Abstract
Phylogeny-based analysis of chitinase and 16S rRNA genes from metagenomic data suggests that salinity is a major driver for the distribution of both chitinolytic and total bacterial communities in aquatic systems. Additionally, more acidic chitinase proteins were observed with increasing salinity. Congruent habitat separation was further observed for both genes according to latitude and proximity to the coastline. However, comparison of chitinase and 16S rRNA genes extracted from different geographic locations showed little congruence in distribution. There was no indication that dispersal limited the global distribution of either gene.
In recent years, progress has been made in elucidating factors that underlie the biogeography of bacterial community assemblages (5, 13, 23, 25). However, function cannot easily be inferred from such studies, since specific functional guilds may be present in low abundances and function is difficult to predict from taxonomic affiliations. To target and analyze the diversity of functional groups, biogeographical studies of protein coding genes is an attractive alternative to studies based on taxonomy markers (e.g., see references 10, 13, 16, 28, and 35). By using a phylogeny-based approach to assess the biogeography of functional guilds, information regarding underlying ecological and evolutionary community assembly processes can be accessed (16).
Marine chitinolytic communities play key roles in global nitrogen and carbon cycles. Chitinases are responsible for the hydrolysis of chitin, which is one of the most abundant biopolymers on earth. It does not accumulate over time and therefore likely has high turnover rates (12, 19). Bacterial degradation of chitin usually involves an initial extracellular hydrolysis of the (1→4)-β-linkage, catalyzed by excreted glycoside hydrolase (GH) family 18 chitinases, which are phylogenetically subdivided into groups A, B, and C. Lateral gene transfer has been discussed as a reason for their widespread but phylogenetically incoherent distribution among bacterial phyla (17). This implies that populations carrying this function may vary greatly in other ecological traits. Environmental factors could consequently act directly on an organism's ability to degrade chitin, independently of the taxonomic background of the organism carrying this trait.
In this study, we used metagenomic data provided by the Global Ocean Sampling expedition (GOS) (32, 33) to test if specific environmental conditions in aquatic habitats shape the chitinase gene assemblages or if limitations in dispersal are important for the distribution of chitinolytic communities. The phylogeography of chitinase genes was also compared to that of the 16S rRNA gene as a universal taxonomic marker. Divergent distribution patterns between the two genes would indicate if there are environmental factors or distribution mechanisms that are specifically relevant for chitinolytic microorganisms. The respective communities were compared among habitats defined by either salinity, latitude, proximity to the coastline, or geographic location.
Sequence extraction and phylogenetic evaluation.
All GOS sequences (32) were retrieved from Cyberinfrastructure for Advanced Microbial Research and Analysis (CAMERA) (33) using the Basic Local Alignment Search Tool (BLAST) (2). The GOS data (all open reading frames [ORFs]) were queried for chitinase genes using short conserved chitinase amino acid sequences (maximum of 24 amino acids) found in 122 genomes (17). All query sequences include the conserved GH family 18 DxxDxDxE motif, which is part of the catalytic domain and corresponds to the following amino acid positions: Serratia marcescens/ChiB (BAA31568), 124 to 148 (group A chitinases); and S. marcescens/ChiC (ABI79318), 124 to 142 (group B chitinases). Parallel tBLASTN searches were performed using the BLOSUM62 matrix (word size 3) and the PAM30 matrix (word size 2). In a second search, 992 chitinase sequences from uncultured bacteria were used as query sequences in a BLASTN search using default settings. All obtained hits were translated to their protein sequence and checked for homology to chitinase genes using a BLASTP search. GOS 16S rRNA gene sequences were extracted using the universal bacterial primer eub27f (21) (BLASTN search, default settings, all metagenomic sequence reads). Hits were confirmed as 16S rRNA genes using the RDP classifier (36) with the confidence threshold set to 95%. We reduced the original 16S rRNA gene set by randomly sampling up to 10 sequences (depending on the number of available reads) with a minimum length of 300 bp from each GOS sampling site with chitinase hits to obtain comparable data set sizes for the two genes. Since the hypersaline and freshwater habitats both consisted of a single site, 30 sequences were sampled from these two sites.
