Abstract
Multidimensional compound-specific stable isotope analysis (CSIA) was applied in combination with RNA-based molecular tools to characterize methyl tertiary (tert-) butyl ether (MTBE) degradation mechanisms occurring in biofilms in an aerated treatment pond used for remediation of MTBE-contaminated groundwater. The main pathway for MTBE oxidation was elucidated by linking the low-level stable isotope fractionation (mean carbon isotopic enrichment factor [ɛC] of −0.37‰ ± 0.05‰ and no significant hydrogen isotopic enrichment factor [ɛH]) observed in microcosm experiments to expression of the ethB gene encoding a cytochrome P450 monooxygenase able to catalyze the oxidation of MTBE in biofilm samples both from the microcosms and directly from the ponds. 16S rRNA-specific primers revealed the presence of a sequence 100% identical to that of Methylibium petroleiphilum PM1, a well-characterized MTBE degrader. However, neither expression of the mdpA genes encoding the alkane hydroxylase-like enzyme responsible for MTBE oxidation in this strain nor the related MTBE isotope fractionation pattern produced by PM1 could be detected, suggesting that this enzyme was not active in this system. Additionally, observed low inverse fractionation of carbon (ɛC of +0.11‰ ± 0.03‰) and low fractionation of hydrogen (ɛH of −5‰ ± 1‰) in laboratory experiments simulating MTBE stripping from an open surface water body suggest that the application of CSIA in field investigations to detect biodegradation may lead to false-negative results when volatilization effects coincide with the activity of low-fractionating enzymes. As shown in this study, complementary examination of expression of specific catabolic genes can be used as additional direct evidence for microbial degradation activity and may overcome this problem.
Due to its physicochemical characteristics (high water solubility, low Henry's law constant, and poor adsorption), the gasoline octane enhancer methyl tertiary (tert-) butyl ether (MTBE) is a particularly challenging compound to use to evaluate the effectiveness of groundwater remediation systems (51). In general, MTBE is more difficult and more expensive to remediate by conventional technologies (aeration, adsorption on activated carbon, ozonation, and advanced oxidation processes) than other typical fuel compounds such as benzene, toluene, ethylbenzene, and xylene (BTEX) isomers (52).
An alternative remedial strategy could be the use of natural treatment systems, including constructed wetlands, ponds, or lagoons originally designed for processing municipal wastewater, which are becoming more popular in recent years for treating a wide variety of pollution sources, including industrial effluents, due to their lower costs and minimal maintenance (24). However, little information is publicly available regarding the removal of specific trace organics (53) or, in particular, petroleum-derived volatile organic contaminants in these systems (4). In a recently published study, MTBE was not degraded in a pilot-scale constructed wetland with subsurface aeration lines operated for the remediation of polluted groundwater from a former refinery site, although aromatic hydrocarbons were effectively removed (4). However, MTBE biodegradation can occur but has been shown to be comparably slower than that of BTEX compounds in contaminated aquifers (43). MTBE biodegradation has also been demonstrated in laboratory studies under a variety of redox conditions, where the highest MTBE degradation rates were observed under aerobic conditions (45). It has thus been suggested that biodegradation of MTBE in the field is often limited by the environmental conditions (12, 37) and, in particular, by the limited concentrations of oxygen typically found in fuel-contaminated aquifers. As a consequence of these findings, in situ bioremediation of MTBE has been successfully enhanced by addition of air, pure oxygen (biostimulation), or previously enriched MTBE-degrading microorganisms (bioaugmentation) (49).
Reliable methods for detecting in situ MTBE biodegradation are crucial for evaluating remediation technologies based on the activities of MTBE-degrading microorganisms. Some of these MTBE-contaminated sites have been characterized recently by compound-specific stable isotope analysis (CSIA) (19, 22, 26, 27, 32, 34, 57). This tool allows for a qualitative and/or quantitative integrative assessment of biological transformation of organic contaminants over a flow path in environmental systems (36, 46), thanks to the reaction-dependent compound-specific isotope enrichment factors (ɛ) obtained under different controlled laboratory conditions employing a modified form of the Rayleigh distillation equation. Due to the significantly different fractionation patterns observed for diverse reaction mechanisms and processes, the MTBE isotopic analysis can be used for (i) discriminating between aerobic and anaerobic biodegradation pathways (14, 26, 50, 57), (ii) understanding the details of the initial reaction mechanisms (oxidation, acidic hydrolysis [SN1], or hydrolysis by [enzymatic] nucleophilic attack [SN2]) (10, 11), (iii) distinguishing degradation mechanisms among different aerobic cultures (33, 41), or even (iv) quantifying the extent of biodegradation when two competing degradation pathways or species are consuming the MTBE simultaneously (3, 42, 55). However, CSIA application at field scale must also account for some uncertainty related to potential fractionation caused by abiotic processes. Although MTBE adsorption on humic substances and the related isotopic effect can be considered negligible (23), other processes, such as diffusion or volatilization, can produce small but measurable normal or inverse isotopic effects (19, 25). This fact becomes even more crucial when considering the diversity of fractionation already found among aerobic MTBE degraders, which possibly correlate with the employment of different MTBE-attacking enzymes (see the following explanation). In the aerobic degradation pathway, the methyl group of the ether is initially hydroxylated in a monooxygenase-catalyzed reaction. High fractionation of carbon and hydrogen was observed for Methylibium sp. (strains PM1 and R8), with ɛC and ɛH ranging from −2.0‰ to −2.4‰ and from −33‰ to −40‰, respectively, while even higher hydrogen fractionation (ɛH = −100‰) for the recently studied Pseudonocardia tetrahydrofuranoxydans K1 has been found, whereas much lower carbon fractionation (ɛC below −0.5‰) and nondetectable hydrogen fractionation was observed for Aquincola tertiaricarbonis L108 or Rhodococcus ruber IFP 2001 (14, 33, 41). This phenomenon might be linked to nonfractionating but rate-limiting steps (e.g., binding of the substrate to the enzyme [30]) masking the carbon and hydrogen isotope fractionation linked to the cleavage of the C-H bond during MTBE oxidation. In the two above-mentioned low fractionating strains, the initial monooxygenase reaction attacking the methyl group of MTBE is catalyzed by a cytochrome P450-type enzyme (CYP249A1) that is encoded by the ethABCD genes, which are also found to be highly conserved in other strains able to grow on the MTBE-related ethyl tert-butyl ether (ETBE) (5, 7, 8, 29). In addition, an alternative and novel enzyme, MdpA, which is closely related to the group of short-chain hydrolyzing alkane hydroxylases (AH1), has been recently discovered to be the only enzyme responsible for MTBE removal in Methylibium petroleiphilum PM1 (44).
In addition to CSIA, molecular tools can also be useful to assess in situ biodegradation of various contaminants (6). The first molecular tools developed to monitor MTBE biodegradation were specific primers and probes designed to PCR amplify partial 16S rRNA gene sequences of this strain (18). When applied to samples from contaminated field sites, these primers could detect native bacterial strains with ≥99% 16S rRNA gene sequence similarity to strain PM1 (17, 49). However, the mere presence of a potential degrader cannot be directly connected to its active participation in the initial MTBE degradation step. On the contrary, the expression of catabolic genes encoding key enzymes of degradation pathways can reveal information on the activity of microorganisms under in situ conditions (6). In light of this situation, multidimensional CSIA might be applied in conjunction with molecular tools in order to identify the activities of MTBE-degrading species in contaminated field sites and provide more reliable assessments of bioremediation effectiveness (3, 32).
In this study, our testing site was an aerated treatment pond system for contaminated groundwater remediation next to a refinery plant in Leuna, Germany. Lower-level removal of MTBE (on average, 38%) versus benzene (99.9%) was observed during the first 14 months of operation (20). MTBE concentration reductions from 4 to 2.5 mg/liter were not sufficient to reach drinking water quality standards (54), and half of the elimination was related to volatilization during the summer of 2008. In addition to field observations, an increasing aerobic biodegradation of MTBE was detected in microcosm experiments, using coconut fiber samples as substrates for biofilms (20). Therefore, increased aeration was thought to enhance MTBE biodegradation in the system, and it was applied in May 2009.
In the present study, we summarize the field data obtained after more than 2 years of continuous operation and compare the removal of MTBE at three different aeration periods. Our aims were to prove MTBE biodegradation in the field, to elucidate the dominant biodegradation mechanism, and to link it to the expression of catabolic genes encoding key enzymes involved in the aerobic MTBE degradation pathway. This goal was achieved by comparing the MTBE carbon and hydrogen isotope enrichment factors obtained in microcosm experiments with the MTBE isotopic compositions observed in the field and by comparing the detection of expressed catabolic genes in both the field and microcosms. For that purpose, biofilms established in the pond system were sampled in October 2009 after almost 5 months of maximum aeration and 2 years of continuous operation. In addition, the potential isotopic effects of the volatilization processes taking place in such an open pond were tested in laboratory experiments in an attempt to better simulate field conditions (bubbling from aqueous solution) in comparison with those shown by previous studies performed in closed systems or sediment columns (19, 25). In summary, a combination of CSIA and RNA-based molecular methods was used in this study to evaluate the removal of MTBE from a complex treatment system.
