Abstract
Diaminodiphenylsulfone (dapsone) has long been used as a first-line drug worldwide for the treatment of leprosy. Diagnosis for dapsone resistance of Mycobacterium leprae by DNA tests would be of great clinical value, but the relationship between the nucleotide substitutions and susceptibility to dapsone must be clarified before use. In this study, we constructed recombinant strains of cultivable Mycobacterium smegmatis carrying the M. leprae folP1 gene with or without a point mutation, disrupting their own folP gene on the chromosome. Dapsone susceptibilities of the recombinant bacteria were measured to examine influence of the mutations. Dapsone MICs for most of the strains with mutations at codon 53 or 55 of M. leprae folP1 were 2 to 16 times as high as the MIC for the strain with the wild-type folP1 sequence, but mutations that changed Thr to Ser at codon 53 showed somewhat lower MIC values than the wild-type sequence. Strains with mutations at codon 48 or 54 showed levels of susceptibility to dapsone comparable to the susceptibility of the strain with the wild-type sequence. This study confirmed that point mutations at codon 53 or 55 of the M. leprae folP1 gene result in dapsone resistance.
The massive use of dapsone for treatment of leprosy led to the isolation of resistant strains of Mycobacterium leprae as early as 1964 (11), only a few years after discovery of the drug. Dapsone is structurally related to the sulfonamides. The mechanism of dapsone resistance in M. leprae is thought to be associated with dihydropteroate synthase (DHPS) in a manner similar to the mechanism of resistance to sulfonamides developed in other bacteria. The sulfonamides are structural analogs of p-aminobenzoate (PABA) and act as antimetabolites by competing with PABA for the active site of DHPS (4). DHPS catalyzes the reaction between dihydropteridine pyrophosphate and PABA as a part of the biosynthetic pathway leading to tetrahydrofolate (5, 12), which acts as a cofactor in the biosynthesis of purines, pyrimidines, and amino acids. Resistance to the sulfonamides has been shown to be mediated by mutations of the chromosomal folP gene encoding DHPS (7, 14, 15). Point mutations in the folP1 gene have been identified in dapsone-resistant strains of M. leprae (9, 10, 16). Because M. leprae cannot be cultivated on any artificial medium and requires 13 days to double in experimentally infected mice, DNA diagnoses to detect dapsone-resistant bacteria would be highly useful. However, not all nucleotide substitutions in the folP1 gene give rise to drug resistance. Therefore, the relationship between drug susceptibility and each nucleotide substitution observed in clinical isolates requires clarification. Dapsone-resistant M. leprae isolates have shown mutation at codon 53 or 55 in the folP1 gene (6, 10, 16). Mutation at codon 48 has also been detected in our clinical specimens (unpublished data). Williams et al. have analyzed two types of mutations at codons 53 and 55 of the M. leprae folP1 gene using a folP-deficient Escherichia coli (16). However, their analysis is as yet insufficient for direct application as molecular diagnosis for dapsone resistance.
In this study, site-directed mutagenesis techniques were used to alter the wild-type M. leprae folP1 gene at codons shown to be mutated in clinical isolates for testing the effects of these mutations on dapsone susceptibility in a folP-disrupted Mycobacterium smegmatis host.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α cells were grown in Luria-Bertani (LB) medium. M. smegmatis mc2155 and its transformants were grown in Middlebrook 7H9 medium (Difco, Detroit, MI) supplemented with 0.5% bovine serum albumin (fraction V), 0.2% glucose, 0.085% NaCl, 0.2% glycerol, and 0.1% Tween 80.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Cloning host | |
| STBL2 | Cloning host | |
| C600 ΔfolP::Kmr | folP mutant | 7 |
| M. smegmatis mc2155 | ||
| Plasmids | ||
| pYUB854 | Cosmid vector | 3 |
| phAE87 | Phasmid vector carrying full length DNA of mycobacteriophage PH101 | 3 |
| pMV261 | E. coli-mycobacteria shuttle plasmid vector (multicopy in mycobacteria) | 13 |
| pNN301a | pMV361-type integrative vector (single copy in mycobacteria) | 13; this study |
pNN301 has an int-attP fragment of mycobacteriophage L5 instead of oriM.
