Abstract
Metallic copper surfaces rapidly and efficiently kill bacteria. Cells exposed to copper surfaces accumulated large amounts of copper ions, and this copper uptake was faster from dry copper than from moist copper. Cells suffered extensive membrane damage within minutes of exposure to dry copper. Further, cells removed from copper showed loss of cell integrity. Acute contact with metallic copper surfaces did not result in increased mutation rates or DNA lesions. These findings are important first steps for revealing the molecular sensitive targets in cells lethally challenged by exposure to copper surfaces and provide a scientific explanation for the use of copper surfaces as antimicrobial agents for supporting public hygiene.
For many organisms, the trace element copper is an essential nutrient. It serves as a cofactor in respiration, and thus copper is required for aerobic metabolism. However, when copper is in excess, it is also highly toxic (29). This is because accumulation of copper ions or intracellular release of free copper ions from proteins causes cell damage. Copper readily catalyzes reactions that result in the production of hydroxyl radicals through the Fenton and Haber-Weiss reactions (13, 14). The highly reactive oxygen intermediates cause lipid peroxidation and oxidation of proteins (14, 16, 33). Free copper ions are able to oxidize sulfhydryl groups, such as cysteine, in proteins or the cellular redox buffer glutathione (15, 34). Specifically, copper ions inactivate proteins by damaging Fe-S clusters in cytoplasmic hydratases. In Escherichia coli, these are dihydroxy-acid dehydratase (IlvD) in the branched-chain amino acid synthesis pathway, isopropylmalate dehydratase (LeuC) in the leucine-specific branch, fumarase A (FumA) in the tricarboxylic acid cycle, and 6-phosphogluconate dehydratase in the pentose phosphate pathway (Edd). All were recently found to be damaged by copper ions (17). Thus, these proteins constitute specific targets for copper-induced toxicity. In Bacillus subtilis, copper ion toxicity was shown to interfere with the biosynthesis of Fe-S clusters and increased production of cluster scaffold and target proteins (5). In vitro exposure of DNA to copper ions causes mutations (36). It has also been thought that in vivo copper ion toxicity in bacteria is mediated by oxidative DNA damage, but this view was challenged because the growth rate of E. coli was found to be more strongly suppressed by copper ions under anaerobic conditions than when oxygen was present (28). Copper ions even decreased oxidative DNA damage when E. coli cells were exposed to hydrogen peroxide (18).
In recent years, it has become evident that copper surfaces with which pathogenic agents may come in contact, i.e., metallic copper touch surfaces, may help diminish surface-related hygiene problems. Dry copper surfaces in laboratory settings and in hospital trials proved to have great killing efficiency against a wide range of microbes (4, 9, 26). In most laboratory studies, cells suspended in buffer were applied to copper surfaces and incubated under ambient conditions. Usually, these cells were killed within hours (8, 25). We recently established a method that mimics contact of microbes with dry copper touch surfaces. Under these conditions, most microbes are killed within minutes (9, 10). Copper ions are released from metallic copper upon contact with bacteria (10) or with buffer alone (25). However, direct copper ion-mediated toxicity, targeting metabolic enzymes such as hydratases involved in amino acid biosynthesis (17), is unlikely to be the reason for contact killing because of the fast killing kinetics. Furthermore, extracellular supplementation with substances known to protect against oxidative stress, such as catalase, superoxide dismutase, or the hydroxyl radical quencher mannitol, delayed the killing of E. coli cells on dry copper surfaces (10). Thus, while we have some insight into the molecular mode of action exerted by copper ions on bacteria, the specific modes of stress exerted by metallic copper surfaces and the identity of sensitive cellular targets have not yet been elucidated. Such knowledge is needed to better understand why surfaces made from copper alloys exhibited efficient antimicrobial properties in recent successfully completed hospital trials (4, 19, 23).
In this study, we investigated the mode of action of dry metallic copper surfaces against E. coli and other bacterial model organisms. Our results demonstrate that exposed cells accumulated copper ions and exhibited membrane and cell envelope damage. It is likely that membrane proteins or the membrane lipids constitute the major targets of copper surface toxicity, but contact killing did not involve lethal damage to the cellular DNA through mutations and lesions.
MATERIALS AND METHODS
Bacterial strains and growth media.
The strains used in this study were Escherichia coli W3110, Bacillus cereus L8, and Deinococcus radiodurans DSM 20539. E. coli was grown in Luria-Bertani (LB) broth (Difco BD) at 37°C for 16 h, Bacillus cereus (9) in LB broth at 30°C, and D. radiodurans (3) in LB broth with 0.5% glucose at 30°C with rotary shaking (250 rpm) until stationary growth phase (approximately 24 h of incubation). Exponential-growth-phase cultures of D. radiodurans and E. coli were grown as described above, but cells were harvested after 16 h for D. radiodurans or after 3 h for E. coli. Bacto agar (Difco BD) was added at 15 g·liter−1 for solid media.
Assay for contact killing on metal surfaces.
Metal surfaces used in this study were 2.5- by 2.5-cm copper coupons (C11000, 99.9% copper) and stainless steel control coupons (AISI 304, approximately 67 to 72% Fe, 17 to 19.5% Cr, and 8 to 10.5% Ni) (C11000 and AISI 304 coupons were supplied by the International Copper Association). All copper alloy coupons were treated prior to each experiment to standardize the surface properties. Coupons were incubated for 30 s in 3% (wt/vol) NaOH solution at 70°C and rinsed in distilled water. After transfer into 10% (vol/vol) sulfuric acid solution for 5 s at room temperature (RT), coupons were immediately washed with distilled water. All coupons were disinfected and cleaned by immersion in ethanol and kept in a sterile container. To prevent surface reoxidation, cleaned coupons were not flamed after immersion in 95% ethanol.
