Abstract
A better understanding of the antimicrobial peptide (AMP) resistance mechanisms of bacteria will facilitate the design of effective and potent AMPs. Therefore, to understand resistance mechanisms and for in vitro assessment, variants of Enterococcus faecalis that are resistant to different doses of the fungal AMP alamethicin (Almr) were selected and characterized. The resistance developed was dose dependent, as both doses of alamethicin and degrees of resistance were colinear. The formation of bacterial cell aggregates observed in resistant cells may be the prime mechanism of resistance because overall, a smaller cell surface in aggregated cells is exposed to AMPs. Increased rigidity of the membranes of Almr variants, because of their altered fatty acids, was correlated with limited membrane penetration by alamethicin. Thus, resistance developed against alamethicin was an adaptation of the bacterial cells through changes in their morphological features and physiological activity and the composition of membrane phospholipids. The Almr variants showed cross-resistance to pediocin, which indicated that resistance developed against both AMPs may share a mechanism, i.e., an alteration in the cell membrane. High percentages of colorimetric response by both AMPs against polydiacetylene/lipid biomimetic membranes of Almr variants confirmed that altered phospholipid and fatty acid compositions were responsible for acquisition of resistance. So far, this is the only report of quantification of resistance and cross-resistance using an in vitro colorimetric approach. Our results imply that a single AMP or AMP analog may be effective against bacterial strains having a common mechanism of resistance. Therefore, an understanding of resistance would contribute to the development of a single efficient, potent AMP against resistant strains that share a mechanism of resistance.
The resistance of most bacterial pathogens to the majority of the available antibiotics/antimicrobial peptides (AMPs) used daily to preserve food or to control disease outbreaks has increased, leading to intensive research efforts of scientists fighting back with alternative antibiotics and combination therapy. Unrestricted overuse of antibiotics has also led to bacterial resistance to nearly all the antibiotics now known. Consequently, there is an urgent need to search for and design efficient and potent AMPs to tackle resistant bacterial strains, the so-called superbugs. Mechanisms of resistance to AMPs in bacteria differ in terms of specificity, efficiency, and occurrence among species (19). By characterizing mechanisms in both naturally resistant variants and those with resistance acquired through extensive exposure to AMPs, several previous reports focused on an understanding of the mechanism of AMP action and the resistance phenomena (6, 8). The mechanism of Listeria monocytogenes resistance to nisin has been ascribed to the altered membrane fatty acid composition, cell wall structure, and requirements for divalent cations (6). However, inadequate information is available for other pathogenic species, such as Enterococcus faecalis, which is an opportunistic pathogen, and there is no report available regarding the mechanism of resistance and cross-resistance against the thoroughly studied model pore-forming (PF) AMPs alamethicin and pediocin. Alamethicin is a small 20-mer peptide produced by the fungus Trichoderma viridae and has served as a model for peptide-membrane interactions (5). Pediocins are 43-mer PF-AMPs with a YGNGV motif at their N termini produced by lactic acid bacteria. Both PF-AMPs act on the cell membrane after diffusion through the cell wall. After initial electrostatic binding, monomers of the peptide can aggregate on the membrane surface to penetrate and ultimately lead to stabilized pore formation in the lipid bilayer (1, 28). One method that effectively helps to tackle the resistance issues is the combinatorial use of AMPs. However, it requires finding new and effective AMPs, either through brute force screening, which is a time-consuming and laborious method, or through AMP design. Understanding the resistance mechanisms is a prerequisite for the AMP design cycle to develop a quantitative structure-activity relationship (QSAR) model for subsequent optimization of peptide design for synthesis and testing. However, the mechanism of resistance and its in vitro assessment remain poorly understood. Therefore, in the present study, the mechanism of resistance and cross-resistance in E. faecalis against PF-AMPs and their in vitro assessment were elucidated. The colorimetric supramolecular assembly of polydiacetylene (PDA)/lipid molecules was used to assess resistance and cross-resistance using an in vitro assay. The conjugated polydiacetylenes used in this assay can undergo blue-to-red color transition in response to AMPs, which can be quantified as the percent colorimetric response (10).
MATERIALS AND METHODS
Bacterial strains and growth media.
E. faecalis NCDC 114 was procured from the National Collection of Dairy Cultures (NCDC), Karnal, Haryana, India, in freeze-dried form. Pediococcus pentosaceus NCDC 273 was used as an AMP producer strain. The cultures were subcultured twice before use to make them physiologically active. Nutrient broth was used for the culturing of E. faecalis NCDC 114, and MRS broth was used for culturing of P. pentosaceus NCDC 273.
Determination of the IC50.
The 50% inhibitory concentration (IC50) was determined using a broth inhibition assay described by Cabo et al. (4). Briefly, 996 μl of nutrient broth (inoculated to 0.5% with E. faecalis) was taken in sterile Eppendorf tubes. To the first Eppendorf tube, 4 μl of alamethicin stock solution was added. To the rest of the Eppendorf tubes, 4 μl of alamethicin stock solution was added after serial double dilutions. A positive control was set up in broth with a test culture, and a negative control was set up with broth alone. All of the tubes were incubated at 37°C for 6 to 8 h, and absorbance was measured at 600 nm. From the A600 data, the percent inhibition (%I) was calculated as follows: %I = 1 − (Ac/Ao) × 100, where Ac is the A600 of the culture at a given concentration of alamethicin and Ao is the A600 of the positive control. The IC50 was then determined as the concentration of alamethicin resulting in 50% inhibition from the log2 value of the alamethicin concentration versus percent inhibition.
Selection of alamethicin-resistant (Almr) variants.
