Abstract
Malic enzyme catalyzes the reversible oxidative decarboxylation of malate to pyruvate and CO2. The Saccharomyces cerevisiae MAE1 gene encodes a mitochondrial malic enzyme whose proposed physiological roles are related to the oxidative, malate-decarboxylating reaction. Hitherto, the inability of pyruvate carboxylase-negative (Pyc−) S. cerevisiae strains to grow on glucose suggested that Mae1p cannot act as a pyruvate-carboxylating, anaplerotic enzyme. In this study, relocation of malic enzyme to the cytosol and creation of thermodynamically favorable conditions for pyruvate carboxylation by metabolic engineering, process design, and adaptive evolution, enabled malic enzyme to act as the sole anaplerotic enzyme in S. cerevisiae. The Escherichia coli NADH-dependent sfcA malic enzyme was expressed in a Pyc− S. cerevisiae background. When PDC2, a transcriptional regulator of pyruvate decarboxylase genes, was deleted to increase intracellular pyruvate levels and cells were grown under a CO2 atmosphere to favor carboxylation, adaptive evolution yielded a strain that grew on glucose (specific growth rate, 0.06 ± 0.01 h−1). Growth of the evolved strain was enabled by a single point mutation (Asp336Gly) that switched the cofactor preference of E. coli malic enzyme from NADH to NADPH. Consistently, cytosolic relocalization of the native Mae1p, which can use both NADH and NADPH, in a pyc1,2Δ pdc2Δ strain grown under a CO2 atmosphere, also enabled slow-growth on glucose. Although growth rates of these strains are still low, the higher ATP efficiency of carboxylation via malic enzyme, compared to the pyruvate carboxylase pathway, may contribute to metabolic engineering of S. cerevisiae for anaerobic, high-yield C4-dicarboxylic acid production.
Malic enzyme is a ubiquitous enzyme that catalyzes the reversible oxidative decarboxylation of malate to pyruvate and CO2, using either NAD+ (EC 1.1.1.38-39) or NADP+ (EC 1.1.1.40) as the redox cofactor. In eukaryotes, malic enzyme has been found in the cytosol and/or in mitochondria and, in plants, in the chloroplasts (20, 35). The reaction has a ΔG0′ of ca. −8 kJ mol−1 in the decarboxylating direction (19) and, although both carboxylation and decarboxylation have been observed in vitro, substrate affinities and maximum reaction rates generally appear to favor the latter (5, 14, 34). Despite decades of study, the physiological function of malic enzyme often remains enigmatic. Proposed physiological roles for the decarboxylating reaction include provision of pyruvate, inorganic carbon, reducing power, and pH regulation (20, 26). Carboxylating roles for malic enzyme have also been suggested (25, 30), although in vivo evidence is sparse. The strongest indications for in vivo malic enzyme activity in the carboxylating direction come from experiments with engineered Escherichia coli, in which overexpression of malic enzyme improved succinate titers (17, 27, 28). However, interpretation of these results was complicated by the fact that these strains still expressed phospho-enol-pyruvate carboxylase, the main carboxylating enzyme in glucose-grown wild-type E. coli (2).
In the yeast Saccharomyces cerevisiae, malic enzyme is encoded by the MAE1 gene (4). The mitochondrial Mae1 protein can use both NAD+ and NADP+ as an electron acceptor and exhibits a relatively high Km toward malate (50 mM, or 17 mM in the presence of 0.5 mM phospho-enol-pyruvate [PEP]) (13). Also in S. cerevisiae, the physiological role of malic enzyme is still uncertain even though the enzyme has been shown to provide the pyruvate required for biosynthesis in pyruvate kinase-negative strains grown on ethanol (4). No kinetic data have been published for the carboxylating reaction, but the inability of pyruvate carboxylase-negative strains of S. cerevisiae to grow on glucose indicates that Mae1p cannot readily take over the anaplerotic role of pyruvate carboxylase (29). Consequently, the in vivo reaction catalyzed by Mae1p is often assumed to be irreversible in the decarboxylating direction (23).
In recent years, interest in the biotechnological production of C4-dicarboxylic acids has intensified, as these acids are envisaged to become key building blocks in an increasingly “bio-based” chemical industry (24, 36). S. cerevisiae is an interesting production host for organic acids (1). Hitherto, attempts to engineer S. cerevisiae for malate (and succinate) production were based on replacement of the native alcoholic fermentation pathway by a malate fermentation pathway. To this end, pyruvate carboxylase, a cytosolically retargeted malate dehydrogenase and a heterologous malate transporter were (over) expressed in a pyruvate decarboxylase-negative strain (38, 39). Although this approach yielded malate titers of up to 59 g liter−1, ATP hydrolysis by pyruvate carboxylase precludes a net synthesis of ATP via this fermentative pathway. In practice, ATP yields are even expected to be negative due to energy requirements for dicarboxylic acid export (32). As a result, part of the glucose fed to the cultures has to be respired to provide ATP, which goes at the expense of the product yield on glucose. Improved understanding of the energetics and thermodynamics of C3-C4 carboxylation reactions is therefore not only of fundamental physiological interest but may also have industrial applications.
