Abstract
The ability to transfer metabolic pathways from the natural producer organisms to the well-characterized cell factory Saccharomyces cerevisiae is well documented. However, as many secondary metabolites are produced by collaborating enzymes assembled in complexes, metabolite production in yeast may be limited by the inability of the heterologous enzymes to collaborate with the native yeast enzymes. This may cause loss of intermediates by diffusion or degradation or due to conversion of the intermediate through competitive pathways. To bypass this problem, we have pursued a strategy in which key enzymes in the pathway are expressed as a physical fusion. As a model system, we have constructed several fusion protein variants in which farnesyl diphosphate synthase (FPPS) of yeast has been coupled to patchoulol synthase (PTS) of plant origin (Pogostemon cablin). Expression of the fusion proteins in S. cerevisiae increased the production of patchoulol, the main sesquiterpene produced by PTS, up to 2-fold. Moreover, we have demonstrated that the fusion strategy can be used in combination with traditional metabolic engineering to further increase the production of patchoulol. This simple test case of synthetic biology demonstrates that engineering the spatial organization of metabolic enzymes around a branch point has great potential for diverting flux toward a desired product.
Nature offers a tremendous repertoire of useful metabolites that are utilized in the food, cosmetic, and pharmaceutical industries. However, many are produced in limiting amounts in organisms that are difficult or expensive to cultivate, and it is therefore often advantageous to transfer pathways from the original producer to a dedicated cell factory, such as the yeast Saccharomyces cerevisiae. Heterologous expression of enzymes in S. cerevisiae poses several layers of complexity, as the foreign proteins need not only to be translated, to be folded, and to work in a nonnatural environment but also to use metabolic intermediates produced by the enzymatic machinery of the host. It is important to consider the latter issue, since in multistep metabolic pathways, the efficiency of forming the final product may be influenced by loss of intermediates through diffusion, degradation, or conversion by competitive enzymes (for reviews, see Conrado et al. [8] and Jørgensen et al. [17]). One way to prevent such losses is to increase the overall turnover rate of intermediate to product. Traditionally, this is optimized by adjusting enzyme levels to outcompete other reactions (32, 10). However, the same goal may also be achieved by coordinating the spatial arrangement of enzymes catalyzing consecutive steps in a metabolic pathway. In the second scenario, close proximity of sequentially acting enzymes speeds up intermediate turnover in the pathway by reducing the transit time required for the intermediate to reach the enzyme that catalyzes the next step in the reaction and also by ensuring that a high local concentration of intermediate exists in the vicinity of the this enzyme. The latter is important, since the global concentration of a metabolic intermediate in most cases is typically lower than the Km of the enzymatic step that converts this intermediate to the next compound (12). Accordingly, enzymes catalyzing sequential steps in a metabolic pathway are often found in close proximity inside the cell, e.g., as the result of being sorted into a single organelle or being physically docked in adjacency (20, 23). One of the goals of metabolic engineering is to optimize metabolic fluxes of pathway intermediates toward a desired product. Employing the benefits of enzyme proximity to efficiently control the flow of intermediates through a pathway is therefore an attractive possibility. To this end, modern genetic techniques allow catalytic sites to be brought into close proximity in several ways. For example, enzymes may be engineered to assemble on supports via docking domains (10, 13) or may be brought to interact via specific communication domains (15). However, the most simple and most studied approach is to simply fuse two open reading frames (ORFs) which encode different enzymatic activities. This allows expression of chimeric proteins with two enzymes fused end-to-end into a single polypeptide (1, 18, 36). A number of observations suggest that this strategy can be used to improve the flux through a metabolic pathway. First, several protein fusions consisting of sequential enzymes have been characterized in vitro and shown to possess kinetic properties superior to those of the individual enzymes (6, 7, 22, 29). Second, experiments carried out with purified fusion proteins in polyethylene glycol solutions to mimic the crowded environment inside the cell indicate that the positive effects of enzyme fusion may be even greater in vivo (38, 25, 5). Last, the production of geranylgeraniol in S. cerevisiae was significantly increased by overexpressing a fusion of the yeast proteins Bts1 and Dpp1 (33). Encouraged by these results, we have explored the possibilities of expressing fusion enzymes, with components originating from different biological kingdoms, in the cell factory Saccharomyces cerevisiae to promote formation of a nonyeast product.
Using this concept, we show that use of a bifunctional enzymatic chimera composed as a fusion between farnesyl diphosphate synthase (FPPS) from yeast and patchoulol synthase (PTS) from patchouli (Pogostemon cablin) improves patchoulol production compared to that with the use of free enzymes. Moreover, the effect is additive to improvements obtained by traditional metabolic engineering, as expression of the FPPS-PTS fusion increased sesquiterpene production 2-fold in an ERG9-repressed strain.
MATERIALS AND METHODS
Plasmid construction.
The plasmids used in this study are listed in Table 1; for the oligonucleotides used in this study, see Table S1 in the supplemental material. All DNA subcloning steps were performed with Escherichia coli DH5α using standard methods as described by Sambrook et al. (28), and sequences of cloned genes were verified by sequencing (MWG-Biotech AG).