MATERIALS AND METHODS
Site description and different setups in the aerated treatment pond.
The aerated treatment pond system next to a refinery plant in Leuna, Germany, was designed to simulate natural transition zones where contaminated groundwater is aerated with ambient air to stimulate aerobic contaminant degradation, and geotextiles provide surface area for biofilm formation and retention of biomass (for a more detailed description, see reference 20). Two different geotextile materials were tested in two parallel basins, coconut fiber textiles (CO) and polypropylene fleece (PO). The minimum oxygen content along the ponds can be regulated by aeration modules based on the feedback from oxygen sensors.
Three different aeration periods had been set up in the system in order to compare and characterize the degradation of the target pollutants ammonium, MTBE, and benzene. Period A1 started in November 2007, and minimum dissolved oxygen concentrations in both ponds were constantly set to a gradient of 0 mg/liter in the inflow section, 0.5 mg/liter in the middle section, and 1 mg/liter in the outflow section to stimulate aerobic degradation processes and at the same time to minimize the loss of contaminants by volatilization and energy costs (20). For period A2, from 18 May 2009 (after 18 months of operation), the aeration was increased in order to reach oxygen saturation levels. For period A3, from 15 October 2009 (after 23 months of operation), the aeration in the PO-amended pond was set back to the adjustment described in period A1. Averages of maximum dissolved oxygen concentrations (measured in the outflow section) and residual MTBE percentages in both basins from more than 2 years of continuous operation (from November 2007 until February 2010) were measured.
Volatilization experiments.
Carbon and hydrogen enrichment factors for volatilization of MTBE from water were determined by a bench-scale experiment. Two 2-liter Duran bottles (Schott AG, Mainz, Germany) were filled with distilled water and MTBE to a final concentration of around 40 g/liter. This maximum dissolved MTBE concentration (close to saturation, as its solubility in water is 44 g/liter at 25°C [13]) was selected with the aim of having a maximal concentration difference within the isotope analysis samples. The bottles were closed with Teflon screw caps and stirred with magnetic stir bars for 2 days at room temperature in order to reach equilibrium and ensure complete dissolution. Subsequently, the bottles were opened, and nitrogen gas was bubbled from the bottoms of the bottles at a constant rate of 500 to 800 ml/min while being stirred at 250 rpm to homogenize the MTBE composition in the liquid volume. MTBE concentration samples were taken in duplicate in regular intervals until complete volatilization. To fit into the linear range of the respective analytical methods (up to 1 g/liter), the samples with expected higher MTBE concentrations were diluted in saturated NaCl aqueous solution to a final volume of 2 ml in 10- or 20-ml vials for concentration or isotopic composition measurements, respectively.
Biodegradation experiments.
The degradation behavior of freshly sampled textile samples was regularly examined in laboratory microcosm experiments in order to evaluate the capability of the developed biofilms to degrade MTBE in the system. Additionally, the isotope enrichment factors and the consumption of oxygen by the MTBE degraders in the biofilms were evaluated. For that purpose, textile samples with attached biofilms were sampled on 5 October 2009 at 120 cm depth and 150 cm distance from the inflow. Samples were transported to the laboratory in 50-ml centrifugation tubes at 4°C for analysis.
The next day, the samples were cultivated in 240-ml serum bottles equipped with oxygen sensor spots and filled with 60 ml phosphate-buffered mineral medium slightly modified from that described in reference 40 (see the details in the supplemental material). Around 0.9 to 1.0 g of CO and 0.5 to 0.7 g of PO (wet weight) was added in duplicate bottles and one control bottle. The average humidity levels of the CO (45% ± 6%) and PO (69% ± 1%) were determined separately by drying triplicate textile samples at 60°C until a constant weight was reached. All bottles were supplemented with 30 μl MTBE (around 400 mg/liter, final concentration) and closed immediately with butyl rubber stoppers and aluminum caps. Control bottles with textiles were autoclaved (121°C, 20 min) before adding vitamins and MTBE, and a single control bottle was prepared with medium but without textiles. Three bottles were monitored for MTBE concentrations, and the last one was monitored for MTBE isotopic composition and oxygen measurements to evaluate abiotic losses. In order to mimic oxic conditions in the field (A2 period) (Fig. 1), high dissolved oxygen concentrations were maintained in the cultures (minimum of 4.7 mg/liter) by respiking sterile air or pure oxygen accordingly (Fig. 2, concentration plots). All the bottles were incubated at room temperature (daily observed average was 23°C ± 1°C, consistent with maximum water temperatures of 22.5°C found in the field) on a horizontal shaker (165 rpm). Samples for MTBE concentration, isotope composition, and pH were taken periodically as described elsewhere (42) until MTBE was completely degraded.
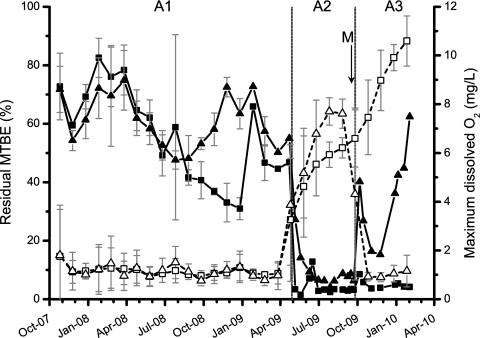
FIG. 1.
Residual MTBE percentages (solid symbols) and maximum dissolved oxygen concentrations (open symbols) at the outflow section in the two parallel basins with CO (squares) and PO (triangles) materials along the three different aeration periods (A1, A2, and A3). The oxygen data and the MTBE residual data are given in monthly averages (± standard deviations [SD]) during the A1 period, as described previously (20). The rest of MTBE sampling times (normally 2 to 3 times a month) are all represented. The biofilms for the studied microcosm experiments (M) and RNA extraction were sampled at the end of period A2 (5 October 2009).
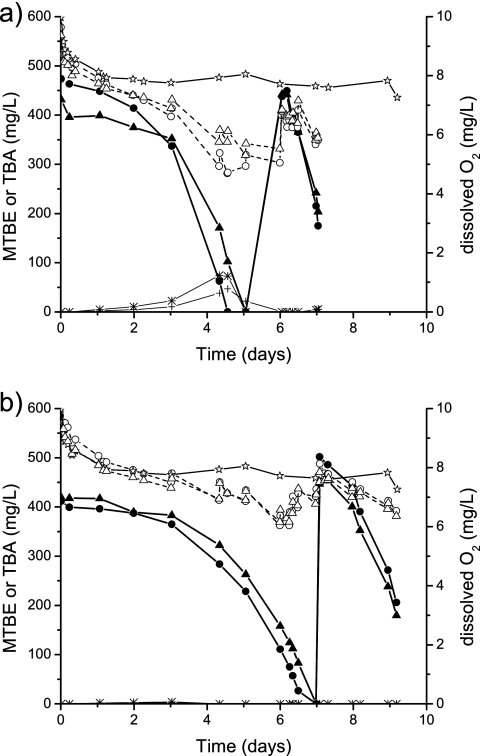
FIG. 2.
Aerobic MTBE biodegradation in replicate microcosm experiments, with biofilms collected on CO (a) and PO (b). High oxygen concentrations (similar to those in field conditions during period A2) were maintained by the addition of air or pure oxygen via the gas phase. MTBE (solid symbols), TBA (asterisks and crosses for first and second replicates, respectively), and dissolved oxygen (open symbols) concentrations in the liquid phase were expressed in mg/liter versus incubation time. Equilibrium between phases and constant dissolved oxygen concentration (8.2 ± 0.6 mg/liter) was attained for the sterile control bottle (open stars). In both graphs, the circles correspond to the first replicate, and the triangles correspond to the second replicate.
After complete degradation of MTBE and the main degradation products, the bottles were opened for 10 min under the sterile bench to renew the oxygen in the headspace, refilled with mineral salt medium and vitamins, and respiked with 30 μl MTBE. Once 50% of MTBE degradation was reached, the textile was taken out for RNA extraction. In parallel, textile samples representative of oxic conditions (end of A2 period) were collected in the pond system at the same location (depth, 120 cm; distance from inflow, 150 cm) where the textiles used in the microcosm experiments were sampled, and additional textiles were collected at a distance of 420 cm from the inflow. All textile samples were immediately deep-frozen in 50-ml centrifugation tubes with a dry ice/ethanol mixture and stored at −80°C until further processing.
Extraction of nucleic acids, RNA quantification, and cDNA synthesis.
