Abstract
Carbapenem-resistant Klebsiella pneumoniae has spread worldwide and throughout the United States. Colistin is used extensively to treat infections with this organism. We describe a cluster of colistin-resistant, carbapenem-resistant K. pneumoniae infection cases involving three institutions in Detroit, MI. A cluster of five cases of colistin-resistant, carbapenem-resistant K. pneumoniae was identified at Detroit Medical Center (DMC) from 27 July to 22 August 2009. Epidemiologic data were collected, and transmission opportunities were analyzed. Isolates were genotyped by using pulsed-field gel electrophoresis and repetitive extragenic palindromic PCR. Data regarding the use of colistin were obtained from pharmacy records. The index case of colistin-resistant, carbapenem-resistant K. pneumoniae was followed 20 days later by four additional cases occurring in a 6-day interval. All of the patients, at some point, had stayed at one particular institution. The mean number of opportunities for transmission between patients was 2.3 ± 0.5, and each patient had at least one opportunity for transmission with one of the other patients. Compared to 60 colistin-susceptible, carbapenem-resistant K. pneumoniae controls isolated in the previous year at DMC, case patients were significantly older (P = 0.05) and the carbapenem-resistant K. pneumoniae organisms isolated from them displayed much higher MICs to imipenem (P < 0.001). Colistin use was not enhanced in the months preceding the outbreak. Genotyping revealed two closely related clones. This report of a colistin-resistant, carbapenem-resistant K. pneumoniae outbreak is strongly linked to patient-to-patient transmission. Controlling the spread and novel emergence of bacteria with this phenotype is of paramount importance.
Carbapenem-resistant members of the family Enterobacteriaceae have spread across the world and within the United States (2, 5, 15). In the United States, carbapenem-resistant Klebsiella pneumoniae constitutes 92% of all carbapenem-resistant Enterobacteriaceae and carbapenemase production mediated by blaKPC is the most prevalent mechanism conferring resistance to carbapenems (5). Carbapenem-resistant Enterobacteriaceae are coresistant to almost all classes of antimicrobials (29), and infections are associated with extremely high rates of morbidity and mortality and high costs (30). Colistimethate sodium, i.e., colistin, is one of the few remaining therapeutic options available to treat infections due to carbapenem-resistant Enterobacteriaceae. Despite the toxicity of this relatively old agent, colistin is frequently used to treat infections due to carbapenem-resistant Enterobacteriaceae because (i) therapeutic alternatives are not yet available and (ii) colistin has a long history and track record in the treatment of invasive infections due to Gram-negative bacilli (28, 31, 35). In most cases, colistin is the last viable effective option for the treatment of invasive bloodstream infections due to carbapenem-resistant Enterobacteriaceae.
In the past few years, there have been sporadic reports of colistin-resistant, carbapenem-resistant Enterobacteriaceae cases from various parts of the world, e.g., Greece (2, 37), Israel (28), South Korea (32), and Singapore (33). In Athens, Greece, an outbreak during the years 2004 and 2005 involving 13 patients in a single hospital unit was reported (2). Colistin-resistant, carbapenem-resistant Enterobacteriaceae cases have never been reported from the United States. There have been case reports of polymyxin B-resistant, carbapenem-resistant Enterobacteriaceae from the New York City area (usually, resistance to polymyxin B confers resistance to polymyxin E, i.e., colistin, and vice versa), but these events remain sporadic and rare, with no outbreaks reported thus far (4, 8, 17). The mechanism(s) of colistin resistance in carbapenem-resistant Enterobacteriaceae is not yet known. The genetic basis of colistin resistance in Acinetobacter baumannii is being investigated (1, 13, 18), but it remains unclear whether the same mechanism(s) is associated with colistin resistance in carbapenem-resistant Enterobacteriaceae. Additionally, the microbiological definitions of colistin resistance are also not well established, as formal Clinical and Laboratory Standards Institute (CLSI) criteria and breakpoints have not been established (7). Lastly, the clinical significance of colistin resistance remains to be defined.
We describe here the first cluster, as far as we know, of colistin-resistant, carbapenem-resistant K. pneumoniae in the United States, which involved two hospitals and a long-term acute-care (LTAC) facility, in the summer of 2009. The epidemiologic and molecular outbreak investigations that followed are detailed.
MATERIALS AND METHODS
Study design and setting.
This outbreak investigation and case-cohort study were conducted in the Detroit Medical Center (DMC) health care system, which consists of eight hospitals and more than 2,000 inpatient beds. The outbreak involved two of these hospitals; both are university hospitals located in Detroit. Hospital A is a 360-bed hospital, and hospital B is a 570-bed hospital. An LTAC facility is housed within one of the study hospitals (hospital A) but is not part of the DMC network. This LTAC facility has 35 beds and is located inside the main inpatient building. Although the LTAC facility is administratively separate from the inpatient hospital, patients from this LTAC facility use, on a routine basis, multiple services of this particular DMC facility, such as radiology, operating rooms, the emergency room, dialysis, respiratory therapy, and laboratory services.
The first part of the outbreak investigation was conducted prospectively during an infection control investigation. Molecular analyses were conducted retrospectively. This study was approved by the Institutional Review Boards at DMC and Wayne State University.
