM−). The closest RSDs between these two methods were observed in the low-fish-meal-fed birds with C. perfringens infections (LFM/Cp), and interestingly, the RSD of the culture-based method in this group was the smallest among the RSDs of all the treatments measured by culture-based method. When the data for day 17 from both methods were compared, it was apparent that the higher the level of fish meal in the diet, the greater the counts of C. perfringens produced by real-time PCR in all the groups with the same Eimeria and/or C. perfringens challenge treatments. However, this trend was observed only in the birds infected with both C. perfringens and Eimeria but not in other two groups for culture-based counts (Fig. 1).
TABLE 2.
Comparison of relative standard deviations for culture-based and quantitative PCR methods for bacterial ileal contents of birds in different treatment groups on day 17a
| Treatment groupb | RSD (%)c |
Ratio of culture RSD to qPCR RSD | |
|---|---|---|---|
| Cp-culture | Cp-qPCR | ||
| 1 (NFM−) | 111.4 | 13.0 | 8.5 |
| 2 (LFM−) | 158.1 | 24.2 | 6.5 |
| 3 (HFM−) | 109.9 | 10.1 | 10.8 |
| 4 (NFM/Cp) | 87.4 | 35.8 | 2.4 |
| 5 (LFM/Cp) | 21.7 | 22.2 | 1.0 |
| 6 (HFM/Cp) | 54.4 | 23.1 | 2.4 |
| 7 (NFM+) | 58.7 | 16.1 | 3.7 |
| 8 (LFM+) | 52.6 | 21.5 | 2.4 |
| 9 (HFM+) | 52.5 | 25.8 | 2.0 |
For all the treatments, the relative standard deviations (RSDs) for real-time PCR analysis were smaller than the RSDs for the culture-based method, suggesting that the real-time PCR was more precise than the culture-based method. qPCR, quantitative PCR.
The treatment group abbreviations shown in parentheses are as follows: NFM−, not fed fish meal and negative for C. perfringens and Eimeria; LFM−, fed low level of fish meal and negative for C. perfringens and Eimeria; HFM−, fed high level of fish meal and negative for C. perfringens and Eimeria; NFM/Cp, not fed fish meal and challenged with C. perfringens only; LFM/Cp, fed low level of fish meal and challenged with C. perfringens only; HFM/Cp, fed high level of fish meal and challenged with C. perfringens only; NFM+, not fed fish meal and challenged by C. perfringens and Eimeria; LFM+, fed low level of fish meal and challenged by C. perfringens and Eimeria; HFM+, fed high level of fish meal and challenged by C. perfringens and Eimeria.
Abbreviations: Cp-culture, C. perfringens culture; Cp-qPCR, C. perfringens quantitative PCR.
High precision of real-time PCR quantification of C. perfringens.
By using the optimized infection-producing necrotic enteritis challenge model (28), we have determined that although both culture-based and real-time PCR methods produced comparable results for broiler ileal C. perfringens counts across nine treatment groups, the precision of real-time PCR methods was substantially better than that of the culture-based method in all treatment groups with only one exception (one group). In addition, the real-time PCR method has been proven to be a reliable, efficient, and cost-effective method in contrast to the tedious and costly culture-based method. Therefore, it is suggested that quantitative real-time PCR could be a good method to replace the traditional culture-based method for the enumeration of C. perfringens in the intestinal tracts of broiler chickens.