TABLE 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant feature(s)a | Source |
|---|---|---|
| Strains | ||
| CEN.PK113-13D | MATα MAL2-8cSUC2 ura3-52 | P. Kötterb |
| CEN.PK113-5D | MATaMAL2-8cSUC2 ura3-52 | P. Kötter |
| YIP-M0-04 | MATaerg9::PMET3-ERG9 MAL2-8cSUC2 ura3-52 | 2 |
| CEN.LA100 | MATa/MATα ERG20/erg20::hph MAL2-8c/MAL2-8cSUC2/SUC2 ura3-52/ura3-52 | This study |
| Plasmids | ||
| pESC-URA | 2μm URA3 | Stratagene |
| pIP029 | 2μm URA3 PGAL1-PatTps177 | 2 |
| pLA001 | 2μm URA3 PGAL1-PatTps177 PGAL10-ERG20 | This study |
| pLA002 | 2μm URA3 PGAL1-ERG20-SF-PatTps177 | This study |
| pLA003 | 2μm URA3 PGAL1-PatTps177-SF-ERG20 | This study |
| pLA004 | 2μm URA3 PGAL1-ERG20-LF-PatTps177 | This study |
| pLA005 | 2μm URA3 PGAL1-ERG20-SR-PatTps177 | This study |
| pLA006 | 2μm URA3 PGAL1-ERG20-LR-PatTps177 | This study |
| pLA007 | 2μm URA3 PGAL1-ERG20-S-PatTps177 | This study |
| pLA009 | 2μm URA3 PGAL1-PGAL10-YFP-ERG20 | This study |
| pLA010 | 2μm URA3 PGAL1-CFP-PatTps177 PGAL10-YFP-ERG20 | This study |
| pLA011 | 2μm URA3 PGAL1-ERG20-CFP-PatTps177 | This study |
| pLA013 | 2μm URA3 PGAL1-CFP-ERG20-PatTps177 | This study |
| pLA014 | 2μm URA3 PGAL1-YFP-ERG20-PatTps177 | This study |
| pWJ1162 | CFP | 26 |
| pWJ1164 | YFP | 26 |
| pJH620 | hph | 16 |
Linker abbreviations are as follows: SF, short flexible; LF, long flexible; SR, short rigid; LR, long rigid; S, stable.
Institut für Mikrobiologie, Johan Wolfgang Goethe-Universität, Frankfurt am Main, Germany.
A plasmid expressing PTS and FPPS as separate enzymes from the GAL1 and GAL10 promoters, respectively, was constructed. An ERG20 fragment was obtained by PCR amplification using genomic DNA from CEN.PK113-5D and the primers E3 and E2. The fragment was digested by EcoRI and ClaI and inserted into an EcoRI-ClaI vector fragment of pIP029 (2) to generate pLA001. To construct a plasmid expressing the fusion protein FPPS-PTS, a fragment encoding FPPS-PTS was constructed in two PCR steps. First, the ERG20 and PatTps177 genes were amplified in two individual PCRs using the primer pair E10 and E12 with genomic yeast DNA as the template or using primer pair P5 and P4 with pIP029 as the template, respectively. The two resulting PCR fragments were fused in a second round of PCR using the primers E10 and P4. Similarly, a fragment containing PTS-FPPS was generated using the primer pair P6 and P7 and primer pair E8 and E11 in the first individual PCRs and using P6 and E11 in the second fusion PCR. The resulting two fusion PCR fragments were cut with BamHI and XhoI and individually inserted into a BamHI-XhoI vector fragment of the 2μm-based pESC-URA plasmid (Stratagene) to generate the plasmids pLA002 and pLA003. Flexible, rigid, and stable linkers were inserted between the ERG20 and PatTps177 genes by inserting PCR fragments of the PatTps177 gene, which were generated by PCR using pIP029 as the template and by using P-FL1, P-RL1, P-RL2, or P-SL as the forward primer and P4 as the reverse primer, into a BspEI-XhoI vector fragment of pLA002 to produce the plasmids pLA004, pLA005, pLA006, and pLA007, respectively. To insert cyan fluorescent protein (CFP) as a linker, CFP was amplified with G15 and G16 using pWJ1162 (26) as the template and inserted into a BspEI vector fragment of pLA002 to generate pLA011.
A plasmid expressing the ERG20 and PatTps177 genes, N-terminally tagged with yellow fluorescent protein (YFP) and CFP, respectively, was constructed in two steps. First, the YFP and ERG20 genes were fused by PCR using primers Y2 and E13 and inserted into an EcoRI-SpeI fragment of pESC-URA to produce pLA009. Second, the CFP and PatTps177 genes were fused by PCR using primers G11 and P4 and inserted into a BamHI-XhoI vector fragment of pLA009 to produce the plasmid pLA010. To N-terminally tag the fusion protein FPPS-PTS with CFP and YFP, PCR fragments of CFP and YFP were generated with the primers G11 and G17, using pWJ1162 and pWJ1164 (26) as templates. The resulting CFP and YFP fragments were inserted into a BamHI vector fragment of pLA002 to produce the plasmids pLA013 and pLA014, respectively.
Media.
All media for genetic manipulations of yeast were prepared as described by Sherman et al. (31) with minor modifications; the synthetic medium contained twice the amount of leucine (60 mg/liter), and the sporulation medium contained galactose (1 g/liter) instead of glucose (1 g/liter). Yeast extract-peptone-galactose (YPGal) medium was prepared the same way as yeast extract-peptone-dextrose (YPD) medium except that it contained 20 g/liter galactose instead of glucose. YPD medium containing hygromycin was prepared by adding hygromycin B to a final concentration of 300 mg/liter. For growth in bioreactors, a defined mineral medium composed of 5 g/liter (NH4)2SO4, 3 g/liter KH2PO4, 0.5 g/liter MgSO4·7H2O, 1 ml/liter trace element solution, 1 ml/liter vitamin solution, 20 g/liter galactose, and 50 μl/liter Synperonic antifoam (Sigma, St. Louis, MO) was prepared as described by Verduyn et al. (35). The trace metal solution was composed of 15 g/liter EDTA, 0.45 g/liter CaCl2·2H2O, 0.45 g/liter ZnSO4·7H2O, 0.3 g/liter FeSO4·7H2O, 100 mg/liter H3BO3, 1g/liter MnCl2·2H2O, 0.3 g/liter CoCl2·6H2O, 0.3 g/liter CuSO4·5H2O, and 0.4 g/liter Na2MoO4·2H2O, whereas the vitamin solution was composed of 50 mg/liter d-biotin, 200 mg/liter para-aminobenzoic acid, 1 g/liter pyridoxine·HCl, 1 g/liter thiamine·HCl, and 25 mg/liter m-inositol. The pH was adjusted to 5.0 by adding KOH. For growth in Erlenmeyer flasks, a similar medium was used, except that this medium contained 2 ml/liter trace element solution and the concentrations of (NH4)2SO4 and KH2PO4 were increased to 7.5 g/liter and 14.4 g/liter, respectively. The pH of the shake flask medium was adjusted to 6.5 by adding NaOH. In all experiments with the strain YIP-M0-04, the ERG9 gene was repressed by supplementing the medium with 2 mM filter-sterilized methionine.