Patients and variables.
Patients were included in the outbreak investigations if they had one or more clinical isolates of K. pneumoniae from 30 July to 22 August 2009 that were resistant to both imipenem and colistin. Imipenem resistance was defined as an MIC of ≥8 μg/ml, and colistin resistance was defined as an MIC of ≥4 μg/ml (7, 11).
Patients were included in the case-cohort study if they had one or more clinical isolates of carbapenem-resistant K. pneumoniae either colistin susceptible or colistin resistant at DMC from 1 September 2008 to 22 August 2009 (from the time testing for carbapenem-resistant Enterobacteriaceae was initiated at DMC to the last day of the outbreak). A cumulative case-cohort analysis was conducted comparing the colistin-resistant, carbapenem-resistant K. pneumoniae case patients (from the outbreak investigation) to the colistin-susceptible, carbapenem-resistant K. pneumoniae controls.
The epidemiologic data collected included demographics, admission source, comorbid conditions, severity-of-illness indices, hospital exposures and procedures, and in-hospital mortality. In addition, opportunities for transmission between patients were analyzed in order to quantify the number of encounters and investigate the possible role of patient-to-patient transmission in the outbreak (19). An opportunity for transmission was defined as two case patients residing at the same hospital ward/unit on the same day before the isolation of colistin-resistant, carbapenem-resistant Enterobacteriaceae from one or both of the patients. Transmission opportunities for each patient were analyzed for the 3 months prior to the initial isolation of colistin-resistant, carbapenem-resistant K. pneumoniae to 2 weeks after the final positive culture.
Cocolonization with non-glucose-fermenting Gram-negative bacilli (such as A. baumannii and Pseudomonas aeruginosa) was assessed in order to examine the possible role of gene transfer from nonfermenters to K. pneumoniae conferring high levels of carbapenem resistance and colistin resistance. Cocolonization was defined as isolation of A. baumannii or P. aeruginosa from a clinical culture on the same day as isolation of the colistin-resistant, carbapenem-resistant K. pneumoniae strain or during the period from 1 week before to 1 week after colistin-resistant, carbapenem-resistant K. pneumoniae isolation.
Data regarding the use of colistin during the outbreak and in the preceding 18 months were obtained from pharmacy records. Colistin defined daily doses (DDDs) were based on the WHO-recommended daily dose of 3 million U/day (i.e., 90 mg colistin base activity) (3, 20, 27, 34).
Microbiology and molecular analyses.
Identification of bacteria and determination of susceptibility to carbapenems (imipenem and meropenem) were initially done by using an automated broth microdilution system (MicroScan, Renton, WA). A modified Hodge test was conducted to assess carbapenemase production according to CLSI criteria (7). Etests (AB Biodisk, Solna, Sweden) were used to determine susceptibility to colistin and determine the MIC to imipenem (the automated panel tests concentrations of 4 and 8 μg/ml only). Etests were conducted according to packet insert instructions, using Mueller-Hinton agar with an inoculum determined by a turbidity of 0.5 in a 0.85% NaCl suspension. The MIC endpoints were defined at the zone of complete inhibition of growth. Quality control tests were performed according to FDA package insert for imipenem Etest strips and included the following organisms and MIC ranges for imipenem: Staphylococcus aureus ATCC 29213, 0.016 to 0.064 μg/ml; Enterococcus faecalis ATCC 29212, 0.5 to 2.0 μg/ml; Escherichia coli ATCC 25922, 0.064 to 0.25 μg/ml; P. aeruginosa ATCC 27853, 1.0 to 4.0 μg/ml. Resistance to colistin was defined as an MIC of ≥4 μg/ml (11).
The presence of a blaKPC gene was determined by PCR as follows. Bacterial DNA was obtained by suspending two to three colonies of each isolate grown on MacConkey agar plates in 500 μl of nuclease-free water (UBS Corporation, Cleveland, OH) and heating them at 90°C for 10 min. Samples were spun at 10,000 rpm for 10 min, and the resulting supernatant was diluted 1:10 with nuclease-free water. Amplification reactions were performed using high-fidelity rTth DNA Polymerase, XL (GeneAmp XL PCR kit; Applied Biosystems, Foster City, CA). The temperature cycling conditions included an initial denaturation at 94°C for 1 min, followed by 30 cycles of 94°C for 30 s, annealing for 30 s at 59°C, and extension at 72°C for 30 s. Cycling was followed by a final extension at 72°C for 10 min. Samples were incubated in the PTC-225 Peltier thermal cycler (MJ Research, Waltham, MA). Each reaction mixture (5 μl) containing the PCR product was mixed with 2 μl of UltraClean gel dye (Mo Bio Laboratories Inc., Carlsbad, CA) and analyzed by electrophoresis in a 1% ultrapure molecular biology grade agarose gel (USB Corporation). For these experiments, previously characterized KPC-producing K. pneumoniae isolates were used as controls (9, 10).
The expression level of K. pneumoniae carbapenemase (KPC) β-lactamase was determined by immunoblotting according to the following procedure. Rabbit polyclonal antibodies were raised against the KPC-2 β-lactamase in order to measure blaKPC expression. The