Resistant variants were selected using the method described by Rekhiff et al. (22). In brief, culture containing 1 × 108 to 5 × 108 CFU was inoculated in nutrient broth containing alamethicin (4, 6, 8, and 10 times the IC50 of the wild-type strain) and incubated at 37°C. After visible growth, the broth culture was serially diluted and plated onto nutrient hard agar without alamethicin. Two colonies from each plate were picked randomly. The IC50 for each of these was determined. Each of the colonies was then propagated through 10 overnight subcultures separately, and the IC50 was determined again.
SEM analysis of Almr cells.
The bacterial cells were Gram stained and examined under a light microscope (magnification, ×1,000). Scanning electron microscopy (SEM) was utilized to visualize both the wild-type strain and the Almr variant (17). Midgrowth log-phase cells were grown for 6 h and collected by centrifugation at 7,000 × g for 10 min. After being washed, the cells were resuspended at 108 CFU/ml in phosphate buffer (pH 7.4) and incubated for 1 h in the presence of alamethicin at 37°C. Then, the cells were washed and fixed with equal volumes of 2.5% glutaraldehyde in phosphate buffer (pH 7.4) for 6 h at 4°C. After centrifugation, the cells were dehydrated by sequential treatments with 50%, 70%, 90%, 95%, and 100% ethanol. After being coated with gold, the samples were examined under SEM at the Sophisticated Analytical Instrumentation Facility (SAIF), Punjab University, Chandigarh, India.
Cell surface hydrophobicity.
The cell surface hydrophobicity of bacterial cells was assayed by the MATH (microbial adhesion to hydrocarbons) assay as described by Reifsteck et al. (21) with slight modifications. The stationary-phase bacterial cells were harvested by centrifugation and washed three times with ice-cold phosphate buffer, followed by resuspension in phosphate buffer to achieve an optical density at 500 nm (OD500) of 0.5. A 4.8-ml volume of each bacterial suspension was mixed with 0.8 ml of n-hexadecane in a glass tube and vigorously shaken for 1 min. After 45 min, the absorbance of the aqueous phase was read at 500 nm. The affinity of bacteria for the solvent, i.e., hexadecane, was evaluated by the following formula: % adherence = (1 − A/A0) × 100, where A0 is the OD500 of the bacterial suspension before being mixed and A is the OD after mixing with solvent.
Growth kinetics.
To different tubes containing 5 ml of sterile nutrient broth, a 1% inoculum of overnight activated culture was added and incubated at 37°C. Bacterial growth was monitored by recording the absorbance at 600 nm from 0 to 24 h at 2-h intervals using sterile nutrient broth as a blank. The specific growth rate (h−1) for both the wild-type strain and Almr variants was determined from A600 data using the following equation: specific growth rate (μ) = ln (X2/X1)/(t2 − t1), where X1 and X2 are equal to the A600 at time 1 (t1) and time 2 (t2) in hours, respectively.
Sensitivity to lysozyme action.
The wild-type strain and Almr variants were subcultured twice before incubation with lysozyme. The Almr variants were then inoculated at 1% (vol/vol) into fresh sterile nutrient broth containing lysozyme at 4 mg ml−1. The cultures were incubated at 37°C, and the absorbance was read at 600 nm.
Analysis of phospholipids.
The bacterial cells were harvested, washed, and resuspended in physiological saline. Total cellular lipids were extracted by the method of Bligh and Dyer (2). The lipid extract was dissolved in chloroform and stored at −20°C. Total lipid extracts of bacterial cells were resolved using chloroform-methanol-distilled water-glacial acetic acid (65:25:4:1 [vol/vol]) as a mobile phase in thin-layer chromatography (TLC) (9), visualized using iodine vapors, and identified using specific stains, viz., ammonium molybdate for phospholipids and ninhydrin for amino group-containing phospholipids (ACPs). The Alpha Imager gel documentation system was used for the quantitation of phospholipids separated in TLC. The pixel areas were determined for each of the standards, and the concentrations of unknown phospholipid spots were determined.
Fatty acid analysis.
Total lipid extracts of bacterial cells were transesterified to obtain fatty acid methyl esters (FAMEs) for further analysis by gas chromatography (23). In brief, lipid extract was transesterified using methanol-hydrochloric acid-chloroform (10:1:1 [vol/vol/vol]) at 90°C for 60 min. The extraction of FAMEs was carried out twice using distilled water (1 ml) and hexane-chloroform (4:1 [vol/vol]). Both aliquots were pooled and allowed to evaporate to 100 μl. The FAMEs were analyzed on a Michro 9100 gas chromatograph using a Supelcowax (Sigma-Aldrich)-fused silica capillary column (60-m by 0.32-mm by 0.25-μm film thickness). Nitrogen gas was used as a carrier gas at a constant flow rate of 1 ml min−1. The initial oven temperature was kept at 50°C, and an isothermal time of 1 min was allowed. Then, a ramp rate of 5°C min−1 was used to bring the temperature to 225°C, and an isothermal time of 15 min was allowed. The injector temperature was kept at 250°C and the flame ionization detector (FID) temperature at 250°C. One microliter of sample containing FAME(s) was injected into the column through the polysilicone diaphragm, and the detector response was recorded.
Purification of YGNGV motif-containing class IIa bacteriocin.
The antibacterial activity of P. pentosaceus NCDC 273 was assessed by the deferred agar spot assay and the spot-on-lawn assay (27). The pedA gene encoding pediocin was detected using PCR, cloned, and sequenced. The AMP produced was purified by three-step purification procedures, which included ammonium sulfate precipitation, cation-exchange chromatography, and reverse-phase high-performance liquid chromatography (HPLC). The purity of the pediocin fraction was checked using SDS-PAGE.