To quantify cell survival on dry metal surfaces, cultures were concentrated 10-fold and tested as described previously (10), with minor changes. Aliquots of 106 cells were streaked out on coupons using sterile cotton swabs. All samples dried completely within 5 s after contact with the surfaces. Unless indicated otherwise, this time point is considered time zero (t0) throughout this study. To avoid contamination from the laboratory environment, coupons were incubated in sterile petri dishes at 23°C for different times. Coupons were transferred into 10 ml ice-cold phosphate-buffered saline (PBS) with approximately 20 glass beads (2 mm; Sigma-Aldrich) (PBSG buffer), and samples were vortexed for 1 min. Samples were diluted in PBS buffer and plated on LB agar. Surviving bacteria were counted as CFU using an automatic counter (Acolyte; Synbiosis) and the associated software (version 2.0.8).
For “moist” copper exposure, 109 cells in 40-μl aliquots in PBS buffer were applied as a standing droplet on coupons, and samples from droplets were removed after specified time intervals for plating on solid agar media. Survivors were counted as CFU.
Mutagenicity assay.
d-Cycloserine is an inhibitor of bacterial cell wall biosynthesis, but mutations in the cycA gene render cells resistant to this antimicrobial agent. CycA is a d-alanine, d-serine, and glycine permease that also transports d-cycloserine. d-Cycloserine uptake leads to cell wall toxicity and finally bacteriostasis but not to cell death. Mutagens increase the overall mutation rate in E. coli, thus leading to the increased appearance of d-cycloserine-resistant clones by inactivation of the cycA gene (11). The advantage of this system is that any inactivation mutation (point, frameshift, deletion, or insertion mutation) will generate resistant cells that can be scored.
A previously described method (11) that tests mutagenesis of growing cells was adapted for use with nongrowing, surface-exposed cells. Cells were applied for 5 s (an exposure period shorter than that required for massive onset of lethal damage) to the surface of the metal coupons, removed as described above, concentrated (6-fold), and spread on solidified minimal medium (21) with glycerol as the sole carbon source for determination of total CFU and on minimal medium containing glycerol and 20 μg·ml−1 d-cycloserine (Sigma-Aldrich) to select for cycA mutants. Colonies were counted after 24 h of incubation. They were assumed to have originated from mutations in the cycA gene. The percentage of cycA mutants was calculated by dividing the number of CFU of cycA mutants by the total number of CFU. As controls, cells were exposed for the same period of time to stainless steel or stainless steel with 0.25% (wt/vol) formaldehyde. This concentration of formaldehyde was used because it did not negatively affect overall survival rate of the challenged cells. To assess statistical significance, a t test was performed with the data from copper-exposed cells, stainless steel-exposed cells, and formaldehyde-exposed cells on stainless steel (positive control). The two-tailed probability (P) values were ≤0.05.
Comet assay.
A comet, or single-cell gel electrophoresis (SCGE), assay was performed with E. coli as described previously (32), with modifications. In short, cells were surface challenged and removed as described above, with minor changes. To prevent any further copper-mediated damage after removal from the surfaces, cells were removed with 10 ml PBSG buffer containing 20 μM EDTA to sequester copper ions. For fixation, cells were treated with 20 mg/ml of lysozyme at 37°C for 15 min, mixed with low-melting 0.8% agarose, and applied to a glass slide precoated with 1.5% agarose. Further steps were performed at 4°C, and ice-cold reagents were used to minimize DNA damage after surface exposure. Complete gelling and solidification of agarose cell suspensions were allowed before addition of lysis buffer (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris-HCl, pH 10, 10% Triton X-100, and 1% dimethyl sulfoxide [DMSO] to prevent oxidation during lysis). Slides were immersed in lysis buffer and carefully agitated (25 rpm). Slides were washed with deionized water, and DNA unwinding was promoted by incubation with denaturation buffer (300 mM NaOH and 1 mM EDTA, pH >13). The samples were neutralized by a short incubation with excess Tris-borate-EDTA (TBE) buffer (30). Slides were subjected to electrophoresis at 25 mV at 10 mA for 3 min at RT, removed, washed with ice-cold deionized water, immersed into absolute ethanol, and air dried overnight. Slides were stained with 1× SYBR Gold (Invitrogen) in TBE and incubated for 1 min in the dark. Fluorescence was then observed (excitation wavelength [λEx] of ∼495 nm, emission wavelength [λEm] of ∼537 nm) with an inverted confocal fluorescence microscope (Olympus IX 81) under oil immersion and with an argon laser at 488 nm (Olympus). The image capture software used was Fluoview 500 (Olympus).
ICP-MS analysis.
Cells were applied as droplets (moist method) or spread directly (dry method) on surfaces of copper coupons as described above. At different time points, cells were removed from the surfaces and excess copper was removed by washing cells with ice-cold PBSG buffer containing 20 μM EDTA. Initial cell numbers were determined by plating as described above, and samples were mineralized with concentrated 70% (vol/vol) nitric acid (trace metal grade; Mallinckrodt) for 2 h at 70°C. Samples were diluted to adjust to a final concentration of 5% (vol/vol) nitric acid. Gallium as Ga(NO3)3 was added at a final concentration of 50 ppb as an internal standard. Inductively coupled plasma mass spectroscopy (ICP-MS) analysis was performed using an Agilent ICP-MS model 7500cx operating with a collision cell with a flow of 3.5 ml·min−1 of H2 and 1.5 ml·min−1 of He. Data for each sample were accumulated in triplicate for 100 ms. An external calibration curve was recorded with gallium in 5% nitric acid. Samples were loaded onto 96-well plates prior to analysis, and an autosampler (Elemental Scientific) was used to inject samples.
General staining methods.