Reference cultures of A. tertiaricarbonis L108 and M. petroleiphilum PM1 were used as controls for testing PCR primers and were cultivated using MTBE as a carbon and energy source as previously described (42), whereas purified DNA from Rhodococcus ruber IFP 2001 and Methylibium sp. strain R8 was available in our laboratory. A total of 10 ml of bacterial culture from the growth phase, with an optical density at 700 nm (OD700) of approximately 0.08 to 0.09 (for strain L108, that meant 0.5 mg dry mass [38]), was filtered using polyethersulfone filters with a 0.2-μm pore size (Pall Corporation). The cells were washed off the filter with 2 ml of RNA-Protection reagent (Qiagen GmbH, Hilden, Germany). The cell suspension was centrifuged for 10 min at 5,900 × g, and the pellet was frozen at −20°C until further RNA extraction. Total RNA was extracted from 0.4 to 0.5 g of textile material and from approximately 109 cells from pure cultures (estimation through OD values) (42) by using the RNeasy minikit (Qiagen GmbH, Germany). Briefly, textile and cell pellets were resuspended in 700 μl RLT buffer (Qiagen GmbH, Germany) containing 7 μl of β-mercaptoethanol and transferred to a bead-beating tube. The samples were mechanically lysed using a FastPrep instrument (FastPrep FP120; Savant Instruments, Inc.) for 20 s at a speed of 6.0. The tubes were incubated on ice for 2 min and centrifuged for 1 min at 15,700 × g. The supernatant was transferred to a new tube, and an equal volume of 70% ethanol was added. The mixture was transferred to an RNeasy column (Qiagen GmbH, Germany). The washing and elution steps were performed by following the manufacturer's instructions. An additional DNase treatment was performed after extraction using the DNA-free kit (Applied Biosystems Inc., Foster City, CA). In order to use an equal amount of total RNA for reverse transcription, quantification of the RNA was performed using RiboGreen as the fluorescent dye (Invitrogen GmbH, Darmstadt, Germany), according to the manufacturer's instructions. Reverse transcription was performed using 200 ng of total RNA and the RevertAid H Minus first-strand cDNA synthesis kit (Fermentas GmbH, St. Leon-Rot, Germany) with random hexamer primers by following the protocol provided by the manufacturer. For each sample, a control reaction without reverse transcriptase was performed in order to detect possible DNA contamination. Samples were stored at −20°C until further processing.
PCR and sequencing reaction.
The primer sets shown in Table 1 were used to amplify genes encoding catabolic enzymes involved in MTBE degradation (ethB, ethR, and mdpA) from prepared cDNA samples and/or for sequencing of the obtained amplification products. Additionally, specific 16S rRNA primers developed by Hristova et al. (17) for detection of bacterial strains similar to the MTBE degrader M. petroleiphilum PM1 were used. PCR was performed in a final volume of 50 μl. A standard PCR mastermix contained 1× HotStar Taq PCR buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 μM each primer, 1.75 U HotStar Taq DNA polymerase (Qiagen GmbH, Germany), and 1 μl of template (cDNA or DNA). The conditions of the PCR were as follows: initial denaturation for 15 min at 95°C, 35 cycles of 94°C for 1 min, annealing (Table 1) for 1 min, and 72°C for 1 min 30 s of extension, followed by a final elongation step of 20 min at 72°C. PCR product size was verified by gel electrophoresis on 1.2% agarose gels.
TABLE 1.
Primer sets used in this study
| Primerb | Primer sequence (5′-3′) | Target gene (GenBank accession no.) | Positiona | Molecular analysis | PCR annealing temp (°C) | Reference |
|---|---|---|---|---|---|---|
| EthB for | AGG AGG AAT CTA TGA CAC TG | ethB, encoding cytochrome P450 monooxygenase of R. ruber IFP 2001 (AF333761) | 12031 | PCR/sequencing | 54 | 7 |
| EthB rev | GGC ATC GGC ATC ACT TCG GGT AG | 13254 | ||||
| EthB for M | TCT ACG ACG ACA CCC | 12586 | Sequencing | |||
| EthR for | ATG GGA ACG TCG ACG ACG AG | ethR, regulator of eth operon of R. ruber IFP 2001 (AF333761) | 9555 | PCR/sequencing | 65 | 7 |
| EthR rev | CTA GGA GCG CAA GGT GTC CG | 10550 | ||||
| EthR rev M | ATC CCG ACT TCC TGC GCG ATC TC | 10318 | Sequencing | |||
| MdpA 1F | CTT ACC GGG CTC AAC TAT GC | mdpA, encoding alkane hydroxylase of M. petroleiphilum PM1 (Mpe_B0606) | 852 | PCR/sequencing | 65 | 44 |
| MdpA 1R | CGC TTC CCT GGA TCG ATG TT | 56 | ||||
| 613F | GTGACTGCAAGGCTGGAGCG | 16S rRNA gene of M. petroleiphilum PM1 (Mpe_A0326a) | 632 | PCR/sequencing | 54 | 17 |
| 988R | TCTGGTAACTTCTAGACA | 1006 |
Positions refer to sequences available under accession numbers provided.
Primer sets used to amplify catabolic genes involved in aerobic MTBE degradation or to detect the presence of bacterial strains similar to the well-known MTBE degrader M. petroleiphilum PM1.
PCR products were purified using the QIAquick PCR purification kit (Qiagen GmbH, Germany) and quantified spectrophotometrically using a NanoDrop apparatus (NanoDrop ND-1000; PeqLab Biotechnologie GmbH, Erlangen, Germany). Purified PCR products were sequenced using a 3130xl genetic analyzer and the BigDye Terminator v1.1 cycle sequencing kit (both from Applied Biosystems Inc., Foster City, CA). The obtained sequences were compared to those in public databases using the Basic Local Alignment Search Tool (BLAST) to identify closely related sequences (http://www.ncbi.nlm.nih.gov/blast/).
MTBE isotope ratio measurements at the field site.
Initially, water samples for MTBE isotopic analysis were taken in duplicate from the in- and outflow of both basins in 240-ml serum bottles prefilled with 70 g of NaCl, leaving a small headspace of around 5 ml. The bottles were closed with Teflon stoppers and aluminum caps for subsequent preheating at 60°C and manual headspace injection similar to the method described elsewhere (41). However, after increasing the aeration in the pond system, the MTBE concentrations decreased to levels below the limits of detection for this methodology. Therefore, 1-liter bottles preserved with 1% (wt/wt) trisodium phosphate dodecahydrate (TSP; Na3PO4·12H2O) or as recommended for MTBE analysis (35) were collected in the field for a purge and trap (P&T) preconcentration step. All bottles were stored at 4°C until measurement. In parallel, MTBE concentrations were measured by a headspace-gas chromatograph-mass spectrometer (HS-GC-MS) method. A detailed description of the different methodologies used is provided in the supplemental material.
Stable isotope calculations and definitions.
Carbon and hydrogen isotopic compositions are reported as δ13C and δ2H values in parts per thousand (‰) relative to the Vienna Pee Dee Belemnite (V-PDB) standard and Vienna Standard Mean Ocean Water (V-SMOW), respectively:
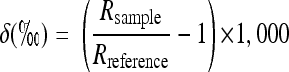 |
(1) |
where Rsample and Rreference are the ratios of the heavy isotope to the light isotope (13C/12C or D/H) in the sample and the international standard, respectively. A simplified Rayleigh equation for a closed system (31) was used to quantify the isotope fractionation,
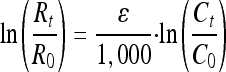 |
(2) |
where the isotopic enrichment factor (ɛ) describes the relationship between changes in isotope composition Rt/R0 = (δt + 1,000)/(δ0 + 1,000) and the concentrations during the course of the experiment.
When possible, two-dimensional CSIA (2D-CSIA) was applied to the data sets. To correct for differences in the initial isotopic composition (δ13C0 and δ2H0) of MTBE, isotopic shifts for hydrogen (Δδ2H) and carbon (Δδ13C) were calculated by subtracting the initial isotopic signature from the value at time t (Δδ = δt − δ0). The slope of a linear regression of Δδ2H versus Δδ13C describes the relationship between carbon and hydrogen fractionation (Λ):
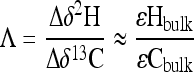 |
(3) |
For the ratio of two competing processes or degradation mechanisms, F (contribution of process 1 expressed as a percentage) has been calculated according to one of the extended Rayleigh-type equations suggested by Van Breukelen (55):
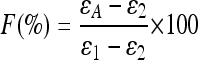 |
(4) |
In equation 4, ɛA is the apparent isotopic enrichment factor as a result of two occurring processes in which individual enrichment factors ɛ1 and ɛ2 were studied separately.
Each sample was measured at least in triplicate, and all linear regression parameters, including the 95% confidence intervals (CI), were obtained by the function “Linear Fit” using errors (standard deviations of measured data sets or corresponding propagated errors in both axes) as weight in OriginPro 7.5.
RESULTS
MTBE removal in the treatment ponds at different dissolved oxygen concentrations.