A seed alignment for chitinase genes was created with sequences from available microbial genomes using the MAFFT v6.626 software program (homolog option) (18). The alignment was refined manually using the secondary structure of a chitinase from Serratia marcescens (Protein Data Bank [PDB] entry 1E15). All sequences were then aligned to the seed alignment using HMMer v2.3 software (9), and the complete alignment was manually corrected for errors. Regions of poor alignment quality were excluded from subsequent analyses. 16S rRNA sequences were imported into the ARB software package (26) and aligned using the ARB integrated aligner, followed by manual curation with the SILVA 100 database as a reference (30). Maximum-likelihood estimation of phylogenies was performed using the GARLI v.0.96b8 software program (38). Selection of the most appropriate substitution models was performed using the Prottest software program (1) for chitinase amino acid and the jModelTest software program (29) for 16S rRNA nucleotide sequences. The WAG+I+Γ model was used for analysis of the chitinase alignment, while the 16S rRNA alignment was analyzed using the GTR+I+Γ substitution model. For each gene, 10 tree search replicates were performed, and the tree with the highest log likelihood score was selected as the final topology. Confidence values at nodes were determined using 500 bootstrap replicates. For the chitinase phylogeny, sequences retrieved from the screening that did not cluster with either of the main groups (A, B, or C) were pruned from the final phylogeny, and the tree was rooted at sequences from chitinase groups B and C. The 16S rRNA gene tree was rooted at archaeal sequences.
We extracted 296 putative chitinase genes from the GOS database (see Fig. S1 and Table S1 in the supplemental material) and detected putative chitinase genes in 58 of the 82 sites. 16S rRNA gene sequences from all major phyla known to inhabit aquatic environments were retrieved from the data set, and the number of sequences per site included in phylogenetic analyses is given in the supplemental material (see Tables S1 and S3).
According to an approach described elsewhere (4, 14) and assuming five chitinase genes per cell (7), we estimated that between 0.2 and 5.8% of total bacterial cells were chitinolytic (see Table S1 in the supplemental material). This agrees with results from a previous metagenomic enzyme screening (7) but contrasts with much higher estimates based on culture-dependent approaches (31). None of the obtained chitinase sequences clustered with group B, and a mere 7 were assigned to group C, whereas the remaining 289 sequences were affiliated with group A (see Fig. S1). Dominance of group A chitinase genes among the GH18 group has been reported before (27, 37), though only in a PCR-based study relying on primers designed from group A sequences (27). Group A chitinase genes are also more frequent than group B or C chitinase genes among cultured organisms (17) (see Fig. S1) but still occur in a far lower proportion than the 98% of the total chitinase pool observed in the GOS data analyzed here. Our study is therefore the first to demonstrate the dominance of group A chitinases with metagenomic data, thereby avoiding bias introduced by either cultivation or PCR. The observed stronger dominance of group A chitinase genes in our study than in cultured chitinolytic organisms could reflect either the composition of chitinase genes specifically in aquatic environments or cultivation bias.
Habitat definition and assessment of diversity coverage.
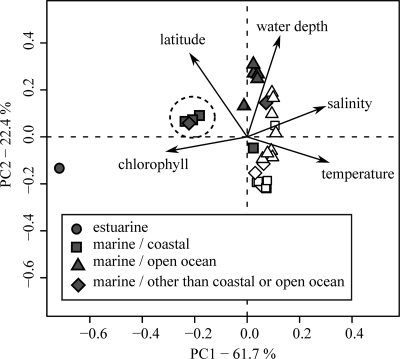
We grouped the obtained sequences into habitats according to the environmental data available for each site. The habitats were grouped into broader categories for subsequent tests on the influence of salinity, latitude, proximity to the coastline, and geographic location (Table 1). To test for hierarchical influence of these broad categories, combinations of categories were used to perform cluster analysis based on UniFrac distance matrices. Combined habitats with fewer than 10 sequences were excluded from the cluster analyses. To assess if sites within the defined habitats share environmental features, contextual data from the GOS expedition (32) were compared with principal components analysis (PCA). Values for latitude were transformed into decimal values, and each parameter was scaled and centered prior to analysis. Sites without contextual data, including the hypersaline and freshwater sites and most of the estuarine sampling sites (see Table S2 in the supplemental material), were excluded from this analysis. Overall, this analysis supported the habitat classification (Fig. 1). Separation of sampling sites was mainly along the second principal component rather than the first, reflecting differences in water depth and latitude. Marine North American East Coast samples grouped separately from the remaining marine samples along the first principal component as a result of differences in chlorophyll concentrations and salinities. This separation was even more apparent for the estuarine North American East Coast sample.
TABLE 1.
