Abstract
Adenoviruses are resistant to monochromatic, low-pressure (LP) UV disinfection—but have been shown to be susceptible to inactivation by polychromatic, medium-pressure (MP) UV—when assayed using cell culture infectivity. One possible explanation for the difference between UV lamp types is that the additional UV wavelengths emitted by MP UV enable it to cause greater damage to viral proteins than LP UV. The objective of this study was to examine protein damage in adenoviruses treated with LP and MP UV. Results show that MP UV is more effective at damaging viral proteins at high UV doses, though LP UV caused some damage as well. To our knowledge, this study is the first to investigate protein damage in UV-treated adenovirus, and the overview presented here is expected to provide a basis for further, more detailed work.
A significant amount of data has been published on UV inactivation of adenovirus and other viruses using monochromatic low-pressure (LP) UV followed by assays of infectivity using cell culture; these studies have shown adenovirus to be highly resistant to LP UV disinfection (2). Low-pressure UV is understood to inactivate pathogens by damaging their genomes (4, 5). In adenovirus, genomic DNA damage may be repaired in host cells, resulting in its apparent UV resistance. When irradiated with medium-pressure (MP) UV, adenoviruses have been shown to be both more sensitive to inactivation than they are upon irradiation with LP UV and as susceptible to UV inactivation as other viruses, even in standard cell culture infectivity assays (1, 6). Medium-pressure UV is polychromatic—it emits a range of wavelengths in the germicidal portion of the UV emission spectrum (200 to 300 nm) which are absorbed by both DNA and proteins, and so MP UV has the potential to damage adenoviral proteins in addition to the genome. Viral proteins are an integral part of every step in the process of infection and enable adenoviruses to successfully infect host cells even if their DNA is damaged (12). Specific adenoviral proteins and UV damage to proteins have been discussed elsewhere (4, 5, 11, 14). Here we describe a study investigating protein damage in LP and MP UV-treated adenovirus using SDS-PAGE. We hypothesize that MP UV is more effective at causing protein damage than LP UV. This work represents an important first step in this field and will help provide a foundation for further, more detailed work.
Preparation of virus, UV irradiation, and dose calculation were carried out as previously described (1). Three independent UV irradiation experiments were conducted for each UV dose; protein precipitation and SDS-PAGE were done twice for each independent experiment. For protein precipitation, 1 ml of irradiated virus was spiked with aprotinin as an internal standard, pretreated with 0.05% sodium deoxycholate, precipitated with 10% trichloroacetic acid (TCA) (9), and resuspended directly in Laemmli sample buffer (Bio-Rad, Hercules, CA). Standard SDS-PAGE was carried out using 4 to 20% gradient Tris-HCl ReadyGel minigels that were fixed and stained overnight using SYPRO Ruby protein gel stain according to the manufacturer's instructions (Bio-Rad, Hercules, CA). Bands were analyzed for molecular weight and protein quantity by using a GelDoc imager and QuantityOne software (Bio-Rad, Hercules, CA). Adenoviral proteins were identified based on molecular weight (8, 11), and the quantity of protein in each sample was determined relative to that of an untreated control. Statistical analyses were carried out using SPSS software (SPSS Inc., Chicago, IL).
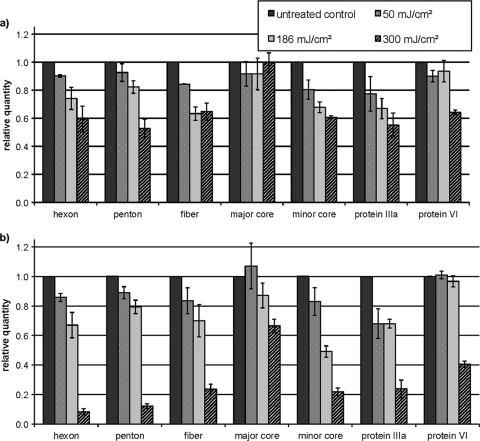
Results are shown in Fig. 1. Seven bands with molecular weights of known adenoviral proteins were identified. Three of these are major capsid proteins, hexon, penton, and fiber proteins; two are minor capsid proteins, protein IIIa and protein VI; and two are core proteins, major core and minor core. Results indicate that MP UV is more effective at damaging adenoviral proteins than LP UV; this is true for all seven viral proteins studied, primarily at a UV dose of 300 mJ/cm2 (Fig. 1). Data (not shown) were similar for a UV dose of 600 mJ/cm2. In general, the major capsid proteins are the most susceptible to UV damage, followed by the minor capsid proteins; the major core protein is least susceptible to MP UV and almost entirely unaffected by LP UV. This suggests that the major core protein may be shielded from UV by the viral DNA within a “nucleosome” structure (7). For MP UV, there is a sharp drop in levels of hexon, penton, and fiber proteins and proteins IIIa and VI between 186 and 300 mJ/cm2, despite the fact that doses up to 186 mJ/cm2 have relatively little effect. This change in kinetics may occur as a result of structural changes in the virus that occur with protein breakdown (9). Preliminary results from our laboratory using transmission electron microscopy suggest that UV does cause structural changes in adenovirus (A. C. Eischeid and K. G. Linden, unpublished results).
