Abstract
The ascomycete fungus Mycosphaerella graminicola is an important pathogen of wheat that causes Septoria tritici blotch. Despite the serious impact of M. graminicola on wheat production worldwide, knowledge about its molecular biology is limited. The velvet gene, veA, is one of the key regulators of diverse cellular processes, including development and secondary metabolism in many fungi. However, the species analyzed to date are not related to the Dothideomycetes, the largest class of plant-pathogenic fungi, and the function of veA in this group is not known. To test the hypothesis that the velvet gene has similar functions in the Dothideomycetes, a veA-homologous gene, MVE1, was identified and gene deletion mutations (Δmve1) were generated in M. graminicola. All of the MVE1 mutants exhibited consistent pleiotropic phenotypes, indicating the involvement of MVE1 in multiple signaling pathways. Δmve1 strains displayed albino phenotypes with significant reductions in melanin biosynthesis and less production of aerial mycelia on agar plates. In liquid culture, Δmve1 strains showed abnormal hyphal swelling, which was suppressed completely by osmotic stress or lower temperature. In addition, MVE1 gene deletion led to hypersensitivity to shaking, reduced hydrophobicity, and blindness to light-dependent stimulation of aerial mycelium production. However, pathogenicity was not altered in Δmve1 strains. Therefore, the light-signaling pathway associated with MVE1 does not appear to be important for Septoria tritici blotch disease. Our data suggest that the MVE1 gene plays crucial roles in multiple key signaling pathways and is associated with light signaling in M. graminicola.
The ascomycete fungus Mycosphaerella graminicola (anamorph Septoria tritici) is the causal agent of Septoria tritici blotch (STB), a serious foliar disease of wheat. This disease has been reported in most wheat-growing regions around the world and can cause yield reductions of 30 to 40% during severe epidemics (26). Moreover, STB is the largest fungicide target on wheat, causing significant economic expenses for disease management worldwide (21, 26). Despite the high economic impact of STB, efficient management strategies against M. graminicola have not been developed fully. Breeding for resistant cultivars and treatments with fungicides are currently used to manage STB. However, neither of those methods provides satisfactory control due to rapid evolution of M. graminicola populations and frequent sexual recombination, which allow the pathogen to overcome available fungicides or resistant cultivars (9, 29). Absence of efficient management strategies for control of STB could be explained in part by our limited knowledge of M. graminicola biology. Therefore, a better understanding of key signaling pathways associated with M. graminicola biology will be essential to facilitate the development of efficient control strategies against STB. Signaling pathways not directly involved in pathogenicity of M. graminicola also are important for a complete understanding of the biology of this economically important pathogen.
The velvet protein encoded by veA has multiple key regulatory roles in fungal development and secondary metabolism (4, 6). Almost half a century ago, a velvet mutant was identified initially in Aspergillus nidulans (14), and since then, veA-homologous genes in several filamentous fungi have been studied intensively. Previous analyses of veA have revealed conserved global regulatory roles influencing fungal phenotypes of tremendous importance, such as morphogenesis and biosynthesis of an array of fungal mycotoxins or antibiotics. In A. nidulans, veA governs both sexual and asexual development by positively regulating formation of cleistothecia while suppressing conidiation (14, 23). As with veA in A. nidulans, gene deletion of veA in Neurospora crassa and Aspergillus flavus increased asexual development (5, 11). However, conidiation is linked to veA in a positive manner in another species, Aspergillus parasiticus, suggesting different functional roles of veA on asexual developments among fungal species (7). In Acremonium chrysogenum, the veA-homologous gene AcveA is associated with hyphal fragmentation and hyperbranching of hyphal tips (10). Besides these effects on fungal morphology, veA-homologous genes also regulate the production of fungal secondary metabolites, including fumonisins and fusarins in Fusarium verticillioides (25), the β-lactam antibiotic cephalosporin C in Acremonium chrysogenum (10), sterigmatocystin and the antibiotic penicillin in A. nidulans (16), and aflatoxin in A. parasiticus (7). Interestingly, most of the phenotypes mediated by veA are light dependent, indicating that this gene may be an important component of fungal light-signaling pathways.
Despite fundamental roles of veA in fungal morphology and secondary metabolite-related chemical development, as well as being an indispensable factor in light-signaling pathway(s), no analyses of veA-homologous genes have been performed on species in the Dothideomycetes, the largest class of plant-pathogenic fungi, such as M. graminicola. In addition, although the signaling pathways associated with veA are well conserved among fungal species, it is clear that there are similarities or differences in veA-mediated signaling among species in different classes of filamentous fungi and even among members of the same genus. Therefore, analysis of veA signaling in the economically important pathogen and model dothideomycete M. graminicola should be performed to test whether there are veA-associated mechanisms that are specific to M. graminicola or whether they are conserved with those in the fungi that have been studied previously.
The purpose of this research was to test the function of the MVE1 gene encoding a veA-homologous protein in M. graminicola. MVE1 gene deletion mutants showed pleiotropic effects on aerial mycelium formation, melanin biosynthesis, hyphal swelling, sensitivity to orbital shaking conditions, and hydrophobicity. Moreover, the MVE1 gene deletion strains did not produce aerial mycelia in response to light. Our results demonstrate that the MVE1 gene plays indispensable roles in multiple key signaling pathways and is associated with the light-signaling pathway(s) in the dothideomycete M. graminicola.
MATERIALS AND METHODS
Fungal strains and culture media.
Highly virulent M. graminicola strain IPO323 was used as the wild type throughout this study and was the subject of all gene disruptions. For MVE1 gene characterization, we analyzed six strains, including the wild type, one ectopic transformant (Ect) as a control, and four independent MVE1 mutants under the same experimental sets or conditions throughout this study. The genome of the wild-type strain has been sequenced completely by the U.S. Department of Energy's Joint Genome Institute (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html). The availability of this sequence allowed identification of the veA-homologous gene in M. graminicola and facilitated primer design for knockout constructs. All of the strains used or generated in this study were stored at −80°C after desiccation on strips of Whatman filter paper (Whatman, Inc., Piscataway, NJ) overnight in a lyophilizer. Cultures grown in yeast-sucrose broth (YSB; 10 g each of sucrose and yeast extract per liter of distilled water) were used for genomic DNA extraction or Agrobacterium tumefaciens-mediated transformation (ATMT). For phenotyping, we used various agar media: yeast-sucrose agar (YSA; YSB with 15 g of agar per liter), potato dextrose agar (PDA; 4%) (Difco Laboratories, Detroit, MI), and water agar (WA; 15 g of agar per liter). For analysis of aerial mycelium, we grew each of the strains on PDA for 10 days at room temperature (∼23 to 28°C) or at 18°C.
Nucleic acid isolation and manipulation.
