Abstract
We screened ∼2,200 compounds known to be safe in people for the ability to reduce the amount of virion-associated hepatitis B virus (HBV) DNA in the culture medium of producer cells. These efforts led to the discovery of an alkylated porphyrin, chlorophyllide, as the compound that achieved the greatest reduction in signal. Here we report that chlorophyllide directly and quantitatively disrupted HBV virions at micromolar concentrations, resulting in the loss of all detectable virion DNA, without detectably affecting cell viability or intracellular viral gene products. Chemophores of chlorophyllide were also tested. Chlorin e6, a metal-free chlorophyllide-like molecule, showed the strongest antiviral activity against HBV as well as profound antiviral effects on other enveloped viruses, such as hepatitis C virus (HCV), human immunodeficiency virus (HIV), dengue virus (DENV), Marburg virus (MARV), Tacaribe virus (TCRV), and Junin viruses (JUNV). Remarkably, chlorin e6 inactivated DENV at subnanomolar-level concentrations. However, the compound had no antiviral effect against encephalomyocarditis virus and adenovirus, suggesting that chlorin e6 may be less active or inactive against nonenveloped viruses. Although other porphyrin derivatives have been previously reported to possess antiviral activity, this is the first analysis of the biochemical impact of chlorophyllide and chlorin e6 against HBV and of the dramatic anti-infectivity impact upon DENV. The possible application of this family of compounds as antiviral agents, as microbicides and systemic virus neutralizing agents, is discussed.
There are currently 7 U.S. FDA-approved medications for the management of chronic hepatitis B (2). These fall into two categories: the interferons (IFNs) and the nucleos(t)ide polymerase inhibitors. Both have medical value, but both have limitations. The IFNs require parenteral injections and are associated with adverse affects that limit their use (15). The polymerase inhibitors are compromised by drug-resistant viruses, and prolonged treatment may be necessary (23). Drugs in both categories are expensive, further limiting use, and are beneficial only in subsets of chronic hepatitis B patients (16). Clearly, alternatives and complements are necessary. However, the pathway to discovery and approval of a new drug entity usually involves years of development and large financial investments.
Drug safety, formulation, and pharmacokinetics are usually the limiting steps in drug development, often being responsible for most of the preclinical cost and failure (1). Therefore, we have been searching for compounds with antiviral activity from libraries of drugs that are already known to be safe in animals or people. Our priority has been hepatitis B virus (HBV). However, since we have been using libraries of compounds that mostly target host functions and these functions could be commonly used by many virus families, we remained open to the possibility of finding broadly active agents.
We have tested approximately 2,200 compounds for the ability to reduce the amount of enveloped viral DNA in the culture medium of HBV-replicating cells, and the assay development and screening work are described in a separate report (X. Lu, J. Lamontagne, A. Cuconati, M. Pinkerton, and T. Block, unpublished data). Our screening effort led to the discovery of chlorophyllide, which noncytotoxically reduced the level of secreted HBV virion DNA by more than 16-fold at micromolar concentrations. This report further describes how chlorophyllide disrupted the intact structure of progeny HBV virion particles secreted in the culture fluid.
Chlorophyllide is an alkylated porphyrin that contains copper and is charged at neutral pH and thus appears green in solution. To determine whether the charge, chelated metal, or porphyrin scaffold was important to the anti-HBV activity, chemophores of chlorophyllide, which lack charge and/or copper binding, replace copper with iron, or were just the porphyrin core ring, were also tested.
The effect upon viral infectivity could not be easily tested with HBV in a cell culture model. Therefore, chlorophyllides were tested for the ability to reduce the infectivity of other medically important enveloped viruses, such as hepatitis C virus (HCV), HIV-1, Dengue virus (DENV-2), Marburg virus (MARV), Junin viruses (JUNV), and herpes simplex virus type 1 (HSV-1). In every case, the chlorin e6 compound was the most potent (the 50% effective concentration [EC50] was approximately 2 to 5 μM), with the exceptions of HSV and DENV. For HSV, the EC50 was 20.93 μM. In the case of DENV, the EC50 was as low as 0.3 nM. To our knowledge, the subnanomolar activity against DENV is unprecedented for an antiviral agent. However, chlorin e6 did not have an antiviral effect against nonenveloped viruses, such as encephalomyocarditis virus (EMCV) and adenovirus (Ad), which suggested that such compounds selectively target enveloped viruses. The activity against multiple enveloped viruses, even as an extracellular acting agent, suggests a use as either a microbicide or a neutralizing antibody-type molecule.
MATERIALS AND METHODS
Materials.
Compound chlorophyllide Cu complex Na salt was purchased from MicroSource Discovery Systems, Inc. (Gaylordsville, CT). Compounds (chlorin e6, porphine, and hemin) were purchased from Frontier Scientific, Inc. (Logan, UT). All the compounds were dissolved in dimethyl sulfoxide (DMSO) and protected from light. Woodchuck sera containing woodchuck hepatitis virus (WHV) and duck sera with duck hepatitis B virus (DHBV) were gifts kindly provided by Pamela Norton (Drexel University College of Medicine) and William Mason (Fox Chase Cancer Center), respectively. Plasmids pE and pS, which express large (L) and middle (M)/small (S) HBV envelope proteins, respectively, were described previously and were kindly provided by Volker Bruss (Helmholtz Center Munich, Neuherberg, Germany) (28).
Cell cultures.
Tetracycline-inducible HBV-expressing HepDE19 cells were maintained in Dulbecco's modified Eagle medium (DMEM)/F12 medium (Mediatech) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin plus 1 μg/ml tetracycline. To initiate HBV replication, tetracycline was withdrawn from the culture medium. To treat the cells, compounds were added to the culture supernatant at indicated concentrations, with DMSO (0.1%) used as a solvent control. Cells and culture medium were harvested at indicated time points. Medium was clarified by centrifugation at 1,000 × g for 10 min, and cells were washed with chilled phosphate-buffered saline (PBS) and stored at −70°C.
Cell transfection.
HepG2 cells were seeded into 35-mm-diameter dishes at a density of 1.2 × 106 cells per dish in antibiotic-free complete DMEM/F12 medium. After 6 h, each well was transfected with a total of 4 μg plasmid with Lipofectamine 2000 (Invitrogen) by following the manufacturer's directions.
HBV particle gel assay.