Habitats and broader categories included in UniFrac P analysesa
| Category | Habitat | No. of sequences |
|
|---|---|---|---|
| Chitinase genes | 16S rRNA genes | ||
| Salinity | Freshwater | 20 | 30 |
| Estuarine | 14 | 34 | |
| Marine | 166 | 310 | |
| Hypersaline | 19 | 30 | |
| Latitude | Tropic climate zone (23.5°N to 23.5°S) | 112 | 137 |
| Nontropic climate zone (>23.5°N, >23.5°S) | 111 | 166 | |
| Coastal/open ocean | Coastal | 46 | 147 |
| Open ocean | 120 | 156 | |
| Geographic | Caribbean Sea | 20 | 50 |
| locationb | Galapagos Islands | 61 | 88 |
| Indian Ocean | 28 | 95 | |
| North American East Coast | 16 | 54 | |
| Sargasso Sea | 86 | 44 | |
With the exception of the category for geographic location, only coastal and open ocean samples were included to represent marine samples in the analyses. The number of sequences included in each habitat is listed for the chitinase and 16S rRNA gene. These habitats were based only on planktonic samples (filter size, <0.8 μm) of group A chitinases. Habitats with fewer than 10 chitinase genes were excluded from further analyses to avoid spurious results. The habitats were further grouped into categories. Samples from a mangrove environment were grouped together with estuarine samples since satellite pictures from the Google Earth software tool (version 4.3.7204.0836 [beta]) show freshwater intrusion at this site. To avoid the influence of special marine habitats, such as reef samples, only coastal and open ocean samples were included in the respective habitats for salinity, latitude, and proximity to the coastline. However, we included sequences from all marine planktonic samples in habitats within the geographic location category, since too few sequences remained for the single categories.
Only sites with at least 10 chitinase sequences were included.
FIG. 1.
Principal components analysis (PCA) of the individual GOS sampling sites grouped according to habitats based on the available environmental data. Sampling sites with missing parameters were excluded from the PCA. Sampling sites derived from the North American East Coast are clustered as indicated by a circle (filled symbols, subtropical; open symbols, tropical).
The covered diversity represented in both the chitinase and 16S rRNA gene data sets was estimated for each habitat by comparison of rarefaction curves against the phylogenetic diversity (PD) (11). An endpoint PD was estimated by fitting the rarefaction curve to an asymptotic model for species accumulation equivalent to the Michaelis-Menten equation (6). For most habitats, sequences included in the UniFrac analyses exceeded 70% of the total estimated PD. The lowest coverage was observed for habitats classified based on salinity, with a mere 31% for the hypersaline habitat and 50% for estuarine waters for both 16S rRNA and chitinase gene sequences (Table 2).
TABLE 2.
Phylogenetic diversity reached by chitinase and 16S rRNA gene sequences included in habitats used for phylogenetic analyses and expected maximal phylogenetic diversity for the respective habitatsa
| Habitat | Diversity of: |
|||||
|---|---|---|---|---|---|---|
| Chitinase genes |
16S rRNA genes |
|||||
| PDreached | PDmax | % coverage | PDreached | PDmax | % coverage | |
| Freshwater | 7.17 | 7.84 | 91.5 | 551.77 | 866.04 | 63.7 |
| Tropics | 19.64 | 22.96 | 85.5 | 1372.38 | 1452.78 | 94.5 |
| Nontropics | 25.78 | 27.79 | 92.7 | 1468.50 | 1730.13 | 84.9 |
| Coastal | 17.77 | 20.73 | 85.7 | 1468.00 | 1678.59 | 87.5 |
| Open ocean | 27.14 | 28.50 | 95.2 | 1500.22 | 1628.94 | 92.1 |
| Hypersaline | 9.27 | 29.96 | 30.9 | 553.65 | 783.61 | 70.7 |
| Marine | 33.82 | 34.90 | 96.9 | 2123.14 | 2271.65 | 93.5 |
| Estuarine | 14.40 | 26.31 | 54.7 | 984.32 | 1778.76 | 55.3 |
| Caribbean Sea | 7.10 | 9.29 | 76.4 | 631.05 | 795.40 | 79.3 |
| Galapagos Islands | 18.39 | 21.50 | 85.5 | 1059.67 | 1244.79 | 85.1 |
| Indian Ocean | 15.99 | 22.05 | 72.5 | 1023.01 | 1189.59 | 86.0 |
| North American East Coast | 14.54 | 23.44 | 62.0 | 830.96 | 1093.74 | 76.0 |
| Sargasso Sea | 18.78 | 21.52 | 87.3 | 854.53 | 1130 | 75.6 |
PD, phylogenetic diversity; PDreached, phylogenetic diversity reached; PDmax, expected maximal PD. PDmax values were obtained by the extrapolation of PD rarefaction curves. The coverage was estimated as the fraction of PDreached to PDmax.
