Abstract
Despite the rapid adoption of crops expressing the insecticidal Cry protein(s) from Bacillus thuringiensis (Bt), public concern continues to mount over the potential environmental impacts. Reduced residue decomposition rates and increased tissue lignin concentrations reported for some Bt corn hybrids have been highlighted recently as they may influence soil carbon dynamics. We assessed the effects of MON863 Bt corn, producing the Cry3Bb protein against the corn rootworm complex, on these aspects and associated decomposer communities by terminal restriction fragment length polymorphism (T-RFLP) analysis. Litterbags containing cobs, roots, or stalks plus leaves from Bt and unmodified corn with (non-Bt+I) or without (non-Bt) insecticide applied were placed on the soil surface and at a 10-cm depth in field plots planted with these crop treatments. The litterbags were recovered and analyzed after 3.5, 15.5, and 25 months. No significant effect of treatment (Bt, non-Bt, and non-Bt+I) was observed on initial tissue lignin concentrations, litter decomposition rate, or bacterial decomposer communities. The effect of treatment on fungal decomposer communities was minor, with only 1 of 16 comparisons yielding separation by treatment. Environmental factors (litterbag recovery year, litterbag placement, and plot history) led to significant differences for most measured variables. Combined, these results indicate that the differences detected were driven primarily by environmental factors rather than by any differences between the corn hybrids or the use of tefluthrin. We conclude that the Cry3Bb corn tested in this study is unlikely to affect carbon residence time or turnover in soils receiving these crop residues.
Transgenic corn variety MON863 that produces the Cry3Bb protein from Bacillus thuringiensis (Bt) with insecticidal activity against the corn rootworm (CRW) complex (Diabrotica spp.) (Coleoptera) was released for commercial use in the United States in 2003 (46). The adoption of Cry3Bb corn in agriculture may achieve some economic, environmental, and social benefits (26), including reduction of the use of chlorpyrifos and other insecticides used to suppress corn rootworm populations, which have serious detrimental effects on the environment (3). However, concerns continue to be raised by the public about the potential environmental impacts of transgenic crops. As is true of other breeding methods, genetic engineering could affect macroscopic plant characteristics (45). Altered lignin concentrations and associated slower residue decomposition rates have been highlighted most recently (8, 17, 18, 32, 38, 50) due to their significance in influencing nutrient cycling, greenhouse gas emissions, and carbon (C) sequestration in soils.
Conflicting results for both the lignin concentration and decomposition rate of Cry1Ab corn residues have been reported, while the information for Cry3Bb corn is still limited (25). A higher lignin concentration (17, 35), slower decomposition rate (8), or both (18) were observed for Cry1Ab corn, but a greater number of studies showed no difference between Bt and non-Bt corn (13, 23, 27, 44, 50). A lower lignin concentration was even found in Cry1Ab Bt corn leaves (X4334-EPR; Novartis) between 2 and 4 weeks after initiating decomposition, albeit without soil (16). These conflicting results may be explained by the use of different Cry1Ab corn hybrids or corresponding isolines tested, or the different soil and environmental conditions encountered in these studies. In the few studies with Cry3Bb1 corn, no differences were observed in decomposition rate and/or lignin concentration (22, 32).
Microorganisms perform many critical ecosystem functions, such as nutrient cycling and residue decomposition, which are controlled by the quality and quantity of plant litter and root exudates (15). Changes in lignin concentration as observed in some Bt crops or Bt protein released from the roots and residues of Bt corn may alter microbial decomposer communities and influence their functions. In laboratory assays, the Cry1Ab protein was released in root exudates (37), bound to clay minerals (41) and humic acids (12), and retained its insecticidal activity (42). This binding likely protects the Cry1Ab protein against microbial degradation (29) and helps it to persist in soils in laboratory incubations (43), plant growth room studies, or field studies (36). The Cry3Bb1 protein was found to be degraded rapidly, and it did not persist in soils in a laboratory study (25) and was not detected in field soils planted with Bt corn for 3 (33) or 4 (24) years. However, it was detected at one of two locations with a 1-year Cry3Bb1 corn planting history in soils near the crop base during the growing season (1). The physicochemical and biological characteristics of soils and locations are likely to influence the persistence of Cry class proteins (25).
To help resolve discrepancies in the literature regarding the environmental effects of transgenic crops, we conducted a field study to evaluate the effects of Cry3Bb Bt corn (MON863), its non-Bt isoline, and the non-Bt isoline treated with the insecticide tefluthrin at planting on residue decomposition rate, initial plant lignin concentration, and composition of the microbial community colonizing the corn residues.
MATERIALS AND METHODS
Field plots.
Field trials were established at Cornell University's Musgrave Farm in Aurora, NY, and continued for 3 consecutive years from 2004 to 2006. The main treatments were as follows: (i) MON863 corn producing the Cry3Bb protein (Bt); (ii) a nontransgenic isoline (non-Bt); and (iii) the non-Bt isoline with the tefluthrin insecticide Force 3G applied to the soil at planting (non-Bt+I) to control rootworms. The field soil is a Lima loam: 43.8% sand, 37.4% silt, and 19% clay, with a pH of 7.4 and 4.6% organic matter.
Seeds of MON863 and its non-Bt isoline were obtained from Monsanto Corp. (St. Louis, MO). The tefluthrin insecticide Force 3G (AstraZeneca Corp., Wayne, PA), a pyrethroid insecticide used commonly to control CRW, was applied to half of the non-Bt corn plots to simulate a typical agricultural practice in the absence of the Bt crop. The treatments were established in plots continuously planted with corn (CC) and duplicated in another location on the farm where alfalfa had been grown previously (AR). Prior to the initial planting of this experiment, the CC plots were found to have CRW present in sufficient numbers to damage corn roots (Leslie Allee, Cornell University, personal communication), while the AR plots had little or no CRW present in the first year. Each treatment had three replicate plots (50 by 50 m). A randomized complete block (RCB) design was used. Blocks established in the AR plots were contiguous, whereas the remaining blocks for the CC plots were not. For blocks established in fields where continuous corn was grown, one was adjacent to the AR blocks and the other two were within 200 m.
The core experiment ended in May 2007. After this time, all plots were planted with oats, except the CC block which was adjacent to the AR blocks. However, litterbags remained in the field until August 2007.
Residue decomposition.