The secreted viral particles in culture medium were assayed as previous described, with modifications (12). Briefly, PEG8000 was added to 1 ml of culture medium to a final concentration of 10%, followed by incubation at 4°C for 1 h. To test the direct effects on HBV viral particles, the compound was incubated with supernatant harvested from HepDE19 cells cultured with tetracycline-free medium at room temperature (RT) for 10 min before polyethylene glycol (PEG) precipitation. To test compound activity on WHV and DHBV, approximately 7 × 107 WHV/DHBV virions were mixed with DMEM/F12 medium to bring the volume to 1 ml, and compound was added to the mixture at 10 μM, incubated at RT for 10 min, and then processed to PEG precipitation. The precipitates were collected by centrifugation at 925 × g for 20 min and dissolved in 40 μl DMEM/F12 medium. The viral particles were fractionated by electrophoresis through nondenaturing 1% agarose gels and transferred to a nitrocellulose filter by blotting with TNE buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 1 mM EDTA). To detect HBV surface and core antigens, membrane was soaked in PBS buffer containing 2.5% formaldehyde at room temperature for 30 min. After a brief rinse with water, the membrane was then fixed with 50% methanol at room temperature for 30 min and washed three times with water. Membranes were blocked with WesternBreeze blocker buffer (Invitrogen) and probed with a monoclonal antibody against HBsAg (Dako) or polyclonal antibody against HBV core (Dako). Bound antibody was revealed by IRDye secondary antibodies and visualized by the Li-COR Odyssey system. For the detection of HBV DNA, the DNA-containing particles on the membrane were denatured with a solution containing 0.5 N NaOH and 1.5 M NaCl, followed by neutralization with a solution containing 1 M Tris-HCl (pH 7.6) and 1.5 M NaCl. HBV DNA was detected by hybridization with an [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus-strand-specific full-length HBV riboprobe. Hybridization was carried out in 5 ml Ekono hybridization buffer (Genotech) with 1 h of prehybridization at 65°C and overnight hybridization at 65°C, followed by a 1-h wash with 0.1× SSC and 0.1% SDS at 65°C. The membrane was exposed to a phosphorimager screen, and hybridization signals were quantified with the QuantityOne software program (Bio-Rad).
Analysis of HBV DNA.
Intracellular viral core DNA was extracted as described previously (13). Briefly, cells from one well of 6-well-plate were lysed with 0.5 ml of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1% NP-40, and 2% sucrose at 37°C for 10 min. Cell debris and nuclei were removed by centrifugation, and the supernatant was mixed with 130 μl of 35% PEG 8000 containing 1.5 M NaCl. After 1 h of incubation in ice, viral nucleocapsids were pelleted by centrifugation at 10,000 rpm for 5 min at 4°C, followed by 1 h of digestion at 37°C in 400 μl of digestion buffer containing 0.5 mg/ml pronase (Calbiochem), 0.5% SDS, 150 mM NaCl, 25 mM Tris-HCl (pH 8.0), and 10 mM EDTA. The digestion mixture was extracted twice with phenol, and DNA was precipitated with ethanol and then dissolved in Tris-EDTA (TE) buffer. One-third of the DNA sample from each plate was resolved by electrophoresis into a 1.2% agarose gel. The gel was then subjected to denaturation in a solution containing 0.5 M NaOH and 1.5 M NaCl, followed by neutralization in a buffer containing 1 M Tris-HCl (pH 7.4) and 1.5 M NaCl. DNA was then blotted onto a Hybond-XL membrane (GE Health care) in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). HBV DNA on the membrane were hybridized with an [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus-strand-specific full-length HBV riboprobe.
Western blot assay.
To detect intracellular protein by immunoblotting, the cells were lysed in 300 μl of 1× Laemmli buffer as used in reference 14, and a total of 30 μl of the cell lysate was resolved on an SDS-12% polyacrylamide gel. For extracellular HBsAg Western blotting, 30 μl of clarified culture supernatant was treated with 10 μl 4× Laemmli buffer and subjected to SDS-PAGE. After electrophoresis, proteins were transferred onto an Immobilon PVDF-FL membrane (Millipore). The membranes were blocked with Western Breeze blocking buffer (Invitrogen) and probed with antibody against HBsAg (Fitzgerald) and β-actin (Chemicon); bound antibodies were revealed by IRDye secondary antibodies and visualized using the Li-COR Odyssey system.
Antiviral activity test with enveloped viruses.
The antiviral activities of chlorin e6 were determined with the following enveloped viruses. (i) For HIV-1, P4-R5 MAGI cells were cultured at a density of 1.2 × 104 cells/well in a 96-well plate 18 h prior to infection. Cells were incubated for 2 h with HIV-1 strains BaL or IIIB (Advanced Biotechnologies, Inc., Columbia, MD) in the absence or presence of chlorin e6. After 2 h, cells were washed, cultured for an additional 46 h, and subsequently assayed for HIV-1 infection using the Galacto-Star β-galactosidase reporter gene assay system for mammalian cells (Applied Biosystems, Bedford, MA) (20). EC50s (concentrations at which exposure to the compound resulted in a 50% decrease in infection relative to results for mock-treated, HIV-1-infected cells) were calculated using the Forecast function of the Microsoft Excel software program. (ii) To test the virucidal activity against HCV and DENV, HCV (Jc-1 strain) or DENV-2 (New Guinea strain) in amounts that would infect Huh7.5 cells at a multiplicity of infection (MOI) of 0.015 or 0.02 was incubated with or without serial concentrations of chlorin e6 at room temperature for 30 min, followed by application of the viruses to Huh7.5 cells and incubation at 37C for 1 h. After removal of the inoculum, cells were fed with fresh medium and cultured for 4 (HCV) or 2 (DENV) days. Cells were then fixed with 3.7% formaldehyde in PBS and permeabilized with 0.25% Triton X-100. Levels of the intracellular HCV NS3 protein or DENV E protein were revealed by sequential incubation with an HCV NS3-specific monoclonal antibody or a monoclonal antibody recognizing the DENV E protein and IRDye 680 goat anti-mouse IgG (LI-COR, Lincoln, NE). The cell viability was determined by DRAQ5 and Sapphire 700 staining. The fluorescence signal intensity was quantified using the LI-COR Odyssey system and normalized to cell viability. (iii) For Tacaribe virus (TCV) (strain TRVL11573; ATCC), virus stock was incubated with or without serial concentrations of the compound chlorin e6 at room temperature for 1 h before being applied to Vero cells for a standard plaque assay. Six days postinfection, plaques were counted after staining of the cells with crystal violet. (iv) For JUNV (strain Romero) and MARV (strain Ci67) virus yield reduction assays, virus stocks were incubated in the presence of serially diluted compound or control medium for 30 min prior to infection. Vero cells were infected for 1 h at a MOI of 1 for MARV and 0.05 for JUNV. Following infection, cells were washed with PBS to remove free virus and compound, and cell culture medium lacking compound was added to cells. Supernatants, harvested after 72 h, were subjected to standard plaque assay using Vero cells. All manipulations of JUNV or MARV were conducted within biosafety level 4 (BSL4) containment facilities located at the U.S. Army Medical Research Institute of Infectious Diseases (Frederick, MD). (v) For HSV, a plaque assay was performed. Briefly, Vero cells (5 × 105 cells/well) were seeded on 6-well plates overnight, followed by infection of HSV-1 (KOS strain) at a MOI of 0.05. Prior to infection, 5 × 104 PFU of HSV-1 was incubated with serial concentrations of chlorin e6 for 20 min at room temperature. After 1 h of infection, 0.5% methylcellulose in the 1% fetal bovine serum (FBS)-containing minimal essential medium (MEM) was added to the cells. Crystal violet staining was performed 2 days thereafter.