B. cereus and E. coli were applied to surfaces of metal coupons as described above, and cells were resuspended on the coupon with 100 μl PBS buffer, transferred onto a glass slide, and air dried. Staining of B. cereus endospores was performed with malachite green and counterstaining with safranin (31). E. coli was stained only with safranin. Glass slides were examined under oil immersion using light microscopy (Olympus AX70 fluorescence microscope).
Live/Dead staining to evaluate membrane damage.
A Live/Dead staining technique was employed to differentiate cells on copper and control surfaces with undamaged and damaged permeable membranes (Live/Dead BacLight bacterial viability kit; Invitrogen). This kit employs two nucleic acid stains: a green-fluorescent SYTO 9 stain and a red-fluorescent propidium iodide stain. These stains differ in their abilities to penetrate healthy bacterial cells. When used alone, the SYTO 9 stain labels DNA of both live and dead bacteria. In contrast, propidium iodide penetrates only bacteria with damaged membranes, reducing SYTO 9 fluorescence when both dyes are present. Thus, live bacteria with intact membranes fluoresce green, while bacteria with damaged membranes fluoresce red. Cells were applied to and removed from surfaces as described above. For staining, cells were suspended in 100 μl of 0.9% NaCl, and 1 μl of the staining mixture (1 part SYTO 9 and 1 part propidium iodide in 60 μl DMSO) was added. Cell suspensions were incubated in the dark for 15 min and then transferred onto glass slides and immediately examined by fluorescence microscopy (λEx of 488/543 nm, λEm of 522/590 nm) under oil immersion using an inverted confocal microscope (Olympus IX 81). For SYTO 9, the laser used was argon at 488 nm, and for propidium iodide, the laser used was HeNe_G at 543 nm. The image capture software used was Fluoview 500 (Olympus).
Visualization of labile intracellular Cu(I) pools.
Coppersensor-1 {CS1; 8-[N,N-bis(3′,6′-dithiaoctyl)-aminomethyl]-2,6-diethyl-4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene} (24, 40) was synthesized and employed to investigate changing intracellular Cu(I) concentrations. CS1 is a membrane-permeable fluorescent dye, which after binding to Cu(I) increases its red fluorescence 10-fold. The dye binds Cu(I) stably and selectively over other metal cations in aqueous solution. The apparent Kd (dissociation constant) for Cu(I) binding to CS1 is 3.6 × 10−12 M (40). CS1 is not a ratiometric dye, but higher copper ion concentrations result in increasing red fluorescence signals as long as the concentration of CS1 is higher than that of Cu(I). As such, CS1 can be used to monitor changes in cellular labile copper levels (24). To visualize intracellular labile Cu(I), cells exposed to a metal surface were removed as described above and stained with CS1 in the dark at RT for 20 min according to a previously described method (40). Copper accumulation within cells was examined (λEx of 543 nm, λEm of 555/600 nm) under oil immersion with an upright fluorescence microscope (Olympus AX70). The laser used was HeNe_G at 543 nm. The image capture software used was Fluoview 500 (Olympus).
RESULTS
E. coli cells release and accumulate copper ions from moist metallic copper surfaces.
Copper ions released from copper surfaces contribute to contact killing (10, 25). However, it is currently not known whether cells exposed to metallic copper actually accumulate copper ions intracellularly. In the present experiments, we found that moist plating of E. coli cells on copper coupons resulted in markedly increased copper ion concentrations over time in the buffer in which the cells were suspended compared to concentrations in buffer alone. At t0, only about (1.2 ± 1.0) × 1016 atoms·ml−1 (4.6 × 10−4 M) copper was detected in buffer-alone samples, and this value increased to (6.4 ± 2.9) × 1017 atoms·ml−1 (0.02 M) after 3 h, with an initial release rate of 9 × 1015 atoms·ml−1·min−1 (3.74 × 10−4 M·min−1). In contrast, we found that the copper content of buffer-alone samples on stainless steel remained constant at (3.8 ± 0.8) × 1014 atoms·ml−1 (1.6 × 10−5 M).
Buffer with suspended cells accumulated (1.2 ± 1) × 1016 atoms·ml−1 (4.6 × 10−4 M) copper at t0 and 3.6 × 1018 (± 7.2 × 1016) atoms·ml−1 (0.15 M) copper after 3 h, yielding an initial release rate of 3 × 1016 atoms·ml−1·min−1 (1.25 × 10−3 M·min−1). Conversely, in samples after contact with stainless steel coupons, the concentration of copper remained constant at (1 ± 0.3) × 1015 atoms·ml−1 (4.3 × 10−5 M) during the course of the 3-h experiment.
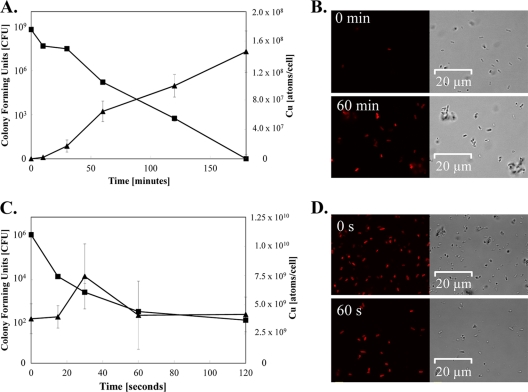
Next, we quantified copper accumulation in cells exposed to moist copper surfaces. At t0, cells contained (1.8 ± 0.5) × 104 Cu atoms/cell. The amount of intracellular copper increased linearly for the next 60 min at a rate of 1.0 × 106 atoms/cell/minute and reached a maximum of (1.5 ± 0.3) × 108 Cu atoms/cell at the end of the experiment at 3 h (Fig. 1A). In contrast, copper contents of cells on stainless steel remained virtually constant during these 3 h, at (7.9 ± 0.5) × 104 atoms/cell.
FIG. 1.