Corn cobs, shoots (stalks plus leaves), and roots were collected separately from the field after harvest in 2004 and stored at 4°C. Residues were oven dried (65°C) to a constant weight and then used in litterbag studies. Separate mesh bags (12.5 by 12.5 cm; 2- by 3-mm-mesh-size ellipse) were filled with 7 g cobs, 10 g shoots, or 5 g roots with two duplicate bags for each retrieval time. In litterbags containing shoots, the ratio of stalks to leaves was 3:2, determined by measuring the ratio of these two components in the field subsamples collected. Litter bags were placed on the soil surface or at a 10-cm depth in the field on 24 June 2005. Each spring when the plots were tilled prior to planting and each fall when the plots were harvested, the litterbags were removed from the field and stored at 4°C for about 2 weeks, until those activities were completed, and then were replaced in the field. Litterbags were sampled on 10 October 2005, 19 October 2006, and 3 August 2007 after 3.5, 15.5, and 25 months in the field, respectively.
In 2005, after the litterbags were recovered from the field, DNA was extracted from residue surface biofilms for one set of litterbags. Arthropods were extracted from the other set of litterbags using the Tullgren funnel method at the New York State Agricultural Experiment Station in Geneva, NY (data not shown). After DNA or arthropods were extracted, litter was dried to a constant weight in a 65°C oven and then ground in an ED-5 Thomas Wiley mill grinder (Arthur H. Thomas Co., Philadelphia, PA) with a sieve size of 2 mm. Approximately 0.5 g of each ground subsample was placed in a borosilicate tube and put into a 450°C muffle furnace for 7 h to determine the ash-free dry weight (AFDW).
After duplicate litterbags were recovered from the field in 2006, AFDW was measured directly for one set of litterbags and DNA was extracted from the surface biofilms for the other set of litterbags. Buried root samples were separated from soil, when needed, using particulate organic matter (POM) extraction (9). A series of nesting sieves with diameters of 12.7 cm (5-in) were stacked in order of 2-mm, 250-μm, and 53-μm openings from top to bottom. Litterbag residues were placed on the top sieves and then rinsed through the sieves with water. The remaining materials in all three sieves were transferred into a container, and POM was collected by density fractionation. POM was placed into a 65°C oven for 1 week, and AFDW was obtained as described above.
AFDW was also measured directly for one set of litterbags in 2007. The POM recovery method was used to recover the remaining residues in buried litterbags for all plant parts.
Initial lignin concentration.
Dry cobs, shoots, and roots, collected at harvest in 2004 and used to prepare the litterbags, were ground and then analyzed for acid detergent lignin (ADL) concentration following the protocol of Goering and van Soest (19). Approximately 1 g of each ground sample was weighed and placed in a 600-ml Pyrex beaker. A 100-ml aliquot of acid detergent solution (1 N sulfuric acid, technical grade cetyl trimethylammonium bromide, and 2% decahydronaphthalene) was added to each beaker, which was then connected to a refluxing apparatus and heated for 1 h. The materials were filtered by a vacuum and transferred into a Gooch crucible. The residues were then washed with acetone to remove all color and break up lumps and put into an oven (100°C) to dry overnight. The acid detergent fiber (ADF) was calculated as follows: ADF = 100[(W0 − Wt)/S], where: W0 is the weight of the oven-dry crucible including fiber, Wt is the tare weight of the oven-dry crucible, and S is the oven-dry sample weight.
Crucibles containing the ADF were half filled with cooled (15°C) 72% H2SO4 and stirred with a glass rod to break up all lumps. The crucible was refilled with 72% H2SO4 three times at hourly intervals and then kept at 20 to 23°C for 3 h. Materials were filtered by vacuum and washed with hot distilled water 5 times to remove the acid. Final filtration was done by adding acetone to the crucible and drying overnight in a 100°C oven. Hot crucibles containing the residues were weighed, ignited for 3 h in a 520°C muffle furnace, cooled to 100°C and then weighed again. The acid detergent lignin (ADL) was calculated as follows: ADL = 100(L/S), where L is the loss upon ignition after 72% H2SO4 treatment and S is the oven-dry sample weight.
Microbial communities colonizing residues in litterbags.
The terminal restriction fragment length polymorphism (T-RFLP) method was used to characterize residue decomposer communities.
Following the method developed in our laboratory, biofilms were detached from residue surfaces by placing residues from litterbags into vials containing 20 ml of 0.5 M sterile potassium phosphate buffer (pH 6.8). The vials were shaken strongly in a mechanical shaker for 1 h. The biofilm suspension was then dispensed into 2-ml tubes and centrifuged until a dense pellet of biomass was obtained. DNA was extracted from approximately 300 μl of this biomass using the PowerSoil DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA). The DNA was then quantified by measuring the fluorescence of ethidium bromide bound to DNA and comparing it against a standard curve prepared with calf thymus DNA (Trevigen Inc., Gaithersburg, MD) using Quantity One Software (Bio-Rad, Hercules, CA).
PCR amplification of the extracted DNA was performed using both bacterial and fungal universal primers. For the bacterial analysis, fluorescently labeled 27f forward primer 5′-6-FAM (6-carboxyfluorescein)-AGA GTT TGA TCC TGG CTC AG-3′ and 1492r reverse primer 5′-GGT TAC CTT GTT ACG ACT T-3′, synthesized by Integrated DNA Technologies (Coralville, IA), were used to amplify 16S rRNA genes, resulting in products of approximately 1,500 bp (30). Each 50-μl PCR mixture contained 7.5 ng of DNA template, 1× PCR buffer, 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.1 mg ml−1 bovine serum albumin (BSA), 0.1 μM each primer, and 0.05 U μl−1 Taq polymerase (all from Promega, Madison, WI). DNA was amplified in an MJ Research thermal cycler using an initial denaturing step of 94°C for 3 min followed by 30 cycles of the following program: denaturation at 94°C for 30 s, primer annealing at 59°C for 45 s, and extension at 72°C for 60 s. A final extension at 72°C for 15 min was performed after the thermal cycling was completed. For the fungal analysis, the internal transcribed spacer (ITS) region between the 18S and 28S rRNA gene regions was amplified using 0.2 μM of both fluorescently labeled ITS1 forward primer 5′-6-FAM-TCC GTA GGT GAA CCT GCG G-3′ and the ITS4 reverse primer 5′-TCC GTA GGT GAA CCT GCG G-3′, resulting in products of approximately 600 bp (48). Each 50-μl PCR mixture contained 15 ng DNA template, 0.05 U μl−1 Taq polymerase, 3.0 mM MgCl2, 1× PCR buffer, 0.6 mM dNTPs, 0.1 g liter−1 BSA, and nuclease-free water. As described above for bacteria, 50-μl reaction volumes were used. The PCR program consisted of an initial denaturing step at 94°C for 5 min, followed by 30 cycles of the following program: denaturation at 94°C for 30 s, annealing at 51°C for 45 s, and extension at 72°C for 45 s. A final extension at 72°C for 10 min was performed. The PCRs for each sample were separated in a 1.5% agarose gel to verify the products, which were then quantified as described above.