Yeast strains.
The genotype and the source of strains used in this study are provided in Table 1. The ERG20 gene was deleted both in a diploid strain resulting from a cross of CEN.PK113-13D and CEN.PK113-5D and in haploid strains derived from CEN.PK113-5D transformed with the plasmids pESC-URA, pLA001, pLA002, and pLA003. To delete the ERG20 gene, all strains were transformed with 300 ng PCR-generated bipartite substrate specifically designed to delete the ERG20 gene according to the method described by Reid et al. (26) and Nielsen et al. (21). More specifically, the bipartite substrate was constructed as follows. First, two targeting fragments composed of sequence homologous to sequence upstream or downstream of the ERG20 gene were amplified by PCR using genomic yeast DNA as the template and the primer pairs E20 and E21 and E22 and E23, respectively. Next, the upper 2/3 and the lower 2/3 of the hph gene were amplified by PCR using pJH620 (16) as the template and the primer pairs H1 and H2 and H3 and H4, respectively. Finally, the bipartite substrate was constructed by fusing the upstream targeting fragment to the upstream hph gene fragment and the downstream targeting fragment to the downstream hph fragment using the primer pairs E20 and H2 and H3 and E23, respectively.
The diploid, heterozygous ERG20/erg20Δ strain resulting from the gene targeting, CEN.LA100, was transformed with plasmids expressing PTS and FPPS as separate enzymes (pLA001) or fused (pLA002 and pLA003) and with a control plasmid (pESC-URA). The resulting transformants were transferred to sporulation medium containing galactose. Tetrads were dissected and allowed to germinate on solid YPGal medium using a TDM50 micromanipulator (Micro Video Instruments) mounted on a Nikon Eclipse 50i microscope.
Shake flask experiments.
Shake flask experiments were carried out essentially as described by Asadollahi et al. (2). Briefly, cultures were grown in 500-ml baffled Erlenmeyer flasks with 100 ml of mineral medium prepared as described above. Shake flasks were incubated in a 30°C shaker running at 150 rpm. When cell densities reached an A600 of 0.5 to 1, an overlay of 10 ml of dodecane was added to the flasks to collect the volatile sesquiterpenes. When cultures entered the stationary phase, the fermentation broth was centrifuged to separate the organic phase from the water phase. To determine the sesquiterpene and farnesol contents, samples from the organic layer were analyzed by using a Finnigan Focus gas chromatography and mass spectrometry system (GC-MS) with a split/splitless injector used in the splitless mode.
Batch fermentations.
Batch fermentations were carried out using well-controlled 1.1-liter DasGip (Jülich, Germany) bioreactors with a 0.6-liter working volume. The pH was controlled at between 4.95 and 5.05 by the automatic addition of 2 M KOH. The temperature was kept constant at 30°C. The airflow was 0.6 liters/min (1 volume of air per volume of medium per min [vvm]), the batch was sterilized by filtration, and the off-gas was passed through a condenser. Agitation was adjusted to maintain the dissolved oxygen tension above 30% of air saturation. Carbon dioxide and oxygen concentrations in the off-gas were determined by a gas analyzer (GA4; DasGip, Jülich, Germany). Mineral medium as described above containing 20 g/liter galactose and 2 mM filter-sterilized methionine was used for all batch fermentations. Batch fermentors were inoculated to an initial A600 of 0.02 from a liquid preculture. Dodecane (45 ml) was added aseptically to the medium at an A600 of 1 ± 0.1.
Fluorescence imaging and quantification.
Fluorescence imaging of CEN.PK113-5D transformed with pLA010, pLA013, or pLA014 was carried out essentially as described by Plate et al. (24). In brief, cells were grown overnight at 23°C in SC-URA medium (synthetic complete medium lacking uracil) with galactose, pelleted by centrifugation, and immobilized on a glass slide by mixing the appropriate amounts of cells with a 1.2% solution of low-temperature-melting agarose (NuSieve 3:1 agarose; FMC). Live-cell images were captured with a cooled Evolution QEi monochrome digital camera (Media Cybernetics, Inc.) mounted on a Nikon Eclipse E1000 camera (Nikon). Images were captured using a Plan-Fluor 100× lens with a 1.3 numerical aperture. The light source was a mercury arc lamp (Osram, Germany). The fluorophores CFP and YFP were visualized with the band-pass filters D436/20, 455DCLP, and D480/40 and HQ500/20, Q515LP, and HQ535/30, respectively (Chroma Technology).
FACS analysis of strains expressing fluorescently tagged FPPS and PTS.
CEN.PK113-5D transformed with pLA010, pLA013, or pLA014 was grown in SC-URA medium (synthetic complete medium lacking uracil) with galactose as a carbon source to an A600 of 1. One hundred microliters of individual cell cultures were transferred to a microtiter plate, and 10,000 cells were analyzed from three independent transfections using a flow cytometer (FACSCanto II; BD Biosciences, San Jose, CA). All transfected cell populations were 100% viable based on forward and side-scatter parameters. YFP fluorescence was detected with the use of a 488-nm solid-state 20-mW laser and band-pass filters 556LP and 585/42, and CFP was detected with a 405-nm solid-state diode with a 30-mW fiber power output and band-pass filters 502LP and 515/50. Data analyses were performed using FCS Express (version 3.00.0611; De Novo Software, Ontario, Canada) and GraphPad Prism software (version 4.03; GraphPad Software) for statistical comparisons. A P value of ≤0.05 was considered significant.