Copper uptake into cells exposed to moist or dry copper surfaces. Cells of E. coli were exposed to moist (A and B) or dry (C and D) metallic copper surfaces for the indicated times, removed, washed, and plated on solidified growth media. Survivors were counted as CFU (▪) (A and C). Parallel samples were mineralized and subjected to ICP-MS analysis for determination of cellular copper content (▴) (A and C) or were stained with the Cu(I)-specific fluorescent dye Coppersensor-1 and subjected to fluorescence microscopy (B and D). Shown are averages and standard deviations (error bars) from triplicate experiments (A and C) and representative phase-contrast (right) and fluorescence (left) microscopy images (B and D).
Copper ion uptake into exposed cells was also followed using the copper-specific dye Coppersensor-1 (CS1), which fluoresces red upon binding to Cu(I) (40). Cells stained with CS1 after 1 h of exposure to moist copper were red, indicating high concentrations of intracellular cuprous copper ions. In contrast, copper-exposed cells at t0 (Fig. 1B), stainless steel-exposed cells, or untreated cells (data not shown) fluoresced only weakly, confirming copper ion uptake from surfaces into exposed cells over the time of the experiment.
Thus, after 3 h on copper surfaces, the concentration of copper in cells was higher than that of the surrounding media. Also, buffer without cells accumulated less copper from the surface than buffer with cells. These results (Fig. 1A and B) indicate that E. coli actively dissolved and accumulated copper ions from moist copper surfaces. Under the conditions tested, cells were inactivated during a 3-h time period, and no live cells could be recovered after 3 h (Fig. 1A). Combining copper accumulation data and killing kinetics clearly demonstrated a correlation between release of copper from the copper coupons and its accumulation by the cells, with lethal consequences.
E. coli cells accumulate large intracellular amounts of copper ions from dry metallic copper surfaces.
Dry copper touch surfaces are more commonly employed to support hygiene in public and health care-related settings than moist copper touch surfaces. Therefore, we also tested copper accumulation in E. coli cells exposed to dry copper coupons. E. coli cells were killed after 1 min on dry copper (10). We repeated earlier experiments (10) to correlate cell inactivation with copper accumulation. However, since EDTA was added to cells after the copper challenge for preparation for ICP-MS analysis, complete killing was not achieved after 1 min. Instead, a 4-log reduction in numbers of live cells was observed (Fig. 1C). This also suggests that addition of a copper chelator postexposure increased the survival rate of cells subjected to contact killing. The intracellular copper content at t0 (which equaled 5 s of exposure, the time needed for the sample to dry) in copper-surface-exposed cells was (3.8 ± 1.3) × 109 Cu atoms/cell (Fig. 1C), which was 200,000 times higher than that in unexposed cells (1.8 × 104 atoms/cell). Remarkably, the intracellular copper content after 1 min still remained at (4.1 ± 2.9) × 109 Cu atoms/cell. By this time, under our test conditions, 99.99% of all cells were lethally damaged. At 30 s, a small but reproducible increase in accumulated copper was observed, probably indicating maximal copper accumulation before cell damage after 1 min countered further accumulation.
To corroborate these findings, cells were stained with CS1 dye after t0 or 1 min of exposure to dry metallic copper. Even the shortest possible exposure time, t0, resulted in bright red cells, indicating very large amounts of intracellular Cu(I) (Fig. 1D). Copper accumulation was also investigated with cells exposed to dry copper for time periods longer than needed for killing (2 min). At that time, cells did not contain more copper than at the time of inactivation (at approximately 1 to 2 min) (data not shown), suggesting that exposure beyond death did not lead to a further increase in intracellular copper concentrations but probably resulted in compromised cell integrity and leakage of previously accumulated copper.
Not only did these results demonstrate that contact killing on dry metallic copper was much faster than that on moist copper, but they also showed that copper ion accumulation was more rapid and resulted in highly elevated intracellular copper concentrations. Unexpectedly, copper accumulation from dry copper surfaces was extremely fast, probably due to the absence of buffering medium, which was present on the moist copper surfaces.
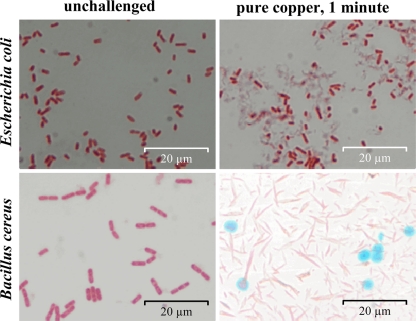
Prolonged exposure to copper surfaces leads to cell disintegration.
Copper accumulation declined when cells were exposed to dry metallic copper for periods exceeding the time needed for killing (Fig. 1C). Thus, we also investigated the structural integrity of copper-surface-exposed cells. After 1 min of exposure, E. coli and B. cereus cells removed from the copper coupons and observed by microscopy had started to disintegrate, and cell debris was detected (Fig. 2). Thus, the contact killing process led to severe structural damage in both Gram-negative and Gram-positive cells. However, Bacillus endospores appeared intact after exposure to copper surfaces and were clearly visible after staining (Fig. 2).
FIG. 2.
Prolonged contact with metallic copper results in cell disintegration. Cells of Gram-negative E. coli and Gram-positive B. cereus were exposed to pure copper for 1 min (right) or unexposed (left), removed, washed, and stained. E. coli was stained red with safranin, and B. cereus was visualized by endospore staining. This process colors endospores green and vegetative cells red after safranin counterstaining. Shown are representative light microscopy images.
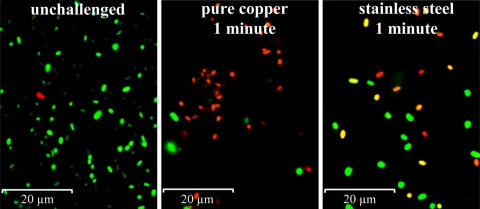
Cells exposed to dry copper surfaces have damaged membranes.