Site-directed mutagenesis.
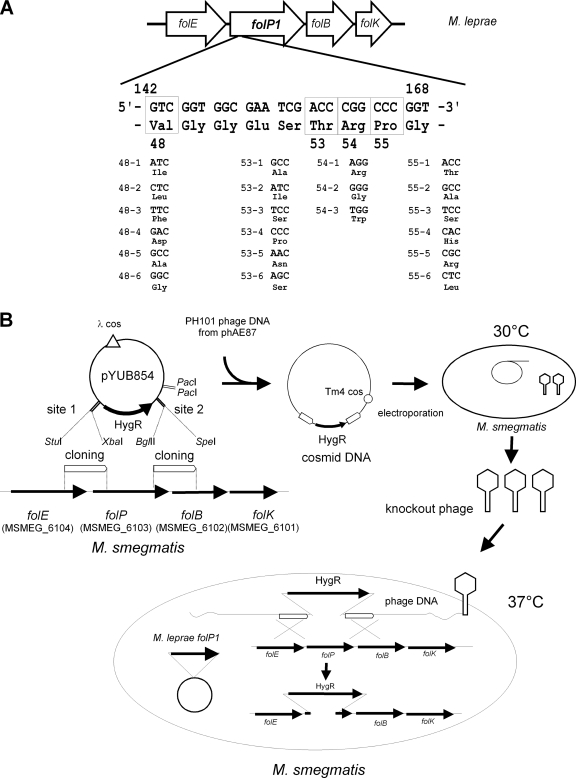
The wild-type M. leprae folP1 gene was amplified by PCR from M. leprae Thai-53 and cloned into pMV261. Site-directed mutagenesis was performed using PCR with KOD DNA polymerase (Toyobo, Osaka, Japan) and the primers listed in Table 2. PCR products were purified and phosphorylated with T4 kinase and ATP and then ligated to become circular. The ligation mixture was used to transform E. coli DH5α, and kanamycin-resistant colonies were isolated. Plasmids were extracted from the transformants, and the mutated sequences were confirmed by sequencing. Mutations introduced in the M. leprae folP1 gene are shown in Fig. 1 A.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Application |
|---|---|---|
| MLFPWTF | GCGAATTCGTGAGTTTGGCGCCAGTGCA | Cloning of M. leprae folP1, forward |
| MLFPWTR | GCAAGCTTTCAGCCATCACATCTAACCT | Cloning of M. leprae folP1, reverse |
| MSFPUF | GCAGGCCTGTATCCTCATCCCGGACAGC | folP disruption, upstream forward |
| MSFPUR | GCTCTAGATGGTGTCGATGCTGATCGTG | folP disruption, upstream reverse |
| MSFPDF | GCAGATCTCGCAAACGTTTCCTCGGTAC | folP disruption, downstream forward |
| MSFPDR | GCACTAGTACTGGTCGATCTCCGACAGC | folP disruption, downstream reverse |
| MSFPF | TCACCGAGTACGGCATGAGC | Detection of folP disruption, forward |
| MSFPR | TAGAGCGCATGGATCAGCAG | Detection of folP disruption, reverse |
| MLFPR1 | CGATTCGCCACCGACGTCGAC | Introduction of point mutations for codons 53, 54, and 55 |
| MLFPR2 | GTCGACAATCGCCGCGCCTT | Introduction of point mutations for codon 48 |
| MLFP48-1 | ATCGGTGGCGAATCGACCCG | Introduction of point mutation 48-1 |
| MLFP48-2 | CTCGGTGGCGAATCGACCCG | Introduction of point mutation 48-2 |
| MLFP48-3 | TTCGGTGGCGAATCGACCCG | Introduction of point mutation 48-3 |
| MLFP48-4 | GACGGTGGCGAATCGACCCG | Introduction of point mutation 48-4 |
| MLFP48-5 | GCCGGTGGCGAATCGACCCG | Introduction of point mutation 48-5 |
| MLFP48-6 | GGCGGTGGCGAATCGACCCG | Introduction of point mutation 48-6 |
| MLFP53-1 | GCCCGGCCCGGTGCCATTAG | Introduction of point mutation 53-1 |
| MLFP53-2 | ATCCGGCCCGGTGCCATTAG | Introduction of point mutation 53-2 |
| MLFP53-3 | TCCCGGCCCGGTGCCATTAG | Introduction of point mutation 53-3 |
| MLFP53-4 | CCCCGGCCCGGTGCCATTAG | Introduction of point mutation 53-4 |
| MLFP53-5 | AACCGGCCCGGTGCCATTAG | Introduction of point mutation 53-5 |
| MLFP53-6 | AGCCGGCCCGGTGCCATTAG | Introduction of point mutation 53-6 |
| MLFP54-1 | ACCAGGCCCGGTGCCATTAG | Introduction of point mutation 54-1 |
| MLFP54-2 | ACCCGGCCCGGTGCCATTAG | Introduction of point mutation 54-2 |
| MLFP54-3 | ACCTGGCCCGGTGCCATTAG | Introduction of point mutation 54-3 |