In the last step of glycolysis, pyruvate kinase transfers the phosphate group of PEP to ADP, producing pyruvate and ATP. In wild-type S. cerevisiae strains grown on glucose, anaplerosis occurs via pyruvate carboxylase, which converts pyruvate to oxaloacetate and consumes 1 ATP per carboxylation. As a result, the overall conversion of PEP to oxaloacetate is ATP neutral. However, if pyruvate kinase can be combined with malic enzyme, which converts pyruvate to malate without consuming ATP, PEP is converted to malate while producing net ATP. The same gain in ATP yield can be achieved by using phospho-enol-pyruvate carboxykinase (PEPCK), which converts PEP directly to oxaloacetate while producing ATP. Any reaction with an improved ATP yield has a lower thermodynamic driving force, since phosphorylation of ADP requires an input of free energy. That this can impose limits on metabolism is illustrated by a previous study, where PEPCK was shown to be able to function as an anaplerotic enzyme in S. cerevisiae, but only when concentrations of the substrates PEP and CO2 were increased (40).
The goal of the present study was to investigate the requirements for an in vivo carboxylating role of malic enzyme in a pyruvate carboxylase-negative S. cerevisiae strain grown on glucose as the sole carbon source. Since in S. cerevisiae endogenous malic enzyme (Mae1p) is mitochondrial, while the pyruvate carboxylases (Pyc1p and Pyc2p) are cytosolic, various malic enzyme expression constructs were tested: (i) overexpression of the endogenous Mae1p, (ii) expression of a truncated form of Mae1p without the mitochondrial targeting sequence, and (iii) expression of the NADH-dependent malic enzyme from E. coli. In addition, the impact of the substrate concentrations of the malic enzyme reaction was tested by varying CO2 concentrations and decreasing the activity of a pyruvate consuming reaction.
MATERIALS AND METHODS
Strains and maintenance.
Strains constructed in the present study (Table 1) were derived from IMK157, an S. cerevisiae strain derived from the CEN.PK family (31), containing targeted deletions of URA3 and the two pyruvate carboxylase genes PYC1 and PYC2 (40). Exceptions were strains IMY030 to IMY032, which originate from S. cerevisiae BY4741 mae1Δ (37). Strains were maintained on YPD (demineralized water; yeast extract [BD Difco], 10 g liter−1; peptone [BD Difco], 20 g liter−1; glucose, 20 g liter−1) or, for strains carrying plasmids, on synthetic stock medium (demineralized water; KH2PO4, 3 g liter−1; MgSO4, 0.5 g liter−1; l-aspartic acid [from a filter-sterilized 7.4-g liter−1 solution set to pH 6 with KOH], 6.6 g liter−1; K2SO4, 6.6 g liter−1; glucose, 20 g liter−1; trace elements and filter-sterilized vitamins) (33). Culture stocks, prepared from shake flask cultures by the addition of glycerol (20% [vol/vol]), were stored at −80°C in 1-ml aliquots. Incubations were performed at 30°C, and shake flasks were kept in orbital shakers at 200 rpm.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| CEN.PK 113-7D | MATa, reference strain |
| IMK157 ura3Δ | MATapyc1::kanMx pyc2::ILV2smr ura3-52 |
| IMK299 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 |
| IMW012 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (cured); derived from IMY017 |
| IMY007 empty vector | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC1) |
| IMY016 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (MB5573) |
| IMY017 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (MB5573*); evolved |
| IMY019 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (MB5573) |
| IMY024 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (MB5573*) |
| IMY025 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC10) |
| IMY026 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC9) |
| IMY027 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (pUDC10) |
| IMY028 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (pUDC9) |
| IMY029 | MATapyc1::kanMx pyc2::ILV2smr pdc2::loxP::hph::loxP ura3-52 (pUDC9); derived from IMY017 |
| BY4741 mae1Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mae1::kanMX |
| IMY030 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mae1::kanMX (pUDC1) |
| IMY031 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mae1::kanMX (MB5573) |
| IMY032 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mae1::kanMX (MB5573*) |
Plasmid construction and transformation.
Plasmid pUG-hphNT1 (Table 2) was constructed by replacing the kanMX marker in pUG6 (15) with the hphNT1 marker (conferring hygromycin B resistance) from pFA6a-hphNT1 (18) using the SacI and BglII restriction sites. The PDC2 deletion cassette was prepared by PCR using pUG-hphNT1 with primers 5′-PDC2-S1 and 3′-PDC2-S2 (Table 3) (10). Transformants of strain IMK157 were selected on YP agar containing ethanol (3% [vol/vol]), glycerol (2% [vol/vol]), and hygromycin B (200 mg liter−1). A single colony isolate, for which deletion of PDC2 was confirmed by PCR, was named IMK299.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristics | Reference |
|---|---|---|
| pFA6a-hphNT1 | Template for hphNT1 | 18 |
| pUG6 | Template for loxP-kanMX-loxP deletion cassette | 15 |
| pUG-hphNT1 | Template for loxP-hphNT1-loxP deletion cassette | This study |
| p416-GPD | Centromeric yeast vector, URA3, PTDH3-TCYC1 | 22 |
| MB5573 | Centromeric yeast vector, URA3, PTDH3-E. coli sfcA-TCYC1 | This study |
| MB5573* | Centromeric yeast vector, URA3, PTDH3-E. coli sfcA-TCYC1, isolated from strain IMY017 | This study |
| pUDC1 | Centromeric yeast vector, URA3, PTDH3-TCYC1 | 40 |
| pUDC9 | Centromeric yeast vector, URA3, PTDH3-sMAE1-TCYC1 | This study |
| pUDC10 | Centromeric yeast vector, URA3, PTDH3-MAE1-TCYC1 | This study |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| 5′-PDC2-S1 | ACGCAACTTGAATTGGCAAAATGGGCTTATGAGACGTTCCCAGCTGAAGCTTCGTACGC |
| 3′-PDC2-S2 | AGCCTGTGTTACCAGGTAAGTGTAAGTTATTAGAGTCTGGGCATAGGCCACTAGTGGATCTG |
| 5′-MAE1-XbaI | CGACGGATTCTAGAGGTTATGCTTAGAACCAGACTATCCG |
| 5′-sMAE1-XbaI | ACCATCTAGAATGTGGCCTATTCAGCAATCGCG |
| 3′-MAE1-SalI | GTTAGTCGACCTACAATTGGTTGGTGTGCACC |
A synthetic gene, consisting of E. coli sfcA malic enzyme (also known as maeA) with flanking SpeI and XhoI restriction sites, was inserted into p416-GPD (22) digested with XbaI and XhoI (synthesis and codon optimization for expression in S. cerevisiae were performed by Blue Heron Biotechnology, Bothell, WA). The resulting plasmid was named MB5573 (Table 2). S. cerevisiae MAE1 and the truncated version sMAE1 (21) were amplified from CEN.PK 113-7D genomic DNA with primers 5′-MAE1-XbaI or 5′-sMAE1-XbaI and 3′-MAE1-SalI (Table 3). After digestion with XbaI and SalI, the PCR products were inserted into p416-GPD via the XbaI and XhoI sites, resulting in plasmids pUDC9 (expressing sMAE1) and pUDC10 (expressing MAE1) (Table 2). Correct PCR amplification was verified by sequencing, performed by BaseClear, Netherlands.