Antiviral activity test with nonenveloped viruses.
The antiviral effect of chlorin e6 on nonenveloped viruses was determined. For EMCV (kindly provided by Christoph Seeger, Fox Chase Cancer Center), virus stock was incubated with serially diluted compound at 37°C for 10 min, and then Huh7.5 cells were infected with compound-treated virus at a MOI of 0.01. A cytopathic effect (CPE) assay was performed at 16 h postinfection. To test the virucidal effect of chlorin e6 against adenovirus (Ad), the recombinant virus Ad-GFP (14) was incubated with serial concentrations of chlorin e6 at RT for 10 min, and then HepG2 cells were infected with virus at a titer of 10 viral genome equivalents per cell. Sixteen hours later, the percentage of GFP-positive cells was quantified by Guava flow cytometry (Millipore).
Cytotoxicity assay.
Cells used in each antiviral assay were seeded in 96-well plates and treated with compound at serial concentrations. The compound treatment was performed for the same duration as the antiviral assay. At the end of treatment, cell viability was determined using the CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega) by following the manufacturer's instructions or using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay of viability (20, 25). The CC50 (concentration at which exposure to the compound resulted in a 50% decrease in cell viability relative to results for mock-treated cells) was calculated using the Forecast function of Microsoft Excel.
RESULTS
Chlorophyllide alters HBV virion particle structure.
We initially identified the anti-HBV activity of chlorophyllide by way of a high-throughput screen. Briefly, compounds were tested for the ability to reduce the amount of real-time-PCR-detectable virion DNA that coimmunoprecipitated from the culture medium of producer cells (HepG2.2.15 cells), using antibodies to the viral envelope protein (Lu et al. unpublished). However, reduction in the amount of enveloped HBV DNA appearing in the culture medium could result from the following: (i) inhibiting HBV nucleic acid or protein synthesis, (ii) inhibiting assembly or secretion of virion, or (iii) the destruction of the secreted virion particles in the culture medium. To confirm the results from the high-throughput screening assay and determine more precisely which step(s) in the viral life cycle was sensitive to chlorophyllide treatment, tetracycline-inducible HepDE19 cells, which produce and secrete HBV following tetracycline withdrawal (12), were incubated with various concentrations of chlorophyllide. After 8 days, the amounts of intracellular HBV DNA and extracellular virion particles were examined.
Chlorophyllide was well tolerated by HepDE19 cells, with no detectable effect on cell viability or function at up to 250 μM, resulting in a concentration necessary to achieve 50% cytotoxicity (CC50) of more than 400 μM (data not shown).
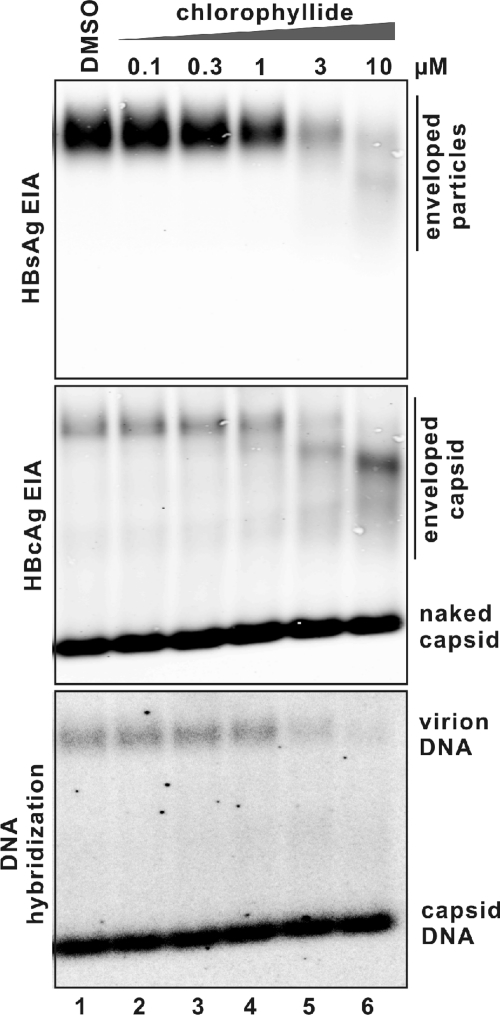
The amount of intact HBV virions was determined by resolving HBV particles present in the culture supernatant through agarose gels, followed by immunostaining for HBV envelope, core, and hybridization to radiolabeled riboprobes for HBV DNA. The mobilities of enveloped particles and nonenveloped capsids (“naked” capsids) are distinct and characteristic in this assay system (12, 21, 32), and the signal intensity can be proportional to the amount of molecule probed.
As Fig. 1 shows, the mobility of envelope particles recovered from the medium of HepDE19 cells incubated with 3 and 10 μM chlorophyllide was much faster and the HBsAg immunostaining intensity was much weaker (Fig. 1, top panel, lanes 5 and 6) than those from control cells (Fig. 1, top panel, lane 1). This implies that the integrity of the enveloped particle was altered intracellularly, extracellularly, or both.
FIG. 1.
Chlorophyllide reduced the amount of intact HBV in the culture medium of producer cells in a dose-dependent manner. HepDE19 cells in a 6-well plate were cultured in tetracycline-free medium and left untreated or treated with the indicated concentration (micromolar) of chlorophyllide. The solvent DMSO was present at 0.1% in every well. Fresh medium with compound was changed every other day. Eight days later, 1 ml of clarified culture fluid was subjected to a particle gel assay as described in Materials and Methods. HBV particles were revealed by enzyme immunoassay (EIA) with anti- HBsAg (top panel) or anti-HBcAg (middle panel) antibodies. HBV particle-associated DNA was hybridized with minus-strand-specific full-length HBV riboprobes (bottom panel). Results represent two experimental trials. No cytoxicity was detectable at any chlorophyllide concentration shown, as determined by MTT assay (see Materials and Methods).