The average MTBE concentration in the inflowing groundwater was relatively constant (3.5 ± 0.8 mg/liter) during the whole study period. As shown in Fig. 1, the MTBE removal in the two aerated treatment ponds was strongly dependent on the oxygen dissolved in the water. During period A1, with maximum oxygen levels of around 1 mg/liter, the residual MTBE in both effluents was on average around 60% of the inflow concentration, whereas during the A2 period, subjected to increased aeration, the levels of oxygen in the water reached saturation levels (4 to 8 mg/liter depending on the water temperature, 11 to 22°C), and the MTBE removal rapidly reached maximums of 98% and 94% for the ponds equipped with CO and PO textiles, respectively. The corresponding lowest MTBE concentrations in the outflows were 56 and 140 μg/liter, respectively. Maximum MTBE removal rates were around 7 g of MTBE per day (1.2 to 1.3 g/m2 surface area/day) in both ponds. These rates are at least double the rates obtained in the first year of operation (20). It should be pointed out that the CO textile-equipped pond showed a faster response to the increased oxygen concentrations caused by aeration than the PO textile-equipped pond, which reacted steadily. In period A3, keeping the same aeration conditions for CO textile-equipped ponds as in period A2, the dissolved oxygen increased accordingly during the low water temperatures registered during the winter (minimum, 2.5°C) without an apparent change in the observed MTBE removal. The aeration levels of the PO textile-equipped pond were changed back to the initial low levels, and the MTBE removal decreased after a certain period of time to values similar to those observed in period A1.
The two main MTBE degradation products, tert-butyl formate (TBF) and tert-butyl alcohol (TBA), were already present in the inflowing groundwater (up to 6 and 100 μg/liter, respectively). TBF concentrations in the outflow were always lower (if detected) than those in the inflow. Fluctuations in the TBA residual fraction in the outflow of both ponds (see Fig. S3 in the supplemental material) showed periods of formation between others of reduction during period A1. This tendency seems to be connected with seasonal low- and high-registered water temperatures, respectively. However, once the aeration was increased, the TBA was removed to an extent similar to that of the MTBE (98%, minimum concentrations of <2 μg/liter), and no significant changes were detected when the temperatures decreased in winter (only in the CO textile-equipped pond in period A3).
Biodegradation and related isotope enrichment factors in microcosm experiments.
Rapid and highly reproducible degradation of MTBE (5 to 7 days) was observed in duplicate oxic microcosms with CO or PO textile samples (Fig. 2). Relatively faster responses in CO microcosms were observed, whereas in PO microcosms, the MTBE biodegradation showed a short lag phase or adaptation period of around 2 days. TBA and TBF were detected temporally and further consumed in all the microcosms but reached higher levels in CO bottles (up to 74 and 2 mg/liter, respectively). However, MTBE degradation rates were comparable when expressed according to dry weights of textiles (90 and 130 ± 10 μmol MTBE/day/g [dry weight] for the experiments with CO and PO textiles, respectively). The same pH decline from 7.5 (as the one measured in both ponds [20]) to 6.6, along with MTBE degradation, in all four microcosms was observed.
Slight differences in the total consumption of oxygen between the two textiles were observed (see Fig. S4 in the supplemental material), which showed average molar ratios differing in one unit (4.2 and 5.3 for CO and PO experiments, respectively) by dividing the slopes of cumulative consumed O2 and degraded MTBE (all with R2 values of >0.9). However, both values were still far from the 7.5 mol of oxygen required for total mineralization of MTBE and close to the range (3.3 to 4.7 mol) previously found for MTBE degraders growing on MTBE (strains PM1 and L108) (42).
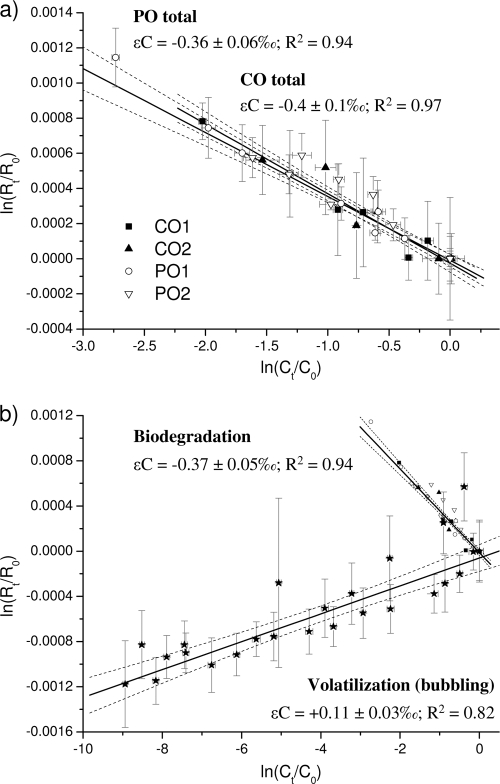
Analogous MTBE isotopic enrichment factors in all the microcosms were obtained in duplicate for each textile, as well as for CO versus PO experiments (see the individual values in Table 2 and Fig. 3 a for the carbon isotope analysis). Low carbon fractionation was observed after 94% of MTBE degradation, resulting in a mean ɛC of −0.37‰ ± 0.05‰ (R2 = 0.94) when plotted together with all the data points obtained in the four microcosms (Fig. 3b). No significant 2H enrichment occurred in any microcosm (not exceeding total analytical uncertainty of 5‰ or associated 95% CI); therefore, the related Λ value could not be determined. These values were similar to the ones observed for A. tertiaricarbonis L108 or R. ruber IFP 2001, with ɛC values below −0.5‰ and no detectable hydrogen fractionation (41, 42).
TABLE 2.
Comparison of MTBE carbon and hydrogen isotopic enrichment factors with 95% confidence intervals and 2D-CSIA slopes caused by abiotic processes and aerobic biodegradation by pure and mixed culturesc
| Process or culture | Carbon |
Hydrogen |
Λ ± 95% CI | Reference(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ɛC (‰) | ±95% CI (‰) | R2 | R (%) | No. of data points | ɛH (‰) | ±95% CI (‰) | R2 | R (%) | No. of data points | |||
| Abiotic processes | ||||||||||||
| Oxidation by permanganate | NA | −109 | 9 | 0.98 | 90 | 16 | NA | 10 | ||||
| Acidic hydrolysis (SN1) | −4.9 | 0.6 | 0.995 | 88 | 6 | −55 | 7 | 0.99 | 88 | 6 | 11 ± 1 | 10 |
| Volatilization | ||||||||||||
| Headspace equilibrium (aqueous, close) | +0.2 | 0.2 | NA | NA | 19 | |||||||
| Diffusive (sand/aqueous) | −1.0 | 0.1 | 0.8 to 0.9 | −5 | 1 | ∼5 | 25 | |||||
| Advective/air sparging (sand/aqueous) | ∼0 | 0.1 | 0.8 to 0.9 | −4 to −12 | NA | 0.88 to 0.99 | NA | 25 | ||||
| Bubbling (aqueous, open, n = 2) | +0.11 | 0.03 | 0.82 | 99.99 | 26 | −5 | 1 | 0.93 | 99.997 | 26 | −27 ± 4 | This study |
| Pure cultures | ||||||||||||
| M. petroleiphilum PM1a | −2.0 to −2.4 | 0.1 to 0.3 | 0.88 to 0.98 | 93 | 39 | −33 to −37 | 4 to 5 | 0.90 to 0.99 | 80 to 90 | 26 | 18 ± 3 | 11, 14 |
| Methylibium sp. strain R8 | −2.3 | 0.1 | 0.98 | 98 | 40 | −40 | 4 | 0.95 | 91 | 36 | 17 ± 1 | 3, 41 |
| A. tertiaricarbonis L108 | −0.48 | 0.05 | 0.94 | 96 | 34 | NS (−0.2) | 8 | 0.00 | 82 | 29 | NA | 41 |
| R. ruber IFP 2001 | −0.28 | 0.06 | 0.81 | 97 | 27 | NS (+5) | 20 | 0.14 | 88 | 24 | NA | 41 |
| P. tetrahydrofuranoxydans K1 | −2.3 | 0.2 | 0.99 | 12 | −100 | 10 | 0.99 | 11 | 48 ± 5 | 33 | ||
| Mixed cultures | ||||||||||||
| Borden enrichment culture | −1.52 to −1.97 | 0.06 | 0.99 | 95 to 97 | 38 | NA | NA | 19 | ||||
| VAFB mixed consortiuma | −1.5 to −1.8 | 0.1 | 0.95 to 0.99 | 99.7 | 55 | −29 to −66 | 3 to 4 | 0.91 to 0.99 | 80 to 90 | 24 | 12 ± 2 to 45 ± 2 | 14, 33 |
| Port Hueneme microcosm | −1.4 | NA | NA | 27 | ||||||||
| Artificial mixture (L108-PM1) | −0.2 to −1.0 | 0.1 to 0.2 | 0.96 to 0.995 | 98.5 | 36 | NS (−4 to +2) | 13 to 15 | 0.04 to 0.09 | 83 | 18 | NA | 42 |
| Biofilms on geotextiles from Leuna, Germanyb | ||||||||||||
| Replicate CO1 | −0.4 | 0.2 | 0.98 | 87 | 7 | NS (−4) | 9 | 0.36 | 87 | 4 | NA | This study |
| Replicate CO2 | −0.4 | 0.4 | 0.94 | 76 | 6 | NS (−9) | 11 | 0.59 | 76 | 5 | NA | This study |
| CO total | −0.4 | 0.1 | 0.97 | 87 | 13 | NS (−5) | 4 | 0.37 | 87 | 9 | NA | This study |
| Replicate PO1 | −0.37 | 0.09 | 0.96 | 94 | 10 | NS (−7) | 18 | 0.81 | 86 | 4 | NA | This study |
| Replicate PO2 | −0.4 | 0.1 | 0.91 | 80 | 9 | NS (−7) | 17 | 0.89 | 80 | 4 | NA | This study |
| PO total | −0.36 | 0.06 | 0.94 | 94 | 19 | NS (−7) | 7 | 0.85 | 86 | 8 | NA | This study |
Original data reevaluated by Elsner et al. (11).