A 300-ng subsample of each PCR product was digested in separate reactions using the restriction endonucleases HhaI and MspI (for fungi) and HhaI and Sau96I (for bacteria) in 30-μl reaction mixtures containing 300 ng of amplified DNA, 1 U of the restriction enzyme, 3 μl of the appropriate 10× buffer, and 0.002 mg μl−1 BSA. The reaction mixtures were incubated at 37°C for 4.5 h, followed by an inactivation step at 68°C for 15 min. The restricted products were purified using Performa DTR Edge plates (Edge BioSystems, Gaithersburg, MD) and then evaporated and resuspended in 10 μl of a mixture containing 9.9 μl of 99% formamide and 0.1 μl of the 500 LIZ size standard. The length of the fluorescently labeled terminal fragments was determined using an ABI 3730 X1 capillary DNA sequencer (Life Sciences Core Facility, Cornell University, Ithaca, NY).
Electropherograms obtained from the ABI 3730 X1 were analyzed using GeneMapper 3.0 software (Applied Biosystems). Only those bands sized between 50 and 500 bp were used for statistical analysis using MATMODEL software (Microcomputer Power, Ithaca, NY) to generate additive main effects and multiplicative interactions (AMMI) analyses.
Statistical analysis.
Analysis of variance (ANOVA) with a linear mixed-effect model was conducted using S-PLUS 8.0 (Tibco Software Inc., Palo Alto, CA). Block was set as the random factor. All other treatment variables were set as fixed factors. Differences between treatments were compared by a post hoc Turkey's honestly significant difference (HSD) test or a post hoc Fisher's least significant difference (LSD) test using R 2.9.1 (The R Foundation for Statistical Computing).
RESULTS
Initial lignin concentration.
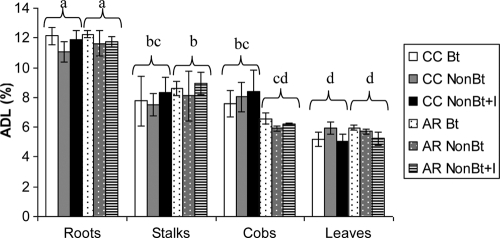
The initial lignin concentration of the 2004 corn residues used to prepare the litterbags is shown in Fig. 1. There was no significant effect of treatment (Bt, non-Bt, or non-Bt+I) on lignin concentration in the residues, but the plant part was a significant factor (P < 0.0001) in the ANOVA analysis (Table 1). The order from highest to lowest lignin concentration in the plant parts was roots > stalks > cobs > leaves. The lignin concentrations in roots and leaves ranged from 9.9 to 13.3% and 4.2 to 6.7%, respectively. Based on the ratio of 3:2 (stalks/leaves), the lignin concentrations of mixed litterbags ranged from 4.7 to 12.5%.
FIG. 1.
Initial acid detergent lignin (ADL) concentration in the corn tissues used to prepare litterbags. Error bars, SE. CC refers to plots continuously planted with corn, while AR refers to plots in which alfalfa was grown previously. Bt, NonBt, and NonBt+I refer to the treatments consisting of planting Bt corn (MON863), its non-Bt isoline, and the non-Bt isoline treated with the insecticide tefluthrin. Different letters indicate statistical differences at P < 0.05 by a post hoc Fisher's least significant difference (LSD) test.
TABLE 1.
ANOVA for initial acid detergent lignin (ADL) concentration
| Source of variance | ADL concentration result |
||
|---|---|---|---|
| dfa | F | P valuee | |
| Tb | 2 | 0.27 | 0.77 |
| Hc | 1 | 0.29 | 0.59 |
| Pd | 3 | 68.55 | <0.0001 |
| T × H | 2 | 0.31 | 0.74 |
| T × P | 6 | 0.43 | 0.86 |
| H × P | 3 | 2.92 | 0.04 |
| T × H × P | 6 | 0.12 | 0.99 |
df, degree of freedom.
Treatment (T) includes Bt (MON863 corn), non-Bt (the isoline), and non-Bt+I (the non-Bt isoline treated with the insecticide tefluthrin).
Plot history (H) includes CC (plots continuously planted with corn) and AR (plots in which alfalfa was grown previously).
Plant part (P) includes roots, stalks, cobs, and leaves.
Bold font indicates a significant difference.
There was a significant interaction between plant part and plot history for lignin concentration (P = 0.04) (Table 1). Only cobs had a higher average lignin concentration in CC plots than in AR plots, while all other parts had a lower lignin concentration in the CC plots. Based on Tukey's HSD test, the lignin concentration in AR stalks was significantly higher than that in AR cobs (P = 0.02), while there was no significant difference between CC stalks and CC cobs (P = 1.00).
Residue decomposition.
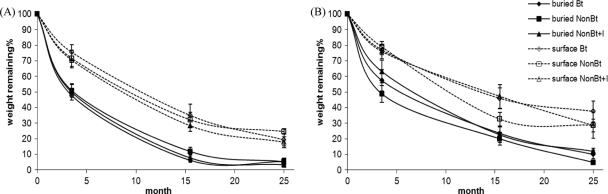
Residue mass loss over time was determined by measuring the ash-free dry weight of litter remaining (%) in the litterbags at each sampling time (Fig. 2). After 25 months in the field, an average of 90.8% of buried cobs and 95.5% of buried shoots were decomposed, while 68.3% of surface-placed cobs and 79.4% of surface-placed shoots were decomposed. For all three sampling times, treatment (Bt, non-Bt, or non-Bt+I) was not a significant factor affecting decomposition in the ANOVA analysis (Table 2) in which root data were not included.
FIG. 2.
Decomposition of litterbags containing shoots (A) or cobs (B). Error bars, SE. Bt, NonBt, and NonBt+I refer to the treatments consisting of planting Bt corn (MON863), its non-Bt isoline, and the non-Bt isoline treated with the insecticide tefluthrin. Regarding placement, litterbags were put on the soil surface (surface) or at a 10-cm depth (buried).
TABLE 2.
