Abstract
Perchlorate is a known health hazard for humans, fish, and other species. Therefore, it is important to assess the response of an ecosystem exposed to perchlorate contamination. The data reported here show that a liquid chromatography-mass spectrometry-based proteomics approach for the detection of perchlorate-reducing enzymes can be used to measure the ability of microorganisms to degrade perchlorate, including determining the current perchlorate degradation status. Signature peptides derived from chlorite dismutase (CD) and perchlorate reductase can be used as biomarkers of perchlorate presence and biodegradation. Four peptides each derived from CD and perchlorate reductase subunit A (PcrA) and seven peptides derived from perchlorate reductase subunit B (PcrB) were identified as signature biomarkers for perchlorate degradation, as these sequences are conserved in the majority of the pure and mixed perchlorate-degrading microbial cultures examined. However, chlorite dismutase signature biomarker peptides from Dechloromonas agitata CKB were found to be different from those in other cultures used and should also be included with selected CD biomarkers. The combination of these peptides derived from the two enzymes represents a promising perchlorate presence/biodegradation biomarker system. The biomarker peptides were detected at perchlorate concentrations as low as 0.1 mM and at different time points both in pure cultures and within perchlorate-reducing environmental enrichment consortia. The peptide biomarkers were also detected in the simultaneous presence of perchlorate and an alternate electron acceptor, nitrate. We believe that this technique can be useful for monitoring bioremediation processes for other anthropogenic environmental contaminants with known metabolic pathways.
An assessment of the physiological state of an ecosystem is a useful tool for evaluating exposure to and effects of an environmental contaminant. The physiological state of an ecosystem can be assessed by measuring the changes in the autochthonous microbial community in response to a disturbance such as contamination by heavy metals, organic and inorganic contaminants, or other xenobiotic substances. For example, changes in the native microbial community's structure and function reflected in changed rates of biodegradation, biotransformation, and bioaccumulation of contaminants by microbes can be early indicators of how an ecosystem is adjusting to a given pollutant. These induced alterations in cellular or biochemical components or processes in a microbial community can be measured and used as biological markers to measure the ability of the ecosystem to respond to a pollutant (13, 22) and can help in making decisions about its extent and the need for human involvement in the cleanup process. Because microbes adjust to environmental changes through modifications of their proteomes, the global analysis of microbial proteins is an effective way to track subtle changes in a microbial community's physiology and structure if the functions of proteins are known and there is a metagenomic scaffold to identify the full-length sequences. For example, it has been shown that perturbation of an ecosystem can modify indigenous microbial protein sequences or induce the expression, repression, degradation, or overproduction of certain proteins. Therefore, analyses of microbial proteins (metaproteomics) can provide insight into the dynamics of both pristine and perturbed biological systems (17, 19).
Proteins as diagnostic biomarkers have been used and researched in the area of medical toxicology for several years (24, 37). The use of proteomics in environmental toxicology is also growing steadily. Environmental toxicology studies thus far have focused mostly on the effects of toxic chemicals on a variety of biological systems, including microorganisms (bacteria and fungi), vertebrates (fish and rodents), invertebrates (insects, worms, and mollusks), and plants (1, 3, 6, 16, 25, 27, 28, 32, 34). The responses of these organisms were determined by proteome comparisons between altered and control states, which led to the identification of toxicity-associated protein biomarkers.
Here, we report a new proteomics-based tool that can be used together with other chemical and biomolecular methods to develop an effective system for monitoring and measuring the exposure of a given environment to perchlorate contamination and/or to determine the current perchlorate degradation status. Perchlorate is a health hazard for humans, fish, and other species. In humans, it competitively inhibits iodine uptake by the thyroid gland and affects thyroid hormone synthesis (9, 29, 31, 35). Thyroid hormones are required for normal development in children and regulation of metabolism in adults. In pregnant females, destruction of thyroid function can affect the fetus and can lead to delayed development and decreased learning capability. Short-term exposure to high perchlorate doses can result in eye and skin irritation, cough, nausea, vomiting, and diarrhea (11, 35). Perchlorate can impair the proper functioning of thyroid glands at concentrations as low as 24 ppb (12). Thus, it is important to assess the ecological responses of environments where perchlorate contamination is known or suspected. We believe that proteomics-based techniques can be useful for developing protein biomarkers for monitoring the dynamics of perchlorate metabolic processes. We also believe that such an approach can be used to develop protein biomarkers to monitor environmental responses to other anthropogenic environmental contaminants with known metabolic pathways, leading to the future creation of peptide sequence libraries that can be used to monitor the fate and degradation dynamics of anthropogenic chemicals in the biosphere.
Several microorganisms are known to grow by anaerobic reductive dissimilation of perchlorate into chloride using two essential enzymes, perchlorate reductase (Pcr) and chlorite dismutase (CD) (14). Perchlorate reductase catalyzes the first step in the perchlorate reduction pathway and reduces perchlorate (ClO4−) to chlorite (ClO2−) and oxygen (O2), which is followed by disproportionation (dismutation) of toxic chlorite into molecular oxygen and chloride by chlorite dismutase. Work to date indicates that perchlorate reductase is an inducible enzyme and is expressed only when an organism capable of perchlorate reduction is grown with perchlorate as the primary electron acceptor (14). The mechanism of expression of chlorite dismutase is more variable and species specific. For example, in D. agitata strain CKB, chlorite dismutase is detectable under aerobic conditions, but its expression is upregulated in the presence of perchlorate; however, in Pseudomonas strain PDA and Pseudomonas strain PK, CD is constitutively expressed (14, 36). CD, encoded by the cld gene, is unique and highly conserved in dissimilatory perchlorate- and chlorate-reducing organisms (4, 21), with low sequence similarity between cld and any other sequence in GenBank (30). Both Pcr and CD satisfy the important attributes of a biological marker. They are unique and specific for perchlorate, quantifiable, and since their synthesis is induced or upregulated in the presence of perchlorate, the concentration of these proteins indicates the state of perchlorate metabolism in a given environment (5). Thus, we suggest that the amino acid sequences of signature peptides from both enzymes can be used to indicate the presence of perchlorate contamination and to detect and quantify perchlorate-degrading enzymes.
As reported here, we used a mass spectrometry (MS)-based proteomics approach to identify signature peptides from known perchlorate-degrading pure bacterial cultures (nine strains) and mixed-culture environmental enrichment samples (three examples) grown under similar conditions. Specific CD and Pcr signature peptides common to all or a majority of the samples were selected as biomarkers to indicate the presence of perchlorate-degrading proteins. Previous research indicated a need to use a number of proteins as biomarkers for reliable results (19, 33). Therefore, multiple signature peptides from both CD and Pcr were chosen as primary biomarkers. To further validate the effectiveness and utility of the selected biomarkers, one of the mixed-culture samples (an aliquot of a biomass-covered activated carbon matrix taken from a bioreactor being used to treat perchlorate-contaminated drinking water) (7) was chosen and the signature peptides were identified under a variety of growth conditions. We also examined this sample to determine the potential for interference (repression) in the activity of perchlorate enzymes by the alternative electron acceptor nitrate.
MATERIALS AND METHODS
Microorganisms used and culture conditions. (i) Pure bacterial cultures and environmental samples.