Susceptibilities of resistant variants to pediocin.
The sensitivities of Almr variants to pediocin were determined using a broth dilution/inhibition assay. The broth only and broth with a test culture were set as negative and positive controls, respectively. After 6 h of incubation, the growth of bacterial cultures was monitored at 600 nm, and the %I was determined using the following formula: %I = 1 − (Am/Ao), where Am is the OD of the culture at different concentrations of pediocin and Ao is the OD of the positive control.
Preparation of PDA-based biomimetic membranes.
The PDA/lipid biomimetic skeletons were prepared using the slightly modified method described by Kolusheva et al. (10). In brief, total lipid extracts (3 mg, from the wild-type strain and Almr variants) and 0.83 mg of 10,12-tricosadiynoic acid (Sigma-Aldrich Pvt. Ltd.) were hydrated by adding 2 ml of triple-distilled water and sonicated at 70°C (or above the melting temperature [Tm] of the phospholipids used) for 5 to 6 min. The vesicle solution was then cooled at room temperature (RT) for 30 min and kept overnight at 4°C. After transfer at RT for 30 min, the vesicles were polymerized using UV irradiation (254 nm) for 45 s, with the resulting solutions exhibiting an intense blue appearance.
In vitro colorimetric estimation of membrane penetration by AMPs.
Samples were prepared by adding 6 μl peptide to 0.03-ml vesicle solutions at 0.2 mM total lipid concentration and 2 mM Tris (pH 8.5). The pH of all components was maintained at 8.5. Alamethicin (Sigma-Aldrich Pvt. Ltd.) solution was prepared in triflouroethanol (TFE). Following addition of the peptides, the solutions were diluted to 1 ml and the absorbances of the solutions were taken at 500 nm and 640 nm. The blue-to-red color transition in vesicle solutions was quantified by calculating the percent colorimetric response (%CR) as follows: %CR = (PB0 − PBI)/PB0 × 100, where PB0 = A640/(A640 + A500) for the control and PBI = A640/(A640 + A500) for samples (PB0 is the blue/red ratio of the control sample, and PBI is the value obtained for the vesicle solution after colorimetric transition occurs).
RESULTS
Selection of alamethicin-resistant variants.
The IC50 of alamethicin for the wild-type strain was found to be 5.0 μg/ml. No growth or turbidity could be seen after 16 to 18 h of coculturing in the presence of alamethicin. After 24 h of incubation, bacterial growth could be seen at alamethicin concentrations 4 and 6 times the IC50 for the wild-type strain. At alamethicin concentrations 8 and 10 times the IC50, growth could be seen after 40 h and 72 h, respectively. The colonies with the same IC50 as before subculturing showed that the resistance developed was a stable phenotypic characteristic, and these colonies were used for further studies. The bacterial colonies that reverted after 10 subculturings were discarded. On the basis of the ratio of the IC50 of alamethicin, the resistant variants were designated as shown in Table 1.
TABLE 1.
Selection of stable resistant variants of E. faecalis using different doses of alamethicina
| Colony no. | [Ala] (μg/ml)b | IC50 before subculturing | IC50 after subculturing | Ratioc | Designation | IC50 of pediocin (μg/ml)d |
|---|---|---|---|---|---|---|
| Wild type | 5.0 | 5.0 | 1.00 | Ef | 6.23 | |
| 1 | 20 | 10.6 | 10.2 | 2.12 | Efv2 | 9.24 |
| 2 | 20 | 9.85 | 5.12e | |||
| 3 | 30 | 15.2 | 15.6 | 3.04 | Efv3 | 18.7 |
| 4 | 30 | 14.7 | 5.27e | |||
| 5 | 40 | 20.1 | 20.4 | 4.02 | Efv4.1 | 22.3 |
| 6 | 40 | 19.6 | 20.2 | 3.92 | Efv4.2 | 21.6 |
| 7 | 50 | 25.8 | 25.4 | 5.16 | Efv5.1 | 24.2 |
| 8 | 50 | 24.75 | 25.1 | 4.95 | Efv5.2 | 26.5 |
Designated on the basis of the ratio of the IC50s of alamethicin for Almr variants and the wild-type (Ef) strain. The cross-resistance of Almr variants to pediocin was checked.
Concentrations of alamethicin at which different resistant variants were selected.
Ratio of the IC50 of alamethicin for resistant variants (with the same IC50 before/after subculturing) to the that of the wild-type strain.
IC50 of pediocin against Almr variants.
Colony was discarded.
Morphologies of the wild-type strain and Almr variants.
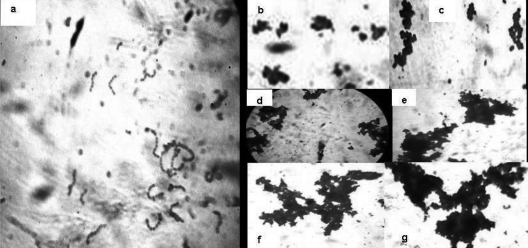
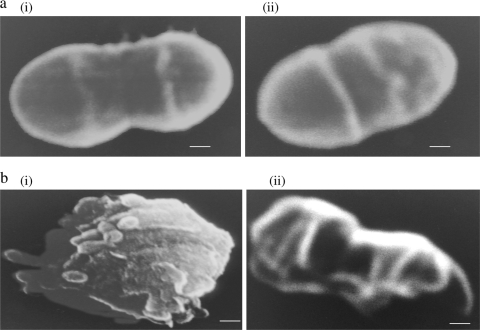
Wild-type cells were found in short, dispersed, straight chains, whereas resistant cells showed aggregation or clump formation (Fig. 1). The aggregation in resistant cells increased with the increase in the degree of resistance (the fold increase in the IC50). Wild-type cells were oval rather than the round or blunt shape of resistant variants (Fig. 2a). The exposure of wild-type cells to alamethicin reduced the cells to a formless mass, whereas abnormal but recognizable cells could be observed after exposure of the resistant cells to alamethicin (Fig. 2b). The resistant cells revealed only contour alterations on the surface.