Yeast transformations were performed as described previously (7). Transformants were selected on synthetic glucose medium agar plates without uracil and with aspartate as the nitrogen source. Histidine (25 mg liter−1), leucine (30 mg liter−1), and methionine (25 mg liter−1) were added as required. Transformations were verified by PCR.
Plate cultivation.
Solid media were prepared by addition of 20 g of Bacto agar liter−1. Apart from the nitrogen source [either 4 g of aspartate or 2 g of (NH4)2SO4 liter−1] and the addition of Tween 80 (0.42 g liter−1) and ethanol-dissolved ergosterol (10 mg liter−1), added to allow for growth under anaerobic conditions, solid synthetic media were identical to the synthetic stock culture medium. Gas-tight jars with 0.2 bar of overpressure were used for incubation of agar plates under CO2 atmosphere.
Batch cultivation.
Batch cultivation was performed at 30°C in 2-liter bioreactors (Applikon, Schiedam, Netherlands) at a working volume of 1 liter. The pH was controlled by automatic addition of 2 M KOH. Bioreactors were sparged with 250 ml of CO2 per min and stirred at 800 rpm. The medium was identical to the synthetic stock culture medium, except for the replacement of the aspartic acid and K2SO4 by 5 g of (NH4)2SO4 liter−1 and the addition of 0.15 ml of silicon antifoam (BDH, Poole, England) liter−1 to control foaming. Tween 80 (0.42 g liter−1) and ethanol-dissolved ergosterol (10 mg liter−1) were added to the medium to allow for growth under anaerobic conditions. For growth rate measurements, the initial optical density at 660 nm (OD660) after inoculation was ≤0.1. Maximum specific growth rates were determined for duplicate cultures (errors are given as deviations from the mean) and based on the OD660. Preculture shake flasks with synthetic stock medium were inoculated with 1-ml aliquots of frozen stock culture. Cells from exponentially growing shake-flask precultures were washed twice with demineralized water and used to inoculate batch cultures.
Selection of IMY017.
Nitrogen-limited medium for the selection of strain IMY017 was based on the synthetic stock culture medium, with the following modifications: medium contained 1 g of (NH4)2SO4 and 5.3 g of K2SO4 liter−1, as well as 30 g of glucose liter−1, 0.15-ml liter−1 of silicon antifoam (BDH, Poole, England) to control foaming, Tween 80 (0.42 g liter−1), and ethanol-dissolved ergosterol (10 mg liter−1) to allow for growth under anaerobic conditions. Medium was added via a peristaltic pump, and the working volume was kept constant by means of a conductive level sensor.
Plasmid isolation and curing.
The plasmid from IMY017 was isolated with a Zymoprep II yeast plasmid miniprep kit and amplified in E. coli. Sequencing of the recovered plasmid, which was designated MB5573* (Table 2), was performed by BaseClear. Plasmid loss in strain IMY017 was achieved by growth under nonselective conditions on liquid YPD. After several serial transfers, the culture was plated on YPD agar. A single colony isolate, which displayed uracil auxotrophy and for which plasmid loss was supported by PCR, was named IMW012 (Table 1).
Metabolite analysis.
Extracellular concentrations of glucose and ethanol were determined by high-performance liquid chromatography, using a Bio-Rad Aminex HPX-87H column eluted with 5 mM H2SO4 at a flow rate of 0.6 ml min−1 and kept at 60°C, coupled to a Waters 2410 refractive index detector.
Preparation of cell extracts.
Samples for enzyme activity measurements were obtained from shake flask cultures on synthetic stock medium, with glucose and aspartate, or 3% (vol/vol) ethanol as the carbon source(s). Cells, harvested in the exponential phase before depletion of either glucose or ethanol, were centrifuged, washed, and resuspended in potassium phosphate buffer (10 mM [pH 7.5], with 2 mM EDTA) and stored at −20°C (each sample contained ∼60 mg biomass dry weight). Cell extracts for isocitrate lyase, pyruvate decarboxylase, and phospho-enol-pyruvate carboxykinase were prepared as described earlier (41). For malic enzyme, cell extracts were prepared in 50 mM Tris-HCl buffer (pH 7.5) containing 1 mM dithiothreitol and 2 mM MgCl2 and dialyzed against this buffer for 4 h at 4°C using 3 ml 10K MWCO Slide-A-Lyzer Dialysis Cassettes (Thermo Scientific).