ANOVA for remaining weight of cob and shoot litterbags
| Source of variance | dfa | Result for samples taken in (yr)f: |
|||||
|---|---|---|---|---|---|---|---|
| 2005 |
2006 |
2007 |
|||||
| F | P value | F | P value | F | P value | ||
| Pb | 1 | 4.39 | 0.04 | 16.64 | 0.0002 | 13.26 | 0.0007 |
| Dc | 1 | 56.79 | <0.001 | 61.28 | <0.0001 | 82.65 | <0.0001 |
| Hd | 1 | 0.10 | 0.75 | 0.65 | 0.42 | 1.38 | 0.25 |
| Te | 2 | 0.23 | 0.80 | 0.60 | 0.56 | 0.50 | 0.61 |
| P × D | 1 | 0.11 | 0.75 | 0.37 | 0.55 | 2.34 | 0.13 |
| P × H | 1 | 2.98 | 0.09 | 5.08 | 0.03 | 5.47 | 0.02 |
| P × T | 2 | 0.48 | 0.62 | 1.80 | 0.18 | 2.05 | 0.14 |
| D × H | 1 | 1.06 | 0.31 | 2.84 | 0.10 | 1.32 | 0.26 |
| D × T | 2 | 0.79 | 0.46 | 1.14 | 0.33 | 0.89 | 0.42 |
| H × T | 2 | 0.91 | 0.40 | 1.69 | 0.20 | 0.19 | 0.82 |
| P × D × H | 1 | 0.58 | 0.45 | 0.14 | 0.71 | 5.58 | 0.02 |
| P × D × T | 2 | 0.92 | 0.40 | 0.40 | 0.67 | 0.66 | 0.52 |
| P × H × T | 2 | 0.31 | 0.73 | 0.04 | 0.96 | 1.04 | 0.36 |
| D × H × T | 2 | 1.12 | 0.33 | 1.85 | 0.17 | 1.44 | 0.25 |
| P × D × H × T | 2 | 0.11 | 0.89 | 0.24 | 0.79 | 2.67 | 0.08 |
df, degree of freedom.
Plant part (P) includes cobs and shoots (stalks plus leaves) made for litterbags.
Depth (D) includes soil surface and 10-cm depth where litterbags were placed.
Plot history (H) includes CC (plots continuously planted with corn) and AR (plots in which alfalfa was grown previously).
Treatment (T) includes Bt (MON863 corn), non-Bt (the isoline), and non-Bt+I (the non-Bt isoline treated with the insecticide tefluthrin).
Bold font indicates a significant difference.
For all sampling times, depth of litterbag placement was a significant factor (P < 0.0001) for the weight of all remaining residues (Table 2). Buried samples decomposed faster than those placed on the soil surface. On average, the remaining weights of surface-placed cobs and shoots were 37% and 46% higher than their corresponding buried samples, respectively, after 3.5 months. After 15.5 months, the remaining weights of surface-placed cobs and shoots were 1.9 and 3.7 times more than those of the corresponding buried samples, respectively. These ratios increased to 3.4 and 4.5 times after 25 months in the field for cobs and shoots, respectively.
As shown in Table 2, the plant part affected residue decomposition significantly for all three sampling times (P = 0.04, P = 0.0002, and P = 0.0007 in 2005, 2006, and 2007, respectively). The remaining weights of cobs were 10.1, 55.1, and 59.2% higher than those of shoots after 3.5, 15.5, and 25 months, respectively.
The interaction between plant part and plot history on remaining weight was significant at P = 0.09 in 2005 and at P < 0.05 in 2006 and 2007 (Table 2). Remaining weights of shoot samples in CC plots were 31.6 and 44.5% lower than those in AR plots in 2006 and 2007, respectively, while those of cob samples in CC plots were 17.9 and 8.8% higher than those in AR plots in 2006 and 2007, respectively.
The weight of surface-placed roots collected in 2005 was heavier than the initial weight, indicating possible soil and/or new roots infiltrating the litterbags. It decreased in 2006 but was still higher than the initial weight. In 2007, the weight of surface-placed roots (± the standard error [SE]) was 85.3% (±8.6%) the initial weight. For buried root samples, although the remaining weight was still higher than the initial weight after 3.5 months, it was lower after 15.5 and 25 months (data not shown). Due to these issues, the ANOVA was performed separately for root data (Table 3), which was excluded from the data set for the ANOVA shown in Table 2. The weight of remaining root residues did not differ significantly by treatment (Bt, non-Bt, or non-Bt+I) but was affected by the depth of litterbag placement (P < 0.0002) for all sampling times. Moreover, the weight of remaining root residues in CC plots was 50% higher than that in AR plots in 2005 (P = 0.0007) (Table 3).
TABLE 3.
ANOVA for remaining weight of root litterbag
| Source of variance | dfa | Result for samples taken in (yr)e: |
|||||
|---|---|---|---|---|---|---|---|
| 2005 |
2006 |
2007 |
|||||
| F | P value | F | P value | F | P value | ||
| Db | 1 | 22.46 | <0.0001 | 19.06 | 0.0002 | 41.40 | <0.0001 |
| Hc | 1 | 12.75 | 0.0007 | 0.16 | 0.69 | 0.04 | 0.85 |
| Td | 2 | 0.27 | 0.76 | 0.03 | 0.97 | 0.18 | 0.84 |
| D × H | 1 | 2.18 | 0.14 | 2.02 | 0.17 | 2.08 | 0.16 |
| D × T | 2 | 0.62 | 0.54 | 0.54 | 0.59 | 0.20 | 0.82 |
| H × T | 2 | 0.03 | 0.97 | 0.10 | 0.91 | 0.15 | 0.86 |
| D × H × T | 2 | 0.25 | 0.78 | 0.21 | 0.81 | 0.88 | 0.43 |
df, degree of freedom.
Depth (D) includes soil surface and 10-cm depth where litterbags were placed.
Plot history (H) includes CC (plots continuously planted with corn) and AR (plots in which alfalfa was grown previously).
Treatment (T) includes Bt (MON863 corn), non-Bt (the isoline), and non-Bt+I (the non-Bt isoline treated with the insecticide tefluthrin).
Bold font indicates a significant difference.
Microbial communities colonizing residues in litterbags.