Enumeration of intestinal bacteria using the culture-based method has its disadvantage, as the processes are extremely laborious (4, 26). Alternative methods including DGGE (10, 24), TGGE (29), conventional PCR (13), and T-RFLP (23) have been applied to analyze bacteria in the environment and animal gastrointestinal tract; however, these approaches are not fully quantitative. Therefore, a quantitative real-time PCR assay has been developed as an alternative to bacterial culture-based counting methods (14, 17, 20, 27). Real-time PCR assays have been established to quantify C. perfringens in chicken gastrointestinal tract or other animal feces by targeting 16S rRNA genes (16, 27), chaperonin 60 gene (6), or genes encoding alpha-toxin (1, 2, 8). The latter assays targeted virulent C. perfringens strains rather than the total amount of the species in the samples. Although culture-based counting results have been compared with quantitative real-time PCR data in these studies, a conclusion on whether real-time PCR achieved higher precision was not reached. The current study applied the RSDs of C. perfringens counts to assess the precision of the results obtained by both methods. It was revealed that real-time PCR enumerations of ileal C. perfringens produced less variation, and thus higher precision and repeatability, than the culture-based method. The reliability of real-time PCR was also reflected by the consistent increase in C. perfringens levels detected by this approach in response to increasing levels of fish meal in the diet in all groups, whereas the culture-based data showed only the trends in the Eimeria and/or C. perfringens challenge groups in this study. The low accuracy and precision of the culture-based method can be attributed to the following. (i) The minimal detectable level of bacteria is higher by the culture-based method, leading to inaccurate counts in replicate samples with small amounts of bacteria and thus higher variation. (ii) The culture-based method is a laborious procedure, which can introduce systematic errors. (iii) Other human errors can be introduced, due to multiple operators required for the culture-based method. In contrast, the real-time PCR method has high sensitivity, as it detects as little as a few copies of the DNA and thus a single bacterial cell. In addition, due to its simplicity, there are fewer chances where systematic or human errors can be introduced. Therefore, the possible between-sample variations that may occur in a culture-based method are largely eliminated in the real-time PCR procedure.
The reliability of real-time PCR enumeration of C. perfringens can be assessed by linear regression analysis against culture-based counting. In an assay of C. perfringens in dog feces, a poor correlation between real-time PCR and agar plate results was found (R2 = 0.0016); however, other reports have demonstrated that the real-time PCR quantification results generally show high correlations with the culture-based counting (R2 values range from 0.386 to 0.990) (6, 16, 27). In this study, a similar result was achieved by linear regression analysis of the results from these two methods (R2 = 0.845). Although the real-time PCR assay resulted in a lower number of C. perfringens than plate counts of bacteria from the intestines of broiler chickens by using the alpha-toxin-encoding gene plc as the PCR target (1), overestimations of the number of bacteria have been suggested in a few other reports in which the cpn-60 gene (6, 12) or 16S rRNA genes (16, 27) were targeted. Our results demonstrated overestimation by real-time PCR compared to culture-based counting, as the culture-based method resulted in an average of approximately 1.3 log units less C. perfringens per gram of digesta (wet weight) (data not shown). This result was largely attributed to the inclusion of DNA from dead or nonculturable cells (16, 27). On the other hand, our data revealed that substantially lower counts of the colonies following agar culture of the samples with less C. perfringens, such as in the unchallenged groups of this study, may also be responsible for this dissociation.
Acknowledgments
This study was funded by The Australian Poultry CRC (project 09-22).
We acknowledge Shuyu Song, Mark Porter, and Gary Taylor for their technical assistance, Rob Moore and Anthony Keyburn of CSIRO Livestock Industries, Geelong, Victoria, Australia, for providing a C. perfringens strain for the challenge experiment, and Rima Youil, Clodualdo Villaflor, and Wayne Woods of Bioproperties Pty Ltd., Glenorie New South Wales, Australia, for providing Eimeria spp.
Footnotes
Published ahead of print on 10 December 2010.