RESULTS
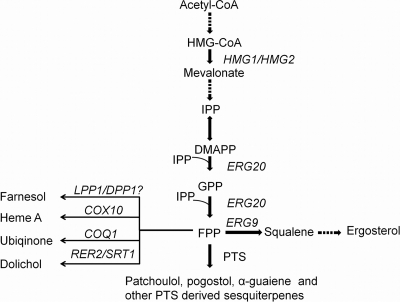
FPPS produces farnesyl diphosphate (FPP), which is the substrate for a number of endogenous S. cerevisiae enzymes, including those that are involved in production of sterols, dolichols, heme A, quinones, farnesol, and farnesylated mating factors (14) (Fig. 1). PTS catalyzes the conversion of FPP to at least 14 different sesquiterpenes, of which patchoulol is the major product (37%) (9). Since FFP is an intermediate in many different yeast pathways, it is possible that a fusion of FPPS and PTS could benefit patchoulol production by alleviating some of the competition for FPP. Accordingly, we fused yeast FPPS to plant PTS as a model system to test whether new metabolites can be advantageously produced by linking enzymes at the branch point between a yeast pathway and a foreign pathway.
FIG. 1.
The mevalonate pathway in S. cerevisiae. Patchoulol synthase (PTS) has been introduced to enable conversion of FPP to 14 different sesquiterpenes, including patchoulol. FPP is also the substrate for several other enzymes, including squalene synthase (encoded by the ERG9 gene), heme A:farnesyltransferase (COX10), hexaprenyl diphosphate synthetase (COQ1), the cis-prenyltransferases (RER2 and SRT1), and possibly the phosphatases encoded by the LPP1 and DPP1 genes (11). The pathway intermediates HMG-CoA, IPP, DMAPP, and GPP are defined as 3-hydroxy-3-methylglutaryl-coenzyme A, isopentenyl pyrophosphate, dimethyl allyl pyrophosphate, and geranyl pyrophosphate, respectively.
FPPS-PTS and PTS-FPPS fusions are functional in vivo.
A successful metabolic process based on fusion proteins requires that the chimeric enzyme is expressed and soluble and that its two enzymatic activities are functional. We therefore determined the in vivo functionality of the two possible fusion configurations, FPPS-PTS and PTS-FPPS.
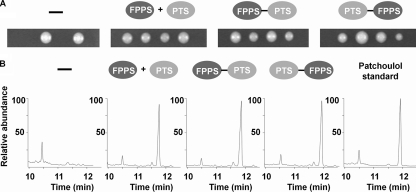
First, we tested whether FPPS could be fused to PTS and retain activity. The ERG20 gene, encoding FPPS, is essential for the viability of S. cerevisiae, and the functionality of FPPS can therefore be evaluated by investigating whether expression of the fusion enzymes can support growth of a strain containing an ERG20 deletion. In one experiment, we individually transformed ERG20 haploid strains with four different versions of a 2μm-based plasmid that contains a bidirectional GAL1/GAL10 promoter: one that serves as an “empty” control vector, one that expresses PTS and FPPS as free enzymes from the GAL1/GAL10 promoter, and two that express a fusion enzyme, either FPPS-PTS or PTS-FPPS, from the GAL1 promoter. In the latter two cases, the enzymes are fused via a short flexible linker, Gly-Ser-Gly. Subsequently, the four different types of transformants were transformed with gene-targeting substrates designed to delete the endogenous ERG20 gene. ERG20 deletions in strains harboring plasmids encoding FFPS, FPPS-PTS, or PTS-FPPS were readily obtained. No deletions were obtained with strains harboring the empty vector (data not shown). In a second experiment, we individually transformed the heterozygous diploid ERG20/erg20Δ strain with the same four plasmids. In agreement with the idea that the ERG20 gene is essential, the strain transformed with the empty plasmid produced tetrads containing two viable spores (Fig. 2A). In contrast, the three other strains occasionally produced tetrads containing four viable spores. Hence, expression of free FPPS, FPPS-PTS, or PTS-FPPS rescues the viability of erg20Δ spore clones. For these strains, we also observed tetrads containing less than four viable spores. In these cases, the erg20Δ spore(s) likely did not inherit a plasmid. Together, the two experiments show that FPPS can be fused both N and C terminally to PTS and retain activity.
FIG. 2.
Functionality of FPPS-PTS and PTS-FPPS when they are expressed in S. cerevisiae. (A) Tetrad analysis of a ERG20/erg20Δ diploid strain harboring an empty plasmid (−) or plasmids expressing FPPS and PTS as free enzymes, an FPPS-PTS fusion, or a PTS-FPPS fusion, as indicated. (B) Gas chromatography spectra for ERG20 strains harboring an empty plasmid (−) or plasmids expressing FPPS and PTS as free enzymes, an FPPS-PTS fusion, or a PTS-FPPS fusion, as indicated. The spectrum of a patchoulol standard is shown to the right. To facilitate comparisons of the shapes of individual chromatograms, they are all shown as relative abundance (%).
Second, it was tested whether PTS remains active after fusion to FPPS. As patchoulol is a nonnative yeast compound, we used patchoulol as a reporter metabolite to demonstrate PTS activity. Accordingly, haploid ERG20 strains were transformed with the four plasmids described above. The four transformed strains were grown in Erlenmeyer shake flasks using a two-phase fermentation system in which dodecane was used as a solvent layer to collect volatile compounds, such as patchoulol. The dodecane phase was analyzed for reaction products using GC-MS. The peak corresponding to patchoulol was identified and confirmed by comparisons to the retention time and mass spectra of a patchoulol standard (Fig. 2B). No production of patchoulol was observed when the strain harbored the empty plasmid (Fig. 2B). In contrast, patchoulol production was observed for strains expressing free FPPS and PTS as well as the fusion enzymes FPPS-PTS and PTS-FPPS. PTS can therefore be extended N- and C-terminally with FPPS and retain PTS activity in vivo. Also noteworthy was that the product profile of PTS in the fusion enzymes appears similar to that of free PTS, as three selected PTS-derived sesquiterpenes, patchoulol, alpha-guaiene, and pogostol, are produced in the same ratios in all cases (data not shown).