Enzyme activity assays.
Activities of isocitrate lyase, pyruvate decarboxylase and phospho-enol-pyruvate carboxykinase were determined as previously described (9, 11). As similar assay mixtures have been used to determine in vitro malic enzyme activities for S. cerevisiae and E. coli (4, 5), malic enzyme activities were determined as described for S. cerevisiae (4) with either 0.4 mM NAD+ or NADP+ as the redox cofactor. Malic enzyme activity was measured in the decarboxylating direction to avoid interference with pyruvate decarboxylase and alcohol dehydrogenase. Values for Vmax and Km were determined with nonlinear fits, assuming Michaelis-Menten kinetics (using GraphPad Prism 4).
RESULTS
Expression of E. coli NAD+-dependent malic enzyme is insufficient to rescue C4 auxotrophy of Pyc− S. cerevisiae.
In wild-type S. cerevisiae grown on glucose, pyruvate carboxylase functions as the sole anaplerotic enzyme (6, 29), replenishing the intracellular pools of C4 acids. To test whether malic enzyme can assume this anaplerotic role in pyruvate carboxylase-negative (Pyc−) S. cerevisiae, a codon-optimized version of the E. coli NAD+-dependent malic enzyme, sfcA (also known as maeA), was overexpressed in the pyc1Δ pyc2Δ strain IMK157, resulting in strain IMY019. Compared to the Pyc− empty vector control strain IMY007, overexpression of the E. coli malic enzyme increased in vitro NAD+-dependent malic enzyme activity by circa 8-fold to 0.49 ± 0.03 mmol min−1 g of protein−1 (Table 4). However, strain IMY019 showed no growth on synthetic medium agar plates with glucose as the sole carbon source, even when cultures were incubated under a CO2 atmosphere to thermodynamically favor pyruvate carboxylation (Fig. 1).
TABLE 4.
In vitro whole-cell enzyme activities
| Strain | μmax (h−1) | Mean enzyme activity (mmol min−1 g of protein−1) ± SDa |
||||
|---|---|---|---|---|---|---|
| ME-NAD | ME-NADP | Pdc | PEPCK | Icl | ||
| CEN.PK 113-7D | 0.30 ± 0.01 | 0.04 ± 0.00 | 0.03 ± 0.00 | 1.5 ± 0.2 | 0.04 ± 0.01 | ND |
| CEN.PK 113-7D on ethanol | NA | 0.09 ± 0.02 | 0.02 ± 0.00 | 0.2 ± 0.0 | 0.44 ± 0.05 | 0.22 ± 0.03 |
| IMY007 (pyc1,2Δ, empty vector) | NG | 0.06 ± 0.00 | 0.03 ± 0.01 | 1.9 ± 0.3 | 0.04 ± 0.00 | ND |
| IMY019 (pyc1,2Δ, E. coli sfcA) | NG | 0.49 ± 0.03 | 0.03 ± 0.00 | 1.6 ± 0.1 | 0.04 ± 0.00 | ND |
| IMY016 (pyc1,2Δ pdc2Δ, E. coli sfcA) | NG | 1.03 ± 0.07 | 0.03 ± 0.00 | 0.7 ± 0.0 | 0.04 ± 0.01 | ND |
| IMY017 (pyc1,2Δ pdc2Δ evolved, mutated E. coli sfcA) | 0.06 ± 0.01 | 0.12 ± 0.01 | 0.39 ± 0.02 | 0.7 ± 0.0 | 0.04 ± 0.02 | ND |
| IMY027 (pyc1,2Δ pdc2Δ, ↑MAE1) | NG | 0.30 ± 0.03 | 0.34 ± 0.04 | 0.5 ± 0.0 | 0.04 ± 0.00 | ND |
| IMY028 (pyc1,2Δ pdc2Δ, ↑sMAE1) | 0.012 ± 0.000 | 0.17 ± 0.01 | 0.18 ± 0.01 | 0.7 ± 0.0 | 0.04 ± 0.00 | ND |
| IMY029 (pyc1,2Δ pdc2Δ, ↑sMAE1), IMY017 background | 0.019 ± 0.003 | 0.18 ± 0.02 | 0.19 ± 0.02 | 1.3 ± 0.0 | 0.04 ± 0.00 | ND |
Abbreviations: ME-NAD; malic enzyme (with NAD+); ME-NADP, malic enzyme (with NADP+); Pdc, pyruvate decarboxylase; PEPCK, phospho-enol-pyruvate carboxykinase; Icl, isocitrate lyase. Yeast strains were grown on synthetic medium with glucose and aspartate unless indicated otherwise. Errors are deviations from mean (for each condition two shake flask cultures were run). Also indicated are the maximum specific growth rates (μmax) on glucose under a CO2 atmosphere in batch cultures. NA, not applicable; ND, not detectable (<0.005 mmol min−1 g of protein−1); NG, no growth (tested on plates).
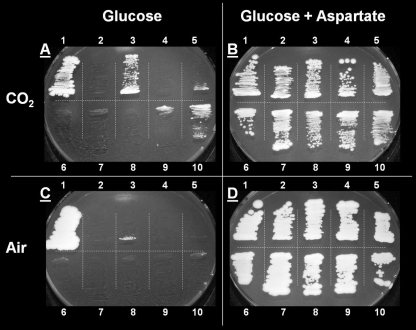
FIG. 1.