All plasmid DNAs used in this study were prepared with the QIAprep spin miniprep kit (Qiagen, Valencia, CA). The MVE1 gene knockout construct, DVE1, was made via double-joint PCR as described previously (31). All primers used for PCR are listed in Table 1. Primer pairs MVE1-dis-A and MVE1-dis-B and MVE1-dis-C and MVE1-dis-D were used to amplify regions of approximately 1 kb flanking the MVE1 gene from M. graminicola genomic DNA based on the genomic sequence. Simultaneously, a hygromycin phosphotransferase (HPH) gene was amplified from plasmid vector pBHT2 (24) with HPH-F and HPH-R primers, using AccuSure DNA polymerase (Bioline, Taunton, MA). Subsequently, all three amplicons were mixed in a single tube in a 1:3:1 5′-fragment-marker-3′-fragment molar ratio and joined by PCR without primers. Finally, nested primers MVE1-nest-F and MVE1-nest-R were used to amplify the 3.5-kb DVE1 construct (Fig. 1). Primers MVE1-nest-F and MVE1-nest-R contain recognition sites for the restriction enzymes XbaΙ and HindIIΙ, respectively. Using these enzyme sites, the DMC1 construct was cloned into vector pBHT2 (24) for ATMT. The resulting vector, designated pMV1, was subsequently transformed into competent cells of Agrobacterium by electroporation following the standard protocols for further use of ATMT.
TABLE 1.
Primers used for generation and analysis of Mycosphaerella graminicola MVE1 deletion mutants
| Primer no. | Primer name | Primer sequence (5′→3′)a |
|---|---|---|
| 1 | HYG-F | GTCCAGGCGGTGAGCACAAAA |
| 2 | HYG-R | TTCCCGGTCGGCATCTACTCTATTC |
| 3 | MVE1-dis-A | CAGTTCCCAACTACTTCGTGCTCC |
| 4 | MVE1-dis-B | TTTTGTGCTCACCGCCTGGACCGGTGGTCAATGAGTATATCCAGCAG |
| 5 | MVE1-dis-C | GAATAGAGTAGATGCCGACCGGGAACTCACACTCCACAGACGGGTTACC |
| 6 | MVE1-dis-D | GACTTCATGCAGCCAGTCGACATC |
| 7 | MVE1-nest-F | GTCTCATTGGTTGGtctagaAAGGCTGCAG |
| 8 | MVE1-nest-R | CGTCGTCGGCAACAGaagcttGCAATGATG |
| 9 | MVE1-che-F | GCCCGCCTTTCATTCGCTTTG |
| 10 | MVE1-che-R | GAGGCTGTGGGTAAGTCTGC |
| 11 | MVE1-neg-F | GTTCATACCACCTTCAAGTTTGTC |
| 12 | MVE1-neg-R1 | CATCCATGTAGCGAGCATTCTG |
Nucleotide sequences from the hygromycin phosphotransferase gene are underlined, and restriction enzyme sites introduced into the primers for cloning purposes are lowercase.
FIG. 1.
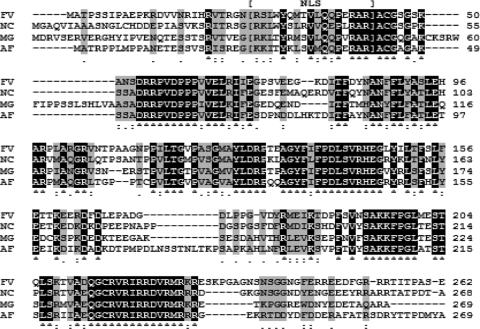
In silico bioinformatic analysis of MVE1. Amino acid alignment of the Mycosphaerella graminicola velvet gene ortholog Mve1 (MG) with those from Fusarium verticillioides (FV), Neurospora crassa (NC), and Aspergillus fumigatus (AF) was performed via ClustalW. Amino acids common to all proteins are indicated by white letters on a black background. Similar amino acids are indicated by a gray background. Mve1 displays significant similarity to all of the velvet proteins included in this analysis. Amino acids corresponding to the nuclear localization signal (NLS) are indicated by brackets.
Agrobacterium tumefaciens-mediated transformation.
An Agrobacterium strain containing the MVE1 knockout construct pVE1 was used for transformation into M. graminicola. Cells for gene disruption were grown in YSB at 18°C for 2 weeks in an incubated orbital shaker. ATMT was carried out using fungal spores adjusted to 1 × 107/ml by essentially following the protocol described previously (32). PDA containing 100 U of hygromycin and 200 μM cefotaxime was used for the selective medium. After 2 to 3 weeks, hygromycin-resistant colonies appeared on the selective medium, and every isolated colony was transferred for subsequent subculture on fresh selective medium for further verification of DVE1 incorporation into the genome.
Melanin assays.
Melanin was extracted following the protocol described previously (1). Fungal mycelia grown on YSA and YSB at room temperature for 15 days were used for the melanin extraction. Briefly, melanin was extracted with 2% NaOH at 100°C for 2 h in a water bath. The cooled extracts were acidified to pH 2.0 with HCl to precipitate the melanin pigments. Subsequently, extracts were centrifuged to separate melanin, and the resulting pellets were dissolved in 2% NaOH. The absorbance of the extract was measured spectrophotometrically at 459 nm, and the amount of melanin was determined by a calibration curve plotted from the results of a correlation between melanin dry weight and absorbance. All assays were biologically replicated at least three times.
Microscopy.
Microscopic observations were made with a dissecting (Olympus America, Inc., Melville, NY) or compound (Olympus America, Inc., Melville, NY) microscope. For images of aerial mycelia and pycnidia, we used a model SZX12 dissecting microscope and Qcapture software. Hyphal growth morphologies in the liquid medium were analyzed with DP controller software and an Olympus BX41 microscope equipped with a DP70 camera.
Aerial mycelium development under different light conditions.
To determine light dependence of MVE1, we used the experimental instruments designed specifically by Flaherty and Dunkle (12) for establishing the ranges of transmitted light. Briefly, boxes with a window containing blue or red cellophane were placed beneath a light-emitting diode (LED) lamp at room temperature. Therefore, only specific wavelengths of light (blue, 465 to 480 nm; or red, 610 to 800 nm) were transmitted to the fungal cultures inside the boxes. Fungal cultures were incubated on water agar with an initial inoculation of 105 cells for 10 days under the indicated light conditions. Two sets of light conditions were included: continuous or temporary. For continuous light, cultures were subjected to 24 h of illumination, whereas for temporary light, we shifted light-inoculated cultures to continuous darkness after 2 consecutive days of illumination. Ten days later, morphological differences under both treatments were analyzed microscopically.
Growth phenotypes in liquid medium.
Cultures grown in YSB were used to analyze phenotype. Cultures were initiated with 1 × 105 cells/ml in 250-ml Erlenmeyer flasks containing 100 ml of medium and were shaken at 0, 50, or 120 rpm for 10 or 15 days at either room temperature or 18°C. To test the effect of osmotic stabilization, we grew cultures in YSB with the addition of 1 M sorbitol under the same conditions described above.
Virulence assays.
The susceptible wheat line Taichung 29 was inoculated by stem or spray inoculations. Briefly, we grew Taichung 29 in a Purdue University greenhouse facility until the wheat seedlings reached the 3- to 5-leaf stage. Fungal inoculum was prepared from 2-week-old cultures of M. graminicola grown in YSB at room temperature in an orbital shaker adjusted to 120 rpm. For inoculations, we followed the protocol described previously (21, 22). Wheat was inoculated by spraying 108 spores suspended in water per pot. After inoculation, the pots were placed in the dark at near 100% humidity for 48 h. Subsequently, infected wheat was incubated in 14-h light/10-h dark cycles. After 21 days, when distinct STB disease symptoms of clear black pycnidia appeared, we analyzed the extent of the infections. We also analyzed pycnidial development with a dissecting microscope. All assays were repeated at least three times.