FIG. 1.
Gram staining of the E. faecalis wild-type strain Ef (a) and resistant variants Efv2 (b), Efv3 (c), Efv4.1 (d), Efv4.2 (e), Efv5.1 (f), and Efv5.2 (g) examined under a light microscope (magnification, ×1,000).
FIG. 2.
(a) Scanning electron micrographs showing single wild-type (i) and resistant (ii) cells (magnification, ×45,000; bar = 100 nm). (b) Scanning electron micrographs of the wild-type strain (magnification, ×27,000; bar = 1 μm) (i) and an Almr variant (Efv5.2) (magnification, ×45,000; bar = 100 nm) (ii) in the presence of alamethicin.
Cell surface hydrophobicity.
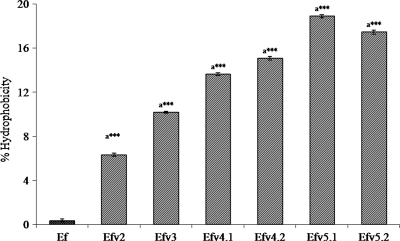
The percent cell surface hydrophobicity of Almr variants was found to be higher than that of the wild type (Fig. 3). One-way analysis of variance (ANOVA) revealed significant differences between the wild-type strain and the Almr variants [F(6,14) = 5,995.380; P < 0.001]. Bonferroni post-hoc analysis revealed a significant increase in the cell surface hydrophobicity of Almr variants compared to the wild-type strain (P < 0.001).
FIG. 3.
Comparison of percent hydrophobicity values of the wild type and Almr variants of E. faecalis. The error bars indicate the standard deviations (SD). a***, significantly different (P < 0.001) from the “EF” value.
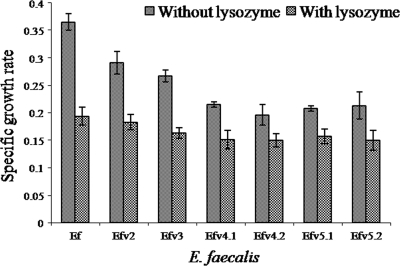
Growth kinetics and lysozyme sensitivity.
A delay in the onset of log phase due to an increase in the lag phase from 1 h in the case of the wild-type strain to 3 h was observed. In the absence of lysozyme, the specific growth rate (h−1) decreased with all Almr variants, whereas in the presence of lysozyme, the rates for the wild-type strain and the Almr variants were similar (Fig. 4).
FIG. 4.
Comparison of the specific growth rates of the wild type (Ef) and Almr variants of E. faecalis before and after lysozyme treatment. The data are the means of three independent experiments, with error bars representing the SD.
Changes in the phospholipid head group.
The phospholipids resolved were identified as either ACPs or amino group-lacking phospholipids (ALPs). In the wild-type strain and the resistant variant Efv2, spots with Rf values of 0.9, 0.79, 0.704, and 0.257 could be identified as ALPs, and a spot with an Rf value of 0.36 was identified as an ACP. In higher-resistance variants (Efv3, Efv4.1, Efv4.2, Efv5.1, and Efv5.2), spots with Rf values of 0.83 and 0.704 could be identified as ALPs, and a spot with an Rf value of 0.36 was identified as an ACP. The ACP/ALP ratio increased in all Almr variants (Table 2).
TABLE 2.
Phospholipid compositions of wild-type (Ef) and Almr variants of E. faecalis
| Rf | Type | % of total phospholipidsa |
||||||
|---|---|---|---|---|---|---|---|---|
| Ef | Efv2 | Efv3 | Efv4.1 | Efv4.2 | Efv5.1 | Efv5.2 | ||
| 0.9 | ALP | 22.94 ± 1.118 | 21.00 ± 0.586 | ND | ND | ND | ND | ND |
| 0.83 | ALP | ND | ND | 42.53 ± 1.394 | 42.61 ± 1.185 | 44.33 ± 1.148 | 41.09 ± 0.307 | 43.87 ± 0.888 |
| 0.79 | ALP | 18.71 ± 1.568 | 17.22 ± 0.790 | ND | ND | ND | ND | ND |
| 0.704 | ALP | 17.69 ± 1.249 | 17.58 ± 0.725 | 21.5 ± 0.775 | 21.10 ± 0.688 | 21.90 ± 0.825 | 22.85 ± 0.811 | 21.56 ± 0.670 |
| 0.36 | ACP | 15.45 ± 0.890 | 21.14 ± 1.275 | 35.92 ± 1.028 | 36.34 ± 0.987 | 33.73 ± 1.150 | 36.04 ± 0.997 | 34.53 ± 0.455 |
| 0.257 | ALP | 25.19 ± 0.864 | 23.03 ± 0.587 | ND | ND | ND | ND | ND |
| ACP/ALP | 0.182 | 0.27 | 0.56 | 0.57 | 0.51 | 0.563 | 0.53 | |
Experiments were conducted in triplicate, and their mean values ±SD are presented. ND, not detected.
Fatty acid analysis.
