Abstract
Anaerobic ammonium-oxidizing (anammox) bacteria have been recognized as an important sink for fixed nitrogen and are detected in many natural environments. However, their presence in terrestrial ecosystems has long been overlooked, and their contribution to the nitrogen cycling in natural and agricultural soils is currently unknown. Here we describe the enrichment and characterization of anammox bacteria from a nitrogen-loaded peat soil. After 8 months of incubation with the natural surface water of the sampling site and increasing ammonium and nitrite concentrations, anammox cells constituted 40 to 50% of the enrichment culture. The two dominant anammox phylotypes were affiliated with “Candidatus Jettenia asiatica” and “Candidatus Brocadia fulgida.” The enrichment culture converted NH4+ and NO2− to N2 with the previously reported stoichiometry (1:1.27) and had a maximum specific anaerobic ammonium oxidation rate of 0.94 mmol NH4+·g (dry weight)−1·h−1 at pH 7.1 and 32°C. The diagnostic anammox-specific lipids were detected at a concentration of 650 ng·g (dry weight)−1, and pentyl-[3]-ladderane was the most abundant ladderane lipid.
Anaerobic ammonium-oxidizing (anammox) bacteria form a deep-branching, monophyletic group within the planctomycetes and anaerobically oxidize ammonium to dinitrogen gas with nitrite as an electron acceptor (14). They are active at redox transition zones, particularly in oceanic oxygen minimum zones (6, 10, 21, 24) and in other marine ecosystems (3, 8, 9, 22). The discovery of anammox bacteria led to the realization that a substantial part of the nitrogen loss that is observed in the marine environment—up to 50% of the total nitrogen turnover—was due to the activity of these bacteria (2). There are also several studies reporting the presence of anammox bacteria in many freshwater and marine sediments (5, 13, 25, 27, 29). However, little is known to date about the distribution, diversity, and activity of anaerobic ammonium oxidation in terrestrial ecosystems. Only this year, anammox bacteria were detected in permafrost and agricultural soils by a 16S rRNA-based molecular approach (12). Nevertheless, anammox bacteria originating from terrestrial ecosystems have not yet been enriched in the laboratory under controlled conditions, and the physiology of these bacteria is still unknown.
Since anammox bacteria depend on the concomitant presence of nitrate/nitrite (NOx) and ammonium under oxygen-limited conditions, oxic-anoxic interfaces in terrestrial ecosystems should provide a suitable habitat for anammox bacteria. It is believed that in the marine environment, the source of ammonium is mineralization and nitrifiers, or dissimilatory nitrate reduction provides the necessary NO2− and NH4+ (16, 23, 24). In many heavily farmed soil ecosystems, agricultural runoff contributes significantly to the flux of inorganic nitrogen in the form of ammonium and nitrate.
Since the 1960s, anthropogenic nitrogen deposition has led to an increased inorganic nitrogen load in the Netherlands (34). At the sampling site in the Hierdense Beek valley, a swampy peat soil is fed by nitrate-enriched local groundwater, which infiltrates the adjacent pine forest. In the pine forest, atmospheric ammonia, originating from nearby agricultural activities, is intercepted by the canopy and is nitrified in the forest soil. As a result, local groundwater is acidified and strongly enriched in nitrate. This nitrate-enriched groundwater flows underneath the peat toward the “Hierdense Beek” (34). The ammonium-rich peat soil comes into contact with nitrate-rich groundwater, and oxygen is rapidly depleted in these organic soils, creating a perfect environment for anammox bacteria.
The aim of the present study was to cultivate bacteria from peat soil and to describe these soil-derived anammox bacteria. In order to enrich microorganisms indigenous to the peaty soil, the in situ niche should be mimicked as closely as possible in the laboratory (18). To this end, we used a sequencing batch reactor (SBR) that provides the biomass retention that is essential for the cultivation of slow-growing bacteria (35). The SBR system, combined with a “standardized” medium (38) rich in inorganic nutrients, was used in previous studies describing enrichment cultures of freshwater anammox bacteria (17, 20, 35). However, the attempts to grow marine anammox species were successful only when not only the salinity but also the micronutrient concentrations of the marine environment were simulated by the inclusion of Red Sea salt (40). Here, with a similar approach, we used the in situ surface water to re-create the field-like conditions that would most suit the anammox bacteria from the soil ecosystem.