RESULTS
MVE1 encodes the velvet protein in Mycosphaerella graminicola.
The M. graminicola genomic database (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html) was searched for a veA-homologous gene with the nucleotide sequence of the Fusarium verticillioides velvet gene, FvVe1 (GenBank accession no. DQ274059) (19), as a query. The search revealed a putative veA gene of 810 bp on chromosome 2, bases 2995014 to 2995823, of the M. graminicola genomic sequence. Analysis of the conceptually translated amino acids revealed significant similarities to various veA genes across different fungal species. For example, it shared high similarity with sexual development activator VeA (XP_752619.1) in A. fumigatus (E value, 1e−65), VeA (ABC02879.1) in F. verticillioides (E value, 9e−57), and a VeA-homologous gene (XP_957154.1) in Neurospora crassa (E value, 1e−59). Based on these similarities, we designated the identified gene in M. graminicola as MVE1 (probable M. graminicola Velvet-like gene 1). Analysis of amino acid alignments showed that the deduced sequence of the MVE1 product corresponded to the N-terminal region of the velvet proteins in other filamentous fungi and is highly conserved with VeA proteins in F. verticillioides, A. fumigatus, and N. crassa (Fig. 1). Further in silico analyses demonstrated that Mve1 has a clear nuclear localization signal (NLS) from amino acids 32 to 48, based on previous studies of N. crassa (5), but no nuclear export signal (NES) was identified in the database search (http://www.cbs.dtu.dk/services/NetNES/).
MVE1 gene deletion mutants were generated via ATMT.
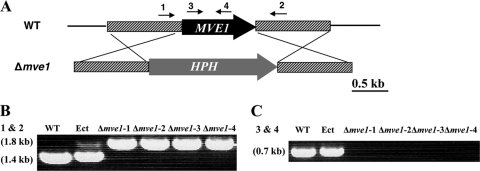
To investigate the functional roles of MVE1 in M. graminicola, we generated MVE1 gene deletion mutants via ATMT (24). Among 72 hygromycin-resistant transformants, 11 independent MVE1 gene deletion mutants were identified in which the entire MVE1 gene was replaced by the gene knockout construct DVE1. PCR screens with the gene-specific primers MVE1-che-F1 and -R1 (Table 1) were performed to confirm the homologous recombination event. The MVE1-che-F1 and -R1 primers are located on the upstream and downstream flanking regions of the MVE1 open reading frame (ORF), and the resultant MVE1 gene deletion mutants should have the entire MVE1 ORF replaced with a hygromycin phosphotransferase gene (HPH) (Fig. 2 A). Based on that, we identified gene deletion mutants of MVE1 by the different sizes of PCR amplicons. The wild-type and ectopic control strains had a 1.4-kb band originating from the MVE1 ORF, whereas MVE1 gene deletion mutants had a 1.8-kb band due to replacement of MVE1 by the HPH gene (Fig. 2B). Transformation was confirmed with another set of primers, MVE1-neg-F1 and -R1 (Table 1), located within the MVE1 ORF. The wild-type and ectopic control strains generated an expected 0.7-kb band from the MVE1 ORF, whereas MVE1 gene disruption strains did not produce any detectable PCR amplicons, confirming MVE1 deletion in the putative mutants (Fig. 2C). We selected four isolates randomly among 11 mutants for further MVE1 gene characterization, as all 11 mutants showed the same morphological phenotypes. We designated the four independent mutants as the Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4 strains. As a second control, we included an ectopic strain containing the DVE1 construct integrated at an unknown location in the genome and designated it Ect. Throughout this study, these six strains (the wild-type strain, Ect control, and four independent MVE1 mutant strains) were analyzed simultaneously for each set of experiments.
FIG. 2.
Replacement construct for the MVE1 gene of Mycosphaerella graminicola and generation of mutants. (A) Schematic drawing of the MVE1 locus and gene replacement event. Agrobacterium tumefaciens-mediated transformation vector pVE1 harbors the hygromycin phosphotransferase gene (HPH) as the selectable marker. A gene replacement event will replace the entire ORF of MVE1 with HPH. The numbered arrows indicate the locations of primers used for positive and negative PCR assays. (B and C) PCR analysis of MVE1 disruption using the primers described in panel 2A: primers 1 and 2 (B) and primers 3 and 4 (C). WT, wild type; Ect, ectopic integration strain; Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4, four independent MVE1 knockout isolates. The sizes of amplification products are indicated within parentheses on the left.
MVE1 gene deletion mutants are albino on agar plates, but do not differ in radial growth.
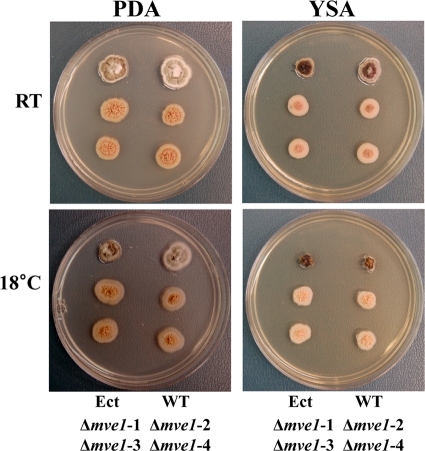
All of the MVE1 gene deletion mutants were defective in the production of melanin-like pigments. Compared to the wild-type and ectopic control strains, all four independent MVE1 gene deletion mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4) showed consistent albino phenotypes, regardless of growth medium (YSA or PDA) or temperature (room temperature or 18°C) (Fig. 3). However, radial growth rates of the M. graminicola Δmve1 mutants did not differ from those of the wild-type progenitor or the ectopic control strain on YSA or PDA plates (data not shown) (Fig. 3). Taken together, we concluded that MVE1 affects melanin-like dark pigmentation on agar plates in a positive manner, but is not associated with radial growth extension in M. graminicola.
FIG. 3.
Morphology on agar plates of Mycosphaerella graminicola wild-type and transformed strains. Growth morphology on agar plates was tested with the wild-type (WT) strain, the ectopic integration (Ect) strain, and four independent MVE1 gene deletion mutants. All strains were grown on potato-dextrose agar (PDA) or yeast-sucrose agar (YSA) for 15 days. Dark pigmentation was completely absent in the four independent MVE1 mutant isolates (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4), regardless of temperature or growth medium. Locations of inoculated strains on the agar plates are indicated at the bottom of each set of images. The growth temperatures are indicated on the left. RT, room temperature.
MVE1 gene deletion mutants are defective in melanin biosynthesis.