Immunostaining with core-specific antibody revealed two major capsid bands (Fig. 1, middle panel). Based upon mobility, DNA and protein content, and previous work (9, 15), the upper band corresponds to enveloped virions (enveloped capsids), and the lower band (with a much stronger signal) represents the nonenveloped capsids (naked capsid). Naked capsids might be an artifact in the cell culture system, since they are not usually detected in the serum of people or animals chronically infected with HBV (12, 21, 32). Nevertheless, as the results show, the virion-associated capsids migrated at the same position as altered enveloped particles did at higher concentrations (3 and 10 μM) of chlorophyllide incubation (Fig. 1, middle panel, lanes 5 and 6), but the signal intensity was not dramatically changed compared with that for the untreated control or cells treated with a lower concentration of chlorophyllide (Fig. 1, middle panel, lanes 1 to 4). Those results suggest that viral capsids were associated with altered envelope structures. However, the migration and abundance of the naked capsids were not detectably affected at any of the chlorophyllide concentrations tested (Fig. 1, middle panel), which implies that nonenveloped capsids are not sensitive to chlorophyllide.
Virion-associated DNA was detected by hybridization (Fig. 1, bottom panel). As the figure shows, although naked capsid associated DNA was not affected, virion associated DNA was eliminated by chlorophyllide treatment at 3 and 10 μM (Fig. 1, bottom panel, lanes 5 and 6), which suggested that chlorophyllide could selectively affect virion DNA.
Chlorophyllide's ability to reduce the amount of intact enveloped virion structures was dose dependent, as shown in Fig. 1, where a range of chlorophyllide concentrations was used. As little as 3 μM chlorophyllide could reduce the HBV DNA signal by 80% compared with untreated control (Fig. 1, bottom panel, compare lane 5 with lane 1), with complete elimination of signal by 10 μM (Fig. 1, bottom panel, lane 6).
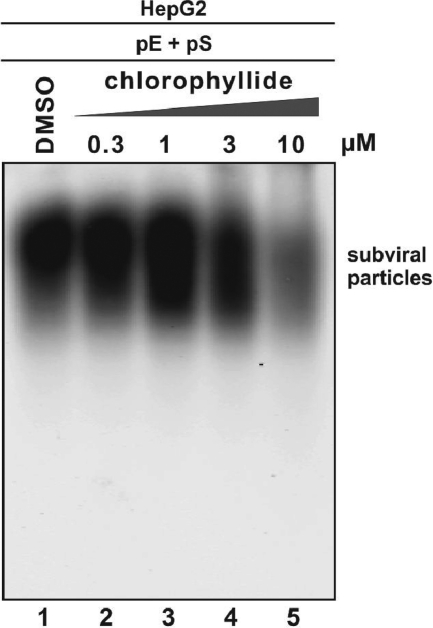
Since the enveloped particle bands showing on the particle gel contain virion particles and subviral particles (2), the data presented above (Fig. 1, top panel) also indicated that chlorophyllide has the ability to affect both populations of HBV enveloped particles. To further support this notion, the compound effect on subviral particles was examined. As shown in Fig. 2, chlorophyllide dose responsively altered the mobility of subviral particles and reduced the HBsAg staining intensity.
FIG. 2.
Chlorophyllide reduced the amount of HBV subviral particles in culture medium. HepG2 cells were transfected with plasmids pE and pS. Compound treatment started at 24 h posttransfection and was repeated every other day. Six days later, culture fluid was subjected to particle gel assay as described in Materials and Methods, and HBV subviral particles were detected by HBsAg EIA.
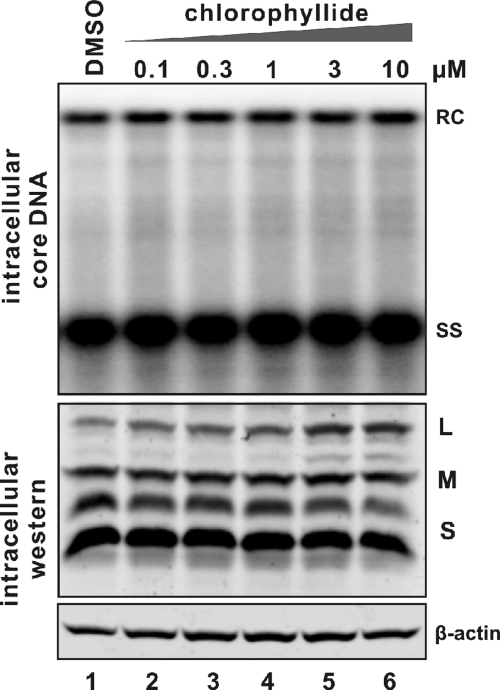
Given chlorophyllide's dramatic effect upon enveloped viral particle structures present in the culture medium, it was of interest to know if intracellular viral DNA or structures had also been affected. The results, as shown in Fig. 3, suggested that the intracellular levels of HBV DNA replication (upper panel) and envelope protein production (bottom panels) did not detectably change following chlorophyllide incubation of cell culture.
FIG. 3.
Intracellular HBV DNA replication and envelope protein expression following cholorophyllide treatment. HepDE19 cell monolayers harvested from the experiment described in the legend to Fig. 1 were processed for intracellular HBV DNA Southern blotting (top panel) and an HBV envelope protein Western blot assay (bottom panel). For DNA analysis, one-half of the HBV capsid-associated DNA extracted from one well of a 6-well plate was loaded. DNA was hybridized with minus-strand-specific full-length HBV riboprobes. Viral relaxed circular (RC) DNA and single strand (SS) were marked. For Western blotting, one-sixth of whole-cell lysates were analyzed for HBV larger (L), middle (M), and small (S) envelope proteins by probing with antibodies against HBsAg. Results represent two experimental trials.
Since chlorophyllide reduced the amount of detectable HBV in the culture medium of HBV-producing cells without detectably affecting intracellular viral DNA, the antiviral mechanism appeared to be due to either (i) an intracellular effect upon nucleocapsid envelopment and/or secretion from the cells or (ii)direct damage to the virion particle in the extracellular medium. The possibility that chlorophyllides directly interacted with virions was examined by incubating cell-free, culture-derived virus with the drug. Briefly, culture fluid from HepDE19 cells cultured under tetracycline-free conditions was collected and incubated with chlorophyllide at the concentration used in Fig. 1, followed by PEG precipitation and particle gel analysis. The results were identical to those of Fig. 1 with concentrations of 3 and 10 μM, with chlorophyllide altering the integrity of virion particle and purging the virion of DNA without affecting the naked capsids and their associated viral DNA (data not shown).