The following perchlorate-degrading pure bacterial strains were used: Dechlorosoma sp. KJ (ATCC BAA-592) (optical density at 600 nm [OD600], 0.53), Dechloromonas hortensis (DSMZ 15637) (OD600, 0.78), Dechloromonas strain MissR (courtesy of J. Coates, University of California—Berkeley) (OD600, 0.76), Dechloromonas agitata CKB (DSMZ 13637) (OD600, 0.52), Dechlorospirillum anomalous (NCCB100047) (OD600, 0.46), Azospira oryzae (DSMZ 11199) (OD600, 0.54), and Azospira oryzae (Dechlorosoma suillum) (DSMZ 13638) (OD600, 0.69). Two unidentified perchlorate-degrading pure strains previously isolated in our laboratory were also used. Isolate “Bior” (OD600, 0.61) was purified from a bioreactor used to treat perchlorate-contaminated drinking water (7), and isolate “Crw” was isolated from the Clearwater River near Lewiston, ID. Bacterial strains were grown and maintained on the culture medium recommended by the culture collections from which the cultures were obtained. Dechloromonas MissR, Bior, and Crw were maintained on R2A medium. The R2A medium (g/liter) consists of 0.5 g proteose peptone, 0.5 g Casamino Acids, 0.5 g yeast extract, 0.5 g dextrose, 0.5 g soluble starch, 0.3 g dipotassium phosphate, 0.3 g sodium pyruvate, and 0.05 g magnesium sulfate. Actively growing bacterial cells from log phase cultures in maintenance medium were used to inoculate ATCC 2106 liquid medium containing 5 mM ammonium perchlorate as the electron acceptor and 20 mM sodium acetate as the electron donor. Three mixed-culture microbiological samples included an aliquot of a biomass-covered activated carbon matrix taken from a bioreactor being used to treat perchlorate-contaminated drinking water (Bior enrichment culture) (OD600, 0.59) (7), a sample of wastewater taken from a paper mill effluent treatment basin (Idaho) (Pt enrichment culture) (OD600, 0.43), and a biofilm sample taken from rock in the Clearwater River near a paper and pulp industry in Lewiston, ID (Crw enrichment culture) (OD600: 0.57). The samples were inoculated and enriched in ATCC 2106 medium supplemented with 5 mM ammonium perchlorate and 20 mM sodium acetate. All the cultures (20 ml) were grown anaerobically in 25-ml gas-tight anaerobic test tubes with 1-cm butyl rubber stoppers secured with aluminum crimp seals and incubated at room temperature without shaking. The growth of cultures was measured by monitoring the OD600. The majority of perchlorate-reducing microbes can also use oxygen as an alternate electron acceptor, and they are also microaerophilic in nature. Therefore, there will not be a significant dissolved oxygen concentration in the medium and it will not affect the growth and activity of the microorganisms.
(ii) Detection of biomarkers in a mixed culture at different growth phases and concentrations of perchlorate.
The enrichment culture from the bioreactor sample was used to inoculate ATCC 2106 medium with 5 mM ammonium perchlorate and 20 mM sodium acetate and incubated anaerobically at room temperature without shaking. Samples for mass spectrometric analysis were drawn from the culture at three different time points (early log, mid-log, and late log phase) during the growth cycle. In a second set of experiments, ATCC 2106 medium was inoculated with enrichment culture from the bioreactor using 20 mM sodium acetate and four different concentrations of ammonium perchlorate (0.1 mM, 0.5 mM, 1 mM, and 2.5 mM). The cultures were also grown anaerobically at room temperature without shaking, and growth was measured by monitoring the OD600. The different perchlorate concentrations were chosen for experimental purposes. There are not many documented sites as yet with perchlorate concentrations higher than 0.1 mM. The U.S. EPA reported ∼10 sites with greater than 0.1 mM perchlorate in 2003 (14). However, our described technique is very sensitive and will be useful for sites with less than 0.1 mM perchlorate concentration due to a lower detection limit (10−15 mole of peptide per 2-μl injection).
(iii) Detection of biomarkers in a mixed culture in the simultaneous presence of perchlorate and nitrate.
ATCC 2106 medium with 20 mM sodium acetate was inoculated with the bioreactor enrichment culture. The following concentration combinations of ammonium perchlorate and sodium nitrate were used: combination a, 1.25 mM perchlorate and 3.75 mM nitrate; combination b, 1.67 mM perchlorate and 3.33 mM nitrate; combination c, 2.5 mM perchlorate and 2.5 mM nitrate; and combination d, 5 mM nitrate (control). The cultures were grown anaerobically at room temperature without shaking, and growth was measured by monitoring the OD600. Perchlorate and nitrate concentrations were monitored at regular time intervals using negative electrospray ionization-MS (ESI-MS). A Micromass Quattro II triple quadrupole mass spectrometer with an electrospray ionization source (Waters Corporation, Milford, MA) was used for these analyses. The instrument was controlled by MassLynx version 4.0 software (Waters Corporation, Milford, MA). The mass spectrometer settings used were the same as described in reference 2. Perchlorate (m/z = 99.45) and nitrate (m/z = 62) concentrations were determined by peak intensities in the total ion spectrum using standard curves (perchlorate and nitrate) prepared by analysis of a range of known concentrations of pure standards between 5 μM and 0.5 mM. All experiments were performed in triplicate.
Proteomics methods. (i) Protein extraction and sequencing.
Late log phase cells, unless specified otherwise, were harvested from pure cultures and enrichment cultures. The volume (range, 0.9 to 1.8 ml) was adjusted to normalize the samples with respect to their optical density depending on the growth of the microorganism. The protein was extracted and digested using the method described in reference 2. Dry peptide samples were solubilized in 20 μl of H2O containing 5% acetonitrile and 0.1% formic acid and spiked with 50 fmol/μl of bovine serum albumin (BSA) peptides as the internal standard (MassPREP BSA digestion standard; Waters Corporation, Milford, MA). Samples were clarified by centrifugation at 10,000 × g for 10 min just before MS analysis. Peptides were then separated and analyzed by a liquid chromatography (LC)-MS method described in reference 2, with some modifications. Peptides in the sample were separated using reverse-phase liquid chromatography on a nanoACQUITY ultra-performance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) and analyzed using a Q-TOF Premier quadrupole time-of-flight-tandem MS (MS-MS) system. Peptides from 2 μl of sample hydrolysate were trapped on an 0.18-mm by 20-mm Symmetry C18 trap column (Waters Corp.) prior to injection into the analytical column [0.075-mm by 200-mm (I) BEH 130 C18 nanoACQUITY UPLC column; Waters Corp.]. The solvents used were 0.1% formic acid in H2O (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Peptides were trapped on the loading column using 100% solvent A at a flow rate of 5 μl min−1 for 3 min. Trapped peptides were then separated at a flow rate of 0.4 μl min−1 using the following conditions: (i) isocratic, 94% solvent A and 6% solvent B for 1 min; (ii) gradient, 1 min after injection, the concentration of solvent A was decreased to 73% and that of solvent B was increased to 27% over the next 70 min; (iii) gradient, the concentration of solvent A was decreased to 43% and that of solvent B was increased to 57% over the next 20 min; (iv) gradient, the concentration of solvent A was decreased to 10% and that of solvent B was increased to 90% over the next 10 min; (v) isocratic, 10% A and 90% B for 10 min; and (vi) gradient, the concentration of solvent A was increased to 94% and that of solvent B was decreased to 6% over the next 10 min. The column effluent was delivered directly to the Q-TOF Premier using a nanospray source inlet. The nanosprayer was fitted with a 20-μm (inner diameter) fused-silica emitter tip. The following mass spectrometer settings were used: 3.80 kV capillary voltage, 25 V cone voltage, 120°C source temperature, 0.45 bar sheath gas pressure, 5.0 V collision energy, and 2,050 V detector voltage. The MS and UPLC were controlled by MassLynx 4.1 software (Waters Corporation, Milford, MA). A high-collision-energy/low-collision-energy switching (MSE)-LC mode of acquisition with alternating 1.5-s scans of low (10 V) or high (15 to 40 V) collision energies was used to obtain both identification and quantification MS-MS information. A reference peptide (or lockmass) was sprayed simultaneously with the LC effluent and was sampled for 1 s every 30 s. The lockmass standard used was human [glu1]-fibrinopeptide B (Sigma-Aldrich, St. Louis, MO).