FIG. 1.
Relative protein quantities after exposure to LP UV (a) and MP UV (b). Results shown are means ± standard errors of the means.
Analysis of variance (ANOVA) results indicated a significant main effect for lamp (P < 0.0001) and a significant main effect for dose (P < 0.0001); there was no significant interaction (P = 0.653), as both lamps cause decreases in the dependent variable. Post hoc tests for UV dose indicate that the protein damage caused by a 300-mJ/cm2 dose is significantly different from that caused by doses of 25 mJ/cm2 (data for 25 mJ/cm2 were not significantly different from data for 50 mJ/cm2; data not shown) and 50 mJ/cm2 (Fig. 1) (P = 0.002 to 0.019). The levels of protein damage caused by 186-mJ/cm2 and 300-mJ/cm2 doses are also significantly different (P = 0.07).
A few bands that do not correspond directly to known adenoviral proteins were also identified. Unknown bands might result from cross-links, aggregation, protein breakdown during handling, translation at alternative start codons, the presence of incomplete virions containing intermediate protein forms, or natural variation in the viral proteins (3, 9, 10, 13). Some of the smallest bands in these gels likely represent protein VIII and protein IX of adenovirus, but the intensity of these bands was very low and they were not used for analysis.
Acknowledgments
This research was supported by the WateReuse Research Foundation, project no. 06-011, and the National Science Foundation, award no. 0832338. The National Water Research Institute Doctoral Fellowship Program provided partial funding for Anne Eischeid while a Ph.D. student at Duke University.
We thank Jeanette Thurston (USDA, Washington, DC) for training on viral stock preparation, Gwy-Am Shin (University of Washington, Seattle, WA) for providing original stock of adenovirus type 2, and Todd Eischeid (IBM, Research Triangle Park, NC) and Amy Eischeid for help with statistical analyses.
Footnotes
Published ahead of print on 3 December 2010.
REFERENCES
- 1.Eischeid, A. C., J. N. Meyer, and K. G. Linden. 2009. UV disinfection of adenovirus: molecular indications of DNA damage efficiency. Appl. Environ. Microbiol. 75:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eischeid, A. C., J. A. Thurston, and K. G. Linden. UV disinfection of adenovirus: current state of the research and future directions. Crit. Rev. Environ. Sci. Technol., in press.
- 3.Everitt, E., B. Sundquist, U. Petersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52:130-147. [DOI] [PubMed] [Google Scholar]
- 4.Harm, W. 1980. Biological effects of ultraviolet radiation. Cambridge University Press, Cambridge, MA.
- 5.Jagger, J. 1967. Introduction to research in ultraviolet photobiology. Prentice-Hall, Inc., Englewood Cliffs, NJ.
- 6.Linden, K. G., J. Thurston, R. Schaefer, and J. P. Malley. 2007. Enhanced UV inactivation of adenoviruses under polychromatic UV lamps. Appl. Environ. Microbiol. 73:7571-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nermut, M. V. 1979. Structural elements in adenovirus cores: evidence for a “core shell” and linear structures in “relaxed” cores. Arch. Virol. 62:101-116. [DOI] [PubMed] [Google Scholar]
- 8.Phillipson, L. 1984. Structure and assembly of adenoviruses. Curr. Top. Microbiol. Immunol. 109:1-52. [DOI] [PubMed] [Google Scholar]
- 9.Rexroad, J., C. M. Wietoff, A. P. Green, T. D. Kierstead, M. O. Scott, and C. R. Middaugh. 2003. Structural stability of adenovirus type 5. J. Pharm. Sci. 92:665-678. [DOI] [PubMed] [Google Scholar]
- 10.Rosenwirth, B., S. Tjia, M. Westphal, and W. Doerfler. 1974. Incomplete particles of adenovirus. II. Kinetics of formation and polypeptide composition of adenovirus type 2. Virology 60:431-437. [DOI] [PubMed] [Google Scholar]
- 11.Rux, J. R., and R. M. Burnett. 1999. Adenovirus capsid proteins, p. 5-16. In P. Seth (ed.), Adenoviruses: basic biology to gene therapy. R. G. Landes Company, Austin, TX.
- 12.Seth, P. (ed.) 1999. Adenoviruses: basic biology to gene therapy. R. G. Landes Company, Austin, TX.
- 13.Van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vellinga, J., S. van der Heijdt, and R. C. Hoeben. 2005. The adenovirus capsid: major progress in minor proteins. J. Gen. Virol. 86:1581-1588. [DOI] [PubMed] [Google Scholar]