Distribution patterns of chitinase and 16S rRNA genes.
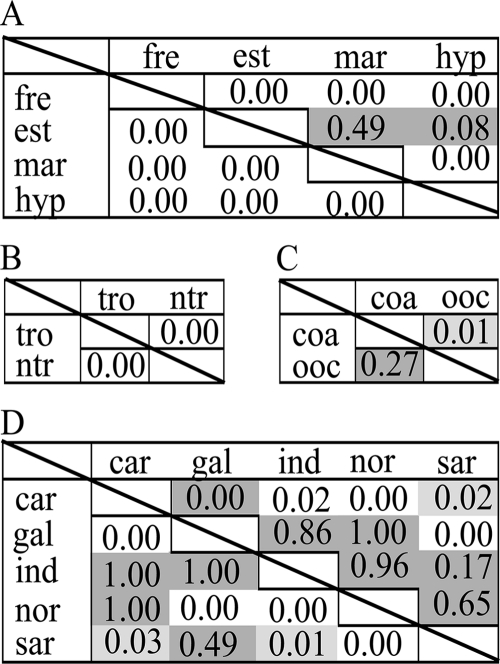
To visualize a possible hierarchy of tested categories (Table 1) on the distribution of the tested genes, cluster analyses were performed while combining categories using the unweighted-pair group method with arithmetic mean (UPGMA), as implemented in the online version of Unifrac. Jackknife support of clusters was determined using 100 permutations (see Fig. 3). Pairwise UniFrac P values (24) were calculated for all habitat pairs within the respective categorical groupings of sites to test the influence of salinity, latitude, proximity to the coastline, and geographic location on the phylogenetic distribution of chitinase and 16S rRNA genes. P values were derived from 10,000 iterations (category geographic location, 50,000 iterations) and were corrected for multiple pairwise comparisons using the Bonferroni correction.
Cluster analyses revealed that chitinolytic organisms follow distribution patterns similar to those of the combined bacterial community, because topology and branch length of the corresponding dendrograms were mostly congruent (Fig. 2). Salinity stands out as a factor strongly influencing the distribution of both genes. This is evident from superior clustering compared to the other categories (Fig. 2). Also, highly significant UniFrac P values between the individual salinity-based habitats illustrate the importance of shifts in salinity for gene distribution patterns (Fig. 3). The striking influence of salinity on the general phylogeographic organization of microbial communities has been demonstrated previously, including studies based on the GOS data set (reviewed in reference 22). However, fewer studies have assessed the influence of salinity on functional guilds (e.g., see references 3 and 16).
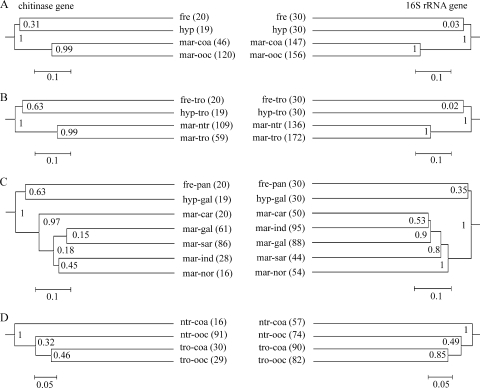
FIG. 2.
Dendrograms with jackknife values derived from the jackknife cluster environments option of the UniFrac metric with estuarine environments excluded to test the hierarchical influence of combined categories. (A) Salinity/proximity of the coastline; (B) salinity/latitude; (C) salinity/geographic location; (D) latitude/proximity of the coastline. Numbers of sequences included in the tested habitats are given in parentheses. Not all possible combinations are displayed, since combined categories containing fewer then 10 sequences were excluded from the analyses (used abbreviations: freshwater, fre; marine, mar; hypersaline, hyp; coastal, coa; open ocean, ooc; tropics, tro; nontropics, ntr; Panama Channel, pan; Caribbean Sea, car; Galapagos Island, gal; Indian Ocean, ind; North American East Coast, nor; Sargasso Sea, sar).
FIG. 3.