(ii) Mass spectrometric data analysis.
Data were analyzed as described in reference 2, with some modifications. ProteinLynx Global Server 2.3 (PLGS) (Waters, Milford, MA) was used for mass spectrum analysis, peptide sequencing, protein identification, and quantification. Partial-protein amino acid sequences were identified by searching a custom database created in PLGS 2.3 using perchlorate reductase and chlorite dismutase amino acid sequences obtained in our laboratory and from a public database (downloaded from ExPASy Proteomics Server, http://ca.expasy.org/). The amino acid sequence of the internal standard protein, bovine serum albumin, was also added to the database. The BSA concentration used was specified in the PLGS program. The PLGS software quantifies the proteins by measuring the average MS signal response of the three most intense tryptic peptides from BSA and comparing them with the MS signal response of other identified proteins. A fixed carbamidomethyl C modification was used, and only 1 missed trypsin cleavage site was allowed.
RESULTS
Selection of protein biomarkers.
The MS spectra obtained for the pure bacterial and environmental consortium samples were searched against the custom database Protein Identity 2.3, created in ProteinLynx Global Server, containing amino acid sequences of known perchlorate-metabolizing enzymes. The amino acid sequences (signature peptides) from chlorite dismutase and perchlorate reductase subunit A and subunit B from all 12 samples (Dechlorosoma sp. KJ, D. hortensis, Dechloromonas MissR, D. agitata CKB, D. anomalous, A. oryzae DSMZ 11199, A. oryzae DSMZ 13638, isolate Bior, isolate Crw, Bior enrichment culture, Crw enrichment culture, and Pt enrichment culture) were compiled and analyzed. The amino acid sequences from CD, PcrA, and PcrB which were detected in the majority of samples were chosen for use as biomarkers (Table 1).
TABLE 1.
Peptide biomarkers from chlorite dismutase (CD) and perchlorate reductase subunit A (PcrA) and subunit B (PcrB)
| Enzyme and samples in which its peptide(s) was detecteda | Signature peptide sequence(s) |
|---|---|
| Chlorite dismutase | |
| All except D. agitata | GLETNSDFFFR, GTILTQPGVFGVFTMFK, HKDNVLVDLYLTR, YVIVIPVKK |
| Perchlorate reductase subunit A | |
| All except strain Bior | MDSTALYSDVVLPSAHWYEK |
| All except D. agitata, strain Crw, and the Pt enrichment culture | DIAPMPNIPEYNPR, EQTDLSYLVR, YIILWGSNPTQTR |
| Perchlorate reductase subunit B | |
| All except strain Crw | NVETAPGLGYPR |
| All except D. agitata and the Pt enrichment culture | IPLAQLEGLFGK |
| All except D. agitata, Crw strain, and the Pt enrichment culture | GKIPPMIDYGIPFEFDYAGR, IEQGVAPACVAQCVGR, IPPMIDYGIPFEFDYAGR, VALPLHPEFGTEPNVFYVPPVLGPR |
| All except D. agitata, strain Bior, and the Pt enrichment culture | SAPNWDEDQGAGEYPNNSFFYLPR |
Samples include Dechlorosoma sp. KJ (OD600, 0.53), Dechloromonas hortensis (OD600, 0.78), Dechloromonas strain MissR (OD600, 0.76), Dechloromonas agitata CKB (OD600, 0.52), Dechlorospirillum anomalous (OD600, 0.46), Azospira oryzae (DSMZ 11199) (OD600, 0.54), Azospira oryzae (DSMZ 13638) (OD600, 0.69), strain Bior (OD600, 0.61), strain Crw, Bior enrichment culture (OD600, 0.59), Pt enrichment culture (OD600, 0.43), and Crw enrichment culture (OD600, 0.57).
None of the CD, PcrA, and PcrB peptide biomarkers detected covered all the pure bacterial and mixed-culture samples used in the study. The chlorite dismutase signature peptides GLETNSDFFFR, GTILTQPGVFGVFTMFK, HKDNVLVDLYLTR, and YVIVIPVKK were detected in all of the samples except D. agitata CKB. Thus, any or all of the chosen CD signature peptides can be used as biomarkers. In the case of the perchlorate reductase A subunit, the amino acid sequence MDSTALYSDVVLPSAHWYEK was universally found in all samples except strain Bior. The signature peptides DIAPMPNIPEYNPR, EQTDLSYLVR, and YIILWGSNPTQTR were found in every sample except D. agitata CKB, strain Crw, and the Pt enrichment culture. For the perchlorate reductase B subunit, the peptide NVETAPGLGYPR was detected in all samples except strain Crw. The PcrB sequence IPLAQLEGLFGK was not found in D. agitata CKB and the Pt enrichment culture. The signature peptides GKIPPMIDYGIPFEFDYAGR, IEQGVAPACVAQCVGR, IPPMIDYGIPFEFDYAGR, VALPLHPEFGTEPNVFYVPPVLGPR, and SAPNWDEDQGAGEYPNNSFFYLPR were detected in nine samples. The first four were absent in D. agitata CKB, strain Crw, and the Pt enrichment culture, while SAPNWDEDQGAGEYPNNSFFYLPR was not found in D. agitata CKB, strain Bior, or the Pt enrichment culture. In the case of perchlorate reductase, a combination of any of the above-mentioned signature peptides from PcrA and PcrB can be used as biomarkers. The signature peptides of chlorite dismutase and perchlorate reductase in D. agitata CKB were found to be different from those in the majority of other perchlorate-degrading microbial cultures. Therefore, a different set of biomarkers for D. agitata (Table 2) should also be included with the selected biomarkers. Any combination of the CD, PcrA, and PcrB signature peptides listed in Table 2 can be used as biomarkers for D. agitata, since all the peptides were detected universally within a similar concentration range.
TABLE 2.