The fatty acid profiles of bacterial cells were identified by comparing the retention times (Rts) of fatty acid methyl ester peaks with the Rts of known bacterial acid methyl esters (Sigma Aldrich Pvt. Ltd.). Further, based on the percentile peak area, the relative quantity of each fatty acid present was determined. A significant decrease and increase were observed in the amounts of total unsaturated fatty acids and total saturated fatty acids of Almr variants, respectively. The saturated/unsaturated (S/U) ratio increased in all Almr variants except Efv4.2 (Table 3 ). Also, an increase in total hydroxy fatty acids (HFA) and a decrease in total branched-chain fatty acids (BCFA) was observed. The cyclopropane fatty acids were detected only in highly resistant variants.
TABLE 3.
Fatty acid compositions of wild-type (Ef) and Almr variants of E. faecalisa
| FAb | % of total fatty acidsc |
Nature of membrane | ||||||
|---|---|---|---|---|---|---|---|---|
| Ef | Efv2 | Efv3 | Efv4.1 | Efv4.2d | Efv5.1 | Efv5.2 | ||
| C12:0 | ND | ND | ND | 0.153 ± 0.020 | ND | 0.223 ± 0.018 | ND | |
| C14:0 | 5.86 ± 0.284 | ND | ND | ND | ND | 0.692 ± 0.022 | 3.84 ± 0.375 | |
| C15:0 | 6.76 ± 0.610 | ND | ND | 0.124 ± 0.082 | ND | 0.543 ± 0.058 | 5.068 ± 0.858 | |
| C16:0 | 1.04 ± 0.496 | 9.097 ± 1.261 | ND | ND | ND | 7.08 ± 1.563 | ND | |
| C17:0 | 6.10 ± 0.998 | ND | 5.69 ± 1.857 | ND | 3.50 ± 0.973 | 36.15 ± 1.456 | 0.89 ± 0.402 | |
| C18:0 | 12.91 ± 1.159 | 9.55 ± 0.965 | 22.83 ± 0.691 | 4.26 ± 0.724 | 3.52 ± 0.778 | 7.32 ± 0.977 | ND | |
| C19:0 | 2.47 ± 1.113 | ND | ND | 29.52 ± 1.446 | ND | 0.16 ± 0.071 | 19.40 ± 1.634 | |
| C20:0 | ND | 10.85 ± 1.039 | ND | 1.368 ± 0.425 | 8.36 ± 1.164 | 2.85 ± 0.948 | ND | |
| Total SFA (S) | 35.14 | 29.5 | 28.5 | 35.4 | 15.38 | 55.02 | 29.19 | |
| C16:19 | 2.23 ± 0.790 | ND | ND | 0.588 ± 0.272 | ND | 1.59 ± 0.501 | 4.398 ± 0.821 | |
| C18:19 | 0.87 ± 0.205 | 9.34 ± 1.266 | 4.70 ± 0.799 | 0.755 ± 0.050 | 8.45 ± 1.487 | ND | ND | |
| C18:29,12 | 17.49 ± 1.486 | ND | ND | 0.863 ± 0.217 | 19.33 ± 2.042 | 1.73 ± 0.342 | ND | |
| Total UFA (U) | 20.59 | 9.34 | 4.70 | 2.21 | 27.78 | 3.32 | 4.398 | Rigid |
| S/U ratio | 1.7 | 3.15 | 6.06 | 16.02 | 0.55 | 16.57 | 6.64 | Rigid |
| 2-OH C12:0 | ND | ND | ND | 4.46 ± 0.749 | 3.06 ± 0.830 | 0.123 ± 0.043 | ND | |
| 3-OH C12:0 | 8.71 ± 1.587 | ND | 19.07 ± 1.250 | 48.10 ± 1.072 | ND | 2.12 ± 0.485 | ND | |
| 3-OH C14:0 | 6.40 ± 0.822 | 4.51 ± 1.005 | 9.465 ± 0.857 | ND | ND | 31.71 ± 0.984 | 34.65 ± 0.834 | |
| 2-OH C16:0 | 1.46 ± 0.711 | 47.43 ± 1.054 | ND | 3.73 ± 0.808 | ND | 5.59 ± 0.655 | 13.85 ± 1.684 | |
| Total HFA | 16.57 | 51.9 | 28.5 | 56.29 | 3.06 | 39.54 | 48.5 | Rigid |
| i-C15:0 | ND | ND | ND | 0.28 ± 0.039 | 2.369 ± 0.578 | 0.397 ± 0.053 | ND | |
| i-C16:0 | 1.52 ± 0.685 | ND | ND | 2.79 ± 0.817 | ND | 1.094 ± 0.329 | ND | |
| i-C17:0 | 6.19 ± 0.346 | ND | ND | 0.694 ± 0.105 | ND | ND | ND | |
| a-C15:0 | 1.22 ± 0.421 | ND | ND | 0.126 ± 0.022 | ND | 0.176 ± 0.028 | 5.00 ± 1.028 | |
| Total BCFA | 8.93 | 3.89 | 2.369 | 1.667 | 5.00 | Rigid | ||
| C17:0Δ9 | ND | ND | ND | 1.092 ± 0.220 | 24.73 ± 0.778 | 0.072 ± 0.008 | 12.83 ± 0.938 | |
| C19:0Δ9 | 18.72 ± 0.772 | 9.20 ± 1.010 | 38.24 ± 0.531 | 1.092 ± 0.036 | 26.66 ± 0.820 | 0.354 ± 0.022 | ND | |
| Total CFA | 18.72 | 9.20 | 38.2 | 2.184 | 51.39 | 0.426 | 12.83 | |
The fatty acid esters of both the wild type and the alamethicin-resistant variants of E. faecalis were separated by gas chromatography and identified by comparing the Rts of their peaks with the Rts of known bacterial acid methyl esters (Sigma Aldrich Pvt. Ltd.). Also, based on the percentile peak area, the relative quantity of each of fatty acid present was determined. The saturated-to-unsaturated fatty acid ratio was determined for the wild type and the alamethicin-resistant variants of E. faecalis. The increase in the S/U ratio, total hydroxy fatty acids (except Efv4.2), and branched-chain fatty acids in resistant variants was responsible for the decreased fluidity of their membranes compared to that of the wild-type strain.