Synthetic medium agar plates, incubated for 14 days under an atmosphere of either 1.2 bar CO2 (plates A and B) or air (plates C and D). Plates contained glucose, Tween 80 and ergosterol and, as the nitrogen source, either ammonium sulfate (plates A and C) or aspartate (plates B and D). Strains: 1, CEN.PK 113-7D (wild-type reference); 2, IMY016 (pyc1,2Δ pdc2Δ, E. coli sfcA); 3, IMY017 (pyc1,2Δ pdc2Δ evolved, mutated E. coli sfcA); 4, IMY019 (pyc1,2Δ, E. coli sfcA); 5, IMY024 (pyc1,2Δ pdc2Δ, mutated E. coli sfcA); 6, IMY025 (pyc1,2Δ, ↑MAE1); 7, IMY026 (pyc1,2Δ, ↑sMAE1); 8, IMY027 (pyc1,2Δ pdc2Δ, ↑MAE1); 9, IMY028 (pyc1,2Δ pdc2Δ, ↑sMAE1); 10, IMY029 (pyc1,2Δ pdc2Δ evolved, ↑sMAE1). sMAE1 is a truncated, cytosolically retargeted version of S. cerevisiae MAE1.
To test whether increased intracellular substrate concentrations would allow for an anaplerotic flux through malic enzyme, an attempt was made to increase intracellular pyruvate levels by deleting PDC2, which encodes a positive regulator of the pyruvate decarboxylase (Pdc) isozymes Pdc1 and Pdc5 (16). Pyruvate decarboxylases convert pyruvate to acetaldehyde, a crucial step in the formation of ethanol. Deletion of PDC2 has previously been shown to result in a 3- to 4-fold reduction of Pdc activity in aerobic glucose-limited chemostat cultures, and in substantial production of pyruvate during batch cultivation on glucose (10). In the present study, PDC2 was deleted in strain IMK157, yielding strain IMK299. Subsequent overexpression of the E. coli sfcA malic enzyme in the pyc1,2Δ pdc2Δ strain IMK299 resulted in strain IMY016 (Table 1). Although a >2-fold reduction in pyruvate decarboxylase activity was observed in cell extracts relative to the parental strain IMY019 (Table 4), strain IMY016 showed no growth on glucose, regardless of whether an air or CO2 atmosphere was applied (Fig. 1).
Evolutionary engineering for increased anaplerotic flux.
To select for mutants with an increased anaplerotic flux through malic enzyme, evolutionary engineering was used, a strategy that previously proved successful for enabling anaplerosis in S. cerevisiae via PEPCK. After precultivation on glucose and aspartate, strain IMY016 was inoculated at an initial OD660 of circa 0.5 in a CO2-sparged batch bioreactor, containing synthetic medium with glucose as the sole carbon source. After a week of incubation without growth, aspartate was pulsed to allow the (viable) biomass concentration to increase. Subsequent nitrogen-limited chemostat cultivation on glucose medium at a dilution rate of 0.05 h−1 resulted in partial washout, and feeding was stopped. During the following batch phase, growth was observed. After two additional washout-batch cycles on medium with glucose as the sole carbon source, a specific growth rate of 0.06 h−1 was reached. At the end of the bioreactor cultivation a culture sample was taken and incubated on synthetic medium agar with glucose as the sole carbon source. Growth on plates was now observed, but only when plates were incubated under a CO2 atmosphere. This was consistent with an anaplerotic role of malic enzyme and indicated that growth did not result from cells with a derepressed glyoxylate cycle (3), for which CO2 is not a substrate. A single colony isolate from the evolved culture, designated IMY017 (Fig. 1), showed a representative maximum specific growth rate of 0.06 ± 0.01 h−1 in CO2-sparged batch cultures on glucose. At a specific growth rate of 0.06 h−1, a single gain-of-function mutant cell would be able to grow to a stationary phase culture in the 1-liter bioreactor (corresponding to approximately 35 generations) within 2 weeks. This time span is well within the 1-month duration of the selective cultivation.
Reverse engineering: a switch in cofactor specificity of E. coli malic enzyme.
To identify the basis of the changed phenotype, the in vitro enzyme activities of malic enzyme, pyruvate decarboxylase, PEPCK, and isocitrate lyase were determined in cell extracts of strain IMY017. Consistent with the strong CO2 dependence observed for growth on glucose (Fig. 1), no activity was found for isocitrate lyase, confirming that the glyoxylate cycle, which is normally repressed during batch cultivation on glucose, did not act as an anaplerotic pathway in the evolved strain. The PEPCK activity remained low at 0.04 ± 0.02 mmol min−1 g of protein−1, while no further decrease in Pdc activity was observed compared to IMY016 (Table 4). However, large changes were observed for malic enzyme. Whereas NAD+-dependent malic enzyme activity had dropped over 8-fold to 0.12 ± 0.01 mmol min−1 g of protein−1, NADP+-dependent activity had increased over 10-fold to 0.39 ± 0.02 mmol min−1 g of protein−1 (Table 4).
The enzyme activity measurements provided strong indications that the new phenotype was primarily due to changes to E. coli malic enzyme. Indeed, when the plasmid from IMY017, named MB5573*, was retransformed to IMK299, the resulting strain immediately showed growth on glucose under a CO2 atmosphere (strain IMY024, Fig. 1). Growth was slower than for IMY017, suggesting that additional beneficial mutations had occurred in IMY017 on a genomic level. No growth was observed when the original MB5573 plasmid or an empty plasmid were retransformed to a cured IMY017, or when the MB5573* plasmid isolated from IMY017 was transformed to strain IMK157, which still carried an intact copy of PDC2 (data not shown).