After 6 months of enrichment, the culture consisted of 40 to 50% anammox bacteria affiliated with two previously unknown species, contained the diagnostic ladderane lipids, and could oxidize ammonium at a rate of 0.94 mmol NH4+·g (dry weight)−1·h−1. The enrichment culture described here presents the first insights into the physiology of anammox bacteria detected in terrestrial environments.
MATERIALS AND METHODS
Sampling site.
The sampling site is located in the Hierdense Beek, Landgoed Staverden, Netherlands (52°16′N, 5°44′E). Samples were taken at 0.1 m, 1 m, and 2 m with a corer. The nitrate and ammonium concentrations, pHs, and organic contents of the samples were measured as described previously (37).
Operation of the bioreactor.
An SBR (working volume, 5 liters) was used for the enrichment and cultivation of anammox bacteria from soil (35). In order to prepare the influent medium, surface water was sterilized by hemofiltration (Fresenius Medical Care, Bad Homburg, Germany). Each SBR cycle consisted of 11 h of filling, 45 min of biomass settling, and 15 min of drawing of the liquid. During each filling period, 0.5 liter of the surface water originating from the sampling site, containing nitrite, ammonium, and nitrate (concentrations are specified in Results), was added continuously to the reactor at a flow rate of 0.5 ml·min−1. To maintain anoxic conditions, the reactor and the medium vessel were flushed continuously with Ar-CO2 (95:5; 10 ml·min−1). The SBR was stirred at 200 rpm and was operated at room temperature (20°C). The pH in the SBR was kept at 7.0 via the CO2 present in the supplied gas and an automated pH control unit supplying KHCO3.
Anammox activity assays.
Biomass (40 ml) from the SBR was transferred to 60-ml serum bottles. The biomass was not washed, because a washing step would have reduced the activity of the anammox bacteria considerably. Bottles were sealed with 5-mm-radius butyl rubber stoppers and were made anoxic by alternately applying underpressure and Ar-CO2 gas at least 7 times. An overpressure of 105 Pa was maintained in the bottles. Soluble substrates were added to the bottles from 10 mM anoxic stock solutions. To measure anaerobic ammonium oxidation activity, final concentrations of 250 μM, 500 μM, or 1,000 μM nitrite were used. Ammonium was not added to the incubated cultures because it was in excess in the reactor. Bottles were incubated at pH 7 and 20°C and were shaken continuously at 200 rpm for 1 day. The activity assays with 15N-labeled nitrogen compounds were conducted as described above except that 15NH4+ and 15NO2− were used at concentrations of 250 μM, 500 μM, or 1,000 μM (99% pure; purchased as chloride and sodium salts, respectively, from Cambridge Isotope Laboratories Inc., Andover, MA).
Optimum temperature and pH assays.
For the optimum temperature analysis, a range of 15 to 35°C with a 5°C increment was used. For the optimum pH determination, HEPES buffer in combination with HCl or NaOH was used to adjust the pH to a range of 6.5 to 8.5 (with 0.5-pH-unit increments). Final concentrations of 250 μM ammonium and nitrite were used in all tests, and the incubations were conducted as described above. The pH of each sample was measured again at the end of the incubation to determine any changes. A highly enriched culture of “Candidatus Brocadia fulgida” was used as a positive control in the assays (19).
Analytical methods.
In the activity tests, nitrite and ammonium were measured as described by Kartal et al. in 2006 (15). The isotopic compositions of dinitrogen gas and nitrous oxide were determined with a gas chromatograph (Agilent 6890) coupled to a quadruple inert mass spectrometer (Agilent 5975c). The total solids (TS) were quantified by drying at 105°C and weighing after cooling in a desiccator.
DNA extraction and PCR.
The original soil sample (1 g) or the biomass from the SBR (1.5 ml) was centrifuged. DNA was extracted with a PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions. DNA was then dissolved in 50 μl ultrapure water and was kept at 4°C until further analysis.