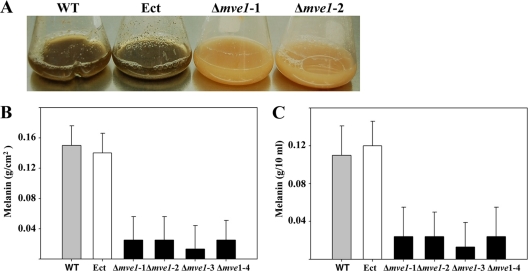
Deletion of the MVE1 gene resulted in complete lack of dark pigmentation on agar and in liquid medium, compared to the wild-type progenitor (Fig. 3 and 4 A). On agar plates and in liquid (YSB) medium, Δmve1 mutants were impaired in the biosynthesis of dark pigmentation compared to the wild-type and ectopic control strains (Fig. 3 and 4A). It has been generally well accepted that the secondary metabolite melanin is primarily responsible for the dark pigmentation in many filamentous fungi. Based on that, we reasoned that the dark pigment in the wild-type and ectopic control strains is melanin. To test this hypothesis, we followed an extraction and measurement protocol specifically designed for melanin (1) to estimate the amounts of melanin produced by each strain with three biological replications on agar plates (YSA) as well as in liquid medium (YSB). We observed more than an 80% decrease in melanin production in Δmve1 mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4) on YSA culture compared to those of the wild-type progenitor or ectopic control (Fig. 4B). In addition, we observed approximately a 70% decrease in production of melanin in the four independent MVE1 disruption mutants in YSB (Fig. 4C). Our results clearly demonstrated that the MVE1 gene in M. graminicola is associated positively with melanin biosynthesis.
FIG. 4.
Assays for melanin production of Mycosphaerella graminicola grown in axenic culture. (A) Yeast-sucrose broth (YSB) cultures of the wild type (WT), an ectopic transformant (Ect), and two independent MVE1 gene deletion mutants (Δmve1-1, and Δmve1-2) were photographed after incubation for 15 days. Black pigmentation was observed in the wild type and ectopic transformant (Ect) but not in two independent MVE1 gene deletion mutants. (B and C) Measurements of melanin production in the wild type strain, Ect transformant, and four independent MVE1 gene deletion mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4) grown on YSA (B) or YSB (C) for 15 days. Data from three biological replications were analyzed to calculate means and standard errors, which are indicated by thin lines above the bars.
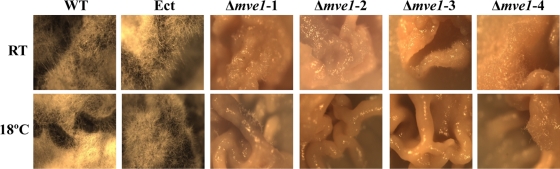
The MVE1 gene deletion mutants are defective in formation of aerial mycelia on agar plates.
Morphological growth patterns on PDA revealed differences between the wild-type or ectopic control strains and Δmve1 mutants. The wild-type and ectopic control strains developed long aerial mycelia, whereas Δmve1 mutants showed significant reduction in the amount of aerial mycelia produced (Fig. 5). The widespread development of thick, white mycelia was observed in the wild-type and ectopic control strains. In contrast, aerial mycelium was almost absent or even nondetectable in Δmve1 mutants (Fig. 5). These phenotypic differences appeared consistently across the four independent MVE1 gene disruption mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4), providing convincing evidence for a functional role of MVE1-mediated signaling in the development of aerial mycelia.
FIG. 5.
Production of aerial mycelia of the wild-type (WT) and transformed strains of Mycosphaerella graminicola. Aerial mycelium was photographed with a dissecting microscope. All strains were grown on potato-dextrose agar (PDA) for 10 days at room temperature (RT) or at 18°C (indicated on the left). Ect, ectopic control; Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4, four independent MVE1 mutants.
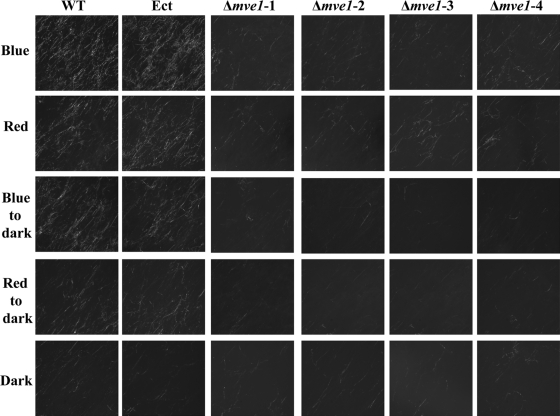
MVE1 gene deletion abolishes the light-stimulated formation of aerial mycelia.
MVE1 is homologous with the fungal velvet gene, which is known for its critical functions in the context of light-signaling pathways (4, 6, 23). Therefore, we tested whether different light conditions known to influence fungal asexual development could affect M. graminicola. To do so, all of the fungal strains used in this study were subjected to five treatments based on the following light conditions: continuous blue or red light, temporary blue or red light, and continuous darkness.
The development of aerial mycelium by wild-type and ectopic control strains was differentiated by the light conditions. Under continuous light, both strains produced more aerial mycelium under blue light than red and thus were more responsive to blue light (Fig. 6). In temporary blue or red light, both the wild-type and ectopic control strains produced more aerial mycelium under the temporary blue light than they did under the temporary red light. Aerial mycelium production in both strains was significantly reduced in complete darkness, indicating the requirement for light, particularly blue light, for its development (Fig. 6). Interestingly, Δmve1 mutants did not respond to environmental light cues for the development of aerial mycelia. Regardless of the light condition, even under blue light, all four independent Δmve1 mutants showed rare, sparse development of aerial mycelia, similar to that produced by the wild-type or ectopic control strains grown in darkness (Fig. 6).
FIG. 6.
Defects in development of aerial mycelia of the Mycosphaerella graminicola MVE1 mutants were independent of light or dark conditions. Aerial mycelium was photographed with a dissecting microscope. All strains were grown on water agar at room temperature for 10 days. Each fungal culture was grown under continuous blue- or red-light conditions as well as under temporary blue- or red-light conditions. For temporary blue or red light, each fungal strain was subjected to continuous blue or red light for 2 days and then shifted to complete darkness. The dark condition was established by wrapping fungal cultures with aluminum foil. A specific spectrum of light was allowed to penetrate into the culture boxes through the use of blue or red cellophane windows under an LED lamp. The light conditions of growth on agar plates are indicated on the left: Blue, fungal cultures grown under continuous blue light; Red, cultures grown under continuous red light; Blue to dark, cultures grown under temporary blue light; Red to dark, cultures grown under temporary red light; Dark, continuous darkness. WT, wild type; Ect, ectopic control; Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4, four independent MVE1 mutants.
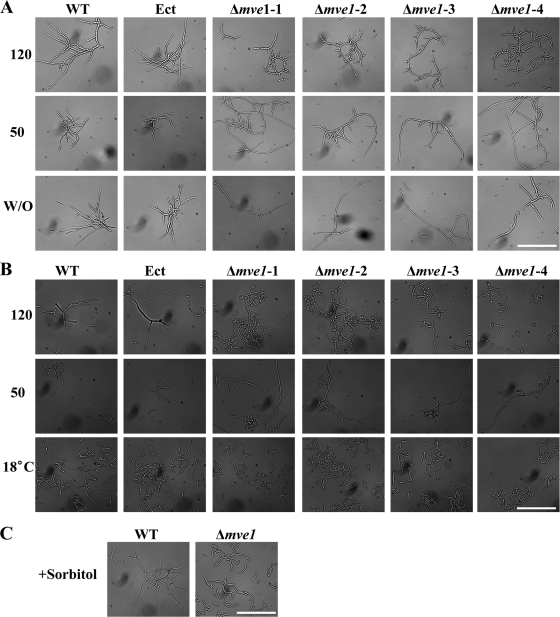
Disruption of MVE1 results in abnormal hyphal swelling and increases sensitivity to shaking force.