| MLFP55-1 | ACCCGGACCGGTGCCATTAG | Introduction of point mutation 55-1 |
| MLFP55-2 | ACCCGGGCCGGTGCCATTAG | Introduction of point mutation 55-2 |
| MLFP55-3 | ACCCGGTCCGGTGCCATTAG | Introduction of point mutation 55-3 |
| MLFP55-4 | ACCCGGCACGGTGCCATTAG | Introduction of point mutation 55-4 |
| MLFP55-5 | ACCCGGCGCGGTGCCATTAG | Introduction of point mutation 55-5 |
| MLFP55-6 | ACCCGGCTCGGTGCCATTAG | Introduction of point mutation 55-6 |
Restriction sites are underlined
FIG. 1.
Construction of recombinant M. smegmatis strains for dapsone susceptibility testing. (A) Point mutations introduced in the M. leprae folP1 gene. Single nucleotide substitutions introduced in the M. leprae folP1 at codons 48, 53, 54, and 55 are shown. Deduced amino acid residues are shown below the triplets. (B) Construction of M. smegmatis recombinants by allelic exchange.
Disruption of the folP gene on the M. smegmatis chromosome.
M. smegmatis mc2155 cells were transformed with plasmids carrying the M. leprae folP1 with or without a point mutation. Recombinants were selected on LB medium containing kanamycin. Allelic exchange mutants were constructed by the temperature-sensitive mycobacteriophage method (3). Using the M. smegmatis mc2155 genome sequence (accession number CP000480), the upstream and downstream flanking DNA sequences were used to generate a deletion mutation in the folP gene (MSMEG_6103). In order to disrupt the folP gene, DNA segments from 736 bp upstream through 286 bp downstream of the initiation codon of M. smegmatis folP and from 198 bp upstream through 832 bp downstream of the termination codon were cloned directionally into the cosmid vector pYUB854, which contains a res-hyg-res cassette and a cos sequence for lambda phage assembly. Plasmids thus produced were digested with PacI and ligated to the PH101 genomic DNA excised from the phasmid phAE87 by PacI digestion. The ligated DNA was packaged using GigaPackIII Gold Packaging Extract (Stratagene, La Jolla, CA), and the resultant mixture was used for transduction of E. coli STBL2 (Life Technologies, Carlsbad, CA) to yield cosmid DNA. After E. coli was transduced and the transductants were plated on hygromycin-containing medium, phasmid DNA was prepared from the pooled antibiotic-resistant transductants and electroporated into M. smegmatis mc2155. Bacterial cells were incubated at 30°C to produce the recombinant phage. The M. smegmatis transformant carrying the M. leprae folP1 gene was infected by the produced temperature-sensitive phage at 37°C for allelic exchange, and kanamycin- and hygromycin-resistant colonies were isolated. Two colonies for each point mutation were subjected to subsequent tests.