Dodecanoate (C12:0), tetradecanoate (C14:0), pentadecanoate (C15:0), hexadecanoate (C16:0), heptadecanoate (C17:0), octadecanoate (C18:0), nonadecanoate (C19:0), eicosanoate (C20:0), cis-9-hexadecenoate (C16:19), cis-9-octadecanoate (C18:19), cis-9,12 octadecadienoate (C18:29,12), 2-hydroxydodecanoate (2OH-C12:0), 3-hydroxydodecanoate (3OH-C12:0), 3-hydroxytetradecanoate (3OH-C14:0), 2-hydroxyhexadecanoate (2OH-C16:0), 13-methyltetradecanoate (i-C15:0), 14 methylpentadecanoate (i-C16:0), 15-methylhexadecanoate (i-C17:0), 12 methyltetradecanoate (a-C15:0), cis-9,10-methylenehexadecanoate (C17:0Δ), cis-9,10 methyleneoctadecanoate (C19:0Δ). FA, fatty acid; SFA, saturated fatty acid; UFA, unsaturated fatty acid.
Experiments were conducted in triplicates and their means ± SD are presented. ND, not detected.
Alar variant Efv4.2, in which the bacterial cell membrane becomes more fluid.
Pediocin cross-resistance.
The presence of a growth-free inhibitory zone confirmed the antimicrobial activity of pediocin. Separation by SDS-PAGE resulted in a band showing antimicrobial activity with a molecular mass of 5.19 kDa. The nucleotide sequence of pediocin showed 100% identity with the other pediocin sequences available at NCBI. The Almr variants showed cross-resistance to pediocin (Table 1). The IC50s of both pediocin and alamethicin against Almr variants were colinear (R2 = 0.973), which indicated that the Almr variants were cross-resistant to pediocin. Further, cross-resistance to pediocin was confirmed using an in vitro lipid assay.
In vitro assessment of resistance and cross-resistance.
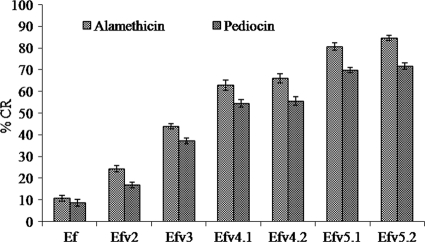
The %CRs of alamethicin and pediocin against PDA/lipid biomimetic membranes of resistant variants were found to be higher than that of the wild-type strain (Fig. 5). The %CRs of both AMPs against the biomimetic membranes of the wild-type strain and the Almr variants differ significantly (Alm, F(6,14) = 815.407, P < 0.001; pediocin, F(6,14) = 290.118, P < 0.001). Bonferroni post-hoc analysis revealed significant increases in the %CRs of alamethicin (P < 0.001) and pediocin (Efv2, P < 0.02, and Efv3, Efv4.1, Efv4.2, Efv5.1, and Efv5.2, P < 0.001) against the membranes of resistant variants compared to the wild-type strain.
FIG. 5.
%CRs of alamethicin and pediocin against biomimetic membranes prepared using total lipid extracts of the wild type (Ef) and Almr variants of E. faecalis. The data are the means of triplicate experiments, with error bars representing the SD. One-way ANOVA and Bonferroni post hoc analysis were done between the wild type and the Almr variants.
DISCUSSION
In this study, variants of E. faecalis that are resistant to different doses of alamethicin were selected and characterized for changes in morphology, cell surface hydrophobicity, growth kinetics, cell wall, membrane surface charge, and membrane fluidity. The IC50 of alamethicin was directly proportional to the concentration used for the selection of resistant variants. The degree of resistance acquired by the wild-type strain was nearly 2-, 3-, 4-, and 5-fold with respect to the 4-, 6-, 8-, and 10-fold concentrations of alamethicin, respectively. Thus, the resistance developed in E. faecalis was dose dependent, as both the doses of alamethicin and degrees of resistance were colinear (R2 = 0.998). To the best of our knowledge, this was the first report of dose-dependent resistance developed against alamethicin in E. faecalis. The clumps or aggregate formation observed in resistant cells may be the prime mechanism of alamethicin resistance and pediocin cross-resistance and could be a very important protective mechanism, because a smaller overall cell surface is exposed to AMPs in clumped or aggregated cells. The inner cells are thus fully protected and survive; therefore, the larger the aggregates, the more cells survive. The bacterial cells can tolerate higher antibiotic/AMP concentrations at higher cell densities, and the ability to aggregate gives bacterial cells an added advantage in overriding the antimicrobial agents. Individuals within the dense clumped cells, those directly exposed to the AMPs, undergo cell death, protecting cells that are not directly exposed (3). Also, the bacterial cell density can induce changes in bacterial regulation, physiology, and metabolic pathways (13). Thus, the aggregation phenomenon has revealed the protective power of large cell densities to withstand exposure to otherwise lethal antibiotic concentrations (3). The increase in cell surface hydrophobicity was colinear with the degree of resistance (R2 = 0.909), which might be responsible for the formation of large clumps of resistant cells. The higher cell surface hydrophobicity and the aggregation observed for resistant cells may be interrelated. However, it is not clear whether aggregation leads to an increase in cell surface hydrophobicity or the increase in cell surface hydrophobicity causes aggregation. Also, the decreased anionic nature of the cell membrane due to an increased percentage of ACP may contribute to the increased cell surface hydrophobicity or aggregation of resistant cells. The reduced anionic nature of the bacterial cell envelope has been exploited by different microbes as a resistance strategy against AMPs. This decrease in anionic charge reduces the interactions of AMPs with the bacterial membrane and contributes to antimicrobial molecule resistance in microbes (18). The decreased growth rate of Almr variants may be seen as a fitness cost associated with the acquisition of resistance. The associated fitness