Each replicate bottle reported individually but also as a total.
NA, not analyzed or not applicable; NS, not significant; CI, confidence interval; R%, maximum percentage of MTBE removal analyzed. Abiotic processes include oxidation, acidic hydrolysis, and different types of volatilization.
FIG. 3.
MTBE carbon fractionation represented, according to a double-logarithmic plot (isotopic composition versus residual concentration), during aerobic degradation in replicate microcosm experiments with biofilms collected on CO and PO (a) and the total enrichment (designed as biodegradation) compared to that obtained in volatilization experiments (b). Solid lines correspond to a linear regression model, and dashed lines correspond to their associated 95% confidential intervals (CI). Related propagated error bars are provided (see Table 2 for more details).
In all sterile controls (with and without textiles), MTBE concentrations remained constant (relative standard deviation of up to 5%) along the whole incubation time, as did the isotopic signatures (bottle without textile, δ13C of −30.4‰ ± 0.3‰ and δ2H of −114‰ ± 3‰), showing that no other abiotic processes affected MTBE in the microcosms.
Volatilization isotope enrichment factors.
Dissolved MTBE concentrations in the two batch experiments decreased exponentially (R2 = 0.98 to 0.99), covering a total range of 4 orders of magnitude. A small but significant inverse carbon fractionation in the water phase (Δδ13C = −1.1 ± 0.3) was observed, corresponding to a mean ɛC value of +0.11‰ ± 0.03‰, according to equation 2 when using all obtained data points (Table 2 and Fig. 3b). In contrast, consistent 2H enrichment in the MTBE fraction in the water phase (Δδ2H = 49 ± 5) during MTBE volatilization was observed, with an average ɛH value of −5‰ ± 1‰. In both cases, the data fit the Rayleigh-type linear fractionation model with R2 values of 0.82 and 0.93, respectively. This led to a negative slope (Λ = −27 ± 4; R2 = 0.8) when plotting Δδ2H versus Δδ13C (equation 3).
MTBE isotope fractionation in the pond system.
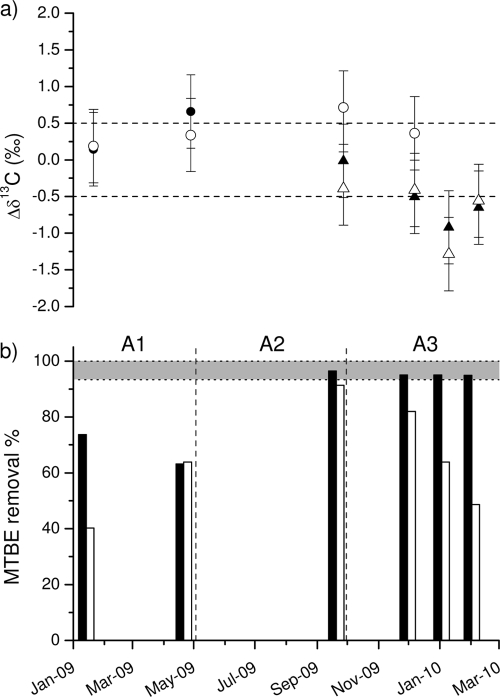
Several water samples obtained for MTBE isotopic measurements were taken after more than 1 year of pond operation, covering the three aeration periods (Fig. 4). δ13C values obtained by two different methodologies (HS versus P&T) were standardized by MTBE aqueous standard injections in order to correct isotopic fractionations caused by the extraction techniques (58). Due to low MTBE concentrations and the available instrumentation (only HS; no P&T system connected to the hydrogen system), δ2H values for the outflows could not be determined. Although the δ13C of the inflow could be considered constant (−29.3‰ ± 0.4‰), a slight tendency to become enriched was observed along the time period (R2 = 0.7). However, no significant fractionation was detected when comparing δ13C values of both outflows versus those of the inflow at any of the aeration periods. As shown in Fig. 4a, the difference of δ13C rarely exceeded the total analytical uncertainty ± 0.5‰ (47) and never left the typical range of δ13C values for MTBE in gasoline (−28.3‰ to −31.6‰) (22, 48).
FIG. 4.
(a) Differences in MTBE carbon isotopic composition in the outflow with respect to the inflow of both ponds, with CO (solid symbols) and PO (open symbols) analyzed by headspace (circles) or purge and trap (triangles) methodology. (a, b) Dashed lines represent the upper and lower limits of total analytical uncertainty (a) (47) and related MTBE removal values (b) for those sampling points and aeration periods. (b) The shaded area represents the required MTBE removal amount (minimum, 93%), which may allow for detection of significant carbon fractionation (absolute Δδ13C ≥ 1) depending on the combination of volatilization and biodegradation processes in the field.
Expression of catabolic genes and detection of strain PM1.
In order to link the isotope fractionation factors obtained in the microcosm experiments with specific microbial activity, previously designed primer sets (Table 1) for the detection of catabolic genes encoding key enzymes involved in aerobic MTBE degradation were used. The results are summarized in Table 3. DNA or RNA (as specified above) from pure reference cultures was used as a control for each primer set, and the expected results were obtained. The ethB gene encoding a cytochrome P450 monooxygenase was detected in R. ruber IFP 2001 and A. tertiaricarbonis L108, whereas the corresponding putative regulator gene ethR was found only in R. ruber IFP 2001, confirming the results from previous studies (7, 8). Although the primer for mdpA was not specifically designed for its detection in other species (44), the positive results obtained for M. petroleiphilum PM1 and for Methylibium sp. strain R8, a closely related MTBE-degrading strain available in our laboratory (99.3% identity to corresponding 16S rRNA gene sequence of strain PM1 and very similar fractionation pattern [41]) indicated its appropriate functioning for tracking the presence of the MdpA enzyme in related bacterial species.
TABLE 3.
Detection of bacterial strains similar to M. petroleiphilum PM1 and catabolic genes encoding enzymes suggested to be involved in aerobic MTBE degradation in textiles from the microcosm experiments or directly from the pondsd
| Culture or textile | Detection of: |
Length of fragment (bp)a | Closest NCBI match | Coverage (%) | Maximum identity (%) | |||
|---|---|---|---|---|---|---|---|---|
| PM1 16S rRNA gene | MdpA 1F/1R | EthB for/for M/rev | EthR for/rev M/rev | |||||
| Reference pure cultures | ||||||||
| A. tertiaricarbonis L108 | − | − | + | − | 1,170 | R. ruber IFP 2001 EthB | 99 | 99 |
| R. ruber IFP 2001 | − | − | + | + | 891 | R. ruber IFP 2001 EthR | 100 | 100 |
| M. petroleiphilum PM1 | + | + | − | − | 684 | M. petroleiphilum PM1 plasmid RPME01 MdpA | 100 | 100 |
| Methylibium sp. strain R8 | + | + | − | − | 659 | M. petroleiphilum PM1 plasmid RPME01 MdpA | 100 | 100 |
| Microcosm expt textiles | ||||||||
| CO1 | + | − | + | − | 1,049 | R. ruber IFP 2007 EthB | 100 | 99 |
| CO2 | + | − | + | − | 1,164 | R. ruber IFP 2001 EthB | 99 | 99 |
| PO1 | +b | − | + | − | 1,093 | R. ruber IFP 2007 EthB | 100 | 99 |
| PO2 | +b | − | + | − | 1,021 | R. zopfii IFP 2005 EthB | 100 | 99 |
| Environmental textilesc | ||||||||
| CO (150 cm) | + | − | + | − | 1,026 | R. ruber IFP 2001 EthB | 99 | 100 |
| CO (420 cm) | + | − | + | − | 1,053 | R. ruber IFP 2001 EthB | 99 | 99 |
| PO (150 cm) | + | − | + | − | 1,166 | R. ruber IFP 2001 EthB | 99 | 99 |
| PO (420 cm) | + | − | + | − | 901 | R. ruber IFP 2007 EthB | 100 | 100 |
Length of sequence obtained from the amplification product of the PCR assay (shown in boldface) used for BLAST analysis.
Successful sequencing; obtained fragment was 100% identical to Methylibium petroleiphilum PM1.
Environmental textiles were all sampled at 120 cm depth. The distance to inflow is given in parentheses.
See details in Table 1.
