Abstract
Yersinia pseudotuberculosis is an enteric human pathogen but is widespread in the environment. Pathogenicity is determined by a number of virulence factors, including the virulence plasmid pYV, the high-pathogenicity island (HPI), and the Y. pseudotuberculosis-derived mitogen (YPM), a superantigen. The presence of the 3 virulence factors varies among Y. pseudotuberculosis isolates. We developed a multilocus sequence typing (MLST) scheme to address the population structure of Y. pseudotuberculosis and the evolution of its pathogenicity. The seven housekeeping genes selected for MLST were mdh, recA, sucA, fumC, aroC, pgi, and gyrB. An MLST analysis of 83 isolates of Y. pseudotuberculosis, representing 19 different serotypes and six different genetic groups, identified 61 sequence types (STs) and 12 clonal complexes. Out of 26 allelic changes that occurred in the 12 clonal complexes, 13 were mutational events while 13 were recombinational events, indicating that recombination and mutation contributed equally to the diversification of the clonal complexes. The isolates were separated into 2 distinctive clusters, A and B. Cluster A is the major cluster, with 53 STs (including Y. pestis strains), and is distributed worldwide, while cluster B is restricted to the Far East. The YPM gene is widely distributed on the phylogenetic tree, with ypmA in cluster A and ypmB in cluster B. pYV is present in cluster A only but is sporadically absent in some cluster A isolates. In contrast, an HPI is present only in a limited number of lineages and must be gained by lateral transfer. Three STs carry all 3 virulence factors and can be regarded as high-pathogenicity clones. Isolates from the same ST may not carry all 3 virulence factors, indicating frequent gain or loss of these factors. The differences in pathogenicity among Y. pseudotuberculosis strains are likely due to the variable presence and instability of the virulence factors.
Yersinia pseudotuberculosis causes human enteric disease in both sporadic and epidemic manners and is widely distributed in countries with cold climates (22, 24, 29-31, 33, 37, 45, 49, 50). Major sources of Y. pseudotuberculosis are animals and the environment. The animals from which Y. pseudotuberculosis has been isolated are mainly warm-blooded, like pigs, rodents, and birds. Contaminated water has also been identified as an important reservoir for Y. pseudotuberculosis (20, 23). Infections caused by Y. pseudotuberculosis are spread through the fecal-oral route, and higher incidences of the infections occur during the cold months of the year (2).
Y. pseudotuberculosis is commonly typed by serotyping based mainly on antigenic differences in the lipopolysaccharide O antigen and is divided into 15 serogroups; serogroups O:1 and O:2 are each divided into subtypes a, b, and c, and serotypes O:4 and O:5 are each divided into subtypes a and b, and thus, there are a total of 21 serotypes (4). Serogroups O:1 to O:5 have been isolated in Europe and the Far East, and most are pathogenic to humans. Serogroups O:6 to O:14 have been isolated from animals and the environment, but not from clinical samples (21).
Pathogenicity in Y. pseudotuberculosis is determined by the virulence plasmid pYV (the plasmid associated with Yersinia virulence), which encodes a type III secretion system and effectors (11). Additional virulence factors include a type IV pilus, which is present in 40% of isolates (10). The presence of the high-pathogenicity island (HPI), which encodes the synthesis of the siderophore yersiniabactin, and the Y. pseudotuberculosis-derived mitogen (YPM), a superantigen, gives rise to more virulent strains (5, 8). The presence of pYV classifies a strain as pathogenic, although pYV− strains have been isolated from human infections (15, 25). In addition, the presence of the HPI further divides strains into high-pathogenicity (HPI+) and low-pathogenicity (HPI−) groups (5).
Y. pseudotuberculosis has been alternatively divided into six genetic groups based on the presence of the pYV, HPI, and YPM (24). The distribution of the genetic groups shows that there are differences, not only in pathogenicity factors, but also in the geographical distribution of Y. pseudotuberculosis strains. Group 1 is positive for all three factors and was seen only in the Far East. Group 2 lacks the toxin (HPI+ YPM− pYV+) and is predominantly found in Europe as serotypes 1a and 1b, but has also been isolated in the Far East as serotypes 1a, 3, 5b, 13, and 14. Group 3 lacks the HPI (HPI− YPM+ pYV+) and is found in 11 serotypes causing systemic infections in the Far East. Group 4 is presumably nonpathogenic, since it has neither the HPI nor the pYV (YPM+ HIP− pYV−). So far, 9 serotypes have been observed in this subgroup. Group 5 has a partial HPI (partial HPI+ YPM+ pYV+) and contains only serotype 3 isolates. Group 6 has neither the HPI nor the toxin (HPI− YPM− pYV+) and is distributed worldwide, with a large number of serotypes (24).
We developed a multilocus sequence typing (MLST) scheme for Y. pseudotuberculosis and used it to study the evolution of Y. pseudotuberculosis, aiming particularly for a phylogenetic analysis of the genetic groups and a better understanding of the heterogeneity of virulence factors and the distribution of these properties in different lineages. This analysis allows us to assess whether pathogenicity classes form natural clades in the phylogeny of Y. pseudotuberculosis.
MATERIALS AND METHODS
Bacterial isolates.