Since cells exposed to copper surfaces for longer time periods acquired structural damage (Fig. 2), it is likely that membrane damage contributes to the mechanism of action of contact killing. Cytoplasmic membrane damage can be assessed using the Live/Dead staining technique, which makes use of a dye, propidium iodide, that enters cells and stains cellular DNA (and thus the cells) only if the membranes are damaged and permeable. To test the extent and rate at which E. coli cells suffer membrane damage, cells were exposed to dry copper coupons for either t0 or 1 min, removed from the coupon surface with PBS buffer, Live/Dead stained, and immediately subjected to fluorescence microscopy. Figure 3 shows that under these conditions most cells turned red, indicating membrane damage. Unchallenged cells and cells exposed to stainless steel for the same time periods remained largely green and thus undamaged (Fig. 3), suggesting that contact with metallic copper and not general desiccation caused the rapid onset of membrane damage. Extended exposure of cells beyond the time needed for killing eventually led to cell disintegration, when cells were removed from surfaces and examined by microscopy (Fig. 2). In contrast, observation of cells directly on copper surfaces by atomic force and scanning electron microscopy did not reveal extensive structural damage, except for a few cells that appeared to have become leaky (data not shown). Thus, it is likely that contact with metallic copper did not puncture cells and result in leakage but rather that cells were permeabilized and destabilized, rendering them more susceptible for subsequent rupture by physical forces.
FIG. 3.
Cells exposed to copper surfaces suffer membrane damage. Cells of E. coli were exposed for 1 min to copper or control surfaces or unchallenged, removed, stained (Live/Dead BacLight bacterial viability kit; Invitrogen), and visualized by fluorescence microscopy. Live bacteria with intact membranes fluoresce green, while those with damaged membranes fluoresce red.
Genomic DNA is not a target of copper-surface-mediated toxicity.
Previously, we showed that bacteria exposed to dry metallic copper surfaces were efficiently killed (10). The molecular cellular targets of metallic copper toxicity, however, are currently not known. We used selection for d-cycloserine resistance to investigate the potential mutagenicity of metallic copper on E. coli cells. Exposure to metallic copper did not increase the mutation rate in E. coli (Fig. 4). Approximately the same percentages of d-cycloserine-resistant mutants arose from sensitive cells when E. coli was exposed to copper and stainless steel surfaces. There was, however, a significant increase in the number of mutants when cells on control surfaces were additionally treated with formaldehyde, a known mutagen. This is the first strong indication that metallic copper is not genotoxic and does not kill exposed cells by generation and accumulation of lethal mutations in the cell DNA.
FIG. 4.
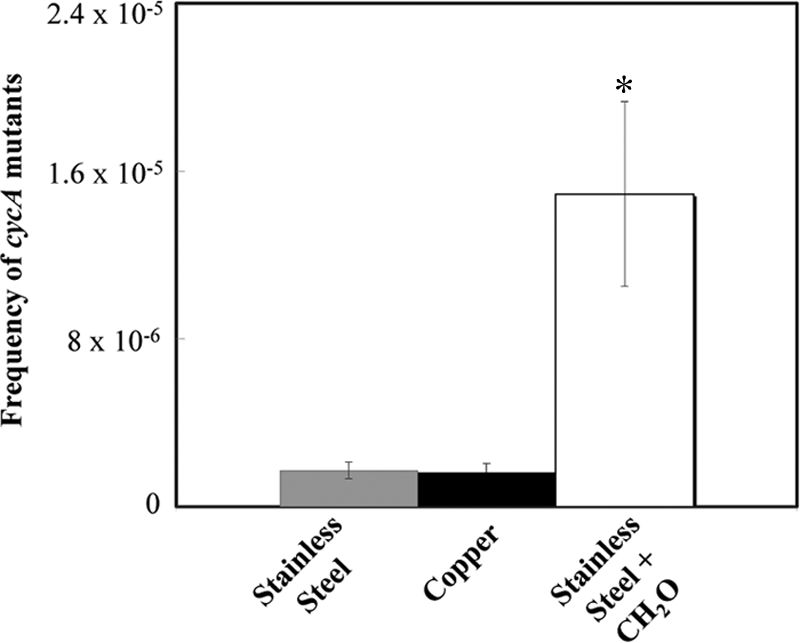
Exposure to metallic copper surfaces does not lead to increased mutations in E. coli. A total of 108 E. coli cells were exposed for 5 s to copper surfaces, stainless steel surfaces, or surfaces containing 0.25% (wt/vol) of the mutagen formaldehyde (CH2O) plus stainless steel, removed, concentrated, and spread on solid medium containing 20 μg·ml−1 of the bacteriostatic compound d-cycloserine. After 24 h of incubation at 37°C, colonies were counted as originating from mutation events leading to resistance via inactivation of CycA, a d-cycloserine uptake permease. Shown are averages from triplicate experiments, with standard deviations (error bars). The asterisk denotes significantly different values (P ≤ 0.05) for formaldehyde-challenged cells.