Enzyme fusion results in an increased production of patchoulol in S. cerevisiae.
To investigate whether enzyme fusions may divert the flux more efficiently toward sesquiterpene production than that with the corresponding free enzymes, ERG20 strains were individually transformed with the three plasmids expressing free FPPS and PTS, FPPS-PTS, and PTS-FPPS. The resulting strains were inoculated into shake flasks in triplicate and grown until the stationary phase. At this point, the organic phase was separated from the medium, and the patchoulol production was determined as described in Materials and Methods. The strain transformed with the plasmid expressing PTS-FPPS produced patchoulol at the same level as that for the strain expressing free FPPS and PTS (Table 2). More interestingly, the strain transformed with the plasmid expressing FPPS-PTS produced patchoulol at an almost 2-fold higher level than that for the strain expressing free FPPS and PTS (Table 2), thus showing that a fusion protein may promote conversion of FPP to patchoulol.
TABLE 2.
Patchoulol production from ERG20 strains expressing FPPS and PTS as individual enzymes or fused
a Cells were grown in 500-ml Erlenmeyer flasks containing 100 ml of mineral medium with 20 g/liter galactose. Final titers are averages from triplicate experiments ± standard deviations.
b FPPS and PTS were expressed from 2 μm-based plasmids under the control of PGAL10 and PGAL1. In all cases, FPPS was also expressed endogenously as a free enzyme. PT, patchoulol.
c The optical density of cell culture measured at 600 nm (OD600) was used to quantify biomass.
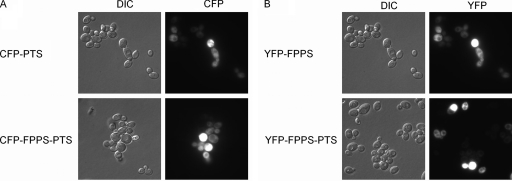
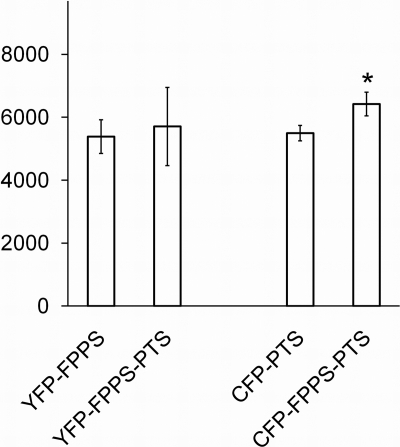
To test whether the increased patchoulol production from the strain expressing FPPS-PTS was due to a generally higher expression level of this protein, the expression levels of free and fused enzymes were assessed by N-terminally fusing FPPS, PTS, or FPPS-PTS to either cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP). This was done by extending the relevant ORFs in the 2μm-based plasmids by the CFP or YFP sequences. The resulting plasmids that expressed YFP-FPPS-PTS, CFP-FPPS-PTS, or YFP-FPPS and CFP-PTS were transformed into an ERG20 strain and inspected by fluorescence microscopy. This analysis showed that all protein species produced fluorescent products that were spread evenly in the cytoplasm (Fig. 3), indicating that all protein species are soluble, folded, and accumulate in the same compartment. However, we noted that the fluorescent enzyme content varied substantially among individual cells. As the large variation among cells makes it hard to assess the expression level using fluorescence imaging, we decided to quantify the signal of a large number of individual cells (10,000 cells) using flow cytometry. The flow cytometry data confirmed that the fluorescent signal varies greatly among cells, as approximately 50% of the cells displayed no fluorescence, and the fluorescence intensity varied more than 100-fold among the cells that did display fluorescence (see Fig. S1 in the supplemental material). However, when the mean fluorescence intensity (MFI) was calculated for three different transformants harboring the same plasmid, it appeared to be similar for the strains expressing YFP-FPPS-PTS, CFP-FPPS-PTS, and YFP-FPPS and CFP-PTS, indicating that the enzyme content in the population of cells is constant in strains transformed with the different constructs (Fig. 4). More noteworthy was the finding that the enzyme content was similar for free and fused enzymes, as there was no significant difference between expression levels of free and fused FPPS and only a slightly higher level of expression for fused PTS than for free PTS. Accordingly, the increased patchoulol production from the strain expressing the fusion protein cannot be accounted for by a higher enzyme concentration in these cells.
FIG. 3.
Cellular localization and expression levels of CFP-PTS, CFP-FPPS-PTS, YFP-FPPS, and YFP-FPPS-PTS fusions. (A) Microscopy of cells expressing PTS-CFP and CFP-FPPS-PTS. (B) Microscopy of cells expressing YFP-FPPS and YFP-FPPS-PTS. DIC, differential interference contrast.
FIG. 4.
Enzyme contents are similar for large populations of cells that express YFP-FPPS or YFP-FPPS-PTS and for those that express CFP-PTS or CFP-FPPS-PTS. Mean fluorescence intensity (MFI) of 10,000 cells expressing YFP-FPPS, YFP-FPPS-PTS, CFP-PTS, or CFP-FPPS-PTS. MFI of cells expressing YFP-FPPS and CFP-PTS was compared to that for YFP-FPPS-PTS and for CFP-FPPS-PTS, respectively, using one-way analysis of variance (ANOVA). *, significant difference (P < 0.05). Error bars represent standard deviations.