In contrast to the wild-type and ectopic controls, the MVE1 gene deletion mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4) developed distinguishable phenotypes when grown in liquid media. During the early stage (10 days after being inoculated into the medium), Δmve1 mutants showed abnormally short, curled cells, whereas the wild-type and ectopic control strains developed normal long hyphae in multiple directions into the liquid medium (Fig. 7 A). During the late stage (15 days), all four independent Δmve1 mutants, but not the wild-type or ectopic control strains, expressed an additional phenotype of swelling of the hyphae (Fig. 7B). Taken together, all of the MVE1 gene deletion mutants had curled as well as abnormal balloon-shaped cells, which were observed during the early and late stages of growth in liquid medium, respectively. Our results clearly demonstrated that MVE1 has critical functional roles in maintaining normal hyphal morphology and suppressing hyphal swelling in M. graminicola.
FIG. 7.
Abnormal morphologies and their conditional suppression of MVE1 mutants of Mycosphaerella graminicola in liquid medium. MVE1 gene deletion resulted in curled cells (A) and eventually the hyphal swelling phenotype (B), which could be suppressed by lowering the shaking condition and lowering the growth temperature or by adding osmotic stabilizer. (A) The curled morphology in MVE1 gene deletion mutants depended on the shaking conditions. Each strain was inoculated into yeast-sucrose broth (YSB) with the same amount of fungal cells (initial concentration of 1 × 105 cells ml−1) and incubated under different shaking conditions by adjusting the rpm. After 10 days of incubation at room temperature, photos were taken under a microscope. WT, wild type; Ect, ectopic control; Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4, four independent MVE1 mutants. 120 and 50, 120 and 50 rpm, respectively; W/O, without shaking. (B) Hyphal swelling in MVE1 gene deletion mutants was suppressed by lowering the growth temperature. The same conditions described in panel A were used for fungal culture, except for different temperatures for fungal growth (room temperature or 18°C). After 15 days, photos were taken with a microscope. 120 and 50, 120 and 50 rpm, respectively; 18°C, growth temperature set to 18°C with shaking at 120 rpm. (C) Hyphal swelling in MVE1 gene deletion mutants reverted completely in the presence of the osmotic stabilizer sorbitol (1 M). The same amounts of cells (initial concentration of 1 ×105 ml−1) of the wild-type strain and the MVE1 gene deletion mutant (Δmve1) were inoculated into YSB medium with 1 M sorbitol and allowed to grow for 15 days with shaking at 120 rpm. After 15 days of incubation at room temperature, photos were taken under the microscope. The white scale bar at the lower right of each panel represents 50 μm.
We suspected that deletion of MVE1 may increase sensitivity to mechanical stress originating from the shaking of liquid culture media, whereby Δmve1 mutants become curled or exhibit hyphal swellings without proper establishment of polarity during germination or by responding too sensitively to the vigorous shaking force. To test this, we examined morphological changes of Δmve1 mutants under different shaking conditions. Consistent with our hypothesis, all four Δmve1 mutants responded drastically to different shaking forces and displayed completely different morphologies, whereas the wild-type and ectopic control strains were not affected by shaking conditions (Fig. 7A). Under high-speed shaking (120 rpm), Δmve1 mutants produced the curled cells. However, hyphae of Δmve1 mutant cells, which were curved or crooked when shaken at 120 rpm, were straight and long when the cultures were shaken at 50 rpm. In stationary culture without shaking, hyphae of the Δmve1 mutants were even straighter and much longer, indicating that the crooked or curled phenotype observed in the Δmve1 mutants was caused by increased sensitivity to the higher shaking force (Fig. 7A). In contrast, the hyphal swelling phenotypes observed in the Δmve1 mutants were not altered regardless of shaking conditions, suggesting that the hyphal swelling phenotype in Δmve1 mutants was not caused by the increased sensitivity to high shaking force (Fig. 7B). Interestingly, hyphal swelling in Δmve1 mutants was completely suppressed by lowering the growth temperature to 18°C or by the addition of sorbitol (1 M) to the medium (Fig. 7B and C). Our results indicate that the hyphal swelling phenotypes in Δmve1 mutants were influenced by temperature and addition of the osmotic stabilizer sorbitol. This is consistent with previous observations made in A. chrysogenum and F. verticillioides that abnormal morphologies of the velvet gene deletion mutants could be rescued by an osmotic stabilizer (10, 19).
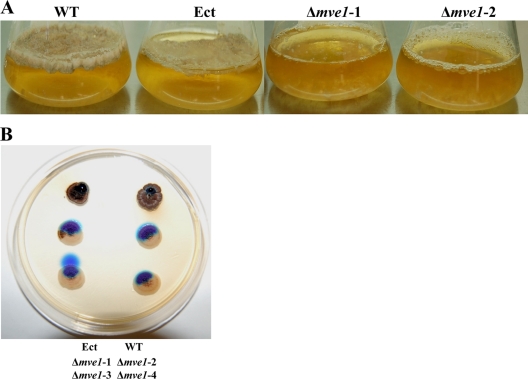
MVE1 disruption significantly reduces hydrophobicity.
A previous study of F. verticillioides showed that deletion of the FvVE1 gene resulted in decreased hydrophobicity of the colony surface (19). Similarly, we observed a significant reduction of hydrophobicity in Δmve1 mutants compared to the wild-type and ectopic control strains (Fig. 8). The wild-type and ectopic control strains produced a dense layer of mycelium on the surface of stationary liquid medium (YSB), presumably due to hydrophobicity of the cellular surfaces. However, Δmve1 mutants produced no detectable fungal mass on the surface by 15 days after inoculation (Fig. 8). Although the Δmve1 mutants eventually produced a thin surface layer, it was thinner and significantly delayed compared to those of the wild-type and ectopic control strains (data not shown). In addition, we tested the hydrophobicity of colony surface between the wild-type strain or ectopic controls and Δmve1 mutants by applying water droplets with dye directly onto fungal cultures, as described previously (19). Water dropped on the wild-type and ectopic control strains formed spherical droplets indicating hydrophobicity of the colony surface. However, on Δmve1 mutants water droplets spread immediately due to reduced hydrophobicity of their colony surfaces. Taken together, we concluded that MVE1 is important for cell surface hydrophobicity in M. graminicola.
FIG. 8.