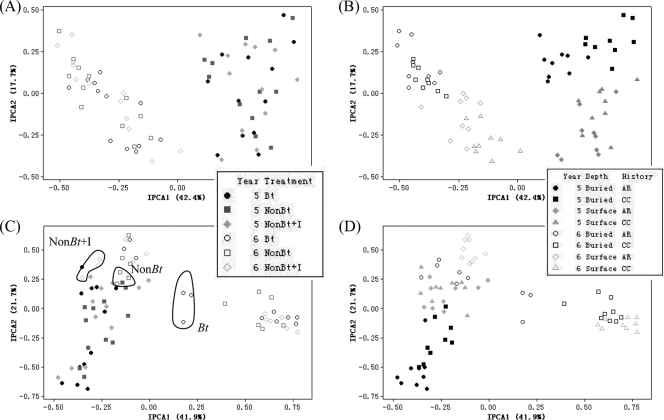
Bacterial and fungal community compositions were determined by T-RFLP analysis for the independent variables of treatment (Bt, non-Bt, or non-Bt+I), litterbag recovery year (2005 or 2006), plot history (CC or AR), plant part (cob, root, shoot), and litterbag placement depth (buried or placed on the surface). In the AMMI analysis, the percentages of predicted interaction signal variation captured in the first interaction principal component (IPCA 1) were all above 41.9% (Fig. 3). For 2006 fungal community terminal restriction fragments (TRFs) resulting from use of the HhaI enzyme (Fig. 4B), this exceeded 100%, indicating that IPCA 1 selectively recovered all predicted signal and contained predicted interaction noise. Results from use of the Sau96I and MspI restriction enzymes (data not shown) show trends similar to those obtained from use of HhaI.
FIG. 3.
Bacterial communities colonizing residues in litterbags, using the HhaI restriction enzyme, labeled by year (5, 2005; 6, 2006) and treatment (A) and year, depth, and plot history (B). Fungal communities labeled by year and treatment (C) and by year, depth, and plot history (D). Regarding treatment, Bt, NonBt, and NonBt+I refer to the treatments consisting of planting Bt corn (MON863), its non-Bt isoline, and the non-Bt isoline treated with the insecticide tefluthrin. For plot history, CC refers to plots continuously planted with corn, while AR refers to plots in which alfalfa was previously grown.
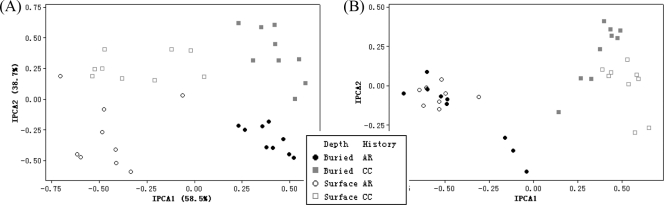
FIG. 4.
Fungal communities colonizing residues in litterbags, using the HhaI restriction enzyme, labeled by depth and plot history in 2005 (A) and 2006 (B). For plot history, CC refers to plots continuously planted with corn, while AR refers to plots in which alfalfa was previously grown.
Bacterial communities.
There was no clear separation of treatment (Fig. 3A) or plant part for either enzyme (HhaI or Sau961), indicating that neither treatment nor plant part was the primary variable controlling residue-colonizing bacterial community composition.
Samples were separated clearly by litterbag recovery year for both enzymes, indicating that bacterial communities differed by this factor. For the HhaI enzyme, the samples were also separated by litterbag placement depth (Fig. 3B). The same trend was observed for the Sau961 digestions but not as clearly as for the HhaI digestions. Litterbag recovery year was the primary main factor affecting bacterial communities, and litterbag placement depth was secondary. These two factors explained most of the variation along IPCA 1 and IPCA 2, respectively (Fig. 3B).
For TRFs resulting from HhaI digestions, 2005 buried samples were separated clearly by plot history (Fig. 3B). The 2006 samples and 2005 surface-placed samples tended to separate by plot history as well, but not as clearly. However, when analyzing the data individually by year, the effect of plot history was evident for both the buried and surface-placed residue samples for the HhaI digests but was less evident for the Sau961 digests (data not shown for both enzymes).
Fungal communities.
Fungal ITS TRFs derived from HhaI digests were separated into three groups along IPCA 1: (i) all 2005 samples, 2006 surface-placed samples, and buried non-Bt and non-Bt+I samples from AR plots; (ii) 2006 buried Bt samples from AR plots; and (iii) 2006 samples from CC plots (Fig. 3C and D). For MspI digests, samples clearly separated into only two groups by litterbag recovery year.
Terminal restriction fragments were separated by treatment (Bt, non-Bt, or non-Bt+I) for only 1 of 16 comparisons across the entire experiment (2 seasons, 2 depths, 2 plot histories, and 2 enzymes). This was for the 2006 buried samples (cobs, shoots, and roots) from the AR plots in the HhaI digests (Fig. 3C). No clustering or separation by treatment was observed for the rest of samples.
There was no clear separation by plant part for either enzyme (HhaI or MspI), indicating that fungal communities colonizing different residue types did not differ strongly. Samples separated clearly by litterbag recovery year for both HhaI and MspI digests, though some overlap existed between 2005 samples and 2006 samples from AR samples for HhaI digests (Fig. 3D). Separations by litterbag placement depth and plot history were also clear for both the HhaI (Fig. 3D) and MspI digests. However, the influence of plot history displayed a single exception, a 2005 surface-placed sample in the HhaI digests. Despite this exception, when analyzing the 2005 HhaI digest data alone (Fig. 4A), surface-placed samples separated clearly by plot history.
The analysis of fungal communities by years (2005 or 2006) is shown in Fig. 4 for HhaI digests. The 2005 residue-colonizing fungal communities were separated clearly by litterbag placement along IPCA 1 and then by plot history along IPCA 2 (Fig. 4A). However, for the 2006 samples, the plot history was the major variable along the IPCA 1 axis (Fig. 4B). Thus, the first dominant factor affecting fungal communities shifted from litterbag placement depth in 2005 to plot history in 2006.
DISCUSSION
Effect of treatment (Bt, non-Bt, and non-Bt+I).
No obvious mechanism would explain a plant lignin change resulting from transgene insertion since the Cry3Bb1 gene transformed into Bt corn is not part of the lignin biosynthesis pathway (27). However, it is theoretically possible that the transformation event causes pleiotropic effects (39). In this study, there was no significant effect of type of treatment (Bt, non-Bt, and non-Bt+I) on initial lignin concentration or litter decomposition rate, which is consistent with the findings of Lehman et al. (32) and Hönemann et al. (22), who studied Cry3Bb corn hybrids and their corresponding controls. Our result is also in accord with a laboratory study (23) and field experiments (27, 44, 50) where Cry1Ab corn was tested. However, a lower (16) or higher lignin concentration of Cry1Ab corn and lower soil respiration rate in response to the addition of finely ground Cry1Ab corn residues have been reported (8, 17, 18, 38).