Dapsone susceptibility testing.
The MIC values for M. smegmatis recombinant clones were determined by culture on Middlebrook 7H10 agar plates containing 2-fold serial dilutions of dapsone (0.25 to 64 μg/ml). The MIC value for each strain was defined as the lowest concentration of dapsone needed to inhibit bacterial growth.
RESULTS
Construction of recombinant M. smegmatis strains.
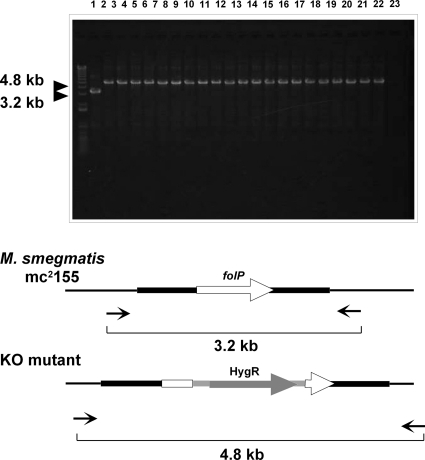
We prepared plasmids with point mutations in the M. leprae folP1 gene. Each plasmid has 1 of 21 single point mutations at codon 48, 53, 54, or 55 (Fig. 1A). The first or second nucleotide at each codon was replaced by another nucleotide to change the amino acid residue. Mutated sequences were confirmed by sequencing. Plasmids carrying the M. leprae folP1 with or without a point mutation were individually introduced into M. smegmatis. The M. smegmatis transformants were subjected to allelic exchange to disrupt the folP gene on their own chromosome (Fig. 1B). PCR analysis confirmed that the folP sequences in the recombinant strains were replaced by hygromycin resistance gene sequences (Fig. 2). Isolation of a folP-disrupted M. smegmatis strain carrying the M. leprae folP1 with mutation 48-4 (mutation 4 at codon 48) was unsuccessful. All the strains except for the strains with mutation 48-5 or 53-4 showed comparable growth rates. The strains with mutation 48-5 or 53-4 grew a little more slowly than the strain with the wild-type sequence. These two mutations may reduce DHPS activity.
FIG. 2.
PCR analysis to confirm the disruption of folP. Black arrows represent primers MSFPF and MSFPR for the PCR amplification. Lane 1, M. smegmatis mc2155; lanes 2 to 22, M. smegmatis strains carrying the M. leprae folP1 without mutation and folP1 with mutations 48-1,48-2, 48-3, 48-5, 48-6, 53-1, 53-2, 53-3, 53-4, 53-5, 53-6, 54-1, 54-2, 54-3, 55-1, 55-2, 55-3, 55-4, 55-5, and 55-6, respectively; lane 23, negative control. KO, knockout.
Dapsone susceptibility.
Dapsone susceptibilities of the recombinant M. smegmatis strains were tested. As shown in Fig. 3, the MIC of dapsone for recombinant M. smegmatis carrying the wild-type M. leprae folP1 gene was 0.5 μg/ml. MIC values for most of the strains with mutations at codon 53 or 55 were 2 to 16 times as high as the MIC for the strain with the wild-type sequence. Interestingly, two strains with alterations in amino acids from threonine to serine (T53S) encoded by different nucleotide sequences (53-3 and 53-6) were more susceptible to dapsone than strains with the wild-type folP1 sequence. MIC values for strains with mutations at codon 48 or 54 were comparable to MICs for strains with the wild-type sequence. MIC values of dapsone for the recombinant M. smegmatis strains are listed in Table 3. Using a multicopy plasmid may affect the expression levels of the M. leprae folP1 and MIC values. Therefore, we tested all the mutations using pNN301, a single-copy integrative vector, instead of pMV261 and obtained MIC values identical to those obtained with pMV261, suggesting that the expression levels did not influence the MIC values.
FIG. 3.