Unifrac statistics and P values for 16S rRNA (lower diagonal) and chitinase (upper diagonal) communities. The level of significance is indicated by shading (white, P < 0.01; light gray, P ≥ 0.01 and < 0.05; dark gray, P ≥ 0.05). (A) Salinity; (B) latitude; (C) proximity of the coastline; (D) geographic location. Abbreviations used: freshwater, fre; estuarine, est; marine, mar; hypersaline, hyp; tropics, tro; nontropics, ntr; coastal, coa; open ocean, ooc; Caribbean Sea, car; Galapagos Island, gal; Indian Ocean, ind; North American East Coast, nor; Sargasso Sea, sar.
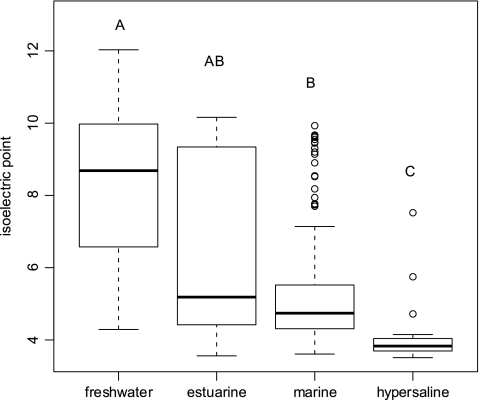
Increased salinity of hypersaline environments shifts entire genomes toward coding for more-acid amino acids (20, 34). We therefore estimated isoelectric point (pI) values for chitinase amino acid sequences from each categorical grouping using EMBOSS pKa values (http://host9.bioinfo3.ifom-ieo-campus.it/sms2/protein_iep.html). The pI is defined as the pH value at which a protein carries no net charge. The obtained pI values were subjected to a reciprocal transformation prior to analysis of variance (ANOVA) analysis, to approximate normal distribution of the residuals. A randomized one-way ANOVA with 10000 iterations was performed (Rundom Pro 3.14 [15]) to assess if significant differences existed in the pI of chitinase genes among the different sets of categories. The pI values were calculated only for protein fragments instead of whole proteins or genomes. This is likely to introduce noise in the analysis since more or less acid regions of a protein will be randomly represented among the extracted fragments. Nevertheless, at a significance level of 0.05, a significant relationship between the predicted pI and habitat was found when habitats were classified based on salinity (F = 22.5; P = 0.0001). Also, pairwise comparisons indicated significant differences between the pI values of sequences found at the extremes of the salinity continuum, whereas no significant differences were observed between the freshwater and estuarine sequences and marine and estuarine sequences (Fig. 4). The influence of salinity or possibly covarying pH values on the pI of chitinase gene fragments reveals a direct impact of these factors on the evolution of chitinase genes, rather than an indirect and random coevolution of the combined gene pool in individual microbial lineages. Such a general impact on protein architecture that probably affects the combined proteome of organisms may explain why salinity appears to be a strong structuring factor for bacteria regardless of their phylogenetic affiliation.
FIG. 4.
Box plot illustrating the distribution of predicted chitinase isoelectric points at different salinity ranges. Values exceeding 1.5 times the interquartile range are indicated individually as circles. The same letters above the box plots indicate no significant difference (P > 0.05) between the tested habitats if identical and significant differences (P < 0.002) if distinct. Differences in the isoelectric points among the habitats were tested for significance by applying a randomized one-way ANOVA (10,000 iterations) with post hoc comparisons (Holm's correction).
The low UniFrac P values when chitinase genes from estuarine waters were compared to those from either freshwater or marine samples suggest that organisms indigenous to both freshwater and marine biomes are present in the estuarine environment. This is in accordance with previous findings (8) and agrees with the large variation in chitinase pI values from estuarine sites (Fig. 4). Subordinate to salinity, other factors associated with the habitat definition influenced the phylogenetic distribution of chitinase genes. A clear congruence of distribution patterns for chitinase and 16S rRNA genes was observed when habitat definitions were based on latitude or proximity to the coastline. This indicates that factors shaping the overall bacterial community composition also control the composition of the chitinolytic community. In contrast, incongruent distributions of chitinase and 16S rRNA genes were observed if habitats were defined by geographic location (Fig. 2C). For example, the separate grouping of North American East Coast samples based on the available environmental data (Fig. 1) was reflected only in the composition of the 16S rRNA assemblages. Thus, the distribution of chitinase genes is influenced by parameters other than those shaping the combined bacterial community. Still, the highly significant UniFrac P values suggest habitat-controlled differences in assemblages of both chitinase and 16S rRNA genes rather than a restriction in dispersal, since communities from geographically distant regions clustered together (Fig. 2C). We suggest that the local availability and crystalline form of chitin shape the assembly of chitinase genes and associated chitinolytic communities in a way that is not reflected at the level of the total bacterial community.