Taken together, these data suggest that chlorophyllide extracellularly disrupts the HBV envelope virion structure, with a loss of viral envelopes and destruction of viral nucleic acid. However, we found that chlorophyllide had no effect on purified viral DNA in FBS-free medium, but purified viral DNA became undetectable in the culture medium containing 10% FBS even without chlorophyllide treatment (data not shown). We thus offer the conclusion that the viral DNA, being exposed after chlorophyllide-mediated virion particle disruption, became degraded by a nuclease(s) in the FBS. The differential effect of chlorophyllide on enveloped capsid DNA and naked capsid DNA may be due to structural differences between these two types of capsid. Since enveloped virions containing DNA appear to be sensitive whereas naked capsid DNA is spared, the primary effects of the drug may be on the envelope, and DNA within the capsids of enveloped particles may be more vulnerable than DNA contained within capsid structures that have not reached the envelopment step yet.
Chemophores of chlorophyllide have antiviral activity.
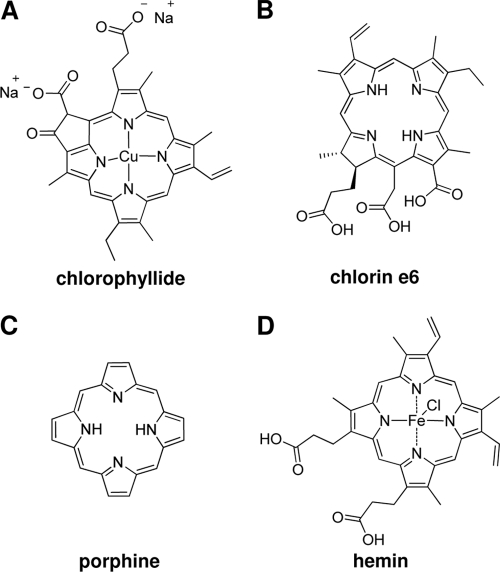
As shown in Fig. 4, chlorophyllide belongs to the family of highly substituted porphyrin analogs, and at neutral pH, it is charged and associated with copper ions chelated in the middle of the porphyrin substructure (Fig. 4A). It was therefore of interest to determine to what extent, if any, these substituents are necessary for antiviral activity. Compounds which vary with respect to key properties of chlorophyllide were selected (see Fig. 4) and were tested for antiviral activity.
FIG. 4.
Structure representations of cholorophyllide, chlorin e6, porphine, and hemin.
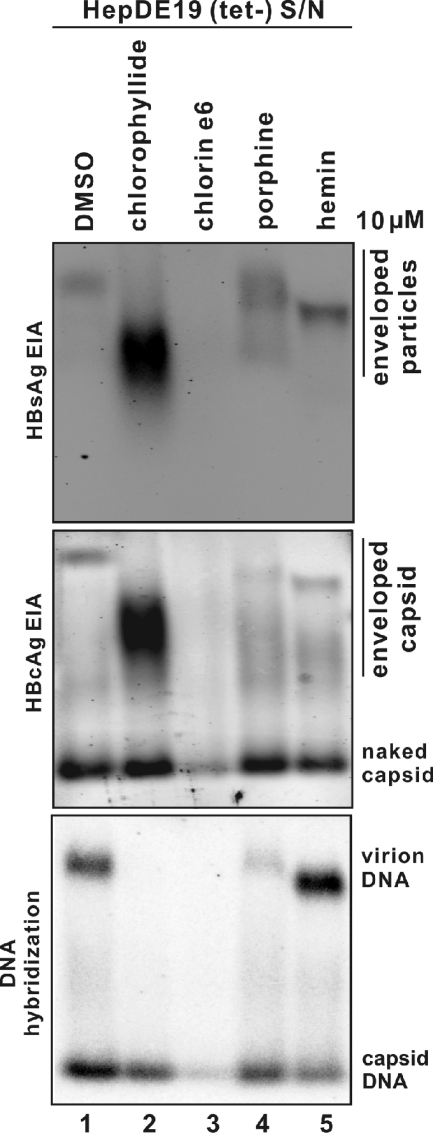
Chlorin e6 (Fig. 4B) retains the key structural features of chlorophyllide (Fig. 4A) but is neither charged nor metal containing. Porphine (Fig. 4C) is strictly a porphyrin, without any substitutions in the core ring structure. Hemin (ferriprotoporphyrin) (Fig. 4D) is chelated with iron rather than copper. These compounds were incubated for 10 min with virus-containing supernatant harvested from HepDE19 cells, and the amount and mobility of viral particles present in the culture medium were analyzed following separation through particle gels.
As shown in Fig. 5, each molecule retains antiviral activity, altering the mobility of enveloped particles and reducing or eliminating the viral DNA from positions associated with enveloped particles compared to results with untreated controls (Fig. 5, lane 1). However, hemin did not cause a reduction in virion DNA, but there was an increased gel mobility of virions (Fig. 5, bottom panel, lane 5), suggesting it might be partially active. Most strikingly, chlorin e6 appeared to have the most dramatic effects, even causing the elimination of all detectable HBV enveloped particles and naked capsids as well as their DNA contents (Fig. 5, lane 3).
FIG. 5.
Impact of chlorophyllide and chemophores upon cell culture-derived HBV particles. One ml of clarified supernatant (S/N) from HepDE19 cells cultured in tetracycline-free medium (days 6 to 8) was incubated at RT for 10 min with 0.1% DMSO or 10 μM (each) indicated compound, followed by PEG precipitation. The pellets were subjected to particle gel assay, and HBV enveloped particles (top panel), capsids (middle panel), and particle-associated DNA (bottom panel) were analyzed as described in the text. Results represent observations conducted with triplicate samples.
The intracellular antiviral effects of these three chlorophyllide derivates were also tested on HepDE19 cells; the results showed that none inhibited intracellular DNA replication and envelope protein expression (data not shown).
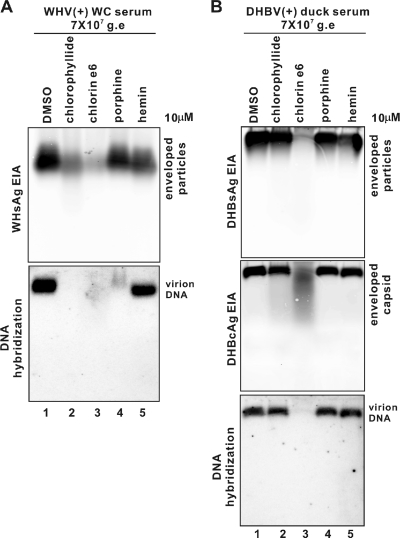
The particle-destroying capabilities of these molecules were also tested on other hepadnaviruses. As Fig. 6 A shows, all chemophores retained activity against woodchuck hepatitis virus (WHV) isolated from animal serum. Chlorophyllide and chlorin e6 dramatically reduced the signals of intact WHV enveloped particles and completely removed virion-associated DNA (Fig. 6A, lanes 2 and 3), with chlorin e6 the most potent. Porphine also greatly reduced the virion DNA level but without changing the gel mobility and intensity of enveloped particles (Fig. 6A, lane 4). Hemin had no ability to affect the level of WHV virion particles and DNA but did slightly increase the mobility of viral particles (Fig. 6A, lane 5).