The primer combination Amx368F-Amx820R was used for the preferential amplification of the 16S rRNA genes of anammox bacteria (31). PCR fragments were cloned directly using the pGEM-T Easy cloning kit (Promega, Leiden, Netherlands) according to the manufacturer's instructions. Plasmid DNA was isolated and purified with the GeneJET plasmid miniprep kit (Fermentas, Burlington, Canada). Plasmids were digested with 5 U EcoRI enzyme in EcoRI buffer for 1.5 h at 37°C. The digestion products were examined for an insert with the expected size by agarose gel (1%) electrophoresis.
Sequencing and phylogenetic analysis.
The sequences of the 16S rRNA gene fragments were determined by using the M13 forward primer, the M13 reverse primer, and an internal primer targeting vector sequences adjacent to the multiple cloning sites. Phylogenetic analyses were performed with the neighbor-joining method using the Tamura-3 algorithm and the pairwise deletion substitution model (36) and were tested by bootstrap analysis with 3,000 replications.
Fluorescence in situ hybridization (FISH).
Biomass (1.5 ml) was harvested from the enrichment culture and was fixed in paraformaldehyde, and hybridizations with fluorescent probes were performed as described previously (31). All probes were purchased as Cy3-, Cy5-, and 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (Fluos)-labeled derivatives from Thermo Electron Corporation (Ulm, Germany). The following probes were used to monitor the enrichment of the anammox population as described by Schmid et al. (31): Amx 368 (S-*-Amx-0368-a-A-18; specific for all known anammox genera [31]), Amx 820 (S-*-Amx-0820-a-A-22; specific for “Candidatus Kuenenia stuttgartiensis” and “Candidatus Brocadia anammoxidans” [30]), EUB 338 (S-D-Bact-0338-a-A-18 [1]), EUB 338 II (S-D-Bact-0338-b-A-18), EUB 338 III (S-D-Bact-0338-c-A-18; together with EUB and EUBII, specific for most bacteria [4]), and Pla46 (S-P-Planc-0046-a-A-18; specific for Planctomycetales [26]).
Lipid analysis.
Biomass (50 ml) was harvested from the SBR and was freeze-dried. It was extracted by a modified Bligh-Dyer method according to the work of Rattray et al. (28). The sample was ultrasonically extracted for 15 min using methanol-dichloromethane (DCM)-phosphate buffer (pH 7.4) at a ratio of 2:1:0.8 (vol/vol/vol). The supernatant was collected, and the residue was reextracted ultrasonically twice. The solvent ratio of the combined supernatants was adjusted to 1:1:0.9 (vol/vol/vol) methanol-dichloromethane-phosphate buffer, and the supernatants were centrifuged. The bottom DCM layer was collected, and the remaining solvent was reextracted twice with DCM. The DCM layers were combined and dried under a rotary evaporator. The extract was then eluted over Na2SO4 and was dried under N2.
An aliquot of the Bligh-Dyer extract was separated over a small column of activated silica (60 mesh; activated for 3 h at 130°C and cooled in a desiccator to room temperature). The aliquot was eluted 3 times with 2:1 (vol/vol) ethyl acetate-hexane and then 3 times with methanol in order to separate the anammox ladderane lipids (methanol fraction) from the soil material (ethyl acetate-hexane fraction). The methanol fraction was saponified by refluxing with aqueous KOH (in 96% methanol) at 100°C for 1 h. Fatty acids were obtained by acidification of the sample to a pH of 3 with 1 N HCl in methanol, followed by extraction using DCM. The fatty acids were converted to their corresponding fatty acid methyl esters (FAMEs) by methylation with diazomethane (CH2N2). Excess CH2N2 was removed by evaporation under N2. Polyunsaturated fatty acids (PUFAs) were removed by elution of the sample over a small AgNO3 (5%)-impregnated silica column with DCM.