A total of 79 Y. pseudotuberculosis isolates were analyzed in this study. The isolates were selected to represent the genetic diversity of Y. pseudotuberculosis based on serotypes, isolate sources (countries and animals/environments), and genetic groups. They were collected from 11 different countries, with the majority isolated from Japan. These isolates were typed previously into their respective genetic groups and serotypes by one of the authors (24). The numbers of isolates for each serotype are as follows: O:1a, 3 isolates; O:1b, 9 isolates; O1c, 1 isolate; O:2a, 5 isolates; O:2b, 5 isolates; O:2c, 3 isolates; O:3, 13 isolates; O:4a, 5 isolates; O:4b, 3 isolates; O:5a, 7 isolates; O:5b, 6 isolates; O:6, 3 isolates; O:7, 3 isolates; O:9, 1 isolate; O:10, 3 isolates; O:11, 1 isolate; O:12, 2 isolates; O:13, 1 isolate; and O:15, 4 isolates. The number of isolates per genetic group are as follows: groups 1, 7 isolates; 2, 11 isolates; 3, 26 isolates; 4, 10 isolates; 5, 4 isolates; and 6, 21 isolates. The details of the strains are shown in Table S1 in the supplemental material. The bacteria were grown on nutrient agar plates at 30°C for 2 days. A single colony from the subculture was inoculated into 10 ml nutrient broth and cultured at 30°C overnight, prior to DNA extraction using the phenol-chloroform method (38).
Design of the MLST scheme.
The 7 housekeeping genes selected were mdh, recA, sucA, fumC, aroC, pgi, and gyrB. The 7 genes were chosen based on several criteria. First, the seven selected genes are involved in essential metabolic pathways and are considered housekeeping genes. Second, they are evenly distributed around the genome to minimize the chance of cotransfer within the same recombination event, such as recombination events involving the O antigen genes, where flanking regions up to 150 kb may be cotransferred (36). The smallest and largest distances between any 2 genes are about 360 kb and 1,235 kb, respectively. Third, the genes chosen were not flanked by genes under high selection pressure, such as outer membrane genes or genes involved in virulence. For each gene of interest, up to 10 flanking genes were examined. Lastly, the genes were selected based on whether they had already been used in MLST schemes for other bacterial species so that comparison of variation between different species of the same gene might be made. Four genes, fumC, mdh, gyrB, and recA, are part of the MLST scheme for Escherichia coli (51), while 2, sucA and aroC, are part of the MLST scheme for Salmonella enterica (32). The seventh gene, pgi, is one of the MLST genes for Haemophilus influenzae (35) but was primarily chosen based on its position on the Y. pseudotuberculosis chromosome. It should be noted that mdh and recA are also used in the H. influenzae MLST scheme. With the exception of pgi, the primers for the amplification of the seven genes were designed in such a way that the fragment used in the other MLST schemes mentioned above were also included within the fragment sequenced in this scheme.
The sequences of the selected seven genes from the Yersinia pestis CO92 genome sequence (40) and the Y. pseudotuberculosis genome sequences (9) were compared with those of Yersinia enterocolitica strain 8081 (accession no. AM286415) (48) in order to design primers that were able to amplify PCR products from potentially divergent strains. The primers were designed using the Y. pestis CO92 genome (no Y. pseudotuberculosis genome sequence was available at the time this study was initiated). The amplicons varied from 750 to 850 bp (Table 1). The primers were manufactured by Sigma Genosys.
TABLE 1.
Details of loci used in the MLST scheme
| Gene | Chromosome locationa | Primer no. | Direction | Size (bp) | Oligonucleotide sequence 5′-3′ | Annealing temp (°C) | Length of sequenced fragment (bp) | No. of alleles | % Sequence diversity [mean (maximum)]b |
|---|---|---|---|---|---|---|---|---|---|
| mdh | 546394-547332 | 9091 | Forward | 18 | CCCAGCTTCCTTCAGGTT | 50-55 | 607 | 8 | 0.95 (3.79) |
| 9171 | Reverse | 18 | AGCACTCAGTTTACCGAT | 50-55 | |||||
| gyrB | 4704462-4706876 | 9169 | Forward | 18 | TGGTATTCGAGGTTGTGG | 50-55 | 627 | 14 | 0.99 (3.19) |
| 9170 | Reverse | 18 | TTCAGGGTACGGGTCATC | 50-55 | |||||
| fumC | 2574258-2575655 | 9167 | Forward | 19 | GATGATGTCAATAAAAGCC | 50-55 | 574 | 13 | 1.53 (6.1) |
| 9168 | Reverse | 18 | GCAGTGCTCATTAAAACC | 50-55 | |||||
| sucA | 1373784-1375007 | 9173 | Forward | 18 | TTCAATGTCGGTTCTTTT | 50-55 | 611 | 13 | 0.88 (2.95) |
| 9174 | Reverse | 18 | CGCGCATCCAGTGGGTTC | 50-55 | |||||
| pgi | 4344997-4346643 | 9144 | Forward | 18 | TTTGCCAAGGACGACCAG | 50-55 | 623 | 10 | 0.69 (2.73) |
| 9145 | Reverse | 18 | CAACCGACAAGGCGATAG | 50-55 | |||||
| recA | 988499-989569 | 9172 | Forward | 18 | TGTTGAAACCATCTCTAC | 50-55 | 584 | 14 | 1.36 (5.48) |
| 9147 | Reverse | 18 | TGGCATTGGCTTTACCCT | 50-55 | |||||
| aroC | 3109434-3110519 | 9148 | Forward | 18 | GGCATTCCGATTACCGAG | 50-55 | 597 | 9 | 1.54 (4.36) |
| 9166 | Reverse | 18 | GCCGATTGATAGTTTGCC | 50-55 |
Positions are based on the Y. pseudotuberculosis strain IP32953 chromosome (9).
Excluding strain Y25.
PCR assay and DNA sequencing.