Subsequent sequencing of the sfcA open reading frame, as well as its TDH3 promoter and CYC1 terminator, in the MB5573* strain uncovered a single A1007G point mutation resulting in an Asp336Gly amino acid change. This mutation lies in the NAD domain of sfcA and is close to the 311- to 319-amino-acid sequence that is predicted to be the NAD(P)-binding site (5). Interestingly, sequencing of the plasmid from an independently evolved single colony isolate, which like strain IMY017 showed growth on glucose but was not further investigated due to a low specific growth rate on glucose of 0.02 ± 0.00 h−1, uncovered a G1006A point mutation, resulting in a change of the same amino acid residue (in this case an Asp336Asn substitution).
To test whether the A1007G mutation had indeed changed the cofactor specificity and to avoid interference of the endogenous Mae1p in enzyme assays, the original and mutated E. coli malic enzyme were introduced in a mae1 knockout strain from the Saccharomyces Genome Deletion Project (37) (Table 1). Although PCR confirmed the deletion of mae1, low background activities were still found in the malic enzyme assay for a mae1 deletion strain carrying an empty vector (strain IMY030). However, at ca. 5% of the activities measured for the strains carrying MB5573 (IMY031) or MB5573* (IMY032), this did not significantly influence the characterization of the E. coli malic enzyme. Interestingly, the A1007G point mutation in sfcA resulted in a 2-fold lower Vmax on NAD+, and a slightly increased Vmax on NADP+ (Table 5). However, a far more striking effect was observed for the cofactor substrate saturation constants. In the mutated E. coli malic enzyme, the Km for NAD+ had decreased ∼30-fold, while the Km for NADP+ was improved by approximately the same factor (Table 5). Combined, these changes effected a drastic switch in cofactor specificity.
TABLE 5.
Kinetic parameters of the original allele of E. coli sfcA NADH-dependent malic enzyme and of a mutated allele obtained by adaptive evolution in yeast, with either NAD+ or NADP+ as the redox cofactor
| Strain | Kinetic enzyme parameters (mean ± SD)a |
|||
|---|---|---|---|---|
| NAD+ |
NADP+ |
|||
| Vmax (mmol min−1 g of protein−1) | Km (mM) | Vmax (mmol min−1 g of protein−1) | Km (mM) | |
| IMY031 (mae1Δ, E. coli sfcA) | 1.8 ± 0.2 | 0.14 ± 0.05 | 0.6 ± 0.3 | 8.7 ± 7.3 |
| IMY032 (mae1Δ, mutated E. coli sfcA) | 1.0 ± 0.2 | 3.9 ± 1.9 | 0.7 ± 0.1 | 0.23 ± 0.08 |
Measurements were performed in whole-cell extracts of S. cerevisiae strains grown on synthetic medium with glucose and aspartate. Ranges indicate 95% confidence intervals (for each strain, enzyme assays were performed on cell extracts from two independent shake flasks).
An anaplerotic role for cytosolically targeted S. cerevisiae Mae1p.
After having established the importance of cofactor specificity in the anaplerotic role of the E. coli enzyme, replacement of the anaplerotic role of pyruvate carboxylase by alleles of the native yeast malic enzyme, encoded by MAE1, was investigated. Interestingly, the endogenous malic enzyme already has the potential to use NADPH as the cofactor, but is, in contrast to the heterologously expressed E. coli enzyme, targeted to the mitochondrion. Therefore, both a full copy of MAE1 and a truncated version lacking the mitochondrial targeting sequence, named sMAE1 (21), were overexpressed in Pyc− S. cerevisiae. No growth was discernible on synthetic glucose medium for the Pyc− strain IMK157 (over)expressing MAE1 (strain IMY025) or sMAE1 (IMY026), and for the pyc1,2Δ pdc2Δ strain IMK299 overexpressing MAE1 (IMY027). However, colonies were obtained for the pyc1,2Δ pdc2Δ strain IMK299 expressing the cytosolic sMAE1 (IMY028, see Fig. 1). This strain showed a maximum specific growth rate of 0.012 ± 0.000 h−1 on glucose in CO2-sparged batch cultures.
The characterization of the evolved strain IMY017, which expresses a mutated copy of the E. coli sfcA gene, suggested that changes in the yeast genetic background, although not essential for growth, contributed to its improved growth. To test whether these unidentified genomic changes would also benefit a strain expressing the truncated S. cerevisiae malic enzyme, sMAE1 was expressed in IMY017 after its original plasmid had been cured. Indeed, growth of the resulting strain IMY029 was faster on glucose plates (Fig. 1) and in CO2-sparged batch cultures, where a specific growth rate of 0.019 ± 0.003 h−1 was observed. The relatively low growth rate of this sMAE1-based strain (the E. coli sfcA-based strain IMY017 showed a maximum specific growth rate of 0.06 ± 0.01 h−1) might in part be explained by a lower NADP+-dependent malic enzyme activity for strain IMY029 compared to strain IMY017 (0.19 ± 0.02 versus 0.39 ± 0.02 mmol min−1 g of protein−1; see Table 4).