Dapsone susceptibility of recombinant M. smegmatis. Results for M. smegmatis strains with point mutations at codon 53 of the M. leprae folP1 are shown. Dapsone concentration is depicted below each plate.
TABLE 3.
Dapsone susceptibility of the recombinant M. smegmatis strains
| Strain or mutation | Dapsone MIC (μg/ml) | Reference of footpad test |
|---|---|---|
| Wild type | 0.5 | |
| 48-1 (Val → Ile) | 0.5 | |
| 48-2 (Val → Leu) | 0.5 | |
| 48-3 (Val → Phe) | 1.0 | |
| 48-4 (Val → Asp) | —a | |
| 48-5 (Val → Ala) | 1.0 | |
| 48-6 (Va → Gly) | 1.0 | |
| 53-1 (Thr → Ala) | 4.0 | 6 |
| 53-2 (Thr → Ile) | 8.0 | 6, 10, 16 |
| 53-3 (Thr → Ser) | 0.25 | |
| 53-4 (Thr → Pro) | 2.0 | |
| 53-5 (Thr → Asn) | 2.0 | |
| 53-6 (Thr → Ser) | 0.25 | |
| 54-1 (Arg → Arg) | 0.5 | |
| 54-2 (Arg → Gly) | 1.0 | |
| 54-3 (Arg → Trp) | 0.5 | |
| 55-1 (Pro → Thr) | 1.0 | |
| 55-2 (Pro → Ala) | 2.0 | |
| 55-3 (Pro → Ser) | 2.0 | |
| 55-4 (Pro → His) | 2.0 | |
| 55-5 (Pro → Arg) | 8.0 | 6, 16 |
| 55-6 (Pro → Leu) | 4.0 | 6, 10 |
Isolation of an folP-disrupted M. smegmatis strain carrying the M. leprae folP1 with mutation 48-4 was unsuccessful.
DISCUSSION
We first attempted using E. coli C600 ΔfolP::Kmr transformants to determine the MIC of dapsone, but susceptibility of the recombinant E. coli strains to dapsone was not stable even in Mueller-Hinton medium. Subsequently, we tried to isolate a folP-deficient M. smegmatis strain by allelic exchange, given the closer association of M. smegmatis to M. leprae than E. coli. The selection held great promise as total-sequence comparison of M. leprae DHPS with M. smegmatis DHPS indicated 83% identity, whereas the identity between M. leprae DHPS and E. coli DHPS is only 41%, indicating the higher potential of M. smegmatis as a host for measuring MIC values of dapsone for M. leprae DHPS. However, isolation of folP-deficient M. smegmatis was unsuccessful. In E. coli, DHPS is not essential for bacterial growth when the cells are cultured with thymidine (7), but DHPS activity may be essential for the growth of M. smegmatis as it could not be replaced by any of the supplemented culture media tested. Hence, we then attempted to disrupt the folP gene on the M. smegmatis chromosome after introducing the M. leprae folP1 gene into the cell to compensate for DHPS activity.
Comparison of the DHPS structures of E. coli, Staphylococcus aureus, and Mycobacterium tuberculosis has suggested that Ser53 and Pro55 of the M. tuberculosis DHPS, which correspond to Thr53 and Pro55 in M. leprae, may be the major sites of interaction with PABA, dapsone, and sulfonamides (1, 2, 8). In the present study, all mutations that cause amino acid substitutions at codon 55 resulted in dapsone resistance. Mutations at codon 53 also gave rise to dapsone resistance except for the T53S substitution, which resulted in less resistance to dapsone than the wild-type sequence (Fig. 3). The results for mutation 53-1, 53-2, 55-5, and 55-6 are consistent with the mouse footpad dapsone susceptibility testing of the M. leprae clinical isolates (6, 10, 16). Mutations at codon 48 or 54 showed comparable levels of susceptibility to dapsone as the wild-type sequence using dapsone susceptibility testing, but the MIC values for mutations 48-3, 48-5, 48-6, and 54-2 were slightly higher than the MIC for the wild-type sequence. Mutation 48-5 for V48A, which has been detected in our clinical samples (unpublished data), might give rise to low-level resistance to dapsone in M. leprae. This level of resistance should be very carefully examined by comparison with the results of footpad testing and clinical data. These data will help the molecular diagnosis of dapsone-resistant M. leprae with the goal of avoiding the wrong choice of drugs for chemotherapy.