The adopted lignin detection methods differed in various studies. For example, the acetyl bromide method was used in some studies (18, 38), while ADL was adopted in this study and some others (17, 32). Regarding the accuracy, there is no “clear winner” (20) among different methods. However, different techniques (ADL, Klason, and acetyl bromide) are unlikely to generate different results or alter the conclusions (27).
The conflicting results for the lignin concentration or decomposition rate for Cry1Ab crop residues might be caused by the various environmental conditions under which these crops were grown. In our study, though AR and CC plots were on the same soil type and within the same farm, the interaction between plant part and plot history was still a significant factor for lignin concentration, indicating the importance of comparing plants grown under identical conditions. Another reason might be the different hybrids used by different researchers. Variations in lignin concentrations or decomposition rates also exist in nontransgenic hybrids, which have not been tested as intensively as transgenic crops. If these variables of Bt corn are within the same range as those of traditional hybrids, it should not be necessary to test them further. This decision should depend more on the agriculture practices that transgenic crops displace.
The mesh size of the litterbags used in this study was 2 by 3 mm (an ellipse). This size allowed the microflora (bacteria, actinomycetes, fungi, and algae), microfauna (Protozoa, Rotatoria, and Tardigrada), and mesofauna (Nematoda, Acari, and Apterygota) to enter the litterbags. As the size of some macrofauna (Enchytraeidae, Lumbricidae, Mollusca, Araneae, Isopoda, Myriapoda, Coleoptera, and Diptera) is approximately 1 to 20 mm (50), most of these would have been excluded from the litterbags in this study. Hence, any effect that the Bt residues have on these populations that in turn may influence residue decomposition would not be detected. Zwahlen et al. (50) tested three different mesh sizes (20, 125, and 5,000 μm) of litterbags and found no significant differences in the remaining weight between the Cry1Ab Bt and non-Bt corn residues at the end of their experiment.
This study and several others show that microbial communities are generally unaffected by transgenic crops. No effect of Cry3Bb corn or insecticide application was reported on soil bacterial communities at another New York field site (14). Icoz et al. (24) did not find any significant effect of Cry3Bb and Cry1Ab crops on microbial diversity during residue decomposition by most-probable-number counts, denaturing gradient gel electrophoresis, and enzyme activity assays. Extracellular enzyme activities in soils (24, 31) and decaying roots (31) in fields planted with Cry3Bb corn have also been assessed, and no significant differences were observed.
In this study, an effect of treatment on fungal communities colonizing residues was observed in only 1 of 16 comparisons. Similarly, in a growth chamber experiment, Blackwood and Buyer (5) reported a significant effect of Bt protein and corn genotype on rhizosphere bacterial communities for only one of three soils tested by community-level physiological profiling, but the amount of variability explained by plant genotype was still minor. In the same study, phospholipid fatty acid analysis indicated no Bt corn effect in any of the three soils (5). Baumgarte and Tebbe (4) used the single-strand conformation polymorphism method to investigate the rhizosphere bacterial community and found the effect of Cry1Ab corn to be less than that of environmental factors, such as plant age or field heterogeneities.
Plant part.
In previous studies, stalks, leaves, or mixed samples (including all plant parts) were used to evaluate the effect of Bt plants on lignin concentration and residue decomposition rate (8, 16, 18, 23, 27, 32, 35, 38, 50). Only one study included root residues (17) and one other included cobs (44). In practice, all roots and some cobs are generally left in the field after harvest. Moreover, Cry3Bb corn was specifically developed to suppress CRW which feeds on corn roots. Thus, cobs and roots need to be assessed in residue decomposition research. In this study, the interaction between plant part and plot history significantly influenced the initial lignin concentration and remaining litterbag weight, due mostly to the distinct patterns of cob decomposition. Roots had the highest lignin concentration and were the most difficult to decompose, yet neither lignin concentration nor its decomposition rate differed by genotype.
The remaining weight of surface-placed root samples after 3.5 and 15.5 months and some buried root samples after 3.5 months was higher than their initial AFDW. This might be attributed to the common limitation of litterbag studies—soil encroachment (34). As root litterbags initially contained the smallest sample weight (5 g) and decomposed the most slowly, soil encroachment could have a greater effect on their decomposition result than on that of other plant parts. Moreover, when litterbags were collected from the field, new roots were often observed growing into or through the root litterbags, but these were rarely seen in cob and shoot litterbags. The new root material was possibly retained in the bags when they were taken from the field. The presence of arthropods and their excretions in litterbags might also be a source of contamination but is unlikely the main reason for observed gains in root litterbag weight over time.
It was surprising that the bacterial and fungal communities' colonizing residues did not differ by plant part in our results. The fragment size, surface-to-volume ratio, lignin concentration, C content/composition, and fragility vary greatly in different plant parts and may have effects on microbial decomposers (7). In this study, such effects might be masked by other environmental factors influencing community composition more strongly, such as litterbag recovery year, placement, or plot history.
Environmental factors.
The environmental factors associated with litterbag recovery year, litterbag placement depth, and plot history clearly affected both the decomposition rate and the microbial communities colonizing the residues.
Litterbag recovery year was a dominant factor controlling microbial community composition. Climate factors, such as temperature and precipitation, vary by year and are often the strongest variables influencing biological populations. Litter quantity and quality changed over time during decomposition (47). All these factors may result in interseasonal differences in microbial communities. In Europe, no field cropping system effect (growing Cry1Ab Bt and non-Bt corn) was observed on wheat straw decomposition, which was affected by climatic conditions such as temperature (11).
In this study, all surface-placed residues decomposed more slowly than buried samples, and bacterial and fungal community compositions were separated clearly by litterbag placement depth. This is consistent with results from other studies (2, 7, 21) and can be explained by differences in key environmental factors between the two locations, such as temperature, moisture, and UV exposure. Surface-placed residues also have less residue-to-soil contact (40).
