Abstract
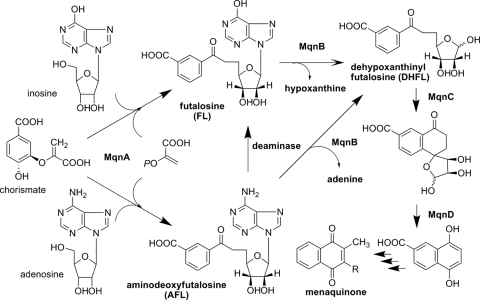
We recently demonstrated that the futalosine pathway was operating in some bacteria for the biosynthesis of menaquinone and that futalosine was converted into dehypoxanthinyl futalosine (DHFL) by an MqnB of Thermus thermophilus. In this study, we found that aminodeoxyfutalosine, which has adenine instead of hypoxanthine in futalosine, was directly converted into DHFL by an MqnB of Helicobacter pylori. Therefore, this step is potentially an attractive target for the development of specific anti-H. pylori drugs.
In prokaryotes, menaquinone (MK) is used for respiration. In Escherichia coli, MK is biosynthesized from chorismate by eight enzymes (4). However, we recently identified an alternative pathway (the futalosine pathway shown in Fig. 1) (2, 3, 5), which operates in bacteria, including some pathogens, such as Helicobacter pylori, Campylobacter jejuni, Chlamydia trachomatis, and Leptospira borgpetersenii. Disruption of this pathway in Streptomyces coelicolor has been shown to lead to the inhibition of bacteriostatic growth. We also showed that the Mqn gene-disrupted S. coelicolor mutants required more than 100 μg/ml menaquinone for their growth. Because we do not consume a diet containing this much menaquinone, the essential amount of MK is not supplied from our diet if the futalosine pathway is blocked. Moreover, humans and commensal intestinal bacteria, including lactobacilli, lack the futalosine pathway. Taking these results together, we think that the futalosine pathway is a potential attractive target for the development of specific anti-H. pylori drugs. However, the details of each of the biosynthetic steps in the futalosine pathway remain unclear. In this study, we investigated the second step of the futalosine pathway, which is the conversion of futalosine (FL) into dehypoxanthinyl futalosine (DHFL) by MqnB (futalosine hydrolase [EC 3.2.2.26]) (3), in Acidothermus cellulolyticus, H. pylori, and S. coelicolor. We found that aminodeoxyfutalosine (AFL), which has adenine instead of hypoxanthine in FL, was an intermediate in these microorganisms.
FIG. 1.
An alternative menaquinone biosynthetic pathway (the futalosine pathway). In A. cellulolyticus and S. coelicolor, dehypoxanthinyl futalosine (DHFL) was perhaps formed from aminodeoxyfutalosine (AFL) via futalosine (FL). In contrast, AFL was directly converted into DHFL by MqnB in H. pylori.
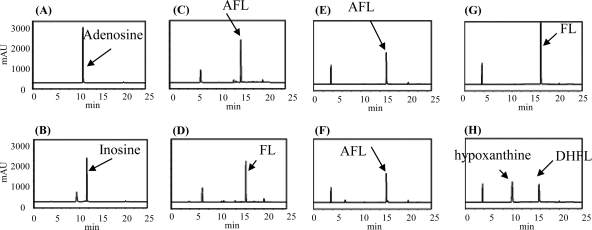
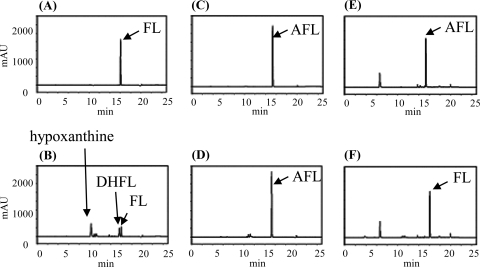
To ascertain the details of each step in the futalosine pathway, we selected the second step catalyzed by MqnB, since FL, the substrate of MqnB, was the only compound that we could readily prepare from the culture broth of the MqnB-disrupted mutant (2). We first investigated the distribution of the futalosine pathway and the organization of the Mqn genes in microorganisms whose genomes had been completely sequenced. In almost all microorganisms, the Mqn genes were scattered throughout the genome. However, in A. cellulolyticus, a thermophilic actinobacterium, the Mqn genes, and other menaquinone-related genes, such as prenylation and methylation genes, were clustered in two loci (1). One cluster contained Acel_0105 and Acel_0106 genes, which encoded orthologs of MqnC and MqnB, respectively. The other was composed of 11 genes (Acel_0255 to Acel_0266) encoding MqnA (Acel_0261) and MqnC (Acel_0263) orthologs. To determine whether Acel_0106 did indeed encode MqnB, we prepared the recombinant Acel_0106 enzyme (see Fig. S1 in the supplemental material) and incubated it with FL. As expected, we detected DHFL under standard conditions (Fig. 2 H). Next, we focused on an adenosine deaminase ortholog (Acel_0264) in one of the clusters. Taking the fact that biosynthetically related genes are usually clustered in microorganism genomes into consideration, we assumed that AFL (Fig. 1) may be formed by MqnA and then converted into FL by Acel_0264. To examine this possibility, we prepared recombinant Acel_0264 (see Fig. S1 in the supplemental material) and incubated it with chemically synthesized AFL (see Fig. S2 in the supplemental material). As shown in Fig. 2D, the enzyme converted AFL (retention time of 15.5 min) into FL (retention time of 16 min). We also examined whether MqnB (Acel_0106) could directly use AFL as a substrate. However, we did not detect the formation of DHFL (Fig. 2F), despite the fact that AFL and FL have very similar structures. This very narrow substrate specificity was in agreement with our previous study with the recombinant MqnB enzyme from Thermus thermophilus (TTHA0556). Of the nucleotide-related compounds examined, the enzyme reacted only with FL (3). Moreover, in this study, we confirmed that recombinant TTHA0556 did not accept AFL as a substrate (Fig. 3 D). Judging from the genome database of T. thermophilus, this species may not have any adenosine deaminases. Therefore, it was quite reasonable that TTHA0556 accepted only FL as a substrate.
FIG. 2.
HPLC analysis of the reaction product without enzymes (A, C, E, and G) and with recombinant Acel_0264 (B and D) and Acel_0106 (F and H). Adenosine (A and B) (positive control), AFL (C, D, E, and F), and FL (G and H) were used as substrates. Time (in minutes) is shown on the x axes, and optical density (in milliabsorbance units [mAU]) is shown on the y axes.
FIG. 3.
HPLC analysis of the reaction product without enzymes (A and C) and with recombinant TTHA0556 (B and D). FL (A and B) and AFL (C and D) were used as the substrates.
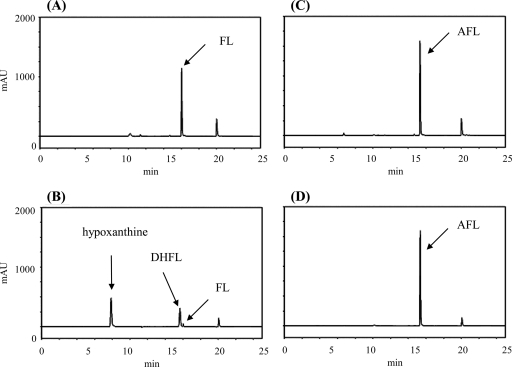
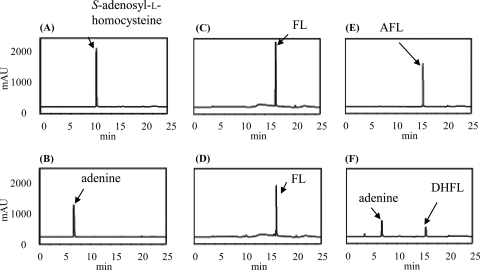
One of our long-term goals is to develop anti-H. pylori drugs by inhibiting the enzymes involved in biosynthesis in the futalosine pathway. To accomplish this, knowledge of the detailed properties of these enzymes will be indispensable. Therefore, we tried to prepare a recombinant enzyme of the MqnB ortholog in H. pylori. We searched for the MqnB ortholog in the genome database of H. pylori strain 26695 with Acel_0106 as the query (6). However, we did not find a typical ortholog with a significant estimated value (E value), although the strain had an MqnC ortholog (HP0656; E value of 4e−81) and an MqnD ortholog (HP0152; E value of 3e−16). Other H. pylori strains, such as J99, HPAG1, Shi470, and G27, had similar results. The most similar ortholog found in the genome of strain 26695 was HP0089 (E value of 0.18), which was annotated as 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (EC 3.2.2.9). Therefore, we prepared a recombinant enzyme of HP0089 (see Fig. S1 in the supplemental material) and incubated it with FL; however, the enzyme did not act on FL (Fig. 4 D). In addition to MqnB, H. pylori strains did not possess typical MqnA orthologs. An orthology search using K07081, which is an ortholog of MqnA, indicated that HP0778 may be MqnA, but it had a low similarity to SCO4506, an MqnA ortholog from S. coelicolor (E value of 7e−4). Taking these results together, we assumed that DHFL might be formed via another route in H. pylori strains. One possibility is a route via AFL, since it may be an intermediate in A. cellulolyticus as described above. Therefore, recombinant HP0089 was incubated with AFL, and the reaction product was analyzed by high-performance liquid chromatography (HPLC). As shown in Fig. 4F, AFL was consumed, and two products were detected. The products were confirmed to be adenine and DHFL by liquid chromatography (LC)-mass spectrometry (MS) analysis (see Fig. S3 in the supplemental material). This result showed that AFL was directly converted into DHFL. This direct conversion without deamination was in agreement with the observation that strain 26695 did not have an ortholog of Acel_0264 (AFL deaminase) (6).