FIG. 6.
Impact of chlorophyllide and chemophores on animal serum-derived hepadnavirus particles. Sera containing approximately 7 × 107 genome equivalent (g.e) virus particles from WHV-infected woodchuck (WC) or DHBV-infected Peking duck were mixed with 0.1% DMSO or 10 μM (each) indicated compound in 1 ml Opti-MEM medium (Invitrogen) and incubated at RT for 10 min, followed by PEG precipitation. The pellets were subjected to a particle gel assay as described in the text. (A) WHV enveloped particles were probed with a monoclonal antibody against the WHV preS2 peptide (top panel); WHV virion DNA was hybridized with minus-strand-specific full-length WHV riboprobes (bottom panel). (B) DHBV enveloped particles were probed with a monoclonal antibody against the DHBV S protein (top panel), DHBV capsids were probed with a polyclonal antibody (11) against full-length DHBV core protein (middle panel), and DHBV virion DNA was hybridized with minus-strand-specific full-length DHBV riboprobes (bottom panel). Results represent two experimental trials.
Chlorin e6 quantitatively eliminated all detectable DHBV virions derived from the duck serum (Fig. 6B, lane 3), including the three major virion-associated components, envelope (Fig. 6B, top panel, lane 3), enveloped capsid (Fig. 6B, middle panel, lane 3), and virion DNA (Fig. 6B, bottom panel, lane 3). Curiously, however, the other three compounds did not show much activity against DHBV particles (Fig. 6B, lanes 2, 4, and 5) beyond the fact that hemin caused virions to migrate slightly faster (Fig. 6B, lane 5).
The above results indicate that the sensitivity of the hepadnaviruses to these chemophores is not an artifact of the tissue culture environment. Moreover, in the same virus family (29), DHBV, WHV, and HBV hepadnaviruses have different sensitivities to chlorophyllide and its derivates. This could be due to species-specific structural properties among these three viruses (7, 17, 30) or a different serum protein(s) in the samples. Having said that, it is worth emphasizing that all viral species were highly sensitive to chlorin e6.
Given its potency and cross-species activity, our attention turned to chlorin e6, which was well tolerated by hepatocyte-derived cells at concentrations of up to 250 μM, as determined in the cell viability assay (data not shown).
HBV envelope proteins are displaced from the virion but remain intact following porphyrin compound exposure.
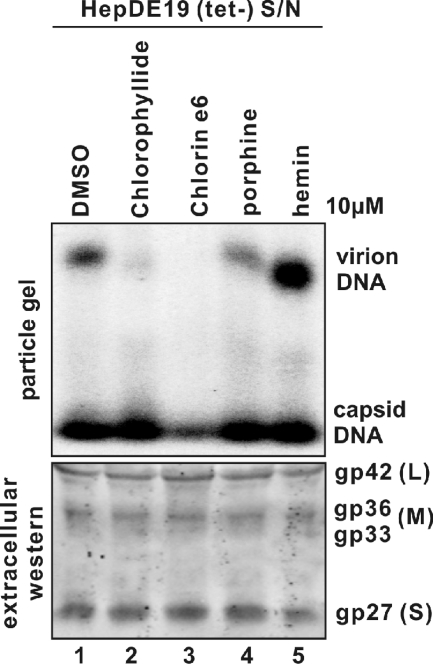
As shown in Fig. 1 and 5, porphyrin-related compounds disrupted the enveloped viral particles, resulting in faster migration of the particles in the particle gel assay. There was a complete loss of envelope protein signal following chlorin e6 incubation (i.e., see Fig. 5, lane 3). The loss of envelope polypeptide signal from the viral particles in the particle gel assay could have either been the result of the degradation of the envelope proteins or of a dissociation of the polypeptides from the structures, causing them to migrate off the particle gel.
Therefore, to determine if HBV envelope proteins remained intact following porphyrin compound treatment, supernatants collected from HepDE19 cells were incubated with each of the four porphyrin compounds. Part of the material was resolved through particle gels for the viral particle assay, and the other part of the material was resolved through a denaturing polyacrylamide gel for detection of envelope protein monomers by analyzing Western blots. The results show that although each of the chlorophyllide compounds affected the mobility of HBV virions in particle gels, with chlorin e6 causing a complete loss of detectable virions (Fig. 7, upper panel), there was no effect on the amount and size of HBs envelope polypeptides, as resolved to monomers in polyacrylamide gels (Fig. 7, lower panel). Thus, the envelope polypeptides themselves remain unaltered.
FIG. 7.
Integrities of HBV envelope proteins following chlorophyllide and chemophores treatment. One milliliter of clarified supernatant (S/N) from HepDE19 cells cultured in tetracycline-free medium (days 6 to 8) was mixed with 0.1% DMSO or 10 μM (each) indicated compound. Ten minutes after incubation at RT, 970 μl of each sample was subjected to particle gel assay, and HBV particle-associated DNA was analyzed by hybridization with minus-strand-specific full-length HBV riboprobes (top panel). Thirty μl of samples were subjected to Western blot analysis (bottom panel), and HBV envelope glycoproteins were probed with antibodies against HBsAg. Results represent observations conducted with triplicate samples.
These data therefore suggest that porphyrin compounds that we have described result in disruption and dissociation of HBV envelope proteins from the enveloped particles without degradation of the envelope polypeptides.
Chlorin e6 is active against other enveloped viruses but less active against nonenveloped viruses.
Since chlorin e6 (and the other chemophores) disrupted intact HBV particles, separating the envelope polypeptides from the virion and causing a loss of viral DNA, it was extremely unlikely that viral infectivity could remain. However, the extent to which infectivity was affected by these compounds was very difficult to assess with the HBV systems available to us. Since chlorin e6 appeared to be acting at the level of the viral envelope, it was also of interest to know if there was selectivity for the viral proteins or if other envelope viruses would be affected.
Thus, the effect of chlorin e6 on infectivity of HCV, DENV-2, HSV-1, HIV-1, MARV, JUNV, and TCRV, all enveloped viruses, was examined. EMCV and adenoviruses were included as examples of nonenveloped viruses.