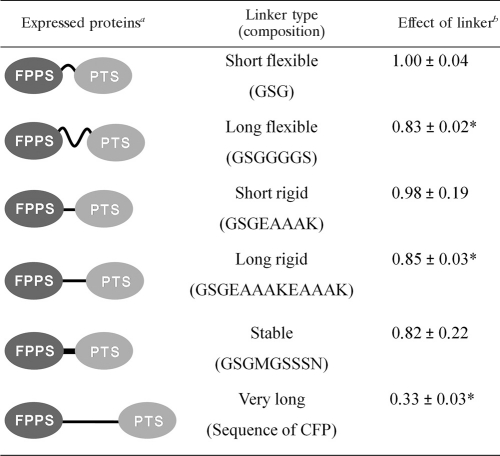
Effect of linker length and composition.
Since specific linkers can presumably favor beneficial conformations of fusion proteins (7, 19, 27, 34), we proceeded to investigate whether the nature of the linker combining FPPS and PTS can influence patchoulol production. To this end, we focused on the configuration FPPS-PTS and examined whether patchoulol production could be further improved by altering the linker composition and length. In addition to the short flexible linker, which we already tested, we also determined patchoulol production of ERG20 strains expressing FPPS-PTS fused by various linker types, namely, short flexible, long flexible, short rigid, long rigid, stable, and very long linkers (Table 3). The stable linker is a short linker that should be resistant to degradation by proteases (37), and the very long linker is an entire protein, CFP. The sequences of the remaining linkers can be found in Table 3. Strains expressing the five new FPPS-PTS variants were analyzed in triplicate for their ability to produce patchoulol. The results of this experiment show that although the FPPS-PTS fusion has a slight preference for short linkers, it does not matter whether these linkers are flexible or rigid in nature; the linker lengths and compositions do not dramatically influence patchoulol production, as they all lead to roughly the same patchoulol production. The exception is when CFP acts as the linker to connect FPPS and PTS. In this case, the amount of patchoulol produced is significantly lower, approximately 2-fold, than that obtained by the free enzymes (Tables 2 and 3).
TABLE 3.
Effect of inserting linkers that varied in length and composition between FPPS and PTS
a Proteins were in all cases expressed from 2μm-based plasmids from PGAL1. In all cases, FPPS was also expressed endogenously as a free enzyme.
b The effect of a linker is given as patchoulol production when using this linker relative to that achieved when using the short flexible linker. *, patchoulol production with this linker was significantly (P < 0.05) different from that with the short flexible linker.
Enzyme fusion can be used in combination with traditional metabolic engineering strategies to increase patchoulol production.
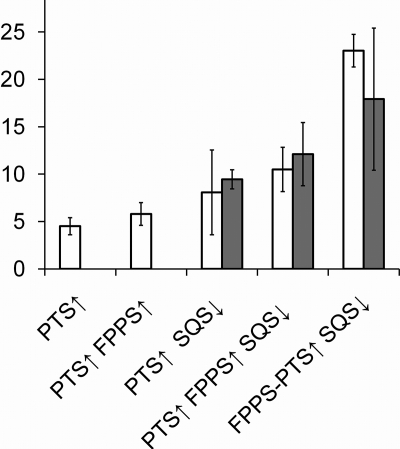
Previously, we have used different metabolic engineering strategies to increase the production of patchoulol and other sesquiterpenes (2, 3, 4). The highest patchoulol production seen by using these strategies was that obtained with a strain for which the flux toward sterol synthesis was reduced by repressing the ERG9 gene, which encodes squalene synthase (SQS) (2). To investigate whether patchoulol production in an ERG9-repressed strain can be further improved by expressing fusion enzymes, we transformed an ERG9-repressed strain with a plasmid expressing FPPS fused to PTS via a short flexible linker. For comparisons, a plasmid expressing FPPS and PTS as separate enzymes and a plasmid that expresses PTS only were each transformed into the ERG9-repressed strain and into an ERG9 strain.
When the fusion protein was expressed in the ERG9-repressed strain, the result was a 2-fold increase in the level of patchoulol production compared to that when the two enzymes were expressed as individual proteins in the same strain and an almost 5-fold increase in the level of patchoulol production compared to that when PTS was expressed alone in the unmodified strain (Fig. 5). The final patchoulol titer was 23 mg/liter when enzyme fusion was combined with ERG9 gene repression, and this is the highest final titer of patchoulol we have achieved for strains grown in shake flasks to date. We also determined the amount of farnesol, another metabolite derived from FPP. Farnesol could be detected only in ERG9-repressed strains (Fig. 5). In the three ERG9-repressed strains, no significant differences in farnesol levels were observed.
FIG. 5.
Production of patchoulol and farnesol (mg/liter) in different S. cerevisiae strains grown in shake flasks. Patchoulol production is shown as white bars and farnesol production as gray bars. ↑, overexpression of a given protein; ↓, downregulation of squalene synthase (SQS). Data shown are averages from experiments performed in triplicate or quadruplicate. Error bars represent standard deviations.
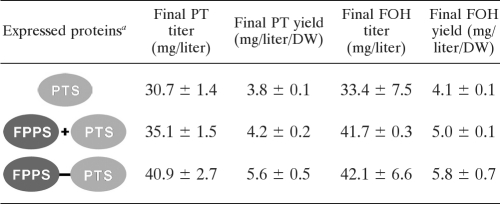
To investigate whether further improvements in the patchoulol titer could be gained under more optimal and controlled growth conditions, the ERG9-repressed strains, containing the same three plasmids as those described above, were grown in 1.1-liter bioreactors. Under these conditions, the strain expressing the fusion enzyme FPPS-PTS produced a 2-fold higher final titer of patchoulol, 41 mg/liter (Table 4), than that produced when it was grown in a shake flask. However, when strains were grown in bioreactors, the effect of enzyme fusion was less pronounced, as the final titer and yield were increased 16% and 33%, respectively, for the strain expressing FPPS-PTS compared to those for the strain expressing the separate counterparts (Table 4).
TABLE 4.
Effect of enzyme fusion and ERG9 repression on patchoulol and farnesol production when strains were grown in 1.1-liter bioreactorsb
a In all cases, free FPPS was also expressed endogenously.