Peptides of chlorite dismutase and perchlorate reductase in late log growth phase of Dechloromonas agitata culture
| Protein (avg sequence coverage [%], no. of peptides matched [range], mean concn of peptides [ng/mg of total protein]a) | Signature peptide sequences |
|---|---|
| Chlorite dismutase (66.3, 39-51, 2,454)b | AAEVVAVVEK, FGNFSTYK, PTLANLVNVK, AEAYLTR, FVIPVKK, QSDFFLR, AEAYLTRGFEAQSDFFLR, GAAAEVVAVVEK, SPNLNAGLTGATYR, AEVVAVVEK, GFEAQSDFFLR, SPNLNAGLTGATYRDATPR, AFLVDFR, IHSYDMAATQAFLVDFR, TAPGVFGNFSTYK, AFVIPVK, ILTAPGVFGNFSTYK, TLANLVNVK, AFVIPVKK, ILTAPGVFGNFSTYKVRPDYYK, TQAFLVDFR, ANLVNVK, KGAAAEVVAVVEK, VKAEAYLTR, AQSDFFLR, KNADWWNLTDEQR, VRPDYYK, DKSPNLNAGLTGATYR, LANLVNVK, VVAVVEK, DLNYITK, LKEMETHTLPTLANLVN, WGSPTVLGTIQSFDSVVNTLSMGR, DWWNLTDEQR, LKEMETHTLPTLANLVNVK, YAFVIPVK, EAQSDFFLR, LSMAER, YAFVIPVKK, EMETHTLPTLANLVNVK, NADWWNLTDEQR, YHVRWGSPTVLGTIQSFDSVVNTLSMGR, EVVAVVEK, NLNAGLTGATYRDATPR, YRDATPR, FGMNAEVTENLVGMTK, PGVFGNFSTYK |
| Perchlorate reductase alpha subunit (66, 85-96, 2,511)c | ACPHFVYTK, GACPHFVYTK, PHFVYTK, ADNNIAK, GAHLINCTGACPHFVYTK, PLPGTAPVSFSAGHR, AMHLLTALVGSEGK, GECAHHYMYGPHR, PPMPNIPELNPR, ANWTSPMK, GGPCVEISPIDATAIGVK, PTHLVGNYGHLVFRPNYYGPAGSQR, ANWTSPMKEGVPYTPFQNYIVDK, GITLQMLR, PVAFMGR, APPVAFMGR, GNWLNQAK, RATWEEALDLISDK, APPVAFMGRNTHTFPK, GNWLNQAKGQK, RYIGATPISL, APVDADKFPFR, GSWGDQPEQK, SISPDFNSSTIK, ASAATLLGNSLTFK, GYIALENLD, SPLPGTAPVSFSAGHR, ATWEEALDLISDK, GYIALENLDPALEGK, SRIEADNNIAK, DGVVIR, GYIALENLDPALEGKFQVK, SVYSPLPGTAPVSFSAGHR, DGVVIREEQSK, HSIHSTFK, TDLPYLIR, DIPPMPNIPELNPR, HSIHSTFKDSVLMLR, TDWQIFLALA, DIPPMPNIPELNPRGCNK, IEADNNIAK, TDWQIFLALAK, DKFYIWDSK, IEADNNIAKAAK, TIIEVAR, DLPYLIR, IIDTIK, TPTHLVGNYGHLVFRPNYYGPAGSQR, DLTNLWNQMTMDGKDNDWVEIWNSHGK, IPDAHFLSEAQLNGAK, TRGAHLINCTGACPHFVYTK, DSVLMLR, ITGVPAK, VDVK, DVRVDVK, ITPTHLVGNYGHLVFRPNYYGPAGSQR, VEIWNSHGK, DWVEIWNSHGK, KPWPTLTGR, VFIVYR, EADNNIAK, LAEDEAAAQYILDTAPHSK, VICR, EADVVAGGSK, LDLNVTAEHTYINMTEPAIKPMWESK, VIREEQSK, EADVVAGGSKDK, LKYPLIR, VSMWHTPELYMDLLEGSTQSVCPVR, EDEAAAQYILDTAPHSK EEQSKDIPPMPNIPELNPR, LQDGNTVEVRPVFEILK, VVSISPDFNSSTIK, EGVPYTPFQNYIVDK, LQRGGPCVEISPIDATAIGVK, VYSPLPGTAPVSFSAGHR, EIWNSHGK, LVGSEGK, WARDLTNLWNQMTMDGK, EQTDLPYLIR, LYDAHNLK, WIHPLPGTDGALALAMAHVIIK, EYATTQPAMIICGGGTMHWYYSDVLLR, LYDAHNLKEQTDLPYLIR, WQIFLALAK, FLREADVVAGGSK, MDSTALYSDVVLPSAHWYEK, WVEIWNSHGK, FLREADVVAGGSKDK, NHSPDCISVYSPLPGTAPVSFSAGHR, YDAHNLK, FNSPHSR, NLWPK, YIGATPISL, FNSPHSRHSIHSTFK, NQWSWDK, YIILWGANPTQTR, FKANWTSPMK, NQWSWDKK, YLMHAIDTR, FPFRFNSPHSR, NWLNQAK, YMDLLEGSTQSVCPVR, FQVK, NWTSPMK, YPLIR, FYDEQFK, PDAHFLSEAQLNGAK, YSPLPGTAPVSFSAGHR, FYIWDSK, PGTAPVSFSAGHR, YVLR, PGTDGALALAMAHVIIK |
| Perchlorate reductase beta subunit (58.5, 26-29, 564)d | AMHVGFIDDQESSVFK, NQPSELMDILIGR, REQDGLVVIHQEK, CIGCFPR, NVETAPGLGYPR, RFGVALPLHPEYGTEPNVFYVPPVLGPR, CIGCQTCTVACK, NVETAPGLGYPRNWQSK, RSADMMISPMT, DDQESSVFK, PFEFDYAGR, RQLAYVADLNK, DQESSVFK, PPVLGPR, SADMMISPMT, EQDGIVVIHQDK, PSELMDILIGR, TAPGLGYPR, ETAPGLGYPR, QLAYVADLNK, TLQAER, EVLK, IEKGVAPACVAECAGR, TLQAEREK, FGVALPLHPEYGTEPNVFYVPPVLGPR, IKNQPSELMDILIGR, TLWTSGPGQDYMYWR, GGGYKDGVLQK, IPPMIDYGVPFEFDYAGR, VEMPNGEHTADPK, GKIPPMIDYGVPFEFDYAGR, ISMTQLEQLFGK, YVADLNK, GVAPACVAECAGR, LFEGKPGR, GYKDGVLQK, MCNHCAKPACLEACPNEAIYK |
| Perchlorate reductase gamma subunit (26.7, 6, 6) | APFTPFSPAVDSK, KPLVDAGQNFDM, LDPK, ASAHAKAFDSLKPNAK, KPLVDAGQNFDMEK, SVTATGKSNPVHTHFK |
| Perchlorate reductase delta subunit (61.3, 9-14, 75) | DFLER, HLCAWLPLVR, QELGVCWGELLEHFNEVLAPGLKLACSR, DFLERHLCAWLPLVR, LAQKDFLER, RPNLLLELK, ELKEELGE, LVLAADK, SDRPNLLLELK, EVFESQFLSAFETNMPSPSASLNEGVHIFK, LVLAADKALCR, SHIYSLLASGFGYPDESGYQGFSDGK, FIDEIR, MNSITDDRLVLAADK, VTTPFFVTLAEFAESFALANLR, GFYSNFGLQVDSK, QAQAHLEGMSADAYK |
Results are the average of three mass spectrometric runs per sample. All the readings have a standard error of less than 12% of the mean.
Peptides also match with (UniProtKB protein database accession numbers in parentheses) Pseudomonas sp. PK (Q673K7), sequence coverage 24%; and Ideonella dechloratans (Q9F437), sequence coverage 2%.
Peptides also match with (UniProtKB protein database accession no. in parentheses) Dechlorosoma sp. KJ (B2D1S4), sequence coverage 19.8%; Dechlorosoma sp. PCC (A7LI70), sequence coverage 18.4%; and Dechloromonas sp. PC1 (A7LI69), sequence coverage 13.4%.
Peptides also match with Dechlorosoma sp. KJ (UniProtKB protein database accession no. B2D1S5), sequence coverage 16.8%.
Validation and quantification of biomarkers.
The presence of the selected CD and Pcr biomarkers when a representative mixed-culture sample (Bior enrichment culture) was grown under different growth conditions (Table 3) was tested as follows: (i) at different time points during the growth cycle, (ii) with different concentrations of perchlorate, and (iii) in the presence of an alternative/competing electron acceptor, nitrate. The chlorite dismutase and perchlorate reductase biomarkers detected under the aforesaid conditions were also quantified using the LC-MSE mode of data acquisition on the Q-TOF Premier mass spectrometer (Table 3).
TABLE 3.