As summarized in Table 1, all enveloped viruses tested were sensitive to chlorin e6 at concentrations well below those that caused cytotoxicity. HSV was the least sensitive enveloped virus tested, and two nonenveloped viruses, EMCV and adenovirus, were strikingly insensitive to chlorin e6. The sensitivity of dengue virus to these compounds deserves particular mention. At a MOI of 0.02, as little as 0.3 nM chlorin e6 was sufficient to reduce DENV infectivity by 50% in this assay. However, the sensitivity of the virus was dependent on the amount of virus in the inoculum. That is, although the EC50 of chlorin e6 was less than 1 nM for a MOI of 0.02 for DENV-2, the EC50 increased to 22 nM when a MOI of 0.2 was used. This suggests that the compound may be working stoichiometrically, and a threshold amount of chlorin e6 may be needed per virion to achieve a virucidal effect. We note also that increased serum concentrations caused a reduction in the antiviral potency of chlorin e6 against dengue virus, with 10% serum leading to an EC50 of closer to 800 nM for a MOI of 0.2 (data not shown).
TABLE 1.
Antiviral activity of chlorin e6 upon enveloped and nonenveloped viruses
| Virus (MOI) | Cell culture | Antiviral activitya |
CC50 (μM) | SIc | |
|---|---|---|---|---|---|
| EC50 (μM) | EC90 (μM) | ||||
| HIV-1 IIIB | P4-R5 MAGI | 15.9 ± 0.2 | 30.3 ± 0.1 | >100 | >6.3 |
| HIV-1 Bal | P4-R5 MAGI | 9.0 ± 1.0 | 25.9 ± 1.4 | >100 | >11.1 |
| HCV Jc-1 | Huh7.5 | 1.6 | >30 | >100 | >62.5 |
| DENV-2 (0.02) | Huh7.5 | 0.0003 | 0.001 | >100 | >333,333.3 |
| DENV-2 (0.2) | Huh7.5 | 0.022 ± 0.02 | 0.098 ± 0.02 | >100 | >4,545.4 |
| TCRV | Vero | 0.8 ± 0.4 | 4.9 ± 4.5 | >100 | >125 |
| JUNV | Vero | <0.01 | 2.7 | >100 | >10,000 |
| MARV | Vero | 4.5 | 9.6 | >100 | >22.2 |
| HSV-1 | Vero | 20.93 ± 4.07 | 45.33 ± 1.71 | >100 | >4.7 |
| Ad-GFP | HepG2 | 50 | NDb | >100 | >2 |
| EMCV | Huh7.5 | >30 | ND | >100 | ND |
Average EC50s and 90% effective concentrations (EC90s) were obtained from at least triplicate repeats in a single experiment. Standalone values represent a single-experiment determination; standard deviations of EC values are provided for experiments performed multiple times.
ND, not determined.
Selectivity index.
DISCUSSION
We have characterized the antiviral activities of chlorophyllide and several chemophores against HBV in tissue culture systems. These compounds, especially chlorophyllide and chlorin e6, appear to disrupt the viral envelope particles and result in the destruction of the viral nucleic acid. However, viral envelope proteins appear to remain intact despite being stripped from the particles.
There is a small body of information regarding the anti-HBV activity of porphyrins. Lin and Hu reported that heme and a few other related porphyrin compounds could inhibit HBV polymerase binding to viral pregenomic RNA in an in vitro binding assay, but there was no intracellular antiviral activity observed when heme was applied to cell cultures (22). We also found little if any intracellular activity of the porphyrin compounds but observed a direct virucidal activity.
Recently Cheng et al. identified a series of anionic tetraphenyl- porphyrins possessing potent antiviral activities against HCV and HIV but with relatively low potency (EC50 > 10 μM) against HBV intracellular replication (5). However, both studies (5, 22) focused only on the ability of the compounds to affect intracellular HBV replication. Thus, the antiviral effects upon extracellular virus of porphyrin compounds may have been underestimated. It is worth noting that substitutions for the tetraphenylporphyrins are located at the bridge carbon of the core porphyrin ring, while the substitutions for the chlorophyllide analogs are directly attached to the aromatic porphine ring.
The molecular mechanism underlying how porphyrins disrupt the HBV enveloped particles is unknown. The idea that chlorophyllide is acting by physically inactivating virus particles in the extracellular environment rather than at an intracellular synthetic step is supported by the observation that there was little effect on viral DNA replication within the cells at concentrations where the extracellular virions were completely destroyed. Therefore, the evidence suggests that chlorophyllide directly disrupts the virus as a virucidal agent which may interact with specific structures (peptide interface, glycosylation, phosphorylation, etc.) on the viral particle surface (27) or lipids within the envelope (3-4, 26, 31). Since all the viral envelopes have a lipid bilayer basal structure similar to that of the cell membrane (2, 29) and these compounds are well tolerated by cells, it is unlikely that porphyrin compounds disrupt envelope through lipid depletion. What's more, their selectivity for viral envelopes over cellular membranes that may contain viral envelope proteins may be due to a relatively lower density of the viral envelopes on the cellular membrane than on the virion, but this is speculative and more work is needed. Nevertheless, the fact that these compounds can inactivate such a diversity of enveloped viruses, but not nonenveloped viruses, implies that some common structural features presented on the surfaces of viral envelope proteins are the most possible targets.
Chlorin e6 was the most potent compound against all the enveloped viruses tested so far. Micromolar EC50s with a selectivity index of more than 100 were also observed against HBV, JUNV, and Tacaribe virus. HSV was the most refractory, with an EC50 of 20.93 μM. The activity of chlorin e6 against DENV is exceptional, where as little as 0.3 nM reduced 50% of the virus infectivity. Curiously, however, the EC50 of chlorin e6 varied with the MOI, which could indicate that at low concentrations and high MOI the amount of compound is limiting and that chlorin e6 is actually acting stoichiometrically. In this case, the number of chlorin e6 molecules needed to inactivate the DENV would be exceeded by the infectious viral particles at low chlorin e6 concentrations and a high MOI of virus. It is also important to note that the chlorophyllides may become heavily protein bound or otherwise lose activity when present in serum, since we observed a loss of anti-dengue virus activity when serum concentrations were raised (preliminary observations).
Since the chlorophyllides have been known to have antiviral activity for more than 40 years and have been shown to have activity against HIV-1 (6, 24) and more recently to be safe in animals (8, 18-19) and, for chlorophyllin, even to be safe in people at 300 mg/day for 4 months (9, 18), it is an open question as to why they have not been developed further. Perhaps the chlorophyllides have not been used against their most sensitive targets, or perhaps it is the undesirable color residues associated with the charged, copper-containing parent compounds and/or formulation reasons. We have found that the copper-free, and hence less colorimetric, chemophore, chlorin e6, is actually the most potent compound against all the viruses we tested.