PT, patchoulol; FOH, farnesol; DW, dry weight of biomass in grams.
DISCUSSION
In the present study, we demonstrate that enzyme fusion can be successfully used to improve the flux through a pathway in the cell factory S. cerevisiae. The process likely benefits from a proximity effect of the two active sites to reduce loss of intermediates to other competing pathways. Previous attempts to use this concept in vivo have generally been unsuccessful (1, 18, 36) despite the fact that fusion enzymes have proven superior to free enzymes in several in vitro studies (6, 7, 22, 29). At least two parameters should be considered in order to obtain successful exploitation of fusion proteins in vivo. First, the cellular concentration of correctly folded chimeric protein should be similar to the concentrations of the corresponding free enzymes. Second, the fusion should not negatively affect the activity of the individual catalytic sites. Unfortunately, with our current knowledge, the impact of a fusion on these two parameters is not easy to predict and the success may depend very much on intrinsic properties of the enzymes to be fused. For example, despite the fact that valencene synthase is approximately 40% identical to PTS at the amino acid residue level, expression of a fusion of FPPS and valencene synthase in S. cerevisiae strains did not increase final valencene titers, compared to the levels obtained for strains expressing the corresponding free enzymes (L.A. and U.H.M., unpublished data). Similarly, Wu and coworkers expressed a fusion of an FPPS of avian origin and the PTS used in this study and obtained a 5- to 6-fold lower production in tobacco plants than that obtained with the free enzymes (36).
The spatial location of the two active sites in the fusion protein is likely key to improved product yields. It is therefore necessary to optimize fusion of the two enzymes for functionality to benefit from this strategy. For example, the configuration of the fusion may play an important role as demonstrated in our study, in which only the fusion of FPPS to the amino-terminal end of PTS (FPPS-PTS) improves patchoulol production (Table 2). Similarly, the spatial positioning of the active site may also depend on the nature of the linker. In our case, strains expressing FPPS-PTS connected via a long linker (CFP) do not produce higher patchoulol yields than those produced by strains expressing free enzymes, as the active sites are likely too far apart to produce a beneficial proximity effect. Moreover, since it is well known that correct folding and activity of a chimeric protein depend on the nature of the linker, it is advisable to create a collection of fusion proteins assembled with different linker types. Alternatively, it may be possible to optimize the positioning of the two active sites in the fusion protein by trimming one or both of the two enzymes before fusion.
We also tested whether the benefits of enzyme fusion were additive to those achieved through traditional strain improvement. This was done by expressing the fusion protein in a strain that had already been optimized for patchoulol production by repression of the ERG9 gene. Two facts suggest that the benefits of this mutation are not fully exploited. First, the main effect of ERG9 gene repression is a substantial increase in farnesol production, indicating that the increased FPP pool is used for farnesol rather than for sesquiterpene production (2). Second, it is known that farnesol mediates product inhibition of an early step in the mevalonate pathway by signaling degradation of Hmg2 (30) (Fig. 1). To overcome these problems, we have previously tried to decrease the flux toward farnesol by deleting the LPP1 and DPP1 genes, which encode the two phosphatases that are believed to be responsible for phosphorylation of FPP to farnesol (11). Furthermore, we have tried to bypass product inhibition of the 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase step by overexpression of a truncated version of the HMG1 gene that contained only the catalytic domain (4). Surprisingly, neither of these modifications increased the sesquiterpene production in the ERG9-repressed strain. In contrast, our strategy of fusing FPPS to PTS increased patchoulol production 2-fold in the ERG9-repressed strain in shake flasks, indicating that it is possible to take advantage of the higher FPP pool in ERG9-repressed strains and redirect the flux toward sesquiterpene production. We also found a positive effect in bioreactor studies, and even though this effect was smaller than that seen with the shake flasks, it is still significant and relevant. The deviation between the effect in the shake flasks and that in the bioreactor can be explained by several factors, e.g., oxygen transfer and the mass transfer between the aqueous and dodecane phases. Importantly, in our continuous efforts to produce patchoulol in the cell factory S. cerevisiae, the 41 mg/liter obtained by expressing the FPPS-PTS fusion in an ERG9-repressed strain represents the highest titer we have obtained to date. Since patchoulol is only one of the products formed by PTS, we estimate that the total titer of sesquiterpene produced in this experiment was 110 mg/liter. The increased titer of patchoulol obtained by using a fusion of FPPS to PTS raises the possibility that heterologous production of other products in a cell factory could also benefit from fusing the first enzyme in the novel pathway to the enzyme that produces the host intermediate, hence ensuring that this intermediate more efficiently enters the new pathway rather than being diverted into the normal metabolism of the host.
In summary, we have used a synthetic biology approach and fused two metabolic enzymes, originating from two different species, to successfully produce a nonyeast product in a yeast cell factory. Physical fusion of the two enzymes optimized the local concentration and spatial organization of the two enzymatic activities to redirect the metabolic flux toward the product more efficiently than that with free enzymes. This may serve as a useful and simple alternative to strategies that employ enzyme-docking stations to achieve similar effects (10). Importantly, we have shown that this enzyme fusion strategy can successfully be used in combination with traditional metabolic engineering strategies to further increase product yields.
Supplementary Material
Acknowledgments
We thank the Danish Council for Independent Research Technology and Production Sciences for financial support (U.H.M.) and Firmenich (J.N.) for financial support and the donation of sesquiterpene synthase genes.
We thank Jesper Mogensen and Kristian Fog Nielsen for invaluable help with GC analysis and Martin Engelhard Kornholt for excellent technical assistance.