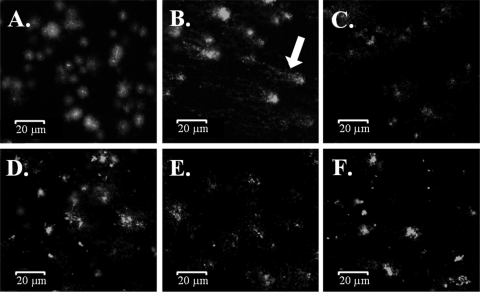
An alternative mode of DNA damage is the generation of double-strand breaks. Recently, it was shown that this mode of action of copper ion toxicity did not apply to E. coli (18). However, it is still possible that the acute toxicity exerted by metallic copper surfaces targets the DNA by this mechanism. To test this, we employed the comet assay (32), in which DNA breaks in genomes are visualized by a trail of stained DNA fragments after gel electrophoresis. E. coli cells exposed to copper or stainless steel surfaces for 1 min, a time needed to kill all cells (106 cells) on copper, were removed and assayed for comets. Unchallenged cells and cells exposed to stainless steel served as negative controls, and cells exposed to ciprofloxacin, a known inducer of DNA breakage, served as a positive control. No comets were observed in cells from challenged or unchallenged cells (Fig. 5). Comets were visible when cells stayed on copper for an extended time, 5 min, a time period 5 times longer than that sufficient to kill all cells. However, picture quality was highly diminished due to the presence of extensive cell debris (data not shown). This indicates that exposure to metallic copper did not cause extensive DNA damage, either through mutation or by fragmentation of double-stranded genomic DNA.
FIG. 5.
Exposure to metallic copper does not cause extensive DNA breakage in E. coli. E. coli cells (106 cells) were challenged on copper (D and F) or stainless steel (C and E) surfaces for t0 (C and D) or 1 min (E and F) and investigated for DNA double-strand breakage by the comet assay (32). Unchallenged (A) and ciprofloxacin-treated (B) cells indicate intact and fragmented DNA, respectively. The arrow indicates characteristic comets, highly fragmented DNA resulting from gyrase inhibition by ciprofloxacin. Images shown are representatives from three independent experiments with similar results.
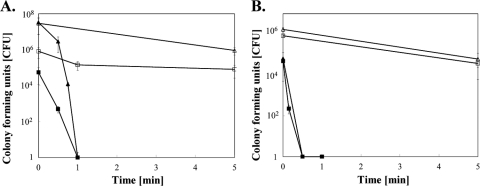
To corroborate these findings, the bacterium Deinococcus radiodurans was also exposed to dry copper surfaces. D. radiodurans possesses sophisticated and very effective DNA repair systems enabling cells to recover from stresses resulting in highly fragmented genomes, damage that is lethal to most microbes (6). Nonetheless, stationary-growth-phase cells of D. radiodurans were inactivated after 1 min of exposure (Fig. 6). This is the same time needed to kill E. coli. We also quantified the killing of exponential-growth-phase cells of D. radiodurans, because cells grown under these conditions were reported to have maximum DNA repair capabilities (35). Here, D. radiodurans cells were completely inactivated after 30 s, 50% faster than stationary-phase cells. Exponential-growth-phase E. coli cells were also tested for comparison. These cells too were 50% more sensitive to metallic copper than the cells in stationary phase. Finally, we tested killing of D. radiodurans on moist copper surfaces. Stationary-phase cells (1.1 × 107) were completely killed after 1 h (E. coli after 3 h) (data not shown). Thus, stationary- and exponential-growth-phase cells from these two species responded similarly to exposure to dry copper surfaces, with exponentially growing cells being more prone to contact killing, but D. radiodurans was even more sensitive to moist copper than E. coli. Stainless steel control surfaces had no antimicrobial activity, but a number of the exposed cells succumbed to desiccation (Fig. 6).
FIG. 6.
Efficient DNA repair provides no protection against toxicity exerted by metallic copper. Contact killing of stationary-phase (A) and exponential-phase (B) cultures of D. radiodurans (squares) or E. coli (triangles) on stainless steel (open symbols) or copper (filled symbols) surfaces. Shown are averages and standard deviations (error bars) from three independent experiments.
Taken together, these results suggest that DNA is not a major target of metallic copper toxicity in Gram-negative E. coli and Gram-positive D. radiodurans. Conversely, cells exhibited vast membrane and envelope damage, which is likely linked to subsequent cell death, and dividing cells are more prone to copper surface toxicity than resting, stationary-growth-phase cells.
DISCUSSION
Cell membranes are primary targets of contact killing through surface-released copper ions.
Significant differences exist between exposure of bacteria to toxic concentrations of copper ions and exposure to metallic copper surfaces. Exposure during growth in media containing copper ions or biofilm growth in copper plumbing systems and colonized medical copper implants is chronic, whereas contact with dry metallic copper is acute. Cells on dry metallic copper surfaces are not in an environment that promotes growth. Thus, these cells face challenges that are different from those of chronically copper ion-challenged cells. Only recently was a major mode of action for the toxicity of elevated intracellular cuprous copper ion concentrations elucidated. In E. coli, this stress causes the inactivation of hydratases, which are necessary for normal cell function. Specifically, cuprous copper ions can damage exposed Fe-S clusters in these proteins, resulting in growth defects of challenged cells (17). Since cells exposed to dry copper surfaces do not proliferate, these sensitive Fe-S clusters within proteins needed for general cellular metabolism do not constitute a likely target of toxicity.
Previously, most studies on the antimicrobial properties of metallic copper touch surfaces described the differences in efficacy caused by various copper contents, temperatures, and other parameters (22, 39). In these studies, the killing kinetics for a wide variety of microbes on copper surfaces were described (9, 20, 38). However, insights into the mechanisms of antimicrobial action are rare. Not surprisingly, copper ions and oxidative stress play a role during contact killing (8, 10, 25), even though contact killing remains efficient and rapid under anaerobiosis (10). The deletion of genes related to cellular copper ion defense was shown to speed up contact killing in Gram-negative E. coli (10) and Pseudomonas aeruginosa (8) and in Gram-positive Enterococcus hirae (25). In our study, Gram-negative E. coli and Gram-positive B. cereus were both damaged after very similar exposure times (Fig. 2), as was D. radiodurans (Fig. 6), with an outer membrane that is different from that of proteobacteria. This suggests that differences in cell wall structure per se are a bad predictor of metallic copper sensitivity. In contrast, it was recently demonstrated that different buffers vary in ability to release copper ions from metallic copper, but the contribution of cells to copper solubilization and uptake was not investigated (25). Here, we show that E. coli cells strongly increased the amount of copper being released from copper surfaces.