Detection of selected chlorite dismutase (CD) and perchlorate reductase subunit A (PcrA) and subunit B (PcrB) peptide biomarkers under different growth conditions in the Bior enrichment culture
| Growth condition | Chlorite dismutase |
Perchlorate reductase subunit A |
Perchlorate reductase subunit B |
|||
|---|---|---|---|---|---|---|
| Peptide sequence(s) | Mean concn (ng of CD/mg of total protein)a | Peptide sequence(s) | Mean concn (ng of Pcr A/mg of total protein)a | Peptide sequences | Mean concn (ng of Pcr B/mg of total protein)a | |
| Phase of growth cycle | ||||||
| Early log phase | GLETNSDFFFR, GTILTQPGVFGVFTMFK, HKDNVLVDLYLTR, YVIVIPVKK | 285 | DIAPMPNIPEYNPR, EQTDLSYLVR, MDSTALYSDVVLPSAHWYEK, YIILWGSNPTQTR | 1,051 | GKIPPMIDYGIPFEFDYAGR, IEQGVAPACVAQCVGR, IPLAQLEGLFGK, IPPMIDYGIPFEFDYAGR, NVETAPGLGYPR, SAPNWDEDQGAGEYPNNSFFYLPR, VALPLHPEFGTEPNVFYVPPVLGPR | 268 |
| Mid-log phase | Same as above | 271 | Same as above | 646 | Same as above | 225 |
| Late log phase | Same as above | 342 | YIILWGSNPTQTR | 317 | Same as above | 243 |
| Perchlorate at indicated concn | ||||||
| 0.1 mM | GTILTQPGVFGVFTMFK | 641 | YIILWGSNPTQTR | 1,203 | GKIPPMIDYGIPFEFDYAGR, IPLAQLEGLFGK, IPPMIDYGIPFEFDYAGR, VALPLHPEFGTEPNVFYVPPVLGPR | 816 |
| 0.5 mM | GLETNSDFFFR, GTILTQPGVFGVFTMFK, YVIVIPVKK | 1,237 | DIAPMPNIPEYNPR, EQTDLSYLVR, YIILWGSNPTQTR | 1,459 | GKIPPMIDYGIPFEFDYAGR, IPLAQLEGLFGK, IPPMIDYGIPFEFDYAGR, SAPNWDEDQGAGEYPNNSFFYLPR, VALPLHPEFGTEPNVFYVPPVLGPR | 774 |
| 1 mM | GLETNSDFFFR, GTILTQPGVFGVFTMFK, HKDNVLVDLYLTR, YVIVIPVKK | 1,353 | DIAPMPNIPEYNPR, EQTDLSYLVR, MDSTALYSDVVLPSAHWYEK, YIILWGSNPTQTR | 1,645 | GKIPPMIDYGIPFEFDYAGR, IEQGVAPACVAQCVGR, IPLAQLEGLFGK, IPPMIDYGIPFEFDYAGR, NVETAPGLGYPR, SAPNWDEDQGAGEYPNNSFFYLPR, VALPLHPEFGTEPNVFYVPPVLGPR | 683 |
| Perchlorate and nitrate at indicated concn | ||||||
| 1.25 mM and 3.75 mM | GLETNSDFFFR, GTILTQPGVFGVFTMFK, HKDNVLVDLYLTR, YVIVIPVKK | 410 | DIAPMPNIPEYNPR, EQTDLSYLVR, MDSTALYSDVVLPSAHWYEK, YIILWGSNPTQTR | 1,213 | IEQGVAPACVAQCVGR,b NVETAPGLGYPR | 655 |
| 1.67 mM and 3.3 mM | Same as above | 725 | Same as above | 1,431 | Same as above | 622 |
| 2.5 mM and 2.5 mM | Same as above | 160 | Same as above | 613 | Same as above | 227 |
| Control | ||||||
| 5 mM nitrate | None of the above | EQTDLSYLVR | 10 | None of the above | ||
Results are the average of three mass spectrometric runs per sample. All the readings have a standard error of less than 12% of the mean.
The rest of the peptide sequences (IPPMIDYGIPFEFDYAGR, IPLAQLEGLFGK; and SAPNWDEDQGAGEYPNNSFFYLPR, VALPLHPEFGTEPNVFYVPPVLGPR) match with peptides from respiratory nitrate reductase beta subunit from Dechloromonas aromatica strain RCB (UniProtKB protein database accession no. Q47CW7).
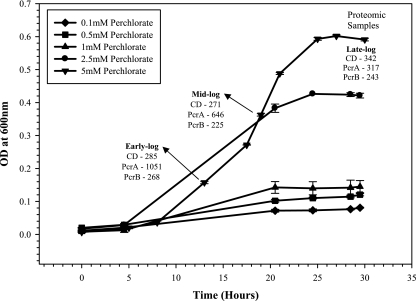
In the early log phase of the growth cycle, we detected all four of the selected CD signature biomarkers, with a mean concentration of 285 ng/mg of total protein. We also detected all the selected PcrA and PcrB subunit biomarkers, with mean concentrations of 1,051 ng/mg and 268 ng/mg of the total protein, respectively. In the mid-log phase of the growth cycle, the mean concentrations for CD (271 ng/mg) and PcrB (225 ng/mg) peptides did not change much; however, the mean concentration for PcrA peptides (646 ng/mg) decreased ∼38% compared to the mean concentration in the early log phase. In the late log phase, all the selected CD and PcrB biomarkers were detected, with mean concentrations of 342 ng/mg and 243 ng/mg of total protein, respectively. Only one peptide for PcrA, YIILWGSNPTQTR, was detected, with a mean concentration of 317 ng/mg, which is ∼50% of the concentration detected at the mid-log phase (Fig. 1).
FIG. 1.
Growth of the bioreactor (Bior) enrichment culture under 0.1, 0.5, 1, 2.5, and 5 mM concentrations of ammonium perchlorate. Concentrations (ng/mg of total protein) of CD, PcrA, and PcrB biomarkers detected at marked time points during growth on 5 mM ammonium perchlorate are indicated. Error bars show standard deviations.
The signature peptides from both CD and PcrA and PcrB were detected at several different perchlorate concentrations (0.1 mM, 0.5 mM, 1 mM, and 2.5 mM) (Table 3). The growth of the Bior enrichment culture on different perchlorate concentrations is shown in Fig. 1. At 0.1 mM perchlorate, we detected one peptide biomarker each of CD (GTILTQPGVFGVFTMFK) and PcrA (YIILWGSNPTQTR), with mean concentrations of 641 ng/mg and 1,203 ng/mg of total protein, respectively. The signature biomarker sequences from PcrB were also found, with a mean concentration of 816 ng/mg of total protein. With a 0.5 mM perchlorate concentration, three biomarkers each of CD and PcrA were identified. The concentrations of CD and PcrA peptides were increased to 1,237 ng/mg and 1,459 ng/mg of total protein, respectively. For PcrB, we detected five peptide biomarkers, with a mean concentration of 774 ng/mg of total protein. At the 1 mM perchlorate concentration, all the above-mentioned selected CD, PcrA, and PcrB signature peptides were detected, with mean concentrations of 1,353 ng/mg, 1,645 ng/mg, and 683 ng/mg of total protein, respectively. Microbial cultures growing on perchlorate concentrations lower than 0.1 mM were not used for analysis as the cellular biomass was not sufficient for proteomic experiments.