The fatty acid fraction was dissolved in acetone, filtered through a 0.45-μm-pore-size, 4-mm-diameter polytetrafluoroethylene (PTFE) filter, and analyzed by high-performance liquid chromatography coupled to positive-ion atmospheric pressure chemical ionization tandem mass spectrometry (HPLC-APCI-MS-MS) in selective reaction monitoring (SRM) mode as described by Hopmans et al. (11) and modified by Rattray et al. (28). Ladderane lipids were quantified using an external calibration curve of standards of isolated methylated ladderane fatty acids containing the [3]- and [5]-ladderane moieties (11, 28, 33). A detection limit of 30 to 35 pg of the injected ladderane lipid was achieved with this technique.
Nucleotide sequence accession numbers.
The sequences determined at the time of inoculation have been deposited in GenBank under accession numbers HQ637487 (cluster 1), HQ637488 (cluster 2), and HQ637489 (cluster 3). Those determined after 8 months of incubation have been deposited in GenBank under accession numbers HQ610833 (cluster 1) and HQ610834 (cluster 2).
RESULTS
Sampling site.
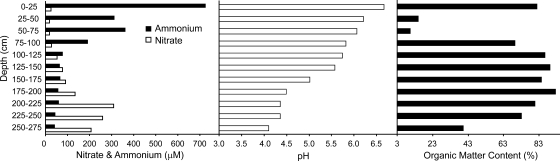
The nitrate and ammonium profiles of the peat soil at the Hierdense Beek valley showed that these nitrogen species co-occurred between 1 and 2 m of the depth profile. The measurements also suggested that there was significant consumption of nitrate and ammonium at these sampling points (Fig. 1). Throughout the depth profile, the O2 concentration was below the detection limit (15 μM).
FIG. 1.
Nitrate and ammonium concentration, pH, and organic matter content profiles of the sampling site.
Screening of the inoculum.
Five samples were taken from Hierdense Beek at two locations and three depths (Table 1). Samples HB1, HB4, and HB5 were taken from the peat soil and samples HB2 and HB3 from the adjacent grassland. PCR amplification with the combination of primer Pla46F and the universal reverse primer 630R (planctomycete specific) did not result in any sequences affiliated with anaerobic ammonium-oxidizing (anammox) bacteria. When the primer set 368F-820R (specific for anammox bacteria) was used, three sequence clusters affiliated with anammox bacteria could be detected in samples HB1, HB4, and HB5. All three clusters were detected in sample HB5; thus, it was decided that sample HB5 would be suitable for inoculating a bioreactor for the enrichment of soil anammox bacteria.
TABLE 1.
Nitrate and ammonium concentrations, depth, and pH of the samples
| Location | Sample site | Concn (μM) of: |
Depth (m) | pH | |
|---|---|---|---|---|---|
| Nitrate | Ammonium | ||||
| HB1 | Peat soil | 12.9 | 740 | 0.1 | 6.8 |
| HB2 | Dry grassland | 14 | 8 | 2 | 4.5 |
| HB3 | Dry grassland | 9 | 7.5 | 1 | 5.8 |
| HB4 | Peat soil | 220 | 1.4 | 2 | 4.6 |
| HB5 | Peat soil | 603 | 43.4 | 1 | 5.8 |
Enrichment of anammox bacteria.
A sequencing batch reactor (SBR) with a working volume of 5 liters was inoculated with 1 kg (wet weight) of soil homogenized with surface water from Hierdense Beek (soil/surface water ratio, 1:4). The influent medium was prepared by filtering surface water from Hierdense Beek with a hemofilter (Fresenius Medical Care, Bad Homburg, Germany). Initially, the medium was supplemented with 0.5 mM nitrite and 1 mM ammonium and nitrate. The substrate concentrations were gradually increased to 4 mM. The effluent concentration of nitrite was always below the detection limit (10 μM). The population diversity of the enrichment culture and the abundance of anammox bacteria were monitored by FISH and 16S rRNA gene clone libraries.
Activity tests.
The anammox activity of the enrichment culture was tested every 2 months during the operation of the reactor. Anammox activity was detected in the batch incubations for the first time after 4 months. This activity increased with a doubling time of 27 days in the following 4 months and reached a pseudo-steady state. The highest specific activity for the enrichment culture was 0.94 (± 0.04) mmol NH4+·h−1·g (dry weight)−1 with a stoichiometry of 1:1.27 (NO2− to NH4+). The optimal temperature and pH of the culture were 32°C and pH 7.1, respectively. These parameters were different from those for the control culture (∼80% enriched “Candidatus Brocadia fulgida” [19]), which had maximum specific activity at 30°C and pH 7.8 with a rate of 9 mmol NH4+·h−1·g (dry weight)−1.