cost may be due to energy-expensive metabolic pathways or lower activity of glucose-transporting enzymes in Almr variants (25). The comparable sensitivities of the wild-type strain and Almr variants toward lysozyme revealed that the cell wall remains unmodified upon acquisition of resistance; this finding was supported by a previous study (26). The increase in the S/U ratio in Almr variants, excluding Efv4.2, might lead to a more rigid cell membrane. The increased rigidity of bacterial cell membranes may prevent the insertion of alamethicin into the phospholipid bilayer. These findings are supported by the previous studies showing that the presence of monolaurin changed the cellular saturated-to-unsaturated fatty acid ratio from 1.4 to 4.35 (7). The decrease in the S/U ratio indicated the increased fluid nature of the cell membrane of Almr variant Efv4.2. This increased fluidity of the bacterial cell membrane did not allow alamethicin to form stable pores in the phospholipid bilayer. A decrease in membrane fluidity has been reported in L. monocytogenes against nisin (11) and in Leuconostoc and Weisela strains against mesentericin 52A and 52B (12). The decrease in the total BCFA of Almr variants led to increased membrane rigidity, as their main role in bacteria is to increase fluidity. The presence of an additional OH group in the fatty acyl chain of a membrane lipid increases H bonding with the neighboring molecules and decreases the membrane permeability. The increased HFA observed in this study increased the membrane rigidity. Thus, either an increase or a decrease in membrane fluidity could affect pore formation by AMPs in the cell membrane. Rigidification of the membrane has been suggested as a mechanism of resistance for membrane-active compounds, such as nisin (14, 15, 24). The ability of the AMPs to generate pores in the membrane is crucial for their antimicrobial potential toward a particular bacterial strain and is governed by the membrane composition and fluidity (19).
The present study correlated limited membrane penetration and poor stability of pore formation by alamethicin due to altered fatty acids of membrane phospholipids. The sensitivity of Almr variants to pediocin was determined to check for the possibility of a common mechanism of resistance for the two AMPs. The Almr variants showed cross-resistance to the YGNGV motif-containing PF-AMP (i.e., pediocin), as the IC50s of the two AMPs were colinear (R2 = 0.97). Cross-resistance in L. monocytogenes strains to class I/II bacteriocins, class IIa bacteriocins, nisin, and class IV leuconocin S has been observed (6, 16). These results indicate that development of resistance against alamethicin and pediocin may share a mechanism, i.e., an alteration in the cell membrane. Thus, the physicochemical properties of the cell envelope can decide the bacterial susceptibility to antimicrobial substances. It has been postulated that bacteria modulate the anionic net charge of the cell envelope to reduce interactions with cationic peptides (20).
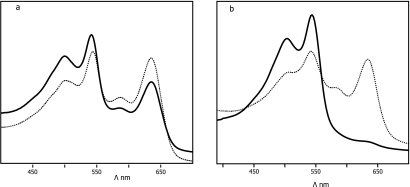
Higher %CRs of AMPs (alamethicin and pediocin) against the biomimetic membrane of Almr variants may be ascribed to their surface interactions at the lipid/water interface. The interactions of AMPs with polar head groups lead to interference in the pendant side chain of PDA and ultimately lead to a stronger blue-to-red color transition, whereas low blue-to-red color transitions occur in the membranes of the wild-type strain due to interactions with fatty acyl chains (Fig. 6). AMPs that preferably interact with the lipid head group region were shown to induce stronger color transitions, while deeper penetration into the hydrophobic lipid core generally gave rise to more moderate blue-to-red transitions. The high %CRs of AMPs indicate the limited insertion of AMPs within the biomimetic membranes of Almr variants. The limited penetration or reduced interactions of AMPs with fatty acyl chains of membrane phospholipids were responsible for the development of resistance. The %CRs of the two AMPs were colinear (R2 = 0.993), and the increase was directly proportional to the level of resistance. Thus, using an in vitro assay, it was confirmed that the altered phospholipid and fatty acid compositions of the Almr variants were responsible for the acquisition of resistance. So, far this is the only report to quantify resistance and cross-resistance phenomena using an in vitro colorimetric approach. In conclusion, this study indicated that the development of resistance against alamethicin was an adaptation of the bacterial cells through changes in their morphological features, physiological activities, cell surface charge, or composition of membrane phospholipids. The resistance developed to an AMP may extend to other AMPs having a common mechanism of action within the same class, or even in other classes. Our results imply that a single AMP or AMP analog may be effective against different bacterial strains having a common mechanism of resistance. Therefore, an understanding of resistance would contribute to the development or design of a single efficient, potent AMP against resistant strains that share a mechanism of resistance.
FIG. 6.
UV-visible light absorption spectra of PDA-biomimetic membranes of the wild type (a) and Almr variants (b) of E. faecalis in the presence (solid lines) and absence (dotted lines) of alamethicin. The blue (640 nm)-to-red (500 nm) color transition in the presence of alamethicin was more pronounced in the biomimetic membranes of the resistant variants.