Each PCR included 2.5 μl of DNA template (approximately 20 ng), 0.5 μl (30 pmol/μl) of each forward and reverse primer, 0.5 μl 10 mM deoxynucleoside triphosphates (dNTPs), 5 μl 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl, pH 9.0, 1% Triton X-100, and 15 mM MgCl2), 0.25 μl (1.25 U) Taq polymerase (Promega), and MilliQ water to a total volume of 50 μl. PCR cycles were performed in a Hybaid PCR Sprint Thermocycler (Thermo Analysis Biocompany, Hybaid Ltd., United Kingdom) under the following conditions: initial DNA denaturation for 2 min at 94°C, followed by DNA denaturation for 15 s at 94°C, primer annealing for 30 s at 50°C, and polymerization for 1 min 30 s at 72°C for 35 cycles, with a final extension of 5 min at 72°C. PCR products were verified on ethidium bromide (EtBr)-stained agarose gels before purification using sodium acetate-ethanol precipitation. The PCR sequencing reaction mixtures contained BigDye and were done as recommended by the manufacturer (Applied Biosystems). We sequenced both the 5′ and 3′ ends of the amplicons. Unincorporated dye terminators were removed by ethanol precipitation. The reaction products were separated and detected by gel electrophoresis using an ABI3730 automated DNA sequence analyzer (Applied Biosystems) at the Ramaciotti Centre (University of New South Wales, Sydney, Australia).
Bioinformatics analysis.
The PHRED-PHRAP-CONSED (26) program package available from the Australian National Genomic Information Service (ANGIS) was utilized for sequence editing. The GCG package (14) and MULTICOMP (43) were used for multiple-sequence alignment and comparison. PHYLIP (19) was used to generate phylogenetic trees and bootstrap values. SplitsTree version 3.2 (3, 27) was used to create a network structure using the uncorrected “p” distance method. STRUCTURE version 2.2 (41), which implements a Bayesian approach for deducing population structure from multilocus data, was used to analyze the population clustering of an isolate, assuming that each isolate has derived all of its ancestry from only one population. The number of populations, K, was determined under the “admixture” model, and in each simulation run, the Markov chain Monte Carlo (MCMC) simulation of 30,000 iterations approximated the posterior probability of K, following a burn-in of 10,000 iterations. Different values of K were run multiple times, and the K value that generated the highest posterior probability was used as the number of possible populations. The assignment of an isolate to a particular population was done under the linkage model. The overall compatibility of informative sites was measured by using the RETICULATE program (28), which gives a measure of phylogenetic concordance between two sites with values ranging from 0% (fully incompatible) to 100% (fully compatible). This method was used to obtain a measure of recombination within and between loci. eBURST (17) was used to cluster sequence types (STs) into clonal complexes (CCs) that consist of STs differing by one of the seven genes typed.
While this study was in progress, an unpublished MLST scheme by M. Achtman's laboratory was released through the Web. However, because their data policy does not allow use of their unpublished data, we could not use that information for comparison with this study.
Accession numbers.
The GenBank accession numbers are HQ622350 to HQ622430. An MLST database has been set up and will be released online when this study is published (http://www.emi.unsw.edu.au/∼lanlab/yersinia/index.html).
RESULTS
Sequence variation.
A total of 79 isolates were sequenced for the 7 housekeeping genes, mdh, recA, sucA, fumC, aroC, pgi, and gyrB. The Y. pseudotuberculosis genome sequenced strains IP32953 (accession no. BX936398), IP31758 (accession no. P000720), YPIII (accession no. CP000950), and PB1 (accession no. CP001048) and the Y. pestis genome sequenced strain CO92 (accession no. AL590842) were included for comparison. The other Y. pestis genome sequenced strains are identical to CO92 in the 7 MLST genes and were excluded. The pairwise percentage differences for all 7 genes are shown in Table 1. The most variable gene is fumC, which has the highest maximum and average pairwise percentage differences of 6.10% and 1.49%, respectively, while the most conserved gene is pgi, with the lowest maximum and average pairwise percentage differences of 2.73% and 0.69%, respectively. Strain Y25 was found to be very divergent for 6 of the 7 genes, with variation up to 21.95%, and was excluded from the percentage calculations. The high level of divergence suggests that Y25 does not belong to Y. pseudotuberculosis. However, the aroC allele (allele 8) of Y25 is very similar to another allele (allele 3), with only a single base difference, and these 2 alleles are not the most divergent alleles among the 9 observed. Therefore, the aroC allele in strain Y25 must have been obtained from Y. pseudotuberculosis in a recent recombination event. Further, 16S rRNA gene sequencing shows that Y25 belongs to Yersinia kristensenii.
Allelic profiles, sequence types, and clonal complexes.
Each isolate is defined by 7 alleles, the combination of which constitutes an allelic profile. Isolates with identical allelic profiles were assigned to the same ST. The allelic profile and STs for the isolates are shown in Table S1 in the supplemental material. For the 84 isolates, including the 4 Y. pseudotuberculosis genome strains and Y. pestis CO92, 62 STs have been identified. Twelve STs are represented by multiple isolates: the largest STs are ST2 with 6 isolates and ST21 with 4 isolates. Four STs (ST3, ST9, ST37, and ST57) contain 3 isolates each. Six STs (ST4, ST6, ST27, ST32, ST36, and ST44) have 2 isolates each. Fifty STs are represented by a single isolate. The genome strains IP32953 and PB1 belong to the largest ST (ST2), while strains YPIII and IP31758 have unique STs (ST61 and ST62, respectively). Y. pestis CO92 also has a unique ST (ST1) with the smallest difference, 4 alleles, from ST29.