Footnotes
Published ahead of print on 10 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.An, J. M., et al. 2005. Evaluation of a novel bifunctional xylanase-cellulase constructed by gene fusion. Enzyme Microb. Technol. 36:989-995. [Google Scholar]
- 2.Asadollahi, M. A., et al. 2008. Production of plant sesquiterpenes in Saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol. Bioeng. 99:666-677. [DOI] [PubMed] [Google Scholar]
- 3.Asadollahi, M. A., et al. 2009. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab. Eng. 11:328-334. [DOI] [PubMed] [Google Scholar]
- 4.Asadollahi, M. A., J. Maury, M. Schalk, A. Clark, and J. Nielsen. 2010. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 106:86-96. [DOI] [PubMed] [Google Scholar]
- 5.Bülow, L. 1987. Characterization of an artificial bifunctional enzyme, beta-galactosidase/galactokinase, prepared by gene fusion. Eur. J. Biochem. 163:443-448. [DOI] [PubMed] [Google Scholar]
- 6.Brodelius, M., A. Lundgren, P. Mercke, and P. E. Brodelius. 2002. Fusion of farnesyldiphosphate synthase and epi-aristolochene synthase, a sesquiterpene cyclase involved in capsidiol biosynthesis in Nicotiana tabacum. Eur. J. Biochem. 269:3570-3577. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, H., S. Ljung, and L. Bülow. 1996. Physical and kinetic effects on introduction of various linker regions in beta-galactosidase/galactose dehydrogenase fusion enzymes. Biochim. Biophys. Acta 1293:154-160. [DOI] [PubMed] [Google Scholar]
- 8.Conrado, R. J., J. D. Varner, and M. P. DeLisa. 2008. Engineering the spatial organization of metabolic enzymes: mimicking nature's synergy. Curr. Opin. Biotechnol. 19:492-499. [DOI] [PubMed] [Google Scholar]
- 9.Deguerry, F., et al. 2006. The diverse sesquiterpene profile of patchouli, Pogostemon cablin, is correlated with a limited number of sesquiterpene synthases. Arch. Biochem. Biophys. 454:123-136. [DOI] [PubMed] [Google Scholar]
- 10.Dueber, J. E., et al. 2009. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27:753-759. [DOI] [PubMed] [Google Scholar]
- 11.Faulkner, A., et al. 1999. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274:14831-14837. [DOI] [PubMed] [Google Scholar]
- 12.Fersht, A. 1984. Enzyme structure and mechanism, 2nd ed. W. H. Freeman and Company, New York, NY.
- 13.Fierobe, H. P., et al. 2001. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 14.Grabinska, K., and G. Palamarczyk. 2002. Dolichol biosynthesis in the yeast Saccharomyces cerevisiae: an insight into the regulatory role of farnesyl diphosphate synthase. FEMS Yeast Res. 2:259-265. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, M., and T. Stachelhaus. 2006. Harnessing the potential of communication-mediating domains for the biocombinatorial synthesis of nonribosomal peptides. Proc. Natl. Acad. Sci. U. S. A. 103:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, E. H., et al. 2009. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker's yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 75:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jørgensen, K., et al. 2005. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8:280-291. [DOI] [PubMed] [Google Scholar]
- 18.Kourtz, L., et al. 2005. A novel thiolase-reductase gene fusion promotes the production of polyhydroxybutyrate in Arabidopsis. Plant Biotechnol. J. 3:435-447. [DOI] [PubMed] [Google Scholar]
- 19.Lu, P., and M. G. Feng. 2008. Bifunctional enhancement of a beta-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl. Microbiol. Biotechnol. 79:579-587. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen, K. A., D. B. Tattersall, P. R. Jones, and B. L. Møller. 2008. Metabolon formation in dhurrin biosynthesis. Phytochemistry 69:88-98. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, M. L., W. A. de Jongh, S. L. Meijer, J. Nielsen, and U. H. Mortensen. 2007. Transient marker system for iterative gene targeting of a prototrophic fungus. Appl. Environ. Microbiol. 73:7240-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orita, I., N. Sakamoto, N. Kato, H. Yurimoto, and Y. Sakai. 2007. Bifunctional enzyme fusion of 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase. Appl. Microbiol. Biotechnol. 76:439-445. [DOI] [PubMed] [Google Scholar]
- 23.Ovadi, J., and P. A. Srere. 1996. Metabolic consequences of enzyme interactions. Cell Biochem. Funct. 14:249-258. [DOI] [PubMed] [Google Scholar]
- 24.Plate, I., et al. 2008. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283:29077-29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prachayasittikul, V., S. Ljung, C. Isarankura-Na-Ayudhya, and L. Bülow. 2006. NAD(H) recycling activity of an engineered bifunctional enzyme galactose dehydrogenase/lactate dehydrogenase. Int. J. Biol. Sci. 2:10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid, R. J. D., M. Lisby, and R. Rothstein. 2002. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350:258-277. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, C. R., and R. T. Sauer. 1998. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 95:5929-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., D. W. Russell, and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Seo, H. S., et al. 2000. Characterization of a bifunctional enzyme fusion of trehalose-6-phosphate synthetase and trehalose-6-phosphate phosphatase of Escherichia coli. Appl. Environ. Microbiol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shearer, A. G., and R. Y. Hampton. 2005. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. EMBO J. 24:149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Stephanopoulos, G. 1999. Metabolic fluxes and metabolic engineering. Metab. Eng. 1:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Tokuhiro, K., et al. 2009. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75:5536-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Leeuwen, H. C., M. J. Strating, M. Rensen, W. de Laat, and P. C. van der Vliet. 1997. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 16:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:395-403. [DOI] [PubMed] [Google Scholar]
- 36.Wu, S., et al. 2006. Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 24:1441-1447. [DOI] [PubMed] [Google Scholar]
- 37.Xue, F., Z. Gu, and J. Feng. 2004. LINKER: a web server to generate peptide sequences with extended conformation. Nucleic Acids Res. 32:W562-W565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz, J. L., and L. Bulow. 2002. Enhanced stress tolerance in Escherichia coli and Nicotiana tabacum expressing a betaine aldehyde dehydrogenase/choline dehydrogenase fusion protein. Biotechnol. Prog. 18:1176-1182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.