16S rRNA analysis and FISH microscopy.
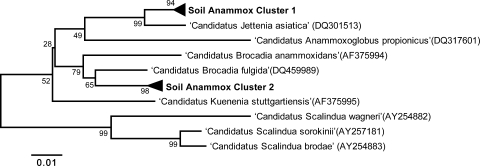
In order to determine the species of anammox bacteria present in the enrichment culture, we applied our anammox-specific 16S rRNA sequence approach. DNA extraction, PCR amplification (with the Amx368F-Amx820R primer set), cloning, and sequencing were conducted at the time of inoculation and after 8 months. Twenty clones were randomly selected for sequencing at each sampling point. Phylogenetic analysis of the clones at the time of inoculation revealed three detectable genera in the Hierdense Beek valley, in contrast to marine sediments, where one genus has been detected to date (see Fig. S1 in the supplemental material). At the end of 8 months, when a pseudo-steady state was reached, two anammox phylotypes, each represented with 9 out of 19 sequences, could be detected in the clone libraries. These clusters (clusters 1 and 2) were unique groups with only 96% sequence similarity to the closest known anammox species, “Candidatus Jettenia asiatica” and “Candidatus Brocadia fulgida,” respectively (Fig. 2).
FIG. 2.
Phylogenetic tree depicting the 16S rRNA gene sequence diversity of the anammox species in the soil enrichment culture after 8 months and the relationship of these sequences to known anammox bacteria.
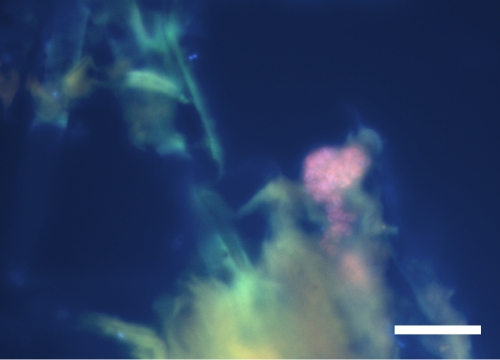
The enriched anammox bacteria could also be visualized by FISH microscopy. Anammox clusters were identified after 6 months of enrichment with the Amx820 probe, specific for “Candidatus Brocadia”-, Candidatus Jettenia”-, and Candidatus Kuenenia”-like bacteria (Fig. 3).
FIG. 3.
Fluorescence in situ hybridization micrograph of the anammox enrichment culture after 6 months. The anammox bacteria were hybridized with the Amx820 (Cy3) (red) and EUB I, II, and III (Fluos) (green) probes and with the DNA stain DAPI; they appear pink. Bar, 10 μm.
Lipid analysis.
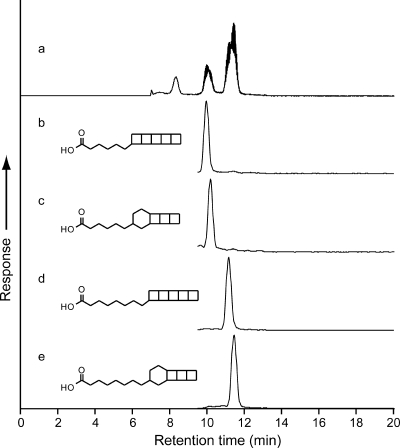
Four ladderane fatty acids (as fatty acid methyl esters [FAMEs]) were analyzed by high-performance liquid chromatography coupled to positive-ion atmospheric pressure chemical ionization tandem mass spectrometry (HPLC-APCI-MS-MS). All scanned ladderane fatty acids were detected in the enrichment culture (Fig. 4b to e). The dominant ladderane fatty acid in the culture was the pentyl-[3]-ladderane FAME (Fig. 4e), which constituted 46% of the total ladderane lipid concentration. The three remaining ladderane fatty acids, the pentyl-[5]-, heptyl-[3]-, and heptyl-[5]-ladderane FAMEs, contributed 14%, 11%, and 29%, respectively. The combined total concentration of ladderane fatty acids in the soil culture was 650 ng·g (dry weight)−1.