FIG. 4.
HPLC analysis of the reaction product without enzymes (A, C, and E) and with recombinant HP0089 (B, D, and F). S-Adenosyl-l-homocysteine (A and B) (positive control), FL (C and D), and AFL (E and F) were used as the substrates.
We previously suggested that SCO4327 from S. coelicolor would be an MqnB ortholog, since a SCO4327 disruptant accumulated FL in the culture broth. However, we did not detect FL hydrolase activity in an in vitro assay using SCO4327 recombinant enzyme and FL (2). As described above, H. pylori strains used AFL as a substrate of MqnB. Therefore, we tested whether S. coelicolor also uses AFL instead of FL. The recombinant enzyme was incubated with AFL, and the reaction product was analyzed by HPLC. However, we still did not detect the formation of DHFL. During the investigation of why the recombinant SCO4327 did not have the predicted enzyme activity, we found that the N-terminal region of the annotated SCO4327 was shorter than those of other MqnB orthologs. We identified an additional start codon that was located 78 nucleotides upstream of the annotated start codon (see Fig. S4 in the supplemental material). The recombinant enzyme with the newly identified start codon was expressed as a maltose binding protein fusion protein (see Fig. S1 in the supplemental material) and incubated with either AFL or FL. As shown in Fig. 5 B and D, DHFL was detected only with FL as the substrate in a manner similar to that with TTHA0556. Next, we examined whether AFL was involved in MK biosynthesis in S. coelicolor. The strain possessed three probable AFL deaminase genes, SCO5662 (E value of 4e−28 with Acel_0264 as the query), SCO2546 (E value of 7e−27), and SCO4644 (E value of 5e−22). Of these three genes, SCO5662 was able to be expressed in a soluble form (Fig. S1) and used for an enzyme assay. As shown in Fig. 5F, the formation of FL was detected.
FIG. 5.
HPLC analysis of the reaction product without enzymes (A, C, and E) and with recombinant SCO4327 (B and D) and SCO5662 (F). FL (A and B) and AFL (C, D, E, and F) were used for the substrates.
In summary, three routes to the formation of DHFL in the futalosine pathway were suggested (Fig. 1). FL may have been directly formed by MqnA in T. thermophilus and converted into DHFL. In A. cellulolyticus, S. coelicolor, and H. pylori, AFL was formed by MqnA. In A. cellulolyticus and S. coelicolor, AFL was converted to FL by the deaminases and then to DHFL. In contrast, AFL was directly converted to DHFL in H. pylori. Therefore, the MqnB in H. pylori is an attractive target for the development of specific anti-H. pylori drugs, since humans and commensal intestinal bacteria, including lactobacilli, lack the alternative pathway. To draw a conclusion, however, detailed studies of the reactions yielding FL and AFL catalyzed by MqnA are essential.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Asahi Glass Foundation, Noda Institute for Scientific Research, and Institute for Fermentation, Osaka, Japan (IFO) to T. Dairi.
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Barabote, R. D., et al. 2009. Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 19:1033-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiratsuka, T., et al. 2008. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321:1670-1673. [DOI] [PubMed] [Google Scholar]
- 3.Hiratsuka, T., N. Itoh, H. Seto, and T. Dairi. 2009. Enzymatic properties of futalosine hydrolase, an enzyme essential for a newly identified menaquinone biosynthetic pathway. Biosci. Biotechnol. Biochem. 73:1137-1141. [DOI] [PubMed] [Google Scholar]
- 4.Meganathan, R. 2001. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitam. Horm. 61:173-218. [DOI] [PubMed] [Google Scholar]
- 5.Seto, H., et al. 2008. Studies on a new biosynthetic pathway for menaquinone. J. Am. Chem. Soc. 130:5614-5615. [DOI] [PubMed] [Google Scholar]
- 6.Tomb, J. F., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.