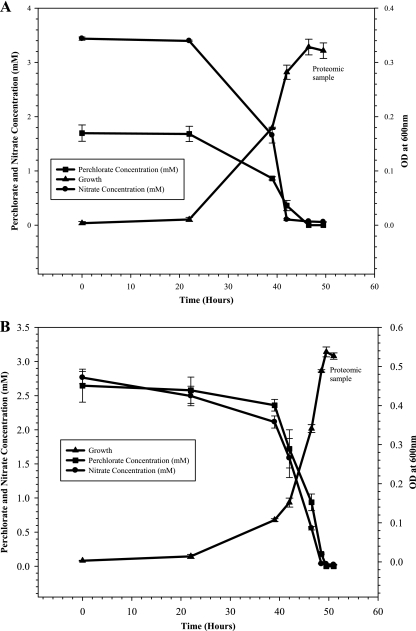
Perchlorate and nitrate degradation by the Bior enrichment culture were monitored along with its growth (Fig. 2 A and B). It was observed that both perchlorate and nitrate were removed simultaneously by the Bior enrichment culture at all the different concentration combinations of these electron acceptors used. The results indicated that the signature biomarkers from CD, PcrA, and PcrB were detected even in the presence of nitrate (Table 3). At 1.25 mM perchlorate and 3.75 mM nitrate (combination a), we detected all selected CD and PcrA and two PcrB biomarkers with mean concentrations of 410 ng/mg, 1,213 ng/mg, and 655 ng/mg of total protein, respectively. Similarly, CD, PcrA and PcrB biomarker peptides were obtained for the 1.67 mM perchlorate and 3.33 mM nitrate (combination b) and 2.5 mM perchlorate and 2.5 mM nitrate (combination c) experimental sets. For the combination b set, the mean concentration for CD was found to be 725 ng/mg. PcrA was 1,431 ng/mg, and PcrB was 622 ng/mg of total protein. And for the combination c set, the mean concentration for CD was found to be 160 ng/mg. PcrA was 613 ng/mg, and PcrB was 227 ng/mg of total protein. The results showed that the PcrB peptides IEQGVAPACVAQCVGR and NVETAPGLGYPR are unique to perchlorate reductase, while other chosen signature peptides from PcrB (GKIPPMIDYGI PFEFDYAGR, IPLAQLEGLFGK, IPPMIDYGIPFEFDYAGR, SAPNWDEDQGAGEYPNNSFFYLPR, and VALPLHPEFGTEPNVFYVPPVLGPR) matched with the beta subunit of respiratory nitrate reductase. At 5 mM nitrate only (control), as expected, no chlorite dismutase or PcrB (IEQGVAPACVAQCVGR and NVETAPGLGYPR) biomarkers were detected. However, one peptide from PcrA was detected with a negligible mean concentration, which could be due to possible cross-contamination of the chromatography column from the previous sample. The signature peptides from respiratory nitrate reductase beta subunit and periplasmic nitrate reductase with their mean concentration are listed in Table S4 in the supplemental material for reference. No similarity was found between the amino acid sequences from the perchlorate reductase beta subunit and periplasmic nitrate reductase.
FIG. 2.
Growth and decomposition of perchlorate and nitrate by the Bior enrichment culture. Concentrations of perchlorate and nitrate were monitored using ESI-MS. The data indicate simultaneous removal of both the electron acceptors irrespective of the concentration ratios used. (A) Results for 1.67 mM ammonium perchlorate and 3.33 mM sodium nitrate. (B) Results for 2.5 mM ammonium perchlorate and 2.5 mM sodium nitrate. Error bars show standard deviations.
DISCUSSION
Microbial proteins are the best indicator of the functional changes in microbial communities or pure cultures because there is a direct association between chemical exposure and changes in microbial cellular components, especially enzymes of biotransformation pathways. Proteins control the real-time metabolic processes within microbial systems and provide a dynamic picture of the systems and how they are adjusting to environmental changes. Protein biomarkers can provide insight into both the occurrence of perchlorate contamination and the current perchlorate degradation status. In the work described here, we used a mass spectrometry-based method to identify signature peptides from the enzymes involved in the bacterial perchlorate degradation pathway for development of the biomarkers that can be used to monitor processes of perchlorate metabolism in cultures and, eventually, contaminated sites. Mass spectrometry, along with an LC-MSE mode of acquisition, is a sensitive method which can detect minute concentrations of analytes (10−15 M) and quantify these even at these very low concentrations. This includes not only pollutants but, also, proteins involved in pollutant degradation.
Chlorite dismutase (CD) and perchlorate reductase (Pcr) are the two enzymes that are known to catalyze perchlorate degradation in bacteria. Both enzymes are unique to perchlorate degradation. Perchlorate reductase consists of four subunits (PcrA, PcrB, PcrC, and PcrD) and is encoded by the pcrABCD operon. Purified Pcr contains two structural subunits, PcrA and PcrB, that are involved in electron transfer. The PcrC subunit functions as a c-type cytochrome and is lost during the purification of the enzyme. The PcrD subunit functions as a chaperone and is not a part of the active enzyme (14). During our analyses, we detected very few signature peptides from PcrC and PcrD. Also, even these few sequences were not found consistently in all the samples. Therefore, we used signature peptides from chlorite dismutase and the perchlorate reductase alpha and beta subunits as biomarkers of the perchlorate reduction pathway. The use of two proteins for the development of biomarkers provides a group of peptides which will act as reliable indicators of the presence of the perchlorate pathway that complement and support each other.
We detected four chlorite dismutase signature peptides which were found to be conserved in all of the microbial cultures examined except D. agitata CKB. The chlorite dismutase sequences obtained from D. agitata CKB were different from those in the other dissimilatory perchlorate-reducing microbial samples used in this study, and no peptide common among the other samples was detected for this bacterium. D. agitata CKB was found to have signature peptides in common with (UniProtKB protein database accession numbers in parentheses) Pseudomonas sp. PK (Q673K7) and Ideonella dechloratans (Q9F437) (Table 2). We compared and aligned the CD amino acid sequence from D. agitata CKB (AAM92878) with other available chlorite dismutase sequences from Dechlorosoma sp. KJ (EU571095), D. hortensis (EU436747), and D. aromatica (CP000089.1) using ClustalX 2.0 (Fig. 3). The sequences were compared without signal peptide sequences, as these are not part of the functional enzyme. The results indicated only 62 to 64% amino acid sequence identity between D. agitata CKB and other sequences compared. Although there is some homology between the compared sequences, the homologous peptides were not detected by mass spectrometry, which could be due to the nature of the peptide, trypsin cleavage specificity, or MS-ESI source ionization efficiency. Therefore, the presence of biomarker peptides from D. agitata CKB listed in Table 2 should be tested for along with the four aforesaid CD biomarkers for a given system. In the case of perchlorate reductase, we detected and selected four PcrA and seven PcrB signature biomarkers that were conserved in the majority of the microbial cultures used. The results indicate that perchlorate reductase peptide sequences are less conserved than those from chlorite dismutase among dissimilatory perchlorate-reducing microorganisms. We found four microbial samples (D. agitata CKB, strain Bior, strain Crw, and the Pt enrichment culture) that were missing one or more of the selected PcrA and PcrB biomarkers.
FIG. 3.
Amino acid alignment of chlorite dismutase, without the signal peptide, from Dechloromonas agitata (D agit), Dechlorosoma sp. KJ (D sp. KJ), Dechloromonas hortensis (D hort), and Dechloromonas aromatica (D arom). The shaded peptide sequences represent regions that are conserved between different organisms and can be used as biomarkers.