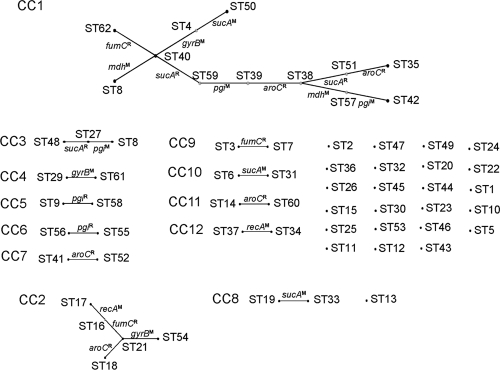
We further analyzed the 62 STs, using eBURST (17), to identify CCs, groups of closely related STs sharing a very recent common ancestor. We used the definition of six out of seven shared alleles for a clonal complex (17) and identified 12 CCs, each of which contains at least 2 member STs. Twenty-four STs are singletons that are unrelated to any other STs (Fig. 1). The 3 largest CCs are CC1 with 12 STs, CC2 with 5 STs, and CC3 with 3 STs. The founders for these CCs are ST40, ST21, and ST27, respectively. The remaining 9 CCs have only 2 member STs, and the founders for these CCs cannot be determined.
FIG. 1.
eBURST analysis of Y. pseudotuberculosis isolates. The 62 STs were analyzed with eBURST (17). Each ST is represented by a dot. STs differing by only one locus were defined as members of a CC. Members of a CC are linked by solid lines, with the single locus difference shown by the gene symbols of the 7 MLST genes. Recombination (differing by 2 or more bases) or mutation (differing by 1 base) is indicated by a superscripted R or M, respectively. CC numbers are shown on the left.
The 83 isolates represented 19 serotypes. There are STs and clonal complexes that contain multiple serotypes, suggesting recent serotyping conversion. The STs and clonal complexes containing multiple serotypes are shown in Table 2. Among the 12 STs with more than 1 isolate, 7 (ST2, ST6, ST9, ST21, ST36, ST37, and ST44) contain isolates of different serotypes. ST21 has the most serotypes, with each of the 4 isolates belonging to a different serotype. ST2, which has the most isolates, contains 3 different serotypes (3 serotype 1a isolates, 1 serotype 1b isolate, and 1 serotype 13 isolate). The remaining 5 STs have 2 serotypes each. Similarly, at the clonal complex level, 9 of the 12 clonal complexes contain 2 or more serotypes, with CC1 and CC2 having 8 and 7 serotypes, respectively.
TABLE 2.
STs and CCs containing more than 1 serotype
| ST or CC | Serotypes |
|---|---|
| ST2 | 1a, 1b, 13 |
| ST6 | 3, 6 |
| ST9 | 3, 12 |
| ST21 | 1b, 5a, 11, 12 |
| ST36 | 2a, 4b |
| ST37 | 2a, 2b |
| ST44 | 2c, 4a |
| CC1 | 1b, 2a, 2b, 2c, 3, 4b, 5a, 5b |
| CC2 | 1b, 5a, 7, 9, 10, 11, 12 |
| CC3 | 1b, 4a |
| CC5 | 3, 12 |
| CC7 | 2b, 5a |
| CC8 | 1c, 10 |
| CC10 | 1b, 3, 6 |
| CC11 | 5b, 7 |
| CC12 | 2a, 2b |
Population structure and detection of recombination.
We first used the Bayesian statistics tool STRUCTURE to divide the 83 isolates (excluding Y25) into subpopulations and to visualize recombination between subpopulations. Three subpopulations were found, with 57, 15, and 11 isolates belonging to subpopulations I, II, and III, respectively (Fig. 2 ). The ancestry of each isolate was then estimated as the sum of probabilities from each subpopulation over all polymorphic nucleotides. Seventeen isolates were found to contain ancestral nucleotides from another subpopulation (Fig. 2). The proportion of nucleotides from one or more subpopulations varied from 5% to 49%.
FIG. 2.
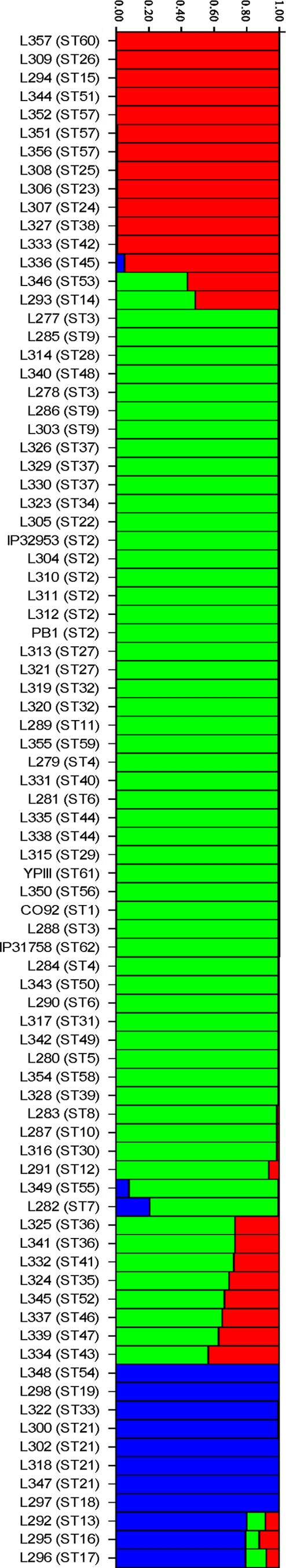
Structure analysis of Y. pseudotuberculosis isolates. The three subpopulations are color coded green, red, and blue for subpopulations I, II, and III, respectively. Each isolate has been allocated to a subpopulation. The isolates were identified by strain name, with the ST in parentheses, on the left. Mosaic colors for an isolate indicate mixed population origins from the respective populations with matching colors. The y axis represents the percentage of population assignment, i.e., the proportions of ancestry from the 3 subpopulations as color coded.