In all textile samples (CO and PO) from the microcosms as well as from the pond system, the ethB gene but not the regulator (ethR) was expressed. Up to now, this pattern has been observed only for A. tertiaricarbonis L108 (7). After sequencing of the obtained PCR products, the closest matches found in the National Center for Biotechnology Information (NCBI) database were, in all cases, ethB genes previously obtained from the genus Rhodococcus, such as the ethB sequence of R. ruber IFP 2001, which was also detected in other ETBE degraders, such as R. ruber IFP 2007 (a constitutive strain derived from strain IFP 2001 [16]), Rhodococcus zopfii IFP 2005 (5, 29), and Rhodococcus sp. strain PEG604 (21). However, a high identity (99 to 100%) was also obtained when comparing directly to the A. tertiaricarbonis L108 ethB sequence (98% identical to that of R. ruber IFP 2001). This is not surprising, considering that this strain was previously isolated from the Leuna, Germany, site (40). Importantly, the ethB sequences found so far form a cluster distinct from other oxygenases and are in all cases related to fuel oxygenates' degradation capacity (29).
Unexpectedly, 16S rRNA gene sequences similar to that of M. petroleiphilum PM1 were detected in all the textile samples (from the microcosms and the pond system) using the previously described specific primer set 613F/988R (17). DNA extracted from M. petroleiphilum PM1 and Methylibium sp. strain R8 was used as a positive control. In all cases, a PCR product with the expected size (375 bp) was obtained. However, in most of the samples, the sequencing of the purified PCR products failed apparently due to a mixture of target sequences. This was not the first time that problems of nonspecificity in the application of these primer sets to complex MTBE cultures were observed (2). When sequencing was successful (both PO samples from the microcosms) (Table 3), the obtained sequence was identical to that of M. petroleiphilum PM1 (100% similarity).
DISCUSSION
In this study, MTBE removal processes in an aerated treatment pond for groundwater remediation were identified by a combination of stable isotope and molecular biological techniques. The removal of MTBE and the formation of potential degradation products were measured in situ while applying different oxygen concentrations as well as in oxic microcosm experiments, with biofilms attached to textile mats sampled in the two parallel ponds.
In contrast to the specific aromatic hydrocarbon metabolites (15), the mere presence of TBA, the main MTBE degradation product, cannot be associated with any particular degradation mechanism or redox condition, as TBA has been detected as a metabolite or end product during MTBE biodegradation under both oxic and anoxic conditions. TBA is also used as a gasoline additive itself or can even originate from other sources (45). On the other hand, TBF was not always detected as an intermediate, with all strains able to degrade MTBE (28). The presence of TBF is evidence that the MTBE degradation pathway is taking place through dehydrogenation rather than by dismutation of the unstable tert-butoxy methanol. However, TBA and TBF can be biodegraded at the same time as MTBE, as observed in our microcosm experiments as well as in numerous studies of the same degrader (with the same or different enzymes used to degrade MTBE) or of other species in mixed cultures (28), and therefore, do not necessarily accumulate in the groundwater. In fact, TBF concentrations were not expected to increase significantly in the ponds, as only 18 μg/liter would be produced at the highest MTBE degradation periods, according to the microcosms, and it can hydrolyze to TBA under a wide range of pHs and temperatures (9).
From a physicochemical point of view, TBA is much less volatile and water miscible in contrast to MTBE (39), so it is less likely to be lost by volatilization. However, its air-water transfer velocity is extremely dependent on temperature, while MTBE is much less influenced (1). This behavior might explain partly why the removal of TBA was temperature dependent (whereas MTBE was not apparently affected) in the ponds during period A1 (see Fig. S3 in the supplemental material). However, if the TBA removal level were due only to volatilization, it should have decreased when the temperature dropped again in the CO-amended pond during high-aeration period A3, but this was not the case. Therefore, this could be an indication of biodegradation, which might still be possible under such cold conditions. Several aerobic MTBE/TBA degraders have shown growth on MTBE in a wide range of temperatures, for example, a cold-adapted mixed culture containing Variovorax paradoxus strain CL-8 (growth range, 3 to 30°C; optimum, 18 to 22°C) (56) or A. tertiaricarbonis L108 (growth range, 4 to 40°C), which had a growth rate of about 15% of its maximum (optimum, 30°C) at 5°C and could even degrade MTBE below this temperature (38).
Similar higher MTBE removals in the two aerated treatment ponds due to the increase of oxygen dissolved in the water (period A2) were obtained. However, in general, the CO textile-equipped pond had a faster response for MTBE removal than the pond equipped with PO textiles, and this could not be directly linked to the stripping/volatilization process, which was equal in both ponds. This same trend was also observed in the microcosm studies performed with the two different textile samples, but no significant differences were found for (i) the total pH decline (probably due to the same formation of CO2 and/or acidic intermediates [3], (ii) oxygen consumption values (indicative of carbon assimilation/biomass formation in both biofilms), (iii) carbon and hydrogen isotope fractionation, or (iv) the expression of ethB genes, which could be detected with both textiles under laboratory and field conditions. Therefore, we might speculate that this behavior is related more to the nature of the material than to the microbial community of MTBE degraders. It is possible that in the less dense CO textile material, the microorganisms of the biofilms might be in closer contact with the bypassing water than the denser PO, in which the attached microorganisms seem to be located deeper inside the textile fleece. In any case, both microcosm experiments showed a great improvement in MTBE biodegradation potential due to aeration of the system compared to that shown in previous microcosm studies at the A1 period (August 2008), with equivalent amounts of textile showing slower degradation of MTBE in CO microcosms (>20 days) and no biodegradation in PO microcosms after 100 days of incubation (20). In fact, estimated maximum MTBE degradation rates of 82 and 115 g MTBE/day in the CO- and PO-equipped ponds, respectively, were extrapolated from the results obtained from the microcosm experiments, considering (i) the average dry weight of the textile samples in the microcosms, (ii) the original density of each material, and (iii) the surface area of the textile used in the treatment ponds (see characteristics elsewhere [20]). However, in the field, the maximum observed MTBE removal rates during the A2 period were only 10 times less effective than the maximal rates predicted above. Possible reasons for this may include the following: (i) competition for oxygen and nutrients by organisms that are degrading other organic matter and pollutants like benzene and ammonium could affect degradation rates by limiting oxygen availability for MTBE degraders; (ii) the hydraulic retention time of 6.3 days may not be enough time to allow for complete biodegradation; and (iii) the lower MTBE concentrations in the water could, at a certain point, not be sufficient enough to enable productive microbial degradation, as calculated threshold concentrations allowing growth on this compound as the sole source of carbon and energy are significantly higher than the MTBE concentrations observed in the ponds (37).
In addition, no significant MTBE isotope fractionation could be detected in any of the ponds during the whole study period. Taking into account the low and divergent carbon enrichment factors obtained for volatilization and biodegradation in the laboratory experiments performed (Fig. 3b), the isotope values observed in the field are likely to be a combination of these two processes. As expected, the predominant process taking place when purging a MTBE aqueous solution is the advective volatilization of the compound, which would produce a 13C depletion in the water phase, leading to an inverse isotope fractionation rather than the passive diffusion which would produce an enrichment (ɛC = −1.0‰) (25). In our volatilization experiments, probably due to the absence of sediments and a broader range of concentrations, the inverse carbon isotope effect predicted for water-air equilibrium was detectable, in contrast to previous air sparging experiments through sand columns (ɛC = ∼0‰ ± 0.1‰) (25), whereas a small normal hydrogen isotope effect with a constant hydrogen fractionation for the whole studied range of concentrations was observed. The normal hydrogen isotope effect of MTBE in the liquid reflects the preferential binding of water to deuterium (2H-MTBE) (25). A theoretical concept was developed by applying the Van Breukelen equation (equation 4) in order to calculate which percentage of total MTBE removal in the field would produce a significant difference of δ13C (±1‰) for increasing contributions of biodegradation (percent F) versus volatilization. For example, a minimum of 93% of MTBE total removal would be necessary to distinguish an MTBE 13C isotope enrichment due to a 100% contribution of biodegradation, and this value would increase to 99.97% for a 50:50 combination and to almost 99.99% if only an inverse isotopic effect were observed (percent F = 0). This range was represented as a shaded area in Fig. 4b. Only the CO samples during the A2 and A3 periods reached high enough MTBE reductions (95 to 96.5%), but significant fractionation was not observed, which therefore means that biodegradation could not be the only removal process occurring.
Although δ2H could not be measured in the outflow pond samples, the low hydrogen fractionation exhibited by the two competing processes (not significant for the biodegradation detected in the microcosms and at −5‰ for the volatilization) would not have improved the interpretation of the results. In contrast, if higher-fractionating aerobic bacteria such as Methylibium sp. strains PM1 and R8 or Pseudonocardia tetrahydrofuranoxydans K1 were dominating the MTBE degradation, lower removal percentages would be necessary to confirm biodegradation and to distinguish biodegradation processes from volatilization.