Copper accumulation within cells on dry copper was extensive and very rapid. However, it could not be determined if this accumulation is the primary cause of lethality or a secondary result caused by compromised membranes, as observed by membrane integrity staining. Clearly, membranes are damaged in cells exposed to copper surfaces. Membrane damage has been observed before (1), but the authors used differential staining techniques only to differentiate live from dead (lethally damaged) cells and did not make the connection to the possibility of membrane damage as the underlying mode of action. Others have used indicators for respiration, such as the fluorescent redox dye 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), to differentiate live (metabolic active) from dead (metabolic inactive) cells (26, 39). However, this is an indirect approach, as it measures cell activity but does not specifically indicate membrane damage. Likewise, an earlier study (2) suggested that E. coli cells embedded in agar overlaid on copper and brass but not stainless steel surfaces were completely disrupted after 24 h as indicated by scanning electron microscopy. During the time periods required for contact killing, we did not observe any widespread gross damage, such as blebs or leakage, to cells exposed to metallic copper, as indicated by high-resolution microscopy (data not shown). Nevertheless, physical cell wall stability after contact killing was compromised in Gram-negative and Gram-positive models (Fig. 2) when cells were examined after contact with copper but not with stainless steel.
In our current working model, the mode of action of antimicrobial copper surfaces comprises cytoplasmic membrane damage and weakening of the cell wall. If free copper ions lead to the damage observed, it is likely that they cause a selective change in membranes, as has been described for Saccharomyces cerevisiae (27). In that study, copper ions were shown to release amino acids, mostly glutamate, and much of the cellular K+, suggesting copper ion-mediated selective lesions of the plasma membrane. Similarly, a study investigating oral streptococci indicated that cytoplasmic membrane-bound F1F0-ATPase was damaged by Cu(I) and Cu(II) ions anaerobically (7). Together with the findings of Keevil and coworkers (26, 39), determined using a respiration indicator, these results demonstrate that molecular targets within membranes might be related to respiration and oxidative energy conservation. Nevertheless, this does not explain why a short, 1-min exposure completely kills cells, preventing them from resynthesizing or repairing damaged target proteins after removal from the surfaces and recovering from this stress.
Lack of a role for DNA damage during copper-surface-mediated contact killing.
Previous work has dismissed the possibility that contact killing on metallic copper surfaces causes cellular DNA damage (10, 22), and for cells challenged with copper ions, DNA damage was disproved (18). However, Warnes et al. recently suggested genotoxicity as a mode of action of copper surfaces (37). In this study, we have provided strong evidence that genotoxicity through mutations and DNA lesions is not the underlying cause for the antimicrobial properties of dry metallic copper, and we offer an alternative explanation for the contact killing mechanism: membrane and envelope damage coupled with extensive copper ion accumulation. In contrast, Warnes et al. observed DNA damage after cells were inactivated on copper (37). It is thus likely that the authors did not identify the primary cause of killing but describe a secondary phenomenon that occurred after the onset of cell death or after lethal damage had accumulated. Further, during the course of our experiments we noticed that fluorescence indicator dyes, such as SYBR Gold, lose their fluorescence upon contact with metallic copper (data not shown). As a consequence, we now routinely remove cells from surfaces prior to staining and fluorescence microscopy.
The genome of D. radiodurans, like those of other bacteria, including E. coli, is highly fragmented after exposure to kGy doses of ionizing radiation (12). However, in contrast to sensitive bacteria, D. radiodurans is able to rejoin these overlapping fragments into complete genomes over a period of 3 to 4 h. After repair, cell division commences normally (6). Thus, radioresistance of D. radiodurans is not due to prevention; instead, this organism relies greatly on a variety of efficient DNA repair functions that have less efficient equivalents in almost all species (12). If contact with metallic copper caused destructive DNA damage in cells, then D. radiodurans would be expected to recover from this stress. The opposite was observed. Cells of D. radiodurans were virtually as sensitive to contact killing as E. coli cells on dry copper surfaces, both in exponential and in stationary growth phase (Fig. 6), and were even more sensitive when exposed to moist copper (data not shown). This and the observation that exposure to copper surfaces did not increase the mutation rate in E. coli make DNA very unlikely as a major target of acute lethal metallic copper stress. Similarly, it was found earlier that chronic, long-term exposure to ionic copper (i.e., growth in media supplemented with toxic concentrations of copper salts) neither increased the mutation rate in E. coli nor increased the numbers of DNA lesions. Conversely, copper ions actually protected the DNA from hydrogen peroxide-mediated oxidative damage (18).
In conclusion, this study proposes cell envelope damage as the mode of action of contact killing mediated by dry metallic copper surfaces. The toxicity exerted does not target the genomic DNA in Gram-negative and Gram-positive organisms tested, even though cells are overloaded with copper ions. Further research will be directed at identifying the molecular targets through which membranes are damaged upon contact with metallic copper. It will be interesting to test whether specific membrane proteins or the lipids themselves constitute the weakest link in cells exposed to this lethal challenge.
Acknowledgments
This research was supported by a pilot grant from NIH, grant P20 RR-017675 from the National Center for Research Resources. We also acknowledge funds from the International Copper Association (ICA) and the Copper Development Association (CDA) to G.G. C.E.S. was supported by a Fundação para a Ciência e Tecnologia, Portugal, graduate fellowship. Equipment for this project was purchased with Nebraska Tobacco Settlement Biomedical Research Development funds and start-up funds from the School of Biological Sciences, University of Nebraska—Lincoln.