We then used the counting method of Feil et al. (18) to determine the ratio of recombination to mutation per locus. The single allele difference between STs within a clonal complex was attributed to mutation if the difference was a single base and otherwise to recombination. Out of a total of 26 allelic changes that occurred in the 12 clonal complexes, 13 each were mutational and recombinational events, with a 1:1 ratio of recombination to mutation, suggesting that the 2 types of events occur at similar frequencies. Two events, both in aroC, involved alleles differing by only 2 bases. These 2 events were assigned as recombinations based on the rule but could also be mutational events. Two genes (aroC and fumC) had only recombinational events, while 3 genes (gyrB, mdh, and recA) had only mutational events. The remaining two genes (pgi and sucA) had both types of events.
We further assessed the level of recombination by compatibility analysis of the 7 genes using the program RETICULATE of Jakobsen and Easteal (28). We calculated both within-gene and between-gene compatibility values, excluding the divergent isolate Y25. sucA has the lowest average within-locus compatibility, at only 71%, while the remaining 6 genes have an average value over 90%. In contrast, compatibility values between genes were much lower than those within genes, ranging from 27% to 69%. The lowest was between fumC and other genes at 27%.
Phylogenetic relationships.
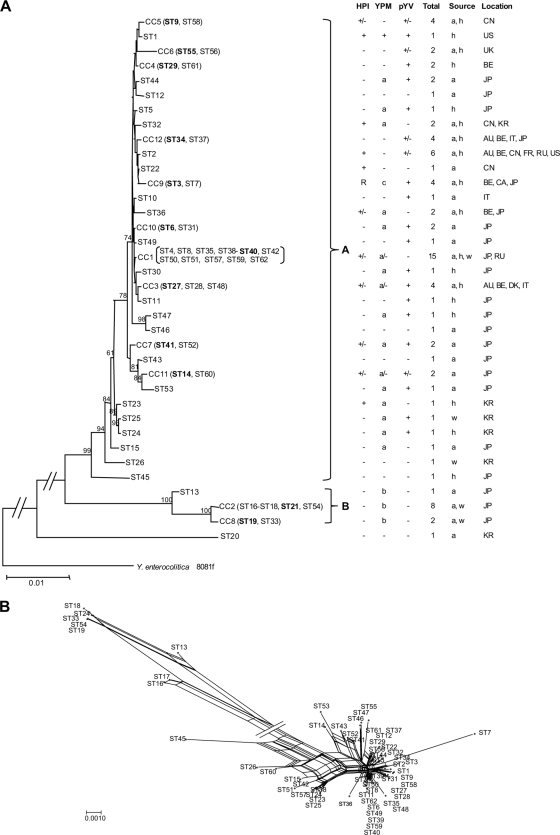
A neighbor-joining tree was constructed using the concatenated sequences of the 7 genes (Fig. 3A). To reduce the size of the tree, clonal complexes were represented by the founder or a randomly selected ST if there was no founder. The Y. enterocolitica genome sequenced strain 8081 (48) was used as an outgroup. The neighbor-joining tree shows two major clusters, A and B, which are supported by bootstrap values of over 90%. The majority of the isolates (72 isolates) are in cluster A, while 11 isolates are in cluster B and 1 (Y25 [ST20]) was not clustered. The divisions are concordant with population divisions based on STRUCTURE analysis. Cluster A is inclusive of subpopulations I and II, while cluster B contains only subpopulation III. We also constructed a neighbor net dendrogram, which revealed an extensive network structure for the STs due to the effect of recombination (Fig. 3B). The isolates studied were previously typed for the presence of the three key virulence factors, the HPI, pYV, and YPM. The presence or absence of these genes was mapped onto the phylogenetic tree. The evolutionary implications are discussed in detail below.
FIG. 3.
Phylogenetic relationships of Y. pseudotuberculosis. (A) Neighbor-joining tree of the 62 STs. STs that form CCs are shown with the CC number followed by the members of the CC in parentheses. The founder of the CC is highlighted in boldface. On the right of the tree are shown the presence and absence of the three virulence factors, the HPI, YPM, and pYV; the number of isolates in each ST; and the source and location of isolation. The sources are h (human), w (water), and a (animal). The locations are AU (Australia), BE (Belgium), CA (Canada), CN (China), DK (Denmark), FR (France), IT (Italy), JP (Japan), KR (South Korea), RU (Russia), UK (England), and US (United States). Y. enterocolitica strain 8081f was used as the outgroup. Bootstrap values greater than 50% are shown at the nodes of the neighbor-joining trees. The name of each cluster is indicated at the right brace. (B) Neighbor net network of the 61 STs, excluding ST20.
DISCUSSION
Y. pseudotuberculosis has an intermediate level of recombination.
In comparison to the other 2 well-known enteric species, E. coli and S. enterica, little was known about the population structure of Y. pseudotuberculosis. From the analysis of 83 representative isolates by MLST, we found that Y. pseudotuberculosis has an intermediate level of recombination. The relative level of recombination in Y. pseudotuberculosis can be compared with those of other bacterial species. By compatibility analysis, Y. pseudotuberculosis has lower between-loci compatibility, averaging 65% over the seven gene comparisons, than those of both S. enterica (90%) and E. coli (76%) but has higher compatibility than that of S. enterica subsp. I (49%) from the data we reported previously (38). Based on the ratio of recombination to mutation that occurred within clonal complexes, Y. pseudotuberculosis has a ratio of 1:1. Although the ratio is quite high, it is much lower than that in Neisseria meningitidis (10:1). On the other hand, it is much higher than those in a number of other species, including Staphylococcus aureus (1:15) (47). Hence, Y. pseudotuberculosis has a moderately clonal population structure.
Cluster A is widely distributed, while cluster B has a restricted geographical distribution.