In summary, we found diverging distribution patterns between chitinase and 16S rRNA genes when habitats were defined by their geographical location, but otherwise the patterns were congruent. Salinity was an important factor shaping chitin-degrading bacterial communities, as well as the general bacterial community. This is likely due to the strong overall physical and chemical influence of the salt concentration on the conformation and tertiary structure of the combined pool of proteins making up the community proteome.
Supplementary Material
Acknowledgments
We thank Michael Chiu for assistance with CAMERA and Ryan Newton and Shalabh Sharma for help in estimating genome equivalents.
Funding was provided by the Swedish Research Council Formas through a grant to Uppsala Microbiomics Center (UMC) and grants to S. Bertilsson and S. Hallin from the Swedish Research Council.
Footnotes
Published ahead of print on 29 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atamna-Ismaeel, N., et al. 2008. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2:656-662. [DOI] [PubMed] [Google Scholar]
- 4.Biers, E. J., S. L. Sun, and E. C. Howard. 2009. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 75:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, J. A., C. Lamanna, H. Morlon, A. J. Kerkhoff, B. J. Enquist, and J. L. Green. 2008. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U. S. A. 105(Suppl. 1):11505-11511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 345:101-118. [DOI] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 10.Elifantz, H., L. A. Waidner, V. K. Michelou, M. T. Cottrell, and D. L. Kirchman. 2008. Diversity and abundance of glycosyl hydrolase family 5 in the North Atlantic Ocean. FEMS Microbiol. Ecol. 63:316-327. [DOI] [PubMed] [Google Scholar]
- 11.Faith, D. P. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61:1-10. [Google Scholar]
- 12.Gooday, G. W. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387-430. [Google Scholar]
- 13.Horner-Devine, M. C., and B. J. M. Bohannan. 2006. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87:S100-S108. [DOI] [PubMed] [Google Scholar]
- 14.Howard, E. C., S. L. Sun, E. J. Biers, and M. A. Moran. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10:2397-2410. [DOI] [PubMed] [Google Scholar]
- 15.Jadwiszczak, P. 2009. Rundom Pro 3.14. Software for classical and computer-intensive statistics. http://pjadw.tripod.com.
- 16.Jones, C. M., and S. Hallin. 2010. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4:633-641. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson, M., and J. Stenlid. 2009. Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. J. Mol. Microbiol. Biotechnol. 16:208-223. [DOI] [PubMed] [Google Scholar]
- 18.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchman, D. L., and J. White. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18:187-196. [Google Scholar]
- 20.Kunin, V., et al. 2008. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol. Syst. Biol. 4:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 22.Logares, R., et al. 2009. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 17:414-422. [DOI] [PubMed] [Google Scholar]
- 23.Logue, J. B., and E. S. Lindstrom. 2010. Species sorting affects bacterioplankton community composition as determined by 16S rDNA and 16S rRNA fingerprints. ISME J. 4:729-738. [DOI] [PubMed] [Google Scholar]
- 24.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, C. A., and R. Knight. 2008. Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32:557-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalfe, A. C., M. Krsek, G. W. Gooday, J. I. Prosser, and E. M. H. Wellington. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakley, B. B., F. Carbonero, C. J. van der Gast, R. J. Hawkins, and K. J. Purdy. 2010. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J. 4:488-497 [DOI] [PubMed] [Google Scholar]
- 29.Posada, D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253-1256. [DOI] [PubMed] [Google Scholar]
- 30.Pruesse, E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramaiah, N., et al. 2000. Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 34:63-71. [DOI] [PubMed] [Google Scholar]
- 32.Rusch, D. B., et al. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seshadri, R., S. A. Kravitz, L. Smarr, P. Gilna, and M. Frazier. 2007. CAMERA: a community resource for metagenomics. PLoS Biol. 5:394-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soppa, J. 2006. From genomes to function: haloarchaea as model organisms. Microbiology 152:585-590. [DOI] [PubMed] [Google Scholar]
- 35.Terahara, T., et al. 2009. Molecular diversity of bacterial chitinases in arable soils and the effects of environmental factors on the chitinolytic bacterial community. Soil Biol. Biochem. 41:473-480. [Google Scholar]
- 36.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson, N., P. Brian, and E. M. H. Wellington. 2000. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek 78:315-321. [DOI] [PubMed] [Google Scholar]
- 38.Zwickl, D. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. thesis. The University of Texas at Austin, Austin, TX.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.