It is known that D. agitata CKB clusters with the Betaproteobacteria group on 16S rRNA gene trees but is placed away from that cluster on the cld tree when all available partial and complete chlorite dismutase nucleotide sequences are compared (1a, 4). Therefore, differences in CD and PcrA and PcrB amino acid sequences between D. agitata CKB and the other samples support the idea of a separate path of evolution of the perchlorate degradation pathway in D. agitata CKB than in other perchlorate-reducing microbes. In future, as more perchlorate-reducing strains, degradation pathways, and enzyme homologs are discovered, there will be a need to introduce and use different sets of biomarkers depending on peptide sequence similarity and divergence. However, the basic premise of using peptide biomarkers will not change. To validate our results, all listed CD, PcrA, and PcrB signature biomarkers were examined in a pure bacterial culture (Dechlorosoma sp. KJ) by targeting the chosen peptide sequences on the basis of the m/z value in the spectra (data not shown). Our conclusion is that the presence of any or a combination of several of the selected microbial peptide biomarkers derived from chlorite dismutase and perchlorate reductase alpha and beta subunits in a given culture and, most likely, ecosystem sample will indicate the presence of perchlorate contamination and microbial reduction of perchlorate. Our results also indicate that the peptide biomarkers should be chosen carefully, as there will be microorganisms that have evolved unique sets of proteins to perform the same metabolic pathway. Hence, known unique sets of biomarker peptides should be included in any environmental study. Similarly, identification of a strain on the basis of unique signature peptides should be done with caution.
To further assess and confirm the utility of the selected CD and Pcr biomarkers, we detected and quantified their presence under different growth conditions. A perchlorate-degrading mixed-culture sample, the Bior enrichment, was selected to be representative of a natural microbial consortium. In the first set of experiments, we tested for the presence of biomarkers at different time points during the growth cycle of the enrichment to make sure that they were detected throughout the cycle and their detection was not affected by or dependent on the time of sample collection. The results indicated that all the chosen CD, PcrA, and PcrB biomarkers were present at all the chosen time points during the growth cycle; however, we detected only one peptide of PcrA in the late log growth phase. This may be because the other perchlorate reductase subunits are turned over faster in an aging culture. This was also evident by the mean concentration of PcrA obtained (Table 3). The concentration of CD biomarkers remained almost constant throughout the log phase of the growth cycle, which indicates that chlorite dismutase was expressed constitutively throughout this phase. In a second experiment, we used different concentrations of perchlorate to determine the lowest concentration at which selected CD, PcrA, and PcrB biomarkers could be detected. Perchlorate concentrations ranging from 0.1 mM to 2.5 mM were used. Microbial growth was observed at perchlorate concentrations lower than 0.1 mM, but sufficient cellular biomass for proteomic analysis was not produced in this experiment. However, the required biomass for environmental proteomics can be obtained by using large culture volumes, as we have demonstrated in our previous work on methanotrophs. We reported that only ∼1,000 cells from a pure culture and ∼10,000 cells from a complex sample are required for the detection of methane monooxygenase peptides using MS-MS (23). The results showed that as the perchlorate concentration increased, microbial growth and the number and concentration of certain signature biomarkers detected also increased; however, the concentrations of PcrB biomarkers showed a reverse trend compared to those of PcrA biomarkers. This could be due to the nature of peptides derived from PcrB, which will affect their stability, extraction, and ionization efficiency during processing and MS-MS analysis, or it could be that the relationship between these two subunits is more complex than the researches of Kengen et al. (15) and Gu and Coates (14) suggest. Their researches indicate that the concentrations of PcrB peptides should follow a trend similar to those of PcrA peptides, as these subunits are closely associated and are translocated together and found as heterodimers (αβ).
To evaluate the effect of the presence of nitrate on the detection of CD and Pcr biomarkers, the Bior enrichment culture was grown with various concentrations of two electron acceptors (ammonium perchlorate and sodium nitrate). It has been shown in previous studies that many of the dissimilatory perchlorate-reducing microorganisms use nitrate as an alternate electron acceptor even in the presence of perchlorate (8, 10). The results show that the Bior enrichment culture, a known perchlorate-degrading consortium, can use both the electron acceptors simultaneously irrespective of their concentrations used in the study. The detection of selected signature biomarkers from CD, PcrA, and PcrB was not affected by the presence of nitrate. However, we found that five out of seven selected PcrB biomarkers were similar to the beta subunit of respiratory nitrate reductase. Both perchlorate and nitrate reductase belong to the dimethyl sulfoxide (DMSO) reductase family of enzymes and require molybdenum as a cofactor. It is known from previous research that enzymes from the DMSO reductase family are highly similar to each other (18). Therefore, the two unique PcrB biomarkers should always be used when nitrate is present in the system. This proteomics-based biomarker approach is thus particularly useful because it can simultaneously detect signature peptides from multiple enzymes in a system where multiple metabolic pathways function simultaneously, as in our example of perchlorate reductase along with nitrate and nitrite reductases.
Previous studies have reported methods to detect and quantify perchlorate-reducing bacteria by targeting either of the two central genes or enzymes (chlorite dismutase, cld, and perchlorate reductase, pcr) involved in the perchlorate degradation pathway. O'Connor and Coates (21) and Bender et al. (4) described detection methods for perchlorate reducers that use an immunoprobe and genetic probes targeting the chlorite dismutase enzyme and gene, respectively. Nozawa-Inoue et al. (20) reported a quantitative detection method using quantitative real-time PCR (qPCR) targeting the perchlorate reductase gene (pcrA). However, our present research indicates that, even though the chlorite dismutase sequence is highly conserved, there will be some exceptions. Also, it is known that chlorite dismutase is not unique to perchlorate reducers and is also found in chlorate-reducing microorganisms. Similarly, the use of perchlorate reductase alone can also lead to biased results, as the Pcr sequence is not highly conserved among perchlorate-reducing microbes and is similar to other DMSO reductases, as described above. Therefore, we highly recommend using peptide biomarkers from both the enzymes simultaneously for reliable interpretation of results. The use of real-time PCR for the detection and quantification of perchlorate-reducing genes offers the advantage of nucleic acid amplification but can lead to biased results. PCR results are affected by the primer design and specificity, amplification reaction efficiency, DNA purity, and cDNA quality (26). On the other hand, the protein biomarker detection system using mass spectrometry overcomes the PCR biases and is a sensitive, direct protein detection technique that provides qualitative and quantitative information about the metabolic processes but is limited by the availability of given protein sequences in the database. Other limitations include knowledge of the key proteins involved in the pathway of interest, the existence of unique and diagnostic peptides, and certain volume and biomass requirements.
The biomarker system developed here shows promise as a tool in the management of environmental restoration activities, such as microbially augmented bioremediation and/or monitored natural attenuation. The biomarker-based monitoring system to this point has only been tested in the laboratory, using both pure cultures and model natural consortia. Thus, it requires validation using samples collected from field locations, such as subsurface biofilms found within perchlorate-contaminated aquifers or streams down-gradient from point source contamination by perchlorate. The system can also be extended to the study of other xenobiotic contaminants of environmental health concern where biodegradation pathways are well understood.
Supplementary Material
Acknowledgments
We thank Lee A. Deobald for assistance with the liquid chromatography-mass spectrometry system. We also thank Stephanie Smith for providing Bior and Crw isolates for this study.
The research results presented are based upon work supported by the U.S. Army Corps of Engineers as Humphreys Engineering Center Support Activity under contract no. W912HQ-07-C-0014.
Views, opinions, and/or findings contained in this report are those of the authors and should not be construed as an official Department of Defense position or decision unless so designated by other official documentation.
Footnotes
Published ahead of print on 29 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bae, H., and R. Sicher. 2004. Changes of soluble protein expression and leaf metabolite levels in Arabidopsis thaliana grown in elevated atmospheric carbon dioxide. Field Crop Res. 90:61-73. [Google Scholar]
- 1a.Bansal, R. 2011. Qualitative and quantitative proteomics analysis of enzymes involved in perchlorate and chlorate metabolism. Ph.D. dissertation. University of Idaho, Moscow, ID.