The current data, therefore, suggest that these compounds could have utility in the following ways, each of which must be rigorously tested: (i) as antiviral agents in the management of HBV, HCV, DENV, Tacaribe virus, and other viruses for which they retain good selectivity; (ii) as microbicides/virucides; disinfection of surfaces that could be contaminated with infectious agents and could transmit infectious diseases is extremely important for the control of sexually transmitted HIV, HBV, and HCV; the chlorophyllide, or chlorin e6, could be an economical, broadly active microbicide/virucide; and (iii) as postexposure “neutralizations”: for HCV, DENV, and HIV, there are currently no effective gamma globulin postexposure treatments; in the case of DENV, gamma globulin (IgG) specific for the virus might actually exacerbate clinical syndromes associated with the infection (10).
Thus, a small molecule that can safely and effectively inactivate circulating viruses could be of great medical value. Further studies with in vivo settings will be carried out to determine whether these molecules have practical value in the control of infectious agents or merely as research tools in exploring an antiviral agent with novel activity.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI061441) and by the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania. Work in Fred Krebs' laboratory was supported by research development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine. Work in Sina Bavari's laboratory was supported by a grant from the Joint Science and Technology Office Transformational Medical Technologies Initiative (TMTI0048_09_RD_T) and Defense Threat Reduction Agency (4.10022_08_RD_B). Haitao Guo is the Bruce Witte Fellow of the Hepatitis B Foundation.
We also thank William Mason, Christoph Seeger, Volker Bruss, and Pamela Norton for providing important reagents and Erica Litschi for help in manuscript preparation.
Footnotes
Published ahead of print on 6 December 2010.
REFERENCES
- 1.Bleicher, K. H., H. J. Bohm, K. Muller, and A. I. Alanine. 2003. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discov. 2:369-378. [DOI] [PubMed] [Google Scholar]
- 2.Block, T. M., H. Guo, and J. T. Guo. 2007. Molecular virology of hepatitis B virus for clinicians. Clin. Liver Dis. 11:685-706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremer, C. M., C. Bung, N. Kott, M. Hardt, and D. Glebe. 2009. Hepatitis B virus infection is dependent on cholesterol in the viral envelope. Cell. Microbiol. 11:249-260. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, G., et al. 2008. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 105:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Y., et al. 2010. A novel class of meso-tetrakis-porphyrin derivatives exhibits potent activities against hepatitis C virus genotype 1b replicons in vitro. Antimicrob. Agents Chemother. 54:197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeCamp, D. L., et al. 1992. Specific inhibition of HIV-1 protease by boronated porphyrins. J. Med. Chem. 35:3426-3428. [DOI] [PubMed] [Google Scholar]
- 7.Dryden, K. A., et al. 2006. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell 22:843-850. [DOI] [PubMed] [Google Scholar]
- 8.Egner, P. A., A. Munoz, and T. W. Kensler. 2003. Chemoprevention with chlorophyllin in individuals exposed to dietary aflatoxin. Mutat. Res. 523-524:209-216. [DOI] [PubMed] [Google Scholar]
- 9.Egner, P. A., et al. 2001. Chlorophyllin intervention reduces aflatoxin-DNA adducts in individuals at high risk for liver cancer. Proc. Natl. Acad. Sci. U. S. A. 98:14601-14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, S., and A. Rothman. 2006. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 19:429-436. [DOI] [PubMed] [Google Scholar]
- 11.Guo, H., C. E. Aldrich, J. Saputelli, C. Xu, and W. S. Mason. 2006. The insertion domain of the duck hepatitis B virus core protein plays a role in nucleocapsid assembly. Virology 353:443-450. [DOI] [PubMed] [Google Scholar]
- 12.Guo, H., et al. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81:12472-12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, H., R. Mao, T. M. Block, and J. T. Guo. 2010. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol. 84:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J. T., T. Zhou, H. Guo, and T. M. Block. 2007. Alpha interferon-induced antiviral response noncytolytically reduces replication defective adenovirus DNA in MDBK cells. Antiviral Res. 76:232-240. [DOI] [PubMed] [Google Scholar]
- 15.Hoofnagle, J. H., and A. M. di Bisceglie. 1997. The treatment of chronic viral hepatitis. N. Engl. J. Med. 336:347-356. [DOI] [PubMed] [Google Scholar]
- 16.Hoofnagle, J. H., E. Doo, T. J. Liang, R. Fleischer, and A. S. Lok. 2007. Management of hepatitis B: summary of a clinical research workshop. Hepatology 45:1056-1075. [DOI] [PubMed] [Google Scholar]
- 17.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 18.Kensler, T. W., et al. 2004. Chemoprevention of hepatocellular carcinoma in aflatoxin endemic areas. Gastroenterology 127:S310-S318. [DOI] [PubMed] [Google Scholar]
- 19.Kostenich, G. A., I. N. Zhuravkin, and E. A. Zhavrid. 1994. Experimental grounds for using chlorin e6 in the photodynamic therapy of malignant tumors. J. Photochem. Photobiol. B 22:211-217. [DOI] [PubMed] [Google Scholar]
- 20.Krebs, F. C., S. R. Miller, D. Malamud, M. K. Howett, and B. Wigdahl. 1999. Inactivation of human immunodeficiency virus type 1 by nonoxynol-9, C31G, or an alkyl sulfate, sodium dodecyl sulfate. Antiviral Res. 43:157-173. [DOI] [PubMed] [Google Scholar]
- 21.Lenhoff, R. J., and J. Summers. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J. Virol. 68:4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, L., and J. Hu. 2008. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J. Virol. 82:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lok, A. S. 2009. Evolution of nucleoside/tide analogues for hepatitis B: is the ideal drug here yet? J. Hepatol. 51:416-418. [DOI] [PubMed] [Google Scholar]
- 24.Mekler, L. B., A. F. Bychovsky, and B. L. Krikun. 1969. Electron microscope study of the viricidal properties of sodium magnesium-chlorophyllin. Nature 222:574-575. [DOI] [PubMed] [Google Scholar]
- 25.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 26.Moss, B. 2006. Poxvirus entry and membrane fusion. Virology 344:48-54. [DOI] [PubMed] [Google Scholar]
- 27.Rando, R. F., et al. 2006. Critical design features of phenyl carboxylate-containing polymer microbicides. Antimicrob. Agents Chemother. 50:3081-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schormann, W., A. Kraft, D. Ponsel, and V. Bruss. 2006. Hepatitis B virus particle formation in the absence of pregenomic RNA and reverse transcriptase. J. Virol. 80:4187-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seitz, S., S. Urban, C. Antoni, and B. Bottcher. 2007. Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions. EMBO J. 26:4160-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf, M. C., et al. 2010. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U. S. A. 107:3157-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, T., et al. 2006. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 72:116-124. [DOI] [PubMed] [Google Scholar]