Although these results should always be initially confirmed by clinical susceptibility testing as well, we believe that the method established in this study should have great utility in further attempts to determine the mutations responsible for giving rise to the dapsone resistance of M. leprae. The advantage of this method lies in the ability to functionally replace an essential gene of fast-growing mycobacteria with the M. leprae counterpart. The method may also be applicable to analysis of the rifampin resistance and quinolone resistance of M. leprae.
Acknowledgments
E. coli C600 ΔfolP::Kmr was kindly given by G. Swedberg (Uppsala University, Uppsala, Sweden). pYUB854 and phAE87 were kindly given by W. R. Jacobs, Jr. (Albert Einstein College of Medicine, New York, NY).
This work was supported by grants from the Ministry of Health, Labor and Welfare (Emerging and Re-Emerging Infectious Diseases) and the Ohyama Health Foundation.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Achari, A., et al. 1997. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4:490-497. [DOI] [PubMed] [Google Scholar]
- 2.Baca, A. M., R. Sirawaraporn, S. Turley, W. Sirawaraporn, and W. G. Hol. 2000. Crystal structure of Mycobacterium tuberculosis 7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J. Mol. Biol. 302:1193-1212. [DOI] [PubMed] [Google Scholar]
- 3.Bardarov, S., et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 4.Baumstark, B. R., L. L. Spremulli, U. L. RajBhandary, and G. M. Brown. 1977. Initiation of protein synthesis without formylation in a mutant of Escherichia coli that grows in the absence of tetrahydrofolate. J. Bacteriol. 129:457-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, G. M. 1962. The biosynthesis of folic acid. II. Inhibition by sulfonamides. J. Biol. Chem. 237:536-540. [PubMed] [Google Scholar]
- 6.Cambau, E., L. Carthagena, A. Chauffour, B. Ji, and V. Jarlier. 2006. Dihydropteroate synthase mutations in the folP1 gene predict dapsone resistance in relapsed cases of leprosy. Clin. Infect. Dis. 42:238-241. [DOI] [PubMed] [Google Scholar]
- 7.Fermer, C., and G. Swedberg. 1997. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J. Bacteriol. 179:831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampele, I. C., et al. 1997. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J. Mol. Biol. 268:21-30. [DOI] [PubMed] [Google Scholar]
- 9.Kai, M., et al. 1999. Diaminodiphenylsulfone resistance of Mycobacterium leprae due to mutations in the dihydropteroate synthase gene. FEMS Microbiol. Lett. 177:231-235. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, S., et al. 2001. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob. Agents Chemother. 45:3635-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees, R. J. 1964. Mycobacterial disease in man and animals. Studies on leprosy bacilli in man and animals. Proc. R. Soc Med. 57:482-483. [PMC free article] [PubMed] [Google Scholar]
- 12.Richey, D. P., and G. M. Brown. 1969. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J. Biol. Chem. 244:1582-1592. [PubMed] [Google Scholar]
- 13.Stover, C. K., et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 14.Swedberg, G., S. Ringertz, and O. Skold. 1998. Sulfonamide resistance in Streptococcus pyogenes is associated with differences in the amino acid sequence of its chromosomal dihydropteroate synthase. Antimicrob. Agents Chemother. 42:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vedantam, G., G. G. Guay, N. E. Austria, S. Z. Doktor, and B. P. Nichols. 1998. Characterization of mutations contributing to sulfathiazole resistance in Escherichia coli. Antimicrob. Agents Chemother. 42:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams, D. L., L. Spring, E. Harris, P. Roche, and T. P. Gillis. 2000. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob. Agents Chemother. 44:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]