FIG. 4.
Base peak chromatogram (a) and SRM traces (b, c, d, and e) of 4 ladderane FAMEs obtained by HPLC-APCI-MS-MS analysis of freeze-dried soil biomass. Traces b, c, d, and e show the pentyl-[5]-, pentyl-[3]-, heptyl-[5]-, and heptyl-[3]-ladderane FAMEs, respectively.
DISCUSSION
The surroundings of the Hierdense Beek valley, Netherlands, have been strongly enriched in anthropogenic nitrogen for more than 40 years due to agricultural activity. Since the 1980s, part of the grasslands in the valley has become progressively wetter, resulting in the development of a swampy peat soil. This can be attributed to the anaerobic decomposition of the deeper peat layer, most likely because the groundwater is enriched in nitrate, which serves as an alternative electron acceptor (34).
For the sampling site, anammox bacteria were detected by PCR cloning in three of the five samples (HB1, -4, and -5) taken from different depths and showed high diversity (3 distinct species) for such a limited set of samples. It is most likely that separate microniches existed in the heterogeneous soil samples, suggesting that the terrestrial ecosystems harbored different anammox species, in contrast to the ubiquitous (and almost exclusive) presence of the “Candidatus Scalindua” genus in the oceans (7, 24, 27, 32). The two samples (HB2 and -3) in which anammox bacteria could not be detected were from a dry grassland in the sampling site, where nitrate/nitrite concentrations in the groundwater were much lower (<10 μM). Nevertheless, the possibility that primer bias may account for the negative result cannot be ruled out.
Our past studies showed that the inorganic medium described by van de Graaf et al. was suitable for the enrichment of wastewater anammox species but was most probably inhibitory to the species originating from natural ecosystems that were adapted for survival under conditions of nutrient limitation (38, 40). In order to enrich the indigenous anammox bacteria, the in situ surface water (hemofilter sterilized) was used as the influent medium, with low initial nitrite and ammonium concentrations (0.5 mM). This resulted in a slow but steady increase in the consumption of the substrates in the reactor. The first detection of anammox activity in batch incubations was followed by an increase in the anaerobic ammonium oxidation rate, and the responsible bacteria could be visualized by FISH microscopy.
The first anammox cells that could be detected by FISH were loosely attached (data not shown). After 6 months of enrichment, as observed for previous enrichment cultures (17, 31, 40), the anammox bacteria were present in tightly packed clusters and constituted about 40 to 50% of the population detectable by FISH microscopy and counterstaining with the DNA dye 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 3). The cells detected seemed to be attached to soil particles, which could facilitate better settling, necessary for growth in a sequencing batch reactor (Fig. 3).
In contrast to the previous reports that show one anammox strain per enrichment culture, there were two clusters of anammox bacteria in the enrichment culture after 8 months (Fig. 2). Neither of these clusters corresponded to the anammox species that have been described. Phylogenetic analysis based on the 16S rRNA gene showed that these clusters had the highest similarities (96%) to “Candidatus Jettenia asiatica” and “Candidatus Brocadia fulgida.” The niche differentiation of anammox bacteria could be defined by micronutrient concentrations or the inclusion of an alternative energy source (16, 17, 40). Compared to the almost exclusively used activated-sludge inoculum, seeding material from the soil environment most likely contained more difficult-to-degrade and slowly released organic compounds (e.g., humic acids). It has been reported that the endogenous electron donors in soils could take as long as several months to be completely degraded anaerobically (39). In the current enrichment culture, there were probably still enough soil-derived compounds to allow the presence of two groups of anammox bacteria (39). Because the enrichment culture was fed with in situ surface water, the compound(s) that affected the selection of a certain species might be constantly supplied to the enrichment culture, leading to the enrichment of two different species.