The plots with two different cropping histories were in different locations within the same farm. Although the soil type was the same and the plots received the same management, the land use history likely resulted in different soil conditions, including the soil biological communities. Though the host range of CRW larvae includes some grass species other than corn (6, 10), no feral western CRW have been found to survive without maize (49). For this reason, the CC plots were found to have CRW present in sufficient numbers to damage corn roots before initiating this experiment, while none were found in the AR plots (Leslie Allee, Cornell University, personal communication). After planting corn in the AR plots, CRW numbers would likely increase over time. The 2006 surface shoot samples, 2007 buried samples, and 2007 surface shoots from CC plots decomposed significantly faster than corresponding samples from AR plots tested by two-tailed t tests. This is likely due to the microbial communities being preadapted to decompose corn residues in CC plots. Consistently, the T-RFLP data also showed that plot history was an important factor controlling the composition of microbial communities colonizing the residues.
Taken together, there was little or no effect of the type of treatment (Bt, non-Bt, and non-Bt+I) on initial residue lignin concentration, residue decomposition rate, or microbial communities colonizing the residues in litterbags. However, environmental factors, including litterbag recovery year, litterbag placement depth, and plot history, resulted in differences for most variables measured. Residue lignin concentration and decomposition rate differed by plant part as well. In short, our results indicate that differences in the variables measured were driven primarily by environmental factors rather than by any inherent differences between the corn hybrids (genotypes) or by the use of the insecticide tefluthrin. The Cry3Bb corn tested in this study did not affect residue decomposition; thus, it is unlikely to affect soil C turnover and have any significant effects on global climate change with respect to changes in CO2 emitted during residue decomposition on a per-unit-mass basis.
Acknowledgments
This work was supported by U.S. Agency for International Development (USAID) Biotechnology (Soil Management-CRSP) (2004-2007) and U.S. Department of Agriculture Biotechnology Risk Assessment (2001-2004).
Footnotes
Published ahead of print on 10 December 2010.
REFERENCES
- 1.Ahmad, A., G. E. Wilde, and K. Y. Zhu. 2005. Detectability of coleopteran-specific Cry3Bb1 protein in soil and its effect on nontarget surface and below-ground arthropods. Environ. Entomol. 34:385-394. [Google Scholar]
- 2.Alva, A. K., H. P. Collins, and R. A. Boydston. 2002. Corn, wheat, and potato crop residue decomposition and nitrogen mineralization in sandy soils under an irrigated potato rotation. Commun. Soil Sci. Plant Anal. 33:2643-2651. [Google Scholar]
- 3.Ando, A. W., and M. Khanna. 2000. Environmental costs and benefits of genetically modified crops—implications for regulatory strategies. Am. Behav. Sci. 44:435-463. [Google Scholar]
- 4.Baumgarte, S., and C. C. Tebbe. 2005. Field studies on the environmental fate of the Cry1Ab Bt-toxin produced by transgenic maize (MON810) and its effect on bacterial communities in the maize rhizosphere. Mol. Ecol. 14:2539-2551. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood, C. B., and J. S. Buyer. 2004. Soil microbial communities associated with Bt and non-Bt corn in three soils. J. Environ. Qual. 33:832-836. [DOI] [PubMed] [Google Scholar]
- 6.Branson, T. F., and E. E. Ortman. 1967. Host range of larvae of western corn rootworm. J. Econ. Entomol. 60:201-203. [Google Scholar]
- 7.Burgess, M. S., G. R. Mehuys, and C. A. Madramootoo. 2002. Decomposition of grain-corn residues (Zea mays L.): a litterbag study under three tillage systems. Can. J. Soil Sci. 82:127-138. [Google Scholar]
- 8.Castaldini, M., et al. 2005. Impact of Bt corn on rhizospheric and on beneficial mycorrhizal symbiosis and soil eubacterial communities in experimental microcosms. Appl. Environ. Microbiol. 71:6719-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, B. T. 1992. Physical fractionation of soil and organic matter in primary particle size and density separates, p. 1-90. In B. A. Stewart (ed.), Advances in soil science, vol. 20. Springer-Verlag, New York, NY. [Google Scholar]
- 10.Clark, T. L., and B. E. Hibbard. 2004. Comparison of nonmaize hosts to support western corn rootworm (Coleoptera: Chrysomelidae) larval biology. Environ. Entomol. 33:681-689. [Google Scholar]
- 11.Cortet, J., et al. 2006. Decomposition processes under Bt (Bacillus thuringiensis) maize: results of a multi-site experiment. Soil Biol. Biochem. 38:195-199. [Google Scholar]
- 12.Crecchio, C., and G. Stotzky. 1998. Insecticidal activity and biodegradation of the toxin from Bacillus thuringiensis subsp. kurstaki bound to humic acids from soil. Soil Biol. Biochem. 30:463-470. [Google Scholar]
- 13.Daudu, C. K., P. Muchaonyerwa, and P. N. S. Mnkeni. 2009. Litterbag decomposition of genetically modified maize residues and their constituent Bacillus thuringiensis protein (Cry1Ab) under field conditions in the central region of the Eastern Cape, South Africa. Agr. Ecosyst. Environ. 134:153-158. [Google Scholar]
- 14.Devare, M. H., C. M. Jones, and J. E. Thies. 2004. Effect of Cry3Bb transgenic corn and tefluthrin on the soil microbial community: biomass, activity, and diversity. J. Environ. Qual. 33:837-843. [DOI] [PubMed] [Google Scholar]
- 15.Dijkstra, F. A., S. E. Hobbie, and P. B. Reich. 2006. Soil processes affected by sixteen grassland species grown under different environmental conditions. Soil Sci. Soc. Am. J. 70:770-777. [Google Scholar]
- 16.Escher, N., B. Kach, and W. Nentwig. 2000. Decomposition of transgenic Bacillus thuringiensis maize by microorganisms and woodlice Porcellio scaber (Crustacea: Isopoda). Basic Appl. Entomol. 1:161-169. [Google Scholar]
- 17.Fang, M., P. P. Motavalli, R. J. Kremer, and K. A. Nelson. 2007. Assessing changes in soil microbial communities and carbon mineralization in Bt and non-Bt corn residue-amended soils. Appl. Soil Ecol. 37:150-160. [Google Scholar]
- 18.Flores, S., D. Saxena, and G. Stotzky. 2005. Transgenic Bt plants decompose less in soil than non-Bt plants. Soil Biol. Biochem. 37:1073-1082. [Google Scholar]
- 19.Goering, H. K., and P. J. van Soest. 1975. Forage fiber analysis—approaches, reagents, procedures, and some applications. Agriculture handbook no. 379. U.S. Government Printing Office, Washington, DC.