We thank You (Joe) Zhou and Terri Fangman for skillful technical assistance with microscopy, Javier Seravalli for performing ICP-MS analysis, Paula V. Morais for helpful discussions, and Filipa Sobreira Pires Solavessa Fontes and Maria Inês Simões Brandão for help with the killing assays.
Contents of this work are solely the responsibility of the authors and do not represent the official views of the NIH.
Footnotes
Published ahead of print on 10 December 2010.
REFERENCES
- 1.Airey, P., and J. Verran. 2007. Potential use of copper as a hygienic surface; problems associated with cumulative soiling and cleaning. J. Hosp. Infect. 67:271-277. [DOI] [PubMed] [Google Scholar]
- 2.Borkow, G., and J. Gabbay. 2005. Copper as a biocidal tool. Curr. Med. Chem. 12:2163-2175. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, B. W., et al. 1980. Red-pigmented micrococci: a basis for taxonomy. Int. J. Syst. Bacteriol. 30:627-646. [Google Scholar]
- 4.Casey, A. L., et al. 2010. Role of copper in reducing hospital environment contamination. J. Hosp. Infect. 74:72-77. [DOI] [PubMed] [Google Scholar]
- 5.Chillappagari, S., et al. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192:2512-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, M. M., J. L. Keck, and J. R. Battista. 2010. Rising from the ashes: DNA repair in Deinococcus radiodurans. PLoS Genet. 6:e1000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunning, J. C., Y. Ma, and R. E. Marquis. 1998. Anaerobic killing of oral streptococci by reduced, transition metal cations. Appl. Environ. Microbiol. 64:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elguindi, J., J. Wagner, and C. Rensing. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 106:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espirito Santo, C., P. V. Morais, and G. Grass. 2010. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espirito Santo, C., N. Taudte, D. H. Nies, and G. Grass. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feher, T., B. Cseh, K. Umenhoffer, I. Karcagi, and G. Posfai. 2006. Characterization of cycA mutants of Escherichia coli. An assay for measuring in vivo mutation rates. Mutat. Res. 595:184-190. [DOI] [PubMed] [Google Scholar]
- 12.Gerard, E., E. Jolivet, D. Prieur, and P. Forterre. 2001. DNA protection mechanisms are not involved in the radioresistance of the hyperthermophilic archaea Pyrococcus abyssi and P. furiosus. Mol. Genet. Genomics 266:72-78. [DOI] [PubMed] [Google Scholar]
- 13.Halliwell, B., and J. M. Gutteridge. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halliwell, B., and J. M. Gutteridge. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186:1-85. [DOI] [PubMed] [Google Scholar]
- 15.Helbig, K., C. Bleuel, G. J. Krauss, and D. H. Nies. 2008. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190:5431-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 17.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marais, F., S. Mehtar, and L. Chalkley. 2009. Antimicrobial efficacy of copper touch surfaces in reducing environmental bioburden in a South African community healthcare facility. J. Hosp. Infect. 74:80-82. [DOI] [PubMed] [Google Scholar]
- 20.Mehtar, S., I. Wiid, and S. D. Todorov. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: an in-vitro study. J. Hosp. Infect. 68:45-51. [DOI] [PubMed] [Google Scholar]
- 21.Mergeay, M., et al. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michels, H. T., J. O. Noyce, and C. W. Keevil. 2009. Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett. Appl. Microbiol. 49:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikolay, A., et al. 2010. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl. Microbiol. Biotechnol. 87:1875-1879. [DOI] [PubMed] [Google Scholar]
- 24.Miller, E. W., L. Zeng, D. W. Domaille, and C. J. Chang. 2006. Preparation and use of Coppersensor-1, a synthetic fluorophore for live-cell copper imaging. Nat. Protoc. 1:824-827. [DOI] [PubMed] [Google Scholar]
- 25.Molteni, C., H. K. Abicht, and M. Solioz. 2010. Killing of bacteria by copper surfaces involves dissolved copper. Appl. Environ. Microbiol. 76:4099-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289-297. [DOI] [PubMed] [Google Scholar]
- 27.Ohsumi, Y., K. Kitamoto, and Y. Anraku. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 170:2676-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 29.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Schaeffer, A. B., and M. D. Fulton. 1933. A simplified method of staining endospores. Science 77:194. [DOI] [PubMed] [Google Scholar]
- 32.Singh, N. P., M. T. McCoy, R. R. Tice, and E. L. Schneider. 1988. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175:184-191. [DOI] [PubMed] [Google Scholar]
- 33.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1224. [DOI] [PubMed] [Google Scholar]
- 34.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 35.Sukhi, S. S., R. Shashidhar, S. A. Kumar, and J. R. Bandekar. 2009. Radiation resistance of Deinococcus radiodurans R1 with respect to growth phase. FEMS Microbiol. Lett. 297:49-53. [DOI] [PubMed] [Google Scholar]
- 36.Tkeshelashvili, L. K., T. McBride, K. Spence, and L. A. Loeb. 1991. Mutation spectrum of copper-induced DNA damage. J. Biol. Chem. 266:6401-6406. [PubMed] [Google Scholar]
- 37.Warnes, S. L., S. M. Green, H. T. Michels, and C. W. Keevil. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76:5390-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver, L., H. T. Michels, and C. W. Keevil. 2010. Potential for preventing spread of fungi in air-conditioning systems constructed using copper instead of aluminium. Lett. Appl. Microbiol. 50:18-23. [DOI] [PubMed] [Google Scholar]
- 39.Wilks, S. A., H. Michels, and C. W. Keevil. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 105:445-454. [DOI] [PubMed] [Google Scholar]
- 40.Zeng, L., E. W. Miller, A. Pralle, E. Y. Isacoff, and C. J. Chang. 2006. A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 128:10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]