DISCUSSION
This study shows that, in addition to a derepressed glyoxylate cycle (3), heterologous PEP carboxylase (12), and PEPCK (40), malic enzyme can assume the anaplerotic role in S. cerevisiae during growth on glucose that is normally fulfilled by pyruvate carboxylase (6, 29). This is substantiated by the fact that isocitrate lyase activity in the Pyc− strains remained below the detection level, growth depended on high CO2 levels, which would only benefit anaplerotic routes involving carbon fixation, and, also in the evolved strains, growth required the overexpression of (cytosolic) malic enzyme. A significant carboxylating flux through PEPCK is not expected, since PEPCK activities were not increased after evolution, and in vitro enzyme activities were only a fraction of those found in PEPCK-anaplerotic strains (0.04 versus 0.29 to 0.87 mmol min−1 g of protein−1; see reference 40). Similarly, the low expression levels of endogenous MAE1 and mitochondrial localization of Mae1p make it unlikely that malic enzyme made a significant contribution to the anaplerotic flux in the earlier-characterized PEPCK-expressing S. cerevisiae strains (40). The present study has shown that a cytosolic localization is crucial for enabling an anaplerotic role of malic enzyme, which might be explained by less favorable conditions for carboxylation in the mitochondrial matrix, an inability to generate mitochondrial NAD(P)H, and/or limitations in the transport of C4 acids from the mitochondria to the cytosol.
NADPH as a preferred cofactor.
The NAD+-specific E. coli sfcA malic enzyme was chosen based on several grounds: (i) as a prokaryotic protein, sfcA lacks an intracellular targeting sequence, and thus can be expected to be cytosolically expressed in S. cerevisiae; (ii) previous studies on malic enzyme as a carboxylating enzyme in E. coli also targeted sfcA (17, 27); and (iii) if malic enzyme would be used for the high-yield production of C4-dicarboxylic acids, the use of NADH instead of NADPH would simplify the balancing of cofactors (e.g., avoiding the need for transhydrogenase activity for the production of malate and fumarate, which can be produced redox neutrally from glucose). However, the Pyc− S. cerevisiae only grew when expressing either the cytosolically relocalized sMAE1p, which can use both NADH and NADPH, or a mutated sfcA allele with a strongly improved affinity for NADPH. This indicates that, in the tested strains, anaplerosis via malic enzyme depended on the use of NADPH for reductive pyruvate carboxylation.
The Asp336Gly mutation clearly resulted in a near complete switch of the cofactor specificity from NAD+ to NADP+ in E. coli malic enzyme. This preferential use of NADPH by malic enzyme in this carboxylating role might be explained by the thermodynamics of the catalyzed reaction. The dependency of growth on deletion of PDC2, which probably increased the cytosolic pyruvate concentration, and on incubation under a CO2 atmosphere, both indicate that this reaction is likely to be close to thermodynamic equilibrium when operating in the carboxylating direction. Although few measurements are available on cytosolic redox factor concentrations in S. cerevisiae (8), whole-cell measurements showed intracellular ratios of NADPH/NADP+ that are higher than those of NADH/NAD+ (21). If this is representative for the cofactor distribution in the cytosol, the use of NADPH would increase the driving force of carboxylation by cytosolic malic enzyme.
Malic enzyme for C4 acid production by S. cerevisiae.
Like PEPCK, malic enzyme offers the prospect of metabolic engineering of S. cerevisiae for C4-dicarboxylic acid production from glucose under anaerobic conditions (40). The higher ATP yield of carboxylation might eliminate the need to respire part of the glucose for ATP generation, thereby increasing product yields. In view of the relatively low growth rates of the current strains (0.06 h−1 with the mutated E. coli malic enzyme versus 0.14 h−1 for PEPCK-anaplerotic strains and 0.30 h−1 for the pyruvate carboxylase-positive reference strain [40]), the first priority will be to increase the in vivo rates of pyruvate carboxylation by malic enzyme. Initially, sequential batch cultivation can be used to select for faster growth and a higher anaplerotic flux. Further increases in the carboxylating fluxes through malic enzyme or PEPCK might be achieved by improving substrate availability and reducing the flux to ethanol. Such an approach previously proved successful in engineering a pyruvate carboxylase-overexpressing S. cerevisiae strain, which was unable to produce ethanol due to deletion of the pyruvate decarboxylase genes, for malate and succinate production (38, 39). The increased availability of pyruvate in pyruvate decarboxylase-negative strains can also improve the carboxylation through malic enzyme. As an additional benefit, when ATP generation becomes dependent on the production (and export) of C4-dicarboxylic acids, this might allow for evolutionary engineering for an improved flux through malic enzyme (40).
Acknowledgments
The Ph.D. research of R.M.Z. was financed by Tate & Lyle Ingredients Americas. This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
We thank our coworkers at the Delft University of Technology, Tate & Lyle, and Microbia for stimulating discussions. We acknowledge Giang Huong Duong for her contribution to the literature review, Erwin Suir for construction of pUG-hphNT1, and Peter Niederberger for granting permission to use strain CEN.JB27, the ancestor of strain IMK157.
Footnotes
Published ahead of print on 3 November 2010.