The MVE1 mutants of Mycosphaerella graminicola exhibited reduced hydrophobicity. (A) Fungal mat formation test. Each strain was inoculated into yeast-sucrose broth (YSB) with the same amount of fungal cells (initial concentration of 1 × 105 cells ml−1). After 15 days of incubation without shaking, YSB cultures of the wild-type strain, an ectopic transformant (Ect), and two independent MVE1 gene deletion mutants (Δmve1-1 and Δmve1-2) were photographed. The wild type and an ectopic transformant generated a fungal mat on the surface of the liquid medium, whereas the two MVE1 mutants did not. (B) On each of the fungal colonies grown on YSA for 30 days, 15 μl of water-based DNA loading dye was pipetted onto the colony surfaces and photographed 10 min later. Spherical water droplets formed on colonies of the wild type and an ectopic transformant, whereas the droplet dispersed immediately on colonies of the four MVE1 gene deletion mutants (Δmve1). The locations of inoculated strains on the agar plate are indicated at the bottom of the image.
Disruption of MVE1 does not affect pathogenicity but decreases pigmentation of pycnidia.
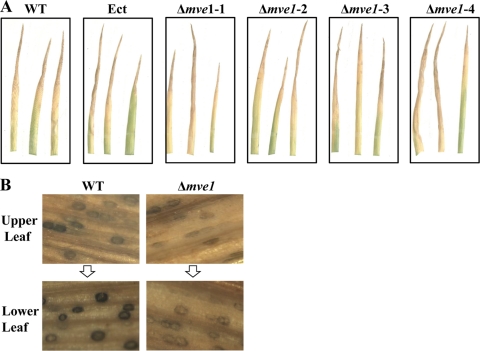
We tested the effect of MVE1 deletion on pathogenicity of M. graminicola by spray inoculations of the highly susceptible wheat cultivar Taichung 29. For these assays, wheat seedlings at the 3- to 5-leaf stage were inoculated with 107 spores of each strain and plants were incubated until the appearance of STB disease symptoms, such as the formation of pycnidia within tan lesions. STB symptom expression by the MVE1 gene deletion mutants was not altered relative to those caused by the wild-type and ectopic control strains (Fig. 9 A). We obtained consistent results from all four independent MVE1 mutants, indicating that MVE1 is not associated with initial plant pathogenicity of M. graminicola.
FIG. 9.
Effect of MVE1 gene deletion on pathogenicity of Mycosphaerella graminicola to wheat. (A) Spray infection assays. Identical quantities of inoculum (108 spores) of the wild type (WT), an ectopic control strain (Ect), and four independent MVE1 gene deletion mutants (Δmve1-1, Δmve1-2, Δmve1-3, and Δmve1-4) were sprayed onto seedling leaves of the highly susceptible wheat cultivar Taichung 29. After inoculation, plants were placed in the dark for 48 h under high humidity. Subsequently, the infected wheat plants were incubated in a greenhouse with a diurnal cycle of 14 h of light/10 h of darkness for 21 days. The inoculation assays were performed with at least three biological replications for each fungal isolate. (B) Effects of the disruption of MVE1 on pycnidial pigmentation. The leaves of wheat cultivar Taichung 29 infected by the wild-type strain or the MVE1 gene deletion mutant (Δmve1) in panel A were harvested and analyzed under a dissecting microscope. The same stages of infected wheat leaves were selected and compared for the development of pycnidia between the wild type and Δmve1 mutant. Upper leaves indicate new development of STB still in progress, whereas the lower leaves were relatively old, with STB showing full development of pycnidia.
However, pigmentation of pycnidia differed significantly between the wild-type or ectopic control strains and Δmve1 mutants. The pycnidia in the wild-type and ectopic control strains became very dark as the infected Taichung 29 leaves aged, indicating the gradual development of pycnidial pigmentation. In contrast, pigmentation of pycnidia produced by the Δmve1 mutants did not display much difference in the intensity of dark coloration of pycnidia between early and late development of STB and the pycnidia remained lightly pigmented (Fig. 9B). Our data demonstrated that MVE1 is dispensable for STB disease development itself, but plays a role in the development of black pigment in pycnidia of M. graminicola in wheat tissue.
DISCUSSION
The velvet gene has attracted a great deal of attention from fungal biologists recently (4, 6). Since the discovery of veA1 in A. nidulans (14), analyses of homologous genes in other filamentous fungi have followed. Although the pathways vary widely, several lines of evidence are consistent with the hypothesis that the velvet gene plays a key regulatory role in development and secondary metabolite production in many fungi (4, 6). However, so far it is not known whether a gene homologous to the velvet gene is present or plays a regulatory function in the wheat pathogen, M. graminicola, or any other member of the Dothideomycetes, the largest class of plant-pathogenic fungi. In this study, we identified MVE1, encoding a velvet-like protein in M. graminicola, and thoroughly characterized its function. Four independent Δmve1 mutants were generated and exhibited consistent multiple phenotypes, suggesting that MVE1 is associated with all of the phenotypes observed in this study and that the importance of the velvet gene in fungal development extends to the Dothideomycetes.
Fungal velvet proteins (veA) consist of conserved N-terminal and variable C-terminal regions. The highly conserved N-terminal region of the velvet protein is responsible for most of its cellular function in fungi (23). However, significant variation in the C-terminal region has been noted among fungi and even within the genus Aspergillus. For example, the velvet gene from F. verticillioides could not rescue a velvet gene mutation in A. nidulans, indicating species-specific functions of this gene in different fungi (19). Therefore, we hypothesized that the functional roles of fungal velvet genes clearly have two aspects: conserved functions due to the N-terminal region of the velvet protein and species-specific functions due to the C-terminal region.
Consistent with conserved N-terminal as well as variable C-terminal functions of the velvet protein, some of the phenotypes in Δmve1 mutants were similar to those reported in other fungi, particularly with FvVE1 (19), while others were novel and unique to M. graminicola. For example, Δmve1 mutants exhibited many common characteristics of the FvVE1 mutants. Reduction of aerial mycelia, alteration of polar growth in liquid culture, swelling and yeast-like growth in liquid shake cultures, and recovery of the wild-type phenotype by osmotic stabilizers presented clear parallelisms of VeA-mediated signaling between these two fungi. On the other hand, Δmve1 mutants showed several phenotypes that were specific to M. graminicola, including melanin biosynthesis and pycnidial pigmentation, possibly due to the C-terminal region. Phylogenetically, M. graminicola belongs to the class Dothideomycetes and is only distantly related to the other filamentous fungi in which the velvet gene has been analyzed (13, 28). To our knowledge, this is the first report on characterization of a velvet-like gene in the class Dothideomycetes or order Capnodiales. Therefore, phenotypes unique to M. graminicola may provide important information on the putative functional roles of the velvet gene in the Capnodiales and closely related fungi. In addition, because some of the phenotypes caused by deletion of the velvet gene in M. graminicola were observed in phylogenetically diverse fungi, some functional roles of the velvet gene appear to be well conserved across divergent lineages of filamentous fungi.