The biomass from the enrichment culture had a substrate (NO2− and NH4+) conversion stoichiometry of 1:1.27, which was almost identical to the previously reported value, 1:1.3 (38). The highest anaerobic ammonium-oxidizing activity occurred at 32°C and pH 7. The optimum pH for the activity reflected the operational conditions of the bioreactor. The maximum specific activity of the biomass from the reactor was 0.94 (± 0.04) mmol NH4+·h−1·g (dry weight)−1. This lower specific activity (approximately 10% that of the control) could be an underrepresentation due to the large amount of nonbiological particles present in the enrichment culture as a result of the very long solids retention time (SRT) required for the enrichment of anammox bacteria.
The long SRT also led to a high abundance of soil- and plant-derived lipids in the fatty acid fraction isolated from the bioreactor. Once these lipids were removed, the ladderane lipids, so far a unique biomarker for anammox bacteria (33), could be detected in relatively high abundances in the enrichment culture. The concentrations of ladderane lipids were lower than those previously reported in highly enriched cultures (75 to 90% [28]), most likely due to the contribution of nonbiological particles to the dry weight measurements. C20 laddrane fatty acids dominated over shorter-chain C18 fatty acids, a lipid distribution comparable to that found in other anammox enrichment cultures. The most abundant lipid type was the C20 fatty acid with 3 cyclobutane rings and 1 cyclohexane ring, as in the previously described enrichment culture of “Candidatus Scalindua”-like bacteria.
In this study, two previously unknown anammox species were enriched from a soil deposited with anthropogenic nitrogen. The physiological parameters and substrate conversion stoichiometry conformed to those for the previously described species. The soil enrichment culture also contained the diagnostic ladderane lipids unique to anammox bacteria, in particular the C20[3] fatty acid. The enrichment of an anammox species from the terrestrial ecosystem is a first step toward better understanding of the contribution of anammox bacteria to nitrogen cycling in natural and agricultural soils.
Supplementary Material
Acknowledgments
This research was financially supported by the Science Foundation of Zhe-jiang Province (Y507227) and the Chinese Universities Scientific Fund (2009QNA6009). M.S.M.J. is supported by ERC advanced grant 232937.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 10 December 2010.
REFERENCES
- 1.Amann, R. I., et al. 1990. Combination of 16S ribosomal RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo, K. R. 2005. Marine microorganisms and global nutrient cycles. Nature 437:349-355. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, N., et al. 2009. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 3:117-123. [DOI] [PubMed] [Google Scholar]
- 4.Daims, H., A. Bruehl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 5.Dale, O. R., C. R. Tobias, and B. K. Song. 2009. Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River Estuary. Environ. Microbiol. 11:1194-1207. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard, T., D. E. Canfield, J. Petersen, B. Thamdrup, and J. Acuna-Gonzalez. 2003. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422:606-608. [DOI] [PubMed] [Google Scholar]
- 7.Engstrom, P., T. Dalsgaard, S. Hulth, and R. C. Aller. 2005. Anaerobic ammonium oxidation by nitrite (anammox): implications for N2 production in coastal marine sediments. Geochim. Cosmochim. Acta 69:2057-2065. [Google Scholar]
- 8.Engstrom, P., C. R. Penton, and A. H. Devol. 2009. Anaerobic ammonium oxidation in deep-sea sediments off the Washington margin. Limnol. Oceanogr. 54:1643-1652. [Google Scholar]
- 9.Glud, R. N., et al. 2009. Nitrogen cycling in a deep ocean margin sediment (Sagami Bay, Japan). Limnol. Oceanogr. 54:723-734. [Google Scholar]
- 10.Hamersley, M. R., et al. 2007. Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol. Oceanogr. 52:923-933. [Google Scholar]
- 11.Hopmans, E. C., et al. 2006. Improved analysis of ladderane lipids in biomass and sediments using high-performance liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20:2099-2103. [DOI] [PubMed] [Google Scholar]
- 12.Humbert, S., et al. 2010. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 4:450-454. [DOI] [PubMed] [Google Scholar]
- 13.Jaeschke, A., et al. 2009. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiol. Ecol. 67:343-350. [DOI] [PubMed] [Google Scholar]
- 14.Jetten, M. S. M., H. J. M. Op den Camp, G. J. Kuenen, and M. Strous. 2010. Family I. “Candidatus Brocadiaceae” fam. nov., p. 596-602. In N. R. Krieg, J. T. Staley, D. R. Brown, B. Hedlund, B. J. Paster, N. Ward, W. Ludwig, and W. B. Whitman (ed.), Bergey's manual of systematic bacteriology, vol. 4. Springer, New York, NY. [Google Scholar]
- 15.Kartal, B., et al. 2006. Adaptation of a freshwater anammox population to high salinity wastewater. J. Biotechnol. 126:546-553. [DOI] [PubMed] [Google Scholar]
- 16.Kartal, B., et al. 2007. Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 9:635-642. [DOI] [PubMed] [Google Scholar]
- 17.Kartal, B., et al. 2007. Candidatus “Anammoxoglobus propionicus,” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30:39-49. [DOI] [PubMed] [Google Scholar]
- 18.Kartal, B., and M. Strous. 2008. Methods to study consortia and mixed cultures, p. 205-219. In K. Zengler (ed.), Accessing uncultivated microorganisms. ASM Press, Washington, DC.