Curiously, even when the presence of organisms closely related to M. petroleiphilum PM1 in some of the samples was proven using 16S rRNA gene-specific primers, its direct participation in the initial MTBE-attacking step could not be demonstrated. First of all, the low enrichment factors observed in the microcosms make very unlikely that higher-fractionating species (or related enzymes) such as M. petroleiphilum PM1 or even potential anaerobes inside the biofilms (see reference 50) contribute significantly to MTBE biodegradation. In fact, if dissolved oxygen concentrations inside the biofilm were lower than those in the surrounding solution, the higher oxygen affinity of the L108-like enzyme would reduce the total observed MTBE fractionation in a mixed culture, as observed previously (42). On the basis of isotopic results (by applying equation 4 and carbon enrichment factors obtained for strains PM1 and L108 under oxic conditions in the above-mentioned study), only 1% of a PM1-like contribution might be possible. Second, the expression of mdpA genes was not observed in any textile sample, suggesting that no active involvement of the AH1-like monooxygenase is responsible for MTBE degradation in M. petroleiphilum PM1. Therefore, the specific roles of the M. petroleiphilum PM1 relatives in the treatment system remain unclear. Their involvement in further degradation of MTBE metabolites or even the possibility that the ethB gene has been horizontally transferred to them from another organism cannot be ruled out. The fact that the eth locus is inserted in a different genomic context for three different ETBE-utilizing strains suggests that it may be transferred horizontally between different species and inserted at different sites in the genome (5).
Indeed, independent of which organism or organisms are degrading MTBE, the key finding of this study is that the expression of a specific catabolic gene such as ethB provides direct evidence for in situ MTBE biodegradation activity in the remediation treatment system. The MTBE biodegradation occurring in the textile biofilms and the observed low MTBE stable isotope fractionation were linked to the expression of this gene.
In conclusion, our work has shown that when low-fractionating enzymes are catalyzing the dominant MTBE biodegradation pathway and volatilization effects are not negligible, the application of CSIA in field investigations to detect biodegradation may lead to false-negative results and should be supplemented by microcosm experiments. For a reliable characterization of MTBE degradation, CSIA should be applied in combination with molecular methods for examining the expression of specific catabolic genes, which constitute direct evidence for the activity of MTBE degraders under in situ conditions in comparison to the mere presence of such organisms.
Supplementary Material
Acknowledgments
This work was supported by the Helmholtz Centre for Environmental Research-UFZ in the scope of the SAFIRA II Research Programme (Revitalization of Contaminated Land and Groundwater at Megasites, subproject “Compartment Transfer-CoTra”). P. M. Martínez-Lavanchy was supported by a collaborative project (BACSIN contract 211684) of the European Commission within its Seventh Framework Program, and M. Rosell was supported by a Beatriu de Pinós postdoctoral grant (2008 BP-A 00054) from the Autonomous Government of Catalonia (Agència de Gestió d'Ajuts Universitaris i de Recerca [AGAUR]).
We thank Francesca Löper (Department of Groundwater Remediation) for technical assistance in the system operation and sampling; Manuela van Pinxteren (Department of Analytical Chemistry) for analyzing the chemical composition of target groundwater from Leuna, Germany; Judith Schuster (Department of Environmental Microbiology) for providing us with purified DNA from Rhodococcus ruber IFP 2001 and EthB/EthR primers; Ursula Günther, Matthias Gehre, and Stephanie Hinke for technical support in our isotope and cultivation laboratories; and last but not least, Kenneth Wasmund for critically editing the manuscript regarding grammar and style.
Footnotes
Published ahead of print on 10 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arp, H. P. H., and T. C. Schmidt. 2004. Air-water transfer of MTBE, its degradation products, and alternative fuel oxygenates: the role of temperature. Environ. Sci. Technol. 38:5405-5412. [DOI] [PubMed] [Google Scholar]
- 2.Babé, A., D. Labbé, F. Monot, C. W. Greer, and F. Fayolle-Guichard. 2007. Biodegradability of oxygenates by microflora from MTBE-contaminated sites: new molecular tools, p. 75-98. In D. Barceló (ed.), Fuel oxygenates, vol. 5, part R. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Bastida, F., et al. 2010. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol. Ecol. 73:370-384. [DOI] [PubMed] [Google Scholar]
- 4.Bedessem, M. E., A. M. Ferro, and T. Hiegel. 2007. Pilot-scale constructed wetlands for petroleum-contaminated groundwater. Water Environ. Res. 79:581-586. [DOI] [PubMed] [Google Scholar]
- 5.Beguin, P., et al. 2003. Genes involved in the degradation of ether fuels by bacteria of the Mycobacterium/Rhodococcus group. Oil Gas Sci. Technol. 58:489-495. [Google Scholar]
- 6.Bombach, P., H. H. Richnow, M. Kästner, and A. Fischer. 2010. Current approaches for the assessment of in situ biodegradation. Appl. Microbiol. Biotechnol. 86:839-852. [DOI] [PubMed] [Google Scholar]
- 7.Breuer, U., C. Bäjen, T. Rohwerder, R. H. Müller, and H. Harms. 2007. MTBE degradation genes of an Ideonella-like bacterium L108, p. 103. In Proceedings of the 3rd European Conference on MTBE and Other Fuel Oxygenates. VITO, Antwerp, Belgium.
- 8.Chauvaux, S., et al. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church, C. D., J. F. Pankow, and P. G. Tratnyek. 1999. Hydrolysis of tert-butyl formate: kinetics, products, and implications for the environmental impact of methyl tert-butyl ether. Environ. Toxicol. Chem. 18:2789-2796. [Google Scholar]
- 10.Elsner, M., J. McKelvie, G. L. Couloume, and B. Sherwood Lollar. 2007. Insight into methyl tert-butyl ether (MTBE) stable isotope fractionation from abiotic reference experiments. Environ. Sci. Technol. 41:5693-5700. [DOI] [PubMed] [Google Scholar]
- 11.Elsner, M., L. Zwank, D. Hunkeler, and R. P. Schwarzenbach. 2005. A new concept linking observable stable isotope fractionation to transformation pathways of organic pollutants. Environ. Sci. Technol. 39:6896-6916. [DOI] [PubMed] [Google Scholar]
- 12.Fayolle, F., et al. 2003. Limitations in MTBE biodegradation. Oil Gas Sci. Technol. 58:497-504. [Google Scholar]
- 13.Gonzalez-Olmos, R., and M. Iglesias. 2008. Study of fuel oxygenates solubility in aqueous media as a function of temperature and tert-butyl alcohol concentration. Chemosphere 71:2098-2105. [DOI] [PubMed] [Google Scholar]
- 14.Gray, J. R., et al. 2002. Carbon and hydrogen isotopic fractionation during biodegradation of methyl tert-butyl ether. Environ. Sci. Technol. 36:1931-1938. [DOI] [PubMed] [Google Scholar]
- 15.Griebler, C., M. Safinowski, A. Vieth, H. H. Richnow, and R. U. Meckenstock. 2004. Combined application of stable carbon isotope analysis and specific metabolites determination for assessing in situ degradation of aromatic hydrocarbons in a tar oil-contaminated aquifer. Environ. Sci. Technol. 38:617-631. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Perez, G., F. Fayolle, and J. P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 17.Hristova, K., B. Gebreyesus, D. Mackay, and K. A. Scow. 2003. Naturally occurring bacteria similar to the methyl tert-butyl ether (MTBE)-degrading strain PM1 are present in MTBE-contaminated groundwater. Appl. Environ. Microbiol. 69:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hristova, K. R., C. M. Lutenegger, and K. M. Scow. 2001. Detection and quantification of methyl tert-butyl ether-degrading strain PM1 by real-time TaqMan PCR. Appl. Environ. Microbiol. 67:5154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunkeler, D., B. J. Butler, R. Aravena, and J. F. Barker. 2001. Monitoring biodegradation of methyl tert-butyl ether (MTBE) using compound-specific carbon isotope analysis. Environ. Sci. Technol. 35:676-681. [DOI] [PubMed] [Google Scholar]
- 20.Jechalke, S., et al. 2010. Aerated treatment pond technology with biofilm promoting mats for the bioremediation of benzene, MTBE and ammonium contaminated groundwater. Water Res. 44:1785. [DOI] [PubMed] [Google Scholar]
- 21.Kim, Y. H., K. H. Engesser, and S. J. Kim. 2007. Physiological, numerical and molecular characterization of alkyl ether-utilizing rhodococci. Environ. Microbiol. 9:1497-1510. [DOI] [PubMed] [Google Scholar]