In M. graminicola, an albino phenotype with a significant reduction of melanin production in Δmve1 mutants showed that MVE1 has a positive regulatory role in melanin biosynthesis (Fig. 3 and 4). This is the first report describing a relationship between the velvet gene and fungal melanin biosynthesis. The fungal velvet gene regulates the biosynthesis of a diverse array of secondary metabolites. For example, veA orthologs have indispensable roles in the biosynthesis of fumonisins and fusarins in Fusarium verticillioides, the β-lactam antibiotic cephalosporin C in Acremonium chrysogenum, sterigmatocystin and the antibiotic penicillin in A. nidulans, and aflatoxin in A. parasiticus (6, 8, 10, 11, 16, 25). Clearly, the MVE1 gene in M. graminicola regulates the biosynthetic pathway of another secondary metabolite, melanin (Fig. 3 and 4). In Alternaria alternata, genes involved in melanin biosynthesis are clustered (18). We speculate that MVE1 might regulate a specific transcription factor for melanin biosynthesis directly, as veA in A. paraciticus is required for the expression of aflR, the key transcription factor for the aflatoxin biosynthetic gene cluster (7, 30). Although we analyzed only melanin production in MVE1 gene deletion mutants, MVE1 also might regulate the production of mycotoxins or secondary metabolites as yet unidentified in M. graminicola.
Deletion of MVE1 also led to significant decreases in development of aerial mycelia (Fig. 5) similar to the functional roles of the velvet gene on aerial mycelium development noted previously in N. crassa and F. verticillioides (5, 19). Interestingly, previous analyses in our laboratory revealed that the MCC1 gene in M. graminicola, encoding a fungal C-type cyclin, influences aerial mycelium positively and melanin biosynthesis negatively (8a). Taken together, MVE1 and MCC1 have a common function in production of aerial mycelia but opposite roles in melanin biosynthesis. We are currently performing microarray experiments to identify genes that are downstream of MVE1 and MCC1. Genes with the same patterns downstream of both MVE1 and MCC1 will be good candidates for aerial mycelium development, whereas downstream genes with opposite trends might be associated with the melanin biosynthetic pathway(s). Development of aerial mycelia in M. graminicola requires light, indicating a pivotal role of the light-signaling pathway (Fig. 6). In A. nidulans, large numbers of conidia are produced only in the light, but neither blue nor red light alone was sufficient for conidial development (25). However, in M. graminicola, we observed a gradual decrease in aerial mycelium development in the sequential order of blue light, red light, and darkness (Fig. 6). The entire visible spectrum of wavelengths affected aerial mycelium development the same as blue light, suggesting that blue light is sufficient to activate the light-signaling pathway regulating development of aerial mycelia (data not shown). The same dependence of aerial mycelium development on blue light also was observed under temporary blue- or red-light conditions. However, aerial mycelium production under temporary blue light was not as high as it was under constant blue light, indicating that in M. graminicola continuous illumination by blue light is necessary for full development of aerial mycelia (Fig. 6). Interestingly, MVE1 gene deletion resulted in complete blindness to light sensing; the Δmve1 mutant did not produce light-dependent aerial mycelia. Our data clearly suggest that MVE1 is one of the key components of the light-signaling pathway in M. graminicola, which is consistent with previous reports for other fungi (4, 6, 23).
In filamentous fungi, several well-known proteins have been identified as components of light-sensing or light-signaling pathways, such as the phototropin-like proteins and White Collar-1 and 2 (WC-1 and WC-2) (2, 3, 20, 27). We believe that the orthologs of WC-1 and WC-2 in M. graminicola should act in similar light-dependent pathways. In addition, the fungal LaeA protein is the master epigenetic regulator of fungal secondary metabolism and physically interacts with VeA in the absence of light (4, 15). Therefore, in M. graminicola, it is necessary to test for molecular linkage or interactions between MVE1 and LaeA as well as WC-1 and WC-2 in the context of a light-signaling pathway; apparent homologs of all of these genes have been identified in the M. graminicola genome sequence (Y.-E. Choi and S. B. Goodwin, unpublished observation). The mRNA expression level of MVE1 was not affected by different light conditions, suggesting that the regulation of Mve1 may occur mostly at the posttranscriptional level for light signaling (data not shown). Our results on MVE1 expression were similar to the constitutive expression of veA in A. nidulans (17). We also detected a nuclear localization signal (NLS) in Mve1, suggesting that nuclear localization of Mve1 protein might be more important than MVE1 transcription (Fig. 1).
Bioinformatic analyses have demonstrated that proteins encoded by velvet genes have few conserved domains and are specific to the filamentous fungi. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe do not contain velvet-like genes, indicating special functional roles of veA in filamentous fungi (6). Considering that the velvet genes are confined to the fungal kingdom, it seemed likely that MVE1 might be involved in fungus-specific developmental processes, such as filamentous growth. Because we observed hyphal swelling phenotypes in Δmve1 mutants (Fig. 7), it is conceivable that MVE1 is associated with filamentous growth by suppressing hyphal swelling. In addition, similar hyphal swelling was observed in FvVE1 gene deletion mutants, indicating conserved functional roles of the velvet gene in filamentous fungi (19). However, the hyphal swelling phenotype in MVE1 gene deletion mutants reverted to normal growth at a lower growth temperature (18°C) or under osmotic stress (Fig. 7B and C). In M. graminicola, filamentous growth was enhanced at room temperature, whereas yeast-like growth was preferred, instead of filamentous growth, at 18°C. Our data suggest that filamentous growth is required for the subsequent hyphal swelling, because it was not detected in Δmve1 mutants at 18°C. In addition, because osmotic stress suppressed hyphal swelling, we suspect that this phenotype associated with MVE1 may involve balancing of the cellular osmotic status during filamentous growth. The similar recovery from morphological defects under osmotic stress of velvet gene deletion mutants in A. chrysogenum and F. verticillioides supports this notion (10, 19). Disruption of MVE1 may result in hyphal swelling phenotypes due to impairment of a putative osmotic balance regulator, Mve1. More studies will be necessary to elucidate the Mve1-associated cellular osmotic balance mechanism. Previous reports on deletion mutants of both MCC1 and MgSlt2, encoding a C-type cyclin and mitogen-activated protein (MAP) kinase, respectively, in M. graminicola, also described large swollen cells (8a, 21). Further study is required to reveal the molecular relationships between MVE1 and MCC1 or MgSlt2 in fungal cell enlargement.
MVE1 gene deletion mutants also displayed increased sensitivity to shaking force and lost their hydrophobicity (Fig. 7 and 8). We suspected that these phenotypes may be linked, because loss of hydrophobicity of the cellular surface in Δmve1 mutants may be associated with increased perturbation due to shaking in the hydrophilic liquid medium. To test that, we increased the hydrophobicity of the medium by adding glycerol and assayed for changes in sensitivity to the shaking force in Δmve1 mutants. However, no differences were observed in the curled cells under vigorous shaking (120 rpm) of Δmve1 mutants even with the addition of glycerol up to 10% (data not shown). Therefore, these two phenotypes in Δmve1 mutants—sensitivity to shaking force and loss of hydrophobicity—do not appear to be related. In F. verticillioides, the veA homolog FvVE1 regulates cell wall formation (19). Therefore, alternatively, we speculate that a change in cell wall composition caused by deletion of MVE1 may explain several phenotypes in M. graminicola, including hyphal swelling, the sensitivity to shaking force, and the loss of hydrophobicity.