REFERENCES
- 1.Abildgaard, L., O. Hojberg, A. Schramm, K. M. Balle, and R. M. Engberg. 2010. The effect of feeding a commercial essential oil product on Clostridium perfringens numbers in the intestine of broiler chickens measured by real-time PCR targeting the α-toxin-encoding gene (plc). Anim. Feed Sci. Technol. 157:181-189. [Google Scholar]
- 2.Albini, S., et al. 2008. Real-time multiplex PCR assays for reliable detection of Clostridium perfringens toxin genes in animal isolates. Vet. Microbiol. 127:179-185. [DOI] [PubMed] [Google Scholar]
- 3.Bonjoch, X., F. Lucena, and A. R. Blanch. 2009. The persistence of bifidobacteria populations in a river measured by molecular and culture techniques. J. Appl. Microbiol. 107:1178-1185. [DOI] [PubMed] [Google Scholar]
- 4.Dahiya, J., D. Hoehler, A. van Kessel, and M. Drew. 2007. Effect of different dietary methionine sources on intestinal microbial populations in broiler chickens. Poult. Sci. 86:2358-2366. [DOI] [PubMed] [Google Scholar]
- 5.Dahiya, J. P., D. C. Wilkie, A. G. van Kessel, and M. D. Drew. 2006. Potential strategies for controlling necrotic enteritis in broiler chickens in post-antibiotic era. Anim. Feed Sci. Technol. 129:60-88. [Google Scholar]
- 6.Dumonceaux, T. J., et al. 2006. Enumeration of specific bacterial populations in complex intestinal communities using quantitative PCR based on the chaperonin-60 target. J. Microbiol. Methods 64:46-62. [DOI] [PubMed] [Google Scholar]
- 7.Fu, C. J., J. N. Carter, Y. Li, J. H. Porter, and M. S. Kerley. 2006. Comparison of agar plate and real-time PCR on enumeration of Lactobacillus, Clostridium perfringens and total anaerobic bacteria in dog faeces. Lett. Appl. Microbiol. 42:490-494. [DOI] [PubMed] [Google Scholar]
- 8.Fukushima, H., K. Katsube, Y. Hata, R. Kishi, and S. Fujiwara. 2007. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl. Environ. Microbiol. 73:92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurjar, A. A., N. V. Hegde, B. C. Love, and B. M. Jayarao. 2008. Real-time multiplex PCR assay for rapid detection and toxin typing of Clostridium perfringens toxin producing strains in feces of dairy cattle. Mol. Cell Probes 22:90-95. [DOI] [PubMed] [Google Scholar]
- 10.Hume, M., et al. 2003. Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult. Sci. 82:1100-1107. [DOI] [PubMed] [Google Scholar]
- 11.Jacangelo, J., and N. Seith. 2001. Investigation of criteria for GWUDI determination. American Water Works Association, Denver, CO.
- 12.Karpowicz, E., A. Novinscak, F. Barlocher, and M. Filion. 2010. qPCR quantification and genetic characterization of Clostridium perfringens populations in biosolids composted for 2 years. J. Appl. Microbiol. 108:571-581. [DOI] [PubMed] [Google Scholar]
- 13.Kato, N., et al. 1993. Identification of enterotoxin-producing Clostridium perfringens by the polymerase chain reaction. J. Jpn. Assoc. Infect. Dis. 67:724-729. [DOI] [PubMed] [Google Scholar]
- 14.Kimura, B., S. Kawasaki, H. Nakano, and T. Fujii. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl. Environ. Microbiol. 67:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, J., et al. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda, K., H. Tsuji, T. Asahara, Y. Kado, and K. Nomoto. 2007. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl. Environ. Microbiol. 73:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuki, T., et al. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonel, J. L. 1980. Clostridium perfringens toxins (type A, B, C, D, E). Pharmacol. Ther. 10:617-655. [DOI] [PubMed] [Google Scholar]
- 19.Peters, F., and H. Maurer. 2005. Bioanalytical method validation and its implications for forensic and clinical toxicology—a review, p. 1-9. In P. Bièvre and H. Günzler (ed.), Validation in chemical measurement. Springer, Berlin, Germany.
- 20.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 21.Shannon, K. E., D. Y. Lee, J. T. Trevors, and L. A. Beaudette. 2007. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 382:121-129. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, J. 1987. Quality assurance of chemical measurements. Lewis Publishers, Boca Raton, FL.
- 23.Torok, V. A., K. Ophel-Keller, M. Loo, and R. J. Hughes. 2008. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 74:783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Wielen, P. W., D. A. Keuzenkamp, L. J. Lipman, F. van Knapen, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 25.Walter, J., et al. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise, M. G., and G. R. Siragusa. 2007. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102:1138-1149. [DOI] [PubMed] [Google Scholar]
- 27.Wise, M. G., and G. R. Siragusa. 2005. Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl. Environ. Microbiol. 71:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, S.-B., N. Rodgers, and M. Choct. 2010. Optimised necrotic enteritis model producing clinical and subclinical infection of Clostridium perfringens in broiler chickens. Avian Dis. 54:1058-1065. [DOI] [PubMed] [Google Scholar]
- 29.Zhu, X., T. Zhong, Y. Pandya, and R. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]