REFERENCES
- 1.Abbott, D. A., R. M. Zelle, J. T. Pronk, and A. J. A. van Maris. 2009. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 9:1123-1136. [DOI] [PubMed] [Google Scholar]
- 2.Ashworth, J. M., and H. L. Kornberg. 1966. The anaplerotic fixation of carbon dioxide by Escherichia coli. Proc. Biol. Sci. 165:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Blazquez, M. A., F. Gamo, and C. Gancedo. 1995. A mutation affecting carbon catabolite repression suppresses growth defects in pyruvate carboxylase mutants from Saccharomyces cerevisiae. FEBS Lett. 377:197-200. [DOI] [PubMed] [Google Scholar]
- 4.Boles, E., P. de Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bologna, F. P., C. S. Andreo, and M. F. Drincovich. 2007. Escherichia coli malic enzymes: two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. J. Bacteriol. 189:5937-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewster, N. K., D. L. Val, M. E. Walker, and J. C. Wallace. 1994. Regulation of pyruvate carboxylase isozyme (PYC1, PYC2) gene expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch. Biochem. Biophys. 311:62-71. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D., D. Dawson, and T. Stearns. 2000. High-efficiency transformation of yeast, p.103-105. In Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 8.Canelas, A. B., W. M. van Gulik, and J. J. Heijnen. 2008. Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol. Bioeng. 100:734-743. [DOI] [PubMed] [Google Scholar]
- 9.de Jong-Gubbels, P., P. Vanrolleghem, S. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 10.Flikweert, M. T., et al. 1999. Steady-state and transient-state analysis of growth and metabolite production in a Saccharomyces cerevisiae strain with reduced pyruvate-decarboxylase activity. Biotechnol. Bioeng. 66:42-50. [DOI] [PubMed] [Google Scholar]
- 11.Flikweert, M. T., et al. 1996. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12:247-257. [DOI] [PubMed] [Google Scholar]
- 12.Flores, C., and C. Gancedo. 1997. Expression of PEP carboxylase from Escherichia coli complements the phenotypic effects of pyruvate carboxylase mutations in Saccharomyces cerevisiae. FEBS Lett. 412:531-534. [DOI] [PubMed] [Google Scholar]
- 13.Fuck, E., G. Stärk, and F. Radler. 1973. Äpfelsäurestoffwechsel bei Saccharomyces. II. Anreicherung und Eigenschaften eines Malatenzyms. Arch. Microbiol. 89:223-231. [PubMed] [Google Scholar]
- 14.Gerrard Wheeler, M., et al. 2008. Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties. Plant Mol. Biol. 67:231-242. [DOI] [PubMed] [Google Scholar]
- 15.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohmann, S. 1993. Characterisation of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 241:657-666. [DOI] [PubMed] [Google Scholar]
- 17.Hong, S. H., and S. Y. Lee. 2001. Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol. Bioeng. 74:89-95. [DOI] [PubMed] [Google Scholar]
- 18.Janke, C., et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers, and promoter substitution cassettes. Yeast 21:947-962. [DOI] [PubMed] [Google Scholar]
- 19.Kunkee, R. E. 1968. Malo-lactic fermentation. Adv. Appl. Microbiol. 9:235-279. [DOI] [PubMed] [Google Scholar]
- 20.Maurino, V. G., M. C. Gerrard Wheeler, C. S. Andreo, and M. F. Drincovich. 2009. Redundancy is sometimes seen only by the uncritical: does Arabidopsis need six malic enzyme isoforms? Plant Sci. 176:715-721. [Google Scholar]
- 21.Moreira dos Santos, M., V. Raghevendran, P. Kötter, L. Olsson, and J. Nielsen. 2004. Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab. Eng. 6:352-363. [DOI] [PubMed] [Google Scholar]
- 22.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 23.Nookaew, I., et al. 2008. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst. Biol. 2:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel, M. K., et al. 2006. Medium and long-term opportunities and risks of the biotechnological production of bulk chemicals from renewable resources. Utrecht University, Utrecht, Netherlands.
- 25.Pound K. M., et al. 2009. Substrate-enzyme competition attenuates upregulated anaplerotic flux through malic enzyme in hypertrophied rat heart and restores triacylglyceride content: attenuating upregulated anaplerosis in hypertrophy. Circ. Res. 104:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 27.Stols, L., and M. I. Donnelly. 1997. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stols, L., G. Kulkarni, B. G. Harris, and M. I. Donnelly. 1997. Expression of Ascaris suum malic enzyme in a mutant Escherichia coli allows production of succinic acid from glucose. Appl. Biochem. Biotechnol. 63-65:153-158. [DOI] [PubMed] [Google Scholar]
- 29.Stucka, R., S. Dequin, J. Salmon, and C. Gancedo. 1991. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 229:307-315. [DOI] [PubMed] [Google Scholar]
- 30.Sundqvist, K. E., J. Heikkilä, I. E. Hassinen, and J. K. Hiltunen. 1987. Role of NADP+-linked malic enzymes as regulators of the pool size of tricarboxylic acid-cycle intermediates in the perfused rat heart. Biochem. J. 243:853-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dijken, J. P., et al. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 32.van Maris, A. J. A., W. N. Konings, J. P. van Dijken, and J. T. Pronk. 2004. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab. Eng. 6:245-255. [DOI] [PubMed] [Google Scholar]
- 33.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 34.Voegele, R. T., M. J. Mitsch, and T. M. Finan. 1999. Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1432:275-285. [DOI] [PubMed] [Google Scholar]
- 35.Volschenk H., H. J. J. van Vuuren, and M. Viljoen-Bloom. 2003. Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr. Genet. 43:379-391. [DOI] [PubMed] [Google Scholar]
- 36.Werpy, T., and G. Petersen. 2004. Top. value added chemicals from biomass: I. The results of screening for potential candidates from sugars and synthesis gas. U.S. Department of Energy, Washington, DC.
- 37.Winzeler, E. A., et al. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 38.Zelle, R. M., E. de Hulster, W. Kloezen, J. T. Pronk, and A. J. A. van Maris. 2010. Key process conditions for production of C4 dicarboxylic acids in bioreactor batch cultures of an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 76:744-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zelle, R. M., et al. 2008. Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 74:2766-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelle, R. M., J. Trueheart, J. C. Harrison, J. T. Pronk, and A. J. A. van Maris. 2010. Phosphoenolpyruvate carboxykinase as the sole anaplerotic enzyme in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 76:5383-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]