- 20.Hatfield, R., and R. S. Fukushima. 2005. Can lignin be accurately measured? Crop Sci. 45:832-839. [Google Scholar]
- 21.Holland, E. A., and D. C. Coleman. 1987. Litter placement effects on microbial and organic-matter dynamics in an agroecosystem. Ecology 68:425-433. [Google Scholar]
- 22.Hönemann, L., C. Zurbrugg, and W. Nentwig. 2008. Effects of Bt-corn decomposition on the composition of the soil meso- and macrofauna. Appl. Soil Ecol. 40:203-209. [Google Scholar]
- 23.Hopkins, D. W., and E. G. Gregorich. 2003. Detection and decay of the Bt endotoxin in soil from a field trial with genetically modified maize. Eur. J. Soil Sci. 54:793-800. [Google Scholar]
- 24.Icoz, I., D. Saxena, D. A. Andow, C. Zwahlen, and G. Stotzky. 2008. Microbial populations and enzyme activities in soil in situ under transgenic corn expressing Cry proteins from Bacillus thuringiensis. J. Environ. Qual. 37:647-662. [DOI] [PubMed] [Google Scholar]
- 25.Icoz, I., and G. Stotzky. 2008. Fate and effects of insect-resistant Bt crops in soil ecosystems. Soil Biol. Biochem. 40:559-586. [Google Scholar]
- 26.James, C. 2008. Global status of commercialized biotech/GM crops: 2008. ISAAA brief no. 39. International Service for the Acquisition of Agri-Biotech Applications, Ithaca, NY.
- 27.Jung, H. G., and C. C. Sheaffer. 2004. Influence of Bt transgenes on cell wall lignification and digestibility of maize stover for silage. Crop Sci. 44:1781-1789. [Google Scholar]
- 28.Reference deleted.
- 29.Koskella, J., and G. Stotzky. 1997. Microbial utilization of free and clay-bound insecticidal toxins from Bacillus thuringiensis and their retention of insecticidal activity after incubation with microbes. Appl. Environ. Microbiol. 63:3561-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 31.Lawhorn, C. N., D. A. Neher, and G. P. Dively. 2009. Impact of coleopteran targeting toxin (Cry3Bb1) of Bt corn on microbially mediated decomposition. Appl. Soil. Ecol. 41:364-368. [Google Scholar]
- 32.Lehman, R. M., S. L. Osborne, and K. A. Rosentrater. 2008. No differences in decomposition rates observed between Bacillus thuringiensis and non-Bacillus thuringiensis corn residue incubated in the field. Agron. J. 100:163-168. [Google Scholar]
- 33.Miethling-Graff, R., S. Dockhorn, and C. C. Tebbe. 2010. Release of the recombinant Cry3Bb1 protein of Bt maize MON88017 into field soil and detection of effects on the diversity of rhizosphere bacteria. Eur. J. Soil Biol. 46:41-48. [Google Scholar]
- 34.Moore-Kucera, J., and R. P. Dick. 2008. Application of 13C labeled litter and root materials for in situ decomposition studies using phospholipid fatty acids. Soil Biol. Biochem. 40:2485-2493. [Google Scholar]
- 35.Poerschmann, J., A. Gathmann, J. Augustin, U. Langer, and T. Gorecki. 2005. Molecular composition of leaves and stems of genetically modified Bt and near-isogenic non-Bt maize—characterization of lignin patterns. J. Environ. Qual. 34:1508-1518. [DOI] [PubMed] [Google Scholar]
- 36.Saxena, D., S. Flores, and G. Stotzky. 2002. Bt toxin is released in root exudates from 12 transgenic corn hybrids representing three transformation events. Soil Biol. Biochem. 34:133-137. [Google Scholar]
- 37.Saxena, D., S. Flores, and G. Stotzky. 1999. Transgenic plants—insecticidal toxin in root exudates from Bt corn. Nature 402:480. [DOI] [PubMed] [Google Scholar]
- 38.Saxena, D., and G. Stotzky. 2001. Bt corn has a higher lignin content than non-Bt corn. Am. J. Bot. 88:1704-1706. [PubMed] [Google Scholar]
- 39.Snow, A. A., et al. 2005. Genetically engineered organisms and the environment: current status and recommendations. Ecol. Appl. 15:377-404. [Google Scholar]
- 40.Summerell, B. A., and L. W. Burgess. 1989. Decomposition and chemical-composition of cereal straw. Soil Biol. Biochem. 21:551-559. [Google Scholar]
- 41.Tapp, H., L. Calamai, and G. Stotzky. 1994. Adsorption and binding of the insecticidal proteins from Bacillus thuringiensis subsp. kurstaki and subsp. tenebrionis on clay minerals. Soil Biol. Biochem. 26:663-679. [Google Scholar]
- 42.Tapp, H., and G. Stotzky. 1995. Insecticidal activity of the toxins from Bacillus thuringiensis subspecies kurstaki and tenebrionis adsorbed and bound on pure and soil clays. Appl. Environ. Microbiol. 61:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tapp, H., and G. Stotzky. 1998. Persistence of the insecticidal toxin from Bacillus thuringiensis subsp. kurstaki in soil. Soil Biol. Biochem. 30:471-476. [Google Scholar]
- 44.Tarkalson, D. D., S. D. Kachman, J. M. N. Knops, J. E. Thies, and C. S. Wortmann. 2008. Decomposition of Bt and non-Bt corn hybrid residues in the field. Nutr. Cycling Agroecosyst. 80:211-222. [Google Scholar]
- 45.Tian, L., and Z. J. Chen. 2001. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. U. S. A. 98:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.U.S. Environmental Protection Agency. 2003. Bacillus thuringiensis Cry3Bb1 protein and the genetic material necessary for its production (vector ZMIR13L) in event MON863 corn fact sheet. U.S. Environmental Protection Agency, Washington, DC.
- 47.Wedin, D. A., L. L. Tieszen, B. Dewey, and J. Pastor. 1995. Carbon-isotope dynamics during grass decomposition and soil organic-matter formation. Ecology 76:1383-1392. [Google Scholar]
- 48.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 49.Wilson, T. A., and B. E. Hibbard. 2004. Host suitability of nonmaize agroecosystem grasses for the western corn rootworm (Coleoptera: Chrysomelidae). Environ. Entomol. 33:1102-1108. [Google Scholar]
- 50.Zwahlen, C., A. Hilbeck, and W. Nentwig. 2007. Field decomposition of transgenic Bt maize residue and the impact on non-target soil invertebrates. Plant Soil 300:245-257. [Google Scholar]