Previous studies have shown geographically restricted distribution of certain genetic groups (24). MLST data revealed that cluster A isolates are distributed on four different continents and in 12 countries. The wide distribution of cluster A isolates is also evident at the clonal complex level. The 72 cluster A isolates are divided into 10 CCs and 22 singletons. CC1, CC3, CC9, and CC12 are present in multiple countries across the globe. However, cluster B is geographically restricted to Japan. As the isolates used in this study were selected from a much larger study of over 2,000 isolates by Fukushima et al. (24), cluster B can now been seen to represent genetic group 4 in that study, as both contain only YPMb isolates. In the study by Fukushima et al. (24), there were 93 isolates belonging to genetic group 4, all isolated in Japan.
Evolutionary dynamics of the three virulence factors, the HPI, YPM, and pYV.
All isolates used in this study have been previously typed for the presence of the 3 virulence factors, the HPI, YPM, and pYV (24). The gain and loss of these virulence factors can be interpreted based on evolutionary relationships established using MLST (Fig. 3A). The HPI is important for systemic disease, as it encodes a system for iron acquisition (5). Sixteen STs on 12 branches in cluster A carry the HPI, while there are no HPI+ isolates in cluster B (Fig. 3). The distribution of the HPI is restricted to a few lineages. It is likely that these lineages gained the HPI independently. It is possible that the ancestral Y. pseudotuberculosis was HPI positive and that lack of the HPI is due to loss, but this scenario is far less likely. Thus, the ability to cause systemic disease is likely to have developed independently in different lineages. However, there are 2 cases where the HPI+ STs are grouped together (ST1 and ST9; ST2, ST22, and ST3), suggesting that in each case the HPI might have been gained by their common ancestors. The R-HPI, in which the 5′ end carrying key iron acquisition genes is deleted, is present in 2 related STs, ST3 and ST7. The deletion must have occurred before ST3 and ST7 diverged. Thus, there are limited independent gains of the HPI. Lesic and Carniel (34) reported that the HPI is transferable in laboratory culture under low-temperature conditions in a RecA-dependent manner, and the recipient strain's genetic background determines whether an HPI can be transferred into it. Such requirements might have restricted the acquisition of the HPI by different strains in the natural environment, since only a limited number of lineages contain the HPI. The gain or loss of the HPI may be quite dynamic, as 3 STs (ST9, ST27, and ST36) contain both HPI+ and HPI− isolates.
In contrast to the restricted distribution of the HPI, YPM is widely distributed on the evolutionary tree (Fig. 3A). Among the 3 known ypm alleles (7, 42), ypmA and ypmC differ by 1 base (7) while ypmA and ypmB differ by 11% at the DNA level (42). ypmA and ypmC are in cluster A only, with the former most widely distributed while the latter is restricted to the R-HPI-containing STs. All ypmC-carrying isolates are closely related. It is clear that ypmA gave rise to ypmC with a single base change. As can be seen from Fig. 3A, some ypmA-carrying isolates are clustered together, while others are interspersed with YPM-negative isolates. Overall, the number of YPM-carrying isolates is more than the number of YPM-negative isolates. The distribution patterns of YPM alleles in the phylogeny suggest that the ancestral Y. pseudotuberculosis was YPM positive and that the negative isolates have lost the ypm gene. It is possible that ypmA transfers frequently within cluster A, leading to the distribution patterns seen. However, the study by Carnoy et al. (6) showed that ypm is located in a highly unstable locus with a high frequency of deletion. Therefore, deletion, rather than gain, is more likely to account for the absence of a YPM in the isolates studied. The ypmB gene is present only in cluster B. The high level of sequence variation between ypmA and ypmB (42) indicates that either they diverged in parallel during the diversification of the 2 clusters or one of them was obtained from another species much earlier by one of the clusters.
pYV is a key virulence factor for Y. pseudotuberculosis (5), and hence, isolates without pYV are likely to be nonpathogenic, although pYV-negative isolates have been isolated from patients (25). The majority of the STs in cluster A carry pYV. However, some pYV-negative isolates may have lost the plasmid in laboratory culture, as loss is frequent (15). For STs containing multiple isolates, 5 STs, ST3, ST4, ST6, ST27, and ST57, are all pYV+ and 3 STs, ST2, ST9, and ST37, contain both pYV-positive and -negative isolates, suggesting recent natural or laboratory loss of pYV in the pYV-negative isolates. It seems that pYV is the least stable among the 3 virulence factors. ST32 contains 2 isolates, both of which are pYV negative. Interestingly, these 2 isolates are positive for both the HPI and YPM. All cluster B isolates are pYV negative but YPM positive, suggesting that this cluster is nonpathogenic, consistent with previous findings by Fukushima et al. (24) that all isolates were obtained from wild-animal or environmental sources. As all cluster B members are positive for ypmB, the gene may play a role in another environment.
Evolution of pathogenicity.
Y. pseudotuberculosis has previously been divided into high- and low-pathogenicity groups based on the presence of the HPI and pYV (5). Three STs (ST8 [CC1], ST52 [CC7], and ST60 [CC11]) belonging to 3 different lineages carry all 3 virulence factors. These STs can be regarded as the most virulent clones, and their high pathogenicity arose independently due to independent gain of the HPI, as discussed above.
Fukushima et al. (24) further divided Y. pseudotuberculosis into six genetic groups based on the presence of the HPI, YPM, and pYV. Two of the genetic groups can now also be seen as phylogenetic groups. Genetic group 4 equates to cluster B and is nonpathogenic (HPI− and pYV−). All isolates were from either wild animals or the environment; none were isolated from a human clinical source (24). Genetic group 5 carries the R-HPI and was referred to as a European low-pathogenicity group (24). As discussed above, the R-HPI isolates studied shared the most recent common ancestry. The 4 remaining genetic groups are not phylogenetic groups.