- 19.Kartal, B., et al. 2010. Effect of nitric oxide on anammox bacteria. Appl. Environ. Microbiol. 76:6304-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kartal, B., et al. 2008. Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 63:46-55. [DOI] [PubMed] [Google Scholar]
- 21.Kuypers, M. M. M., et al. 2005. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl. Acad. Sci. U. S. A. 102:6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuypers, M. M. M., et al. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608-611. [DOI] [PubMed] [Google Scholar]
- 23.Lam, P., et al. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam, P., et al. 2009. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U. S. A. 106:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, R. L., N. Risgaard-Petersen, and D. E. Allen. 2005. Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl. Environ. Microbiol. 71:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neef, A., R. Amann, H. Schlesner, and K.-H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 27.Penton, C. R., A. H. Devol, and J. M. Tiedje. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72:6829-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rattray, J. E., et al. 2008. Ladderane lipid distribution in four genera of anammox bacteria. Arch. Microbiol. 190:51-66. [DOI] [PubMed] [Google Scholar]
- 29.Rich, J. J., O. R. Dale, B. Song, and B. B. Ward. 2008. Anaerobic ammonium oxidation (anammox) in Chesapeake Bay sediments. Microb. Ecol. 55:311-320. [DOI] [PubMed] [Google Scholar]
- 30.Schmid, M., et al. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 31.Schmid, M., et al. 2003. Candidatus “Scalindua brodae,” sp. nov., Candidatus “Scalindua wagneri,” sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 32.Schmid, M. C., et al. 2007. Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 9:1476-1484. [DOI] [PubMed] [Google Scholar]
- 33.Sinninghe Damsté, J. S., et al. 2002. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature 419:708-712. [DOI] [PubMed] [Google Scholar]
- 34.Smolders, A. J. P., E. C. H. E. T. Lucassen, R. Bobbink, J. G. M. Roelofs, and L. P. M. Lamers. 2010. How nitrate leaching from agricultural lands provokes phosphate eutrophication in groundwater fed wetlands: the sulphur bridge. Biogeochemistry 98:1-7. [Google Scholar]
- 35.Strous, M., J. J. Heijnen, J. G. Kuenen, and M. S. M. Jetten. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50:589-596. [Google Scholar]
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Tomassen, H. B. M., A. J. P. Smolders, J. Limpens, L. P. M. Lamers, and J. G. M. Roelofs. 2004. Expansion of invasive species on ombrotrophic bogs: desiccation or high N deposition? J. Appl. Ecol. 41:139-150. [Google Scholar]
- 38.van de Graaf, A. A., P. de Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1996. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142:2187-2196. [Google Scholar]
- 39.Van Den Heuvel, R. N., E. Van Der Biezen, M. S. Jetten, M. M. Hefting, and B. Kartal. 2010. Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 12:3264-3271. [DOI] [PubMed] [Google Scholar]
- 40.van de Vossenberg, J., et al. 2008. Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environ. Microbiol. 10:3120-3129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.