Acknowledgments
We thank the Director, Regional Sophisticated Instrumentation Centre, Panjab University, Chandigarh, India, for providing the SEM facility and Ranjan Gupta, Department of Biochemistry, Kurukshetra University, India, for her critical review of the manuscript.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Béven, L., O. Helluin, G. Molle, H. Duclohier, and H. Wroblewski. 1999. Correlation between antibacterial activity and pore sizes of two classes of voltage-dependent channel forming peptides. Biochim. Biophys. Acta 1421:53-63. [DOI] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and E. J. Dyer. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Butler, M. T., W. Qingfeng, and R. M. Harshey. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107:3776-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabo, M. L., M. A. Murado, M. P. González, and L. Pastoriza. 1999. A method for bacteriocin quantification. J. Appl. Microbiol. 87:907-914. [DOI] [PubMed] [Google Scholar]
- 5.Cafiso, D. S. 1994. Alamethicin: a peptide model for voltage-gating and protein membrane electrostatic interactions. Annu. Rev. Biophys. Biomol. Struct. 23:141-165. [DOI] [PubMed] [Google Scholar]
- 6.Crandall, A. D., and T. J. Montville. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl. Environ. Microbiol. 64:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dufour, M., et al. 2007. Characterization of monolaurin resistance in Enterococcus faecalis. Appl. Environ. Microbiol. 73:5507-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravesen, A., M. Ramnath, B. Rechinger, N. Andersen, L. Jänsch, Y. Héchard, J. W. Hastings, and S. Knøchel. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361-2369. [DOI] [PubMed] [Google Scholar]
- 9.Katz, S. S., Y. Weinrauch, R. S. Munford, and E. P. Weiss. 1999. Deacylation of LPS in whole E. coli during destruction by cellular and extracellular components of a rabbit peritoneal inflammatory exudate. J. Biol. Chem. 274:36579-36584. [DOI] [PubMed] [Google Scholar]
- 10.Kolusheva, S., L. Boyer, and R. Jelinek. 2000. A colorimetric assay for rapid screening of antimicrobial peptides. Nat. Biotechnol. 18:225-227. [DOI] [PubMed] [Google Scholar]
- 11.Li, J., M. L. Chikindas, R. D. Ludescher, and T. J. Montville. 2002. Temperature and surfactant induced membrane modifications that alter Listeria monocytogenes nisin sensitivity by different mechanisms. Appl. Environ. Microbiol. 68:5904-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limonet, M., A. M. Junelles, and J. B. Milliere. 2002. Variations In the membrane fatty acid composition of resistant or susceptible Leuconostoc or Weissela strains in presence or absence of mesenterocin 52A and mesenterocin 52B produced by Leuconostoc mesenteroides subsp. mesenteroides FR52. Appl. Environ. Microbiol. 68:2910-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, X., C. Ng, and T. Ferenci. 2000. Global adaptations resulting from high population densities in Escherichia coli cultures. J. Bacteriol. 182:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loughlin, M. F., M. V. Jones, and P. A. Lambert. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631-639. [DOI] [PubMed] [Google Scholar]
- 15.Ming, X., and M. A. Daeschel. 1993. Nisin resistance of foodborne bacteria and the specific resistance responses of Listeria monocytogenes. J. Food Prot. 56:944-948. [DOI] [PubMed] [Google Scholar]
- 16.Naghmouchi, K., E. Kheadr, C. Lacroix, and I. Fliss. 2007. Class I/class IIa bacteriocin cross resistance phenomenon in Listeria monocytogenes. Food Microbiol. 24:718-727. [DOI] [PubMed] [Google Scholar]
- 17.Park, S. C., et al. 2006. Investigation of toroidal pore and oligomerization by melittin using transmission electron microscopy. Biochem. Biophys. Res. Commun. 43:222-228. [DOI] [PubMed] [Google Scholar]
- 18.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 19.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 20.Peschel, A., and L. V. Collins. 2001. Staphylococcal resistance to antimicrobial peptides of mammalian and bacterial origin. Peptides 22:1651-1659. [DOI] [PubMed] [Google Scholar]
- 21.Reifsteck, F., S. Wee, and B. J. Wilkinson. 1987. Hydrophobicity-hydrophilicity of staphylococci. J. Med. Microbiol. 24:65-73. [DOI] [PubMed] [Google Scholar]
- 22.Rekhiff, N., A. Atrih, and G. Lefebvre. 1994. Characterization and partial purification of plantaricin LC74, a bacteriocin produced by Lactobacillus plantarum LC74. Biotechnol. Lett. 16:771-776. [Google Scholar]
- 23.Sakayori, Y., et al. 2003. Characterization of Enterococcus faecium mutants resistant to mundticin KS, a class Ila bacteriocin. Microbiology 149:2901-2908. [DOI] [PubMed] [Google Scholar]
- 24.To, M. S., S. Favrin, N. Romanova, and M. W. Griffiths. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5258-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vadyvaloo, V., J. L. Snoep, J. W. Hastings, and M. Rautenbach. 2004. Physiological implications of class IIa bacteriocin resistance in Listeria monocytogenes strains. Microbiology 150:335-340. [DOI] [PubMed] [Google Scholar]
- 26.Verheul, A., N. J. Russel, R. V. T. Hof, F. M. Rombouts, and T. Abee. 1997. Modifications of membrane phospholipid composition in nisin resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 63:3451-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, C. W., L. J. Yin, and S. T. Jiang. 2004. Purification and characterization of bacteriocin form Pediococcus pentosaceus ACCEL. J. Agric. Food Chem. 52:1146-1154. [DOI] [PubMed] [Google Scholar]
- 28.Yang, L., T. A. Harroum, and T. M. Weiss. 2001. Barrel stave model or toroidal model? A case study on melittin pores. Biophys. J. 81:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]