We could not differentiate the severity of STB development caused by the wild-type and Δmve1 mutants, indicating that the velvet gene in M. graminicola does not impact pathogenicity directly (Fig. 9A). Clearly, MVE1 is associated with light signaling and impacts the production of melanin as a secondary metabolite, similar to the velvet genes in other fungal genera. Therefore, we concluded that Mve1-associated light signaling and the biosynthesis of at least some secondary metabolites are dispensable for STB development in M. graminicola. Instead, we observed a significant reduction of pigmentation of pycnidia in Δmve1 mutants (Fig. 9B). Reduced pigmentation may affect the viability and survival of pycnidiospores. However, we saw no differences in the survival rates of pycnidiospores between the wild-type strain and Δmve1 mutants in preliminary experiments (Choi and Goodwin, unpublished). Additional work is required to test the hypothesis that MVE1 affects the viability of lightly pigmented pycnidia and pycnidiospores in M. graminicola.
Our results demonstrated that MVE1 is not necessary for STB development, but is important for pigmentation of pycnidia. However, because deletion of the MVE1 gene did not completely block the development of pigment in pycnidia, we believe there might be one or more additional pathways for production of dark pigments in the pycnidia of M. graminicola. The leaky phenotype of black color development in the pycnidia contrasted with the almost complete blockage of melanin production in the Δmve1 mutants on artificial media (Fig. 3 and 4). Therefore, the molecular signaling pathways for black color development of pycnidia and melanin biosynthesis may be related in part via MVE1, but appear to differ in terms of partial or absolute dependence on Mve1 as a regulator for the biosyntheses of pycnidia pigment or melanin, respectively.
In summary, we characterized MVE1, a velvet gene homolog in M. graminicola, and demonstrated that it plays important roles in multiple phenotypes: melanin biosynthesis, development of aerial mycelia, hyphal swellings, sensitivity to shaking, and hydrophobicity. MVE1 also was implicated in the light-signaling pathway, but was dispensable for STB disease pathogenicity. Further research will be necessary to expand our knowledge on each of the mechanisms associated with Mve1 in M. graminicola.
Acknowledgments
We thank Larry Dunkle and Charles P. Woloshuk for critical reading of the manuscript. We also thank Larry Dunkle, Tesfaye Mengiste, and Richard Latin for use of microscopes and experimental equipment. DNA sequencing of M. graminicola was performed at the U.S. Department of Energy's Joint Genome Institute through the Community Sequencing Program (www.jgi.doe.gov/csp/), and all sequence data are publicly available.
This work was supported by USDA-ARS CRIS project 3602-22000-015-00D and by a USDA-ARS Administrator-Funded Postdoctoral position to Y.-E. Choi.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Babitskaya, V. G., V. V. Shcherba, T. V. Filimonova, and E. A. Grigorchuk. 2000. Melanin pigments from the fungi Paecilomyces variotii and Aspergillus carbonarius. Appl. Biochem. Microbiol. 36:128-133. [PubMed] [Google Scholar]
- 2.Ballario, P., and G. Macino. 1997. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 5:458-462. [DOI] [PubMed] [Google Scholar]
- 3.Ballario, P., et al. 1996. White collar-1, a central regulator of blue light responses in Neurospora crassa, is a zinc finger protein. EMBO J. 15:1650-1657. [PMC free article] [PubMed] [Google Scholar]
- 4.Bayram, Ö., et al. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504-1506. [DOI] [PubMed] [Google Scholar]
- 5.Bayram, Ö., S. Krappmann, S. Seiler, N. Vogt, and G. H. Braus. 2008. Neurospora crassa ve-1 affects asexual conidiation. Fungal Genet. Biol. 45:127-138. [DOI] [PubMed] [Google Scholar]
- 6.Calvo, A. M. 2008. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45:1053-1061. [DOI] [PubMed] [Google Scholar]
- 7.Calvo, A. M., J. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cary, J. W., et al. 2007. Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl. Microbiol. Biotechnol. 76:1107-1118. [DOI] [PubMed] [Google Scholar]
- 8a.Choi, Y.-E. and S. B. Goodwin. 2010. Gene encoding a c-type cyclin in Mycosphaerella graminicola is involved in aerial mycelium formation, filamentous growth, hyphal swelling, melanin biosynthesis, stress response, and pathogenicity. Mol Plant-Microbe Interact. [Epub ahead of print.] doi: 10.1094/MPMI-04-10-0090. [DOI] [PubMed]
- 9.Cools, H. J., and B. A. Fraaije. 2008. Azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola. Pest Manag. Sci. 64:681-684. [DOI] [PubMed] [Google Scholar]
- 10.Dreyer, J., H. Eichhorn, E. Friedlin, H. Kürnsteiner, and U. Kück. 2007. A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl. Environ. Microbiol. 73:3412-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran, R. M., J. W. Cary, and A. M. Calvo. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73:1158-1168. [DOI] [PubMed] [Google Scholar]
- 12.Flaherty, J. E., and L. D. Dunkle. 2005. Identification and expression analysis of regulatory genes induced during conidiation in Exserohilum turcicum. Fungal Genet. Biol. 42:471-481. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin, S. B. 2004. Minimum phylogenetic coverage: an additional criterion to guide the selection of microbial pathogens for initial genomics sequencing efforts. Phytopathology 94:800-804. [DOI] [PubMed] [Google Scholar]
- 14.Käfer, E. 1965. The origin of translocations in Aspergillus nidulans. Genetics 52:217-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kale, S. P., et al. 2008. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45:1422-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, N., W. Brooks, and A. M. Calvo. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H.-S., et al. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37:72-80. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, N., and T. Tsuge. 1993. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175:4427-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, S., et al. 2006. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol. Microbiol. 62:1418-1432. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., Q. He, and P. Cheng. 2003. Photoreception in Neurospora: a tale of two white collar proteins. Cell. Mol. Life Sci. 60:2131-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehrabi, R., T. van der Lee, C. Waalwijk, and G. H. J. Kema. 2006. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant Microbe Interact. 19:389-398. [DOI] [PubMed] [Google Scholar]
- 22.Mehrabi, R., L.-H. Zwiers, M. A. de Waard, and G. H. J. Kema. 2006. MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19:1262-1269. [DOI] [PubMed] [Google Scholar]
- 23.Mooney, J. L., and L. N. Yager. 1990. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 4:1473-1482. [DOI] [PubMed] [Google Scholar]
- 24.Mullins, E. D., et al. 2001. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173-180. [DOI] [PubMed] [Google Scholar]
- 25.Myung, K., et al. 2009. FvVE1 regulates biosynthesis of the mycotoxins fumonisins and fusarins in Fusarium verticillioides. J. Agric. Food. Chem. 57:5089-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polley, R. W., and M. R. Thomas. 1991. Surveys of diseases of winter wheat in England and Wales 1976-88. Ann. Appl. Biol. 119:1-20. [Google Scholar]
- 27.Purschwitz, J., et al. 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18:255-259. [DOI] [PubMed] [Google Scholar]
- 28.Schoch, C. L., et al. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98:1041-1052. [DOI] [PubMed] [Google Scholar]
- 29.Wittenberg, A. H. J., et al. 2009. Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS One 4:e5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woloshuk, C. P., et al. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, J.-H., et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 32.Zwiers, L.-H., and M. A. de Waard. 2001. Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 39:388-393. [DOI] [PubMed] [Google Scholar]