It is interesting that ST26 and ST45, both at the base of cluster A, do not contain any of these three virulence factors, suggesting that the ancestor of cluster A may not be pathogenic. However, the only ST45 isolate was obtained from a human and thus presumably is pathogenic. Further studies are needed to shed light on the node point where Y. pseudotuberculosis became pathogenic to humans.
Apart from gastroenteritis and systemic infection, Y. pseudotuberculosis can cause a particular clinical disease called Far East scarlet-like fever (FESLS). Strain IP31758, isolated from a FESLS patient, has been sequenced. IP31758 contains the HPI and ypmA, but no pYV, although a majority of FESLS isolates carry pYV. IP31758 has a unique ST, ST62, which belongs to CC1, the largest clonal complex. It would be interesting to analyze more FESLS isolates by MLST to determine whether they all belong to ST62 or are closely related to ST62 and thus whether FESLS pathogenicity was developed within CC1. Genome sequencing of IP31758 did not reveal FESLS-specific virulence factors (15).
O antigen diversity within sequence types and clonal complexes.
Serotype diversity in Y. pseudotuberculosis is determined by the O antigen gene cluster (44). Most serotypes are distributed in multiple lineages, suggesting extensive transfer of O antigen genes in Y. pseudotuberculosis. This high frequency of transfer has occurred at both ST and clonal complex levels (Table 2). Therefore, Y. pseudotuberculosis changes its O antigens quite rapidly. The switching in serotypes seems to be mediated by recombination of part of the O antigen gene cluster rather than a complete replacement of the O antigen gene cluster, based on known gene cluster structures (44). The O antigen gene clusters in O:2a and O:4b (present in ST36) are nearly identical throughout, except for one gene in the middle (39). Those in O:2c and O:4a (present in ST44) parallel those in O:2a and O:4b, again with one gene in the middle replaced (44). Those in O:2a and O:2b, present in ST37, have a common 5′ end. Those in O:1b and O:5a, present in ST21, share the same genes at both ends but differ in the middle by 6 or 7 genes (44). That in O:11, also present in ST21, shares the same backbone with O:1b (12). For the other serotypes present in the same ST or CC, the O antigen gene clusters are likely to share considerable homology. Thus, apart from the frequent recombination in housekeeping genes, O antigen genes also recombine frequently in Y. pseudotuberculosis, presumably under selection to generate serotype diversity.
The closest relative of Y. pestis so far identified.
Y. pestis is known to be a clone of Y. pseudotuberculosis (1). All genome-sequenced Y. pestis strains of different biovars, including CO92, KIM, and Angola (13, 16, 40), belong to a single ST, and its closest relative is ST29 (isolate H722-36/88) in cluster A. The 2 STs differ by 4 genes, with 1 base difference in three genes, aroC, fumC, and gyrB, and 2 differences in pgi. Considering that there are only 5 base changes in the total 4,223 bp of the 7 MLST gene fragments, these 2 STs are very closely related. Genome sequencing of H722-36/88 will provide the closest non-pestis relative to aid in understanding the earlier evolution of Y. pestis. Interestingly, H722-36/88, a human clinical isolate from Belgium, has the same serotype, 1b, as the ancestral but now nonfunctional Y. pestis serotype (46). Both H722-36/88 and Y. pestis are pYV positive but YPM negative. However, Y. pestis is also HPI positive (6), suggesting that the ancestral Y. pestis gained the HPI.
Conclusions.
An MLST scheme has been established for characterization of Y. pseudotuberculosis isolates. The population of Y. pseudotuberculosis was divided into 3 distinct subpopulations by STRUCTURE analysis and into 2 clusters by phylogenetic analysis. The two clusters differ in virulence and geographic distribution. Cluster A isolates have worldwide distributions and have been isolated from both human infections and animal and environmental sources. Cluster B contains only isolates from the Far East. Cluster A is pathogenic to humans in that most isolates have pYV and YPMa, with a smaller proportion carrying the HPI, while cluster B is nonpathogenic, with only YPMb present. The absence of a ypm gene and pYV in some cluster A isolates is most likely due to loss, while the HPI has been gained independently in several lineages. The genetic groups defined previously are not phylogenetic clades, with the exception of genetic groups 4 and 5. There are a limited number of lineages carrying all three virulence factors. The differences in pathogenicity among Y. pseudotuberculosis strains can be explained by the variable presence and instability of the virulence factors.
Supplementary Material
Acknowledgments
This study was supported by a faculty research grant and an Australian Research Council discovery project grant. Q.X. was supported by a Chinese Government visiting scholarship.