- 22.Kolhatkar, R., T. Kuder, P. Philp, J. Allen, and J. T. Wilson. 2002. Use of compound-specific stable carbon isotope analyses to demonstrate anaerobic biodegradation of MTBE in groundwater at a gasoline release site. Environ. Sci. Technol. 36:5139-5146. [DOI] [PubMed] [Google Scholar]
- 23.Kopinke, F. D., A. Georgi, M. Voskamp, and H. H. Richnow. 2005. Carbon isotope fractionation of organic contaminants due to retardation on humic substances: implications for natural attenuation studies in aquifers. Environ. Sci. Technol. 39:6052-6062. [DOI] [PubMed] [Google Scholar]
- 24.Kruzic, A. P., and J. F. Kreissl. 2009. Natural treatment and onsite systems. Water Environ. Res. 81:1346-1360. [Google Scholar]
- 25.Kuder, T., P. Philp, and J. Allen. 2009. Effects of volatilization on carbon and hydrogen isotope ratios of MTBE. Environ. Sci. Technol. 43:1763-1768. [DOI] [PubMed] [Google Scholar]
- 26.Kuder, T., et al. 2005. Enrichment of stable carbon and hydrogen isotopes during anaerobic biodegradation of MTBE: microcosm and field evidence. Environ. Sci. Technol. 39:213-220. [DOI] [PubMed] [Google Scholar]
- 27.Lesser, L. E., et al. 2008. An evaluation of compound-specific isotope analyses for assessing the biodegradation of MTBE at Port Hueneme, CA. Environ. Sci. Technol. 42:6637-6643. [DOI] [PubMed] [Google Scholar]
- 28.Lopes Ferreira, N., C. Malandain, and F. Fayolle-Guichard. 2006. Enzymes and genes involved in the aerobic biodegradation of methyl tert-butyl ether (MTBE). Appl. Microbiol. Biotechnol. 72:252-262. [DOI] [PubMed] [Google Scholar]
- 29.Malandain, C., F. Fayolle-Guichard, and T. M. Vogel. 2010. Cytochromes P450-mediated degradation of fuel oxygenates by environmental isolates. FEMS Microbiol. Ecol. 72:289-296. [DOI] [PubMed] [Google Scholar]
- 30.Mancini, S. A., et al. 2006. Effects of trace element concentration on enzyme controlled stable isotope fractionation during aerobic biodegradation of toluene. Environ. Sci. Technol. 40:7675-7681. [DOI] [PubMed] [Google Scholar]
- 31.Mariotti, A., et al. 1981. Experimental-determination of nitrogen kinetic isotope fractionation: some principles. Illustration for the denitrification and nitrification processes. Plant Soil 62:413-430. [Google Scholar]
- 32.McKelvie, J. R., et al. 2007. Evaluation of TCE and MTBE in situ biodegradation: integrating stable isotope, metabolic intermediate and microbial lines of evidence. Ground Water Monit. Remediat. 27:63-73. [Google Scholar]
- 33.McKelvie, J. R., et al. 2009. Isotopic fractionation of MTBE suggests different initial reaction mechanisms during aerobic biodegradation. Environ. Sci. Technol. 43:2793-2799. [DOI] [PubMed] [Google Scholar]
- 34.McKelvie, J. R., D. M. Mackay, N. R. de Sieyes, G. Lacrampe-Couloume, and B. Sherwood Lollar. 2007. Quantifying MTBE biodegradation in the Vandenberg Air Force Base ethanol release study using stable carbon isotopes. J. Contam. Hydrol. 94:157-165. [DOI] [PubMed] [Google Scholar]
- 35.McLoughlin, P. W., R. J. Pirkle, D. Fine, and J. T. Wilson. 2004. TBA production by acid hydrolysis of MTBE during heated headspace analysis and evaluation of a base as a preservative. Ground Water Monit. Remediat. 24:57-66. [Google Scholar]
- 36.Meckenstock, R. U., B. Morasch, C. Griebler, and H. H. Richnow. 2004. Stable isotope fractionation analysis as a tool to monitor biodegradation in contaminated aquifers. J. Contam. Hydrol. 75:215-255. [DOI] [PubMed] [Google Scholar]
- 37.Müller, R. H., T. Rohwerder, and H. Harms. 2007. Carbon conversion efficiency and limits of productive bacterial degradation of methyl tert-butyl ether and related compounds. Appl. Environ. Microbiol. 73:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller, R. H., T. Rohwerder, and H. Harms. 2008. Degradation of fuel oxygenates and their main intermediates by Aquincola tertiaricarbonis L108. Microbiology 154:1414-1421. [DOI] [PubMed] [Google Scholar]
- 39.Pankow, J. F., R. E. Rathbun, and J. S. Zogorski. 1996. Calculated volatilization rates of fuel oxygenate compounds and other gasoline-related compounds from rivers and streams. Chemosphere 33:921-937. [Google Scholar]
- 40.Rohwerder, T., U. Breuer, D. Benndorf, U. Lechner, and R. H. Müller. 2006. The alkyl tertiary butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72:4128-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosell, M., et al. 2007. Variations in 13C/12C and D/H enrichment factors of aerobic bacterial fuel oxygenate degradation. Environ. Sci. Technol. 41:2036-2043. [DOI] [PubMed] [Google Scholar]
- 42.Rosell, M., et al. 2010. Evaluation of the effects of low oxygen concentration on stable isotope fractionation during aerobic MTBE biodegradation. Environ. Sci. Technol. 44:309-315. [DOI] [PubMed] [Google Scholar]
- 43.Schirmer, M., and J. F. Barker. 1998. A study of long-term MTBE attenuation in the borden aquifer, Ontario, Canada. Ground Water Monit. Remediat. 18:113-122. [Google Scholar]
- 44.Schmidt, R., V. Battaglia, K. Scow, S. Kane, and K. R. Hristova. 2008. Involvement of a novel enzyme, MdpA, in methyl tert-butyl ether degradation in Methylibium petroleiphilum PM1. Appl. Environ. Microbiol. 74:6631-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, T. C., M. Schirmer, H. Weiss, and S. B. Haderlein. 2004. Microbial degradation of methyl tert-butyl ether and tert-butyl alcohol in the subsurface. J. Contam. Hydrol. 70:173-203. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, T. C., et al. 2004. Compound-specific stable isotope analysis of organic contaminants in natural environments: a critical review of the state of the art, prospects, and future challenges. Anal. Bioanal. Chem. 378:283-300. [DOI] [PubMed] [Google Scholar]
- 47.Sherwood Lollar, B., S. K. Hirschorn, M. M. G. Chartrand, and G. Lacrampe-Couloume. 2007. An approach for assessing total instrumental uncertainty in compound-specific carbon isotope analysis: implications for environmental remediation studies. Anal. Chem. 79:3469-3475. [DOI] [PubMed] [Google Scholar]
- 48.Smallwood, B. J., R. P. Philp, and J. D. Allen. 2002. Stable carbon isotopic composition of gasolines determined by isotope ratio monitoring gas chromatography mass spectrometry. Org. Geochem. 33:149-159. [Google Scholar]
- 49.Smith, A. E., et al. 2005. Comparison of biostimulation versus bioaugmentation with bacterial strain PM1 for treatment of groundwater contaminated with methyl tertiary butyl ether (MTBE). Environ. Health Perspect. 113:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somsamak, P., H. H. Richnow, and M. M. Häggblom. 2006. Carbon isotope fractionation during anaerobic degradation of methyl tert-butyl ether under sulfate-reducing and methanogenic conditions. Appl. Environ. Microbiol. 72:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Squillace, P. J., J. F. Pankow, N. E. Korte, and J. S. Zogorski. 1997. Review of the environmental behavior and fate of methyl tert-butyl ether. Environ. Toxicol. Chem. 16:1836-1844. [Google Scholar]
- 52.Stupp, H. D. 2007. Remediation technologies and costs for cleaning MTBE contaminated groundwater, p. 249-273. In D. Barceló (ed.), Fuel oxygenates, vol. 5, part R. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 53.Thörneby, L., L. Mathiasson, L. Martensson, and W. Hogland. 2006. The performance of a natural treatment system for landfill leachate with special emphasis on the fate of organic pollutants. Waste Manage. Res. 24:183-194. [DOI] [PubMed] [Google Scholar]
- 54.U.S. Environmental Protection Agency. 1997. Drinking water advisory: consumer acceptability advice and health effects analysis on methyl tertiary-butyl ether (MTBE) EPA-822-F-97-009. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 55.Van Breukelen, B. M. 2007. Extending the Rayleigh equation to allow competing isotope fractionating pathways to improve quantification of biodegradation. Environ. Sci. Technol. 41:4004-4010. [DOI] [PubMed] [Google Scholar]
- 56.Zaitsev, G. M., J. S. Uotila, and M. M. Häggblom. 2007. Biodegradation of methyl tert-butyl ether by cold-adapted mixed and pure bacterial cultures. Appl. Microbiol. Biotechnol. 74:1092-1102. [DOI] [PubMed] [Google Scholar]
- 57.Zwank, L., et al. 2005. New evaluation scheme for two-dimensional isotope analysis to decipher biodegradation processes: application to groundwater contamination by MTBE. Environ. Sci. Technol. 39:1018-1029. [DOI] [PubMed] [Google Scholar]
- 58.Zwank, L., M. Berg, T. C. Schmidt, and S. B. Haderlein. 2003. Compound-specific carbon isotope analysis of volatile organic compounds in the low-microgram per liter range. Anal. Chem. 75:5575-5583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.