- 2.Bansal, R., L. A. Deobald, R. L. Crawford, and A. J. Paszczynski. 2009. Proteomic detection of proteins involved in perchlorate and chlorate metabolism. Biodegradation 20:603-620. [DOI] [PubMed] [Google Scholar]
- 3.Bar, C., R. Patil, J. Doshi, M. J. Kulkarni, and W. N. Gade. 2007. Characterization of the proteins of bacterial strain isolated from contaminated site involved in heavy metal resistance—a proteomic approach. J. Biotechnol. 128:444-451. [DOI] [PubMed] [Google Scholar]
- 4.Bender, K. S., M. R. Rice, W. H. Fugate, J. D. Coates, and L. A. Achenbach. 2004. Metabolic primers for detection of (Per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl. Environ. Microbiol. 70:5651-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benninghoff, A. D. 2007. Toxicoproteomics—the next step in the evolution of environmental biomarkers. Toxicol. Sci. 95:1-4. [DOI] [PubMed] [Google Scholar]
- 6.Bestel-Corre, G., S. Gianinazzi, and E. Dumas-Gaudot. 2004. Impact of sewage sludges on Medicago truncatula symbiotic proteome. Phytochemistry 65:1651-1659. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J., C. Lauderdale, E. Morgenroth, and L. Raskin. 2007. Direct fixed-bed biological perchlorate destruction demonstration, poster F-99, p. 118. SERDP ESTCP Partners Environ. Technol. Tech. Symp. Workshop. Washington, DC, 4 to 6 December 2007.
- 8.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)- chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 9.Clark, J. J. 2000. Toxicology of perchlorate, p. 15-29. In E. T. Urbansky (ed.), Perchlorate in the environment. Kluwer Academic, New York, NY.
- 10.Coates, J. D., et al. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Environmental Protection Agency. 2010. Perchlorate Toxicology. Contaminated Site Clean-Up Information (CLU-IN), Technology Innovation and Field Services Division, U.S. Environmental Protection Agency, Washington, DC. http://www.clu-in.org/contaminantfocus/default.focus/sec/perchlorate/cat/toxicology/.
- 12.Environmental Working Group. 2008. Is toxic perchlorate in Utah's food? http://www.ewg.org/node/25973.
- 13.Gil, F., and A. Pla. 2001. Biomarkers as biological indicators of xenobiotic exposure. J. Appl. Toxicol. 21:245-255. [DOI] [PubMed] [Google Scholar]
- 14.Gu, B., and J. D. Coates. 2006. Perchlorate environmental occurrence, interactions and treatment. Springer Science and Business Media, Inc., New York, NY.
- 15.Kengen, S. W. M., G. B. Rikken, W. R. Hagen, C. G. van Ginkel, and A. J. M. Stams. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser, and M. Hecker. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686-697. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Barea, J., and J. L. Gomez-Ariza. 2006. Environmental proteomics and metallomics. Proteomics 6(Suppl. 1):S51-S62. [DOI] [PubMed] [Google Scholar]
- 18.McEwan, A. G., J. P. Ridge, C. A. McDevitt, and P. Hugenholtz. 2002. The DMSO reductase family of microbial molybdenum enzymes: molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19:3-21. [Google Scholar]
- 19.Nesatyy, V. J., and M. J. F. Suter. 2007. Proteomics for the analysis of environmental stress responses in organisms. Environ. Sci. Technol. 41:6891-6900. [DOI] [PubMed] [Google Scholar]
- 20.Nozawa-Inoue, M., et al. 2008. Quantitative detection of perchlorate-reducing bacteria by real-time PCR targeting the perchlorate reductase gene. Appl. Environ. Microbiol. 74:1941-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connor, S. M., and J. D. Coates. 2002. Universal immunoprobe for (per)- chlorate-reducing bacteria. Appl. Environ. Microbiol. 68:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogunseitan, O. A. 2000. Microbial proteins as biomarkers of ecosystem health, p. 207-222. In K. M. Scow, G. E. Fogg, D. E. Hinton, and M. L. Johnson (ed.), Integrated assessment of ecosystem health. CRC Press LLC, Boca Raton, FL.
- 23.Paidisetti, R. 2007. Ecoproteomic and ecogenomic approaches to monitor microbial activity in a subsurface aquifer and effect of compost infusion enrichment on bacterial community structure of polycylic aromatic hydrocarbon contaminated soils. M.S. thesis, Department of Microbiology, Molecular Biology and Biochemistry, University of Idaho, Moscow, ID.
- 24.Petricoin, E. F., et al. 2002. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359:572-577. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Ortega, M. J., B. E. Grosvik, A. Rodriguez-Ariza, A. Goksoyr, and J. Lopez-Barea. 2003. Changes in protein expression profiles in bivalve molluscs (Chamelea gallina) exposed to four model environmental pollutants. Proteomics 3:1535-1543. [DOI] [PubMed] [Google Scholar]
- 26.SABiosciences. 2008. Designing and validating real-time PCR primers: systematic guidelines. SABiosciences, Frederick, MD. http://www.sabiosciences.com/newsletter/PATHWAYS07_PCRprimers.pdf.
- 27.Shepard, J. L., B. Olsson, M. Tedengren, and B. P. Bradley. 2000. Protein expression signatures identified in Mytilus edulis exposed to PCBs, copper and salinity stress. Mar. Environ. Res. 50:337-340. [DOI] [PubMed] [Google Scholar]
- 28.Shrader, E. A., T. R. Henry, M. S. Greeley, Jr., and B. P. Bradley. 2003. Proteomics in zebrafish exposed to endocrine disrupting chemicals. Ecotoxicology 12:485-488. [DOI] [PubMed] [Google Scholar]
- 29.Smith, P. N., C. W. Theodorakis, T. A. Anderson, and R. J. Kendall. 2001. Preliminary assessment of perchlorate in ecological receptors at the Longhorn Army Ammunition Plant (LHAAP), Karnack, Texas. Ecotoxicology 10:305-313. [DOI] [PubMed] [Google Scholar]
- 30.Streit, B. R., and J. L. DuBois. 2008. Chemical and steady-state kinetic analyses of a heterologously expressed heme dependent chlorite dismutase. Biochemistry 47:5271-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theodorakis, C., et al. 2006. Perchlorate in fish from a contaminated site in east-central Texas. Environ. Pollut. 139:59-69. [DOI] [PubMed] [Google Scholar]
- 32.Vido, K., et al. 2001. A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276:8469-8474. [DOI] [PubMed] [Google Scholar]
- 33.Wang, P., F. G. Bouwman, and E. C. Mariman. 2009. Generally detected proteins in comparative proteomics—a matter of cellular stress response? Proteomics 9:2955-2966. [DOI] [PubMed] [Google Scholar]
- 34.Witzmann, F. A., et al. 1999. Regional protein alterations in rat kidneys induced by lead exposure. Electrophoresis 20:943-951. [DOI] [PubMed] [Google Scholar]
- 35.Wolff, J. 1998. Perchlorate and the thyroid gland. Pharmacol. Rev. 50:89-105. [PubMed] [Google Scholar]
- 36.Xu, J., J. J. Trimble, L. Steinberg, and B. E. Logan. 2004. Chlorate and nitrate reduction pathways are separately induced in the perchlorate-respiring bacterium Dechlorosoma sp. KJ and the chlorate-respiring bacterium Pseudomonas sp. PDA. Water Res. 38:673-680. [DOI] [PubMed] [Google Scholar]
- 37.Yang, S. Y., et al. 2005. Application of serum SELDI proteomic patterns in diagnosis of lung cancer. BMC Cancer 5:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.