Footnotes
Published ahead of print on 3 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtman, M., et al. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleksić, S., J. Bockemuhl, and H. H. Wuthe. 1995. Epidemiology of Y. pseudotuberculosis in Germany, 1983-1993. Contrib. Microbiol. Immunol. 13:55-58. [PubMed] [Google Scholar]
- 3.Bandelt, H. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanovich, T., E. Carniel, H. Fukushima, and M. Skurnik. 2003. Use of O-antigen gene cluster-specific PCRs for the identification and O-genotyping of Yersinia pseudotuberculosis and Yersinia pestis. J. Clin. Microbiol. 41:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 6.Carnoy, C., et al. 2002. The superantigen gene ypm is located in an unstable chromosomal locus of Yersinia pseudotuberculosis. J. Bacteriol. 184:4489-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnoy, C., H. Müller-Alouf, S. Haentjens, and M. Simonet. 1998. Polymorphism of ypm, Yersinia pseudotuberculosis superantigen-encoding gene. Zentralbl. Bakteriol. 29:397-398. [Google Scholar]
- 8.Carnoy, C., C. Mullet, H. Muller-Alouf, E. Leteurtre, and M. Simonet. 2000. Superantigen YPMa exacerbates the virulence of Yersinia pseudotuberculosis in mice. Infect. Immun. 68:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P. S., et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collyn, F., et al. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 12.Cunneen, M. M., et al. 2009. The O-specific polysaccharide structure and biosynthetic gene cluster of Yersinia pseudotuberculosis serotype O:11. Carbohydr. Res. 344:1533-1540. [DOI] [PubMed] [Google Scholar]
- 13.Deng, W., et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolz, R. 1994. GCG, p. 9-17. In A. M. Griffin and H. G. Griffin (ed.), Computer analysis of sequence data, methods in molecular biology, vol. 25. Humana, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 15.Eppinger, M., et al. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eppinger, M., et al. 2010. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J. Bacteriol. 192:1685-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 20.Fukushima, H. 1992. Direct isolation of Yersinia pseudotuberculosis from fresh water in Japan. Appl. Environ. Microbiol. 58:2688-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukushima, H. 2003. Molecular epidemiology of Yersinia pseudotuberculosis. Adv. Exp. Med. Biol. 529:357-358. [DOI] [PubMed] [Google Scholar]
- 22.Fukushima, H., et al. 1994. Restriction endonuclease analysis of virulence plasmids for molecular epidemiology of Yersinia pseudotuberculosis infections. J. Clin. Microbiol. 32:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukushima, H., M. Gomyoda, K. Shiozawa, S. Kaneko, and M. Tsubokura. 1988. Yersinia pseudotuberculosis infection contracted through water contaminated by a wild animal. J. Clin. Microbiol. 26:584-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukushima, H., et al. 2001. Geographical heterogeneity between Far Eastern and Western countries in prevalence of the virulence plasmid, the superantigen Yersinia pseudotuberculosis-derived mitogen, and the high-pathogenicity island among Yersinia pseudotuberculosis strains. J. Clin. Microbiol. 39:3541-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukushima, H., T. Sato, R. Nagasako, and I. Takeda. 1991. Acute mesenteric lymphadenitis due to Yersinia pseudotuberculosis lacking a virulence plasmid. J. Clin. Microbiol. 29:1271-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon, D., C. Abajian, and P. Green. 1998. CONSED—a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 27.Huson, D. H. 1998. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsen, I. B., and S. Easteal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. 12:291-295. [DOI] [PubMed] [Google Scholar]
- 29.Jalava, K., et al. 2006. An outbreak of gastrointestinal illness and erythema nodosum from grated carrots contaminated with Yersinia pseudotuberculosis. J. Infect. Dis. 194:1209-1216. [DOI] [PubMed] [Google Scholar]
- 30.Jalava, K., et al. 2004. Multiple outbreaks of Yersinia pseudotuberculosis infections in Finland. J. Clin. Microbiol. 42:2789-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kangas, S., et al. 2008. Yersinia pseudotuberculosis O:1 traced to raw carrots, Finland. Emerg. Infect. Dis. 14:1959-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidgell, C., et al. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 33.Laukkanen, R., et al. 2008. Transmission of Yersinia pseudotuberculosis in the pork production chain from farm to slaughterhouse. Appl. Environ. Microbiol. 74:5444-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesic, B., and E. Carniel. 2005. Horizontal transfer of the high-pathogenicity island of Yersinia pseudotuberculosis. J. Bacteriol. 187:3352-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meats, E., et al. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milkman, R., E. Jaeger, and R. D. McBride. 2003. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics 163:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuorti, J. P., et al. 2004. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J. Infect. Dis. 189:766-774. [DOI] [PubMed] [Google Scholar]
- 38.Octavia, S., and R. Lan. 2006. Frequent recombination and low level of clonality within Salmonella enterica subspecies I. Microbiology 152:1099-1108. [DOI] [PubMed] [Google Scholar]
- 39.Pacinelli, E., L. Wang, and P. R. Reeves. 2002. Relationship of Yersinia pseudotuberculosis O antigens IA, IIA, and IVB: the IIA gene cluster was derived from that of IVB. Infect. Immun. 70:3271-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkhill, J., et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramamurthy, T., K. Yoshino, J. Abe, N. Ikeda, and T. Takeda. 1997. Purification, characterization and cloning of a novel variant of the superantigen Yersinia pseudotuberculosis-derived mitogen. FEBS Lett. 413:174-176. [DOI] [PubMed] [Google Scholar]
- 43.Reeves, P. R., L. Farnell, and R. Lan. 1994. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. Comput. Appl. Biosci. 10:281-284. [DOI] [PubMed] [Google Scholar]
- 44.Reeves, P. R., E. Pacinelli, and L. Wang. 2003. O antigen gene clusters of Yersinia pseudotuberculosis. Adv. Exp. Med. Biol. 529:199-206. [DOI] [PubMed] [Google Scholar]
- 45.Rimhanen-Finne, R., et al. 2009. Yersinia pseudotuberculosis causing a large outbreak associated with carrots in Finland, 2006. Epidemiol. Infect. 137:342-347. [DOI] [PubMed] [Google Scholar]
- 46.Skurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37:316-330. [DOI] [PubMed] [Google Scholar]
- 47.Spratt, B. G. 2004. Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266:323-352. [DOI] [PubMed] [Google Scholar]
- 48.Thomson, N. R., et al. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsubokura, M., and S. Aleksi. 1995. A simplified antigenic scheme for serotyping of Yersinia pseudotuberculosis: phenotypic characterization of reference strains and preparation of O and H factor sera. Contrib. Microbiol. Immunol. 13:99-105. [PubMed] [Google Scholar]
- 50.Vincent, P., et al. 2008. Sudden onset of pseudotuberculosis in humans, France, 2004-05. Emerg. Infect. Dis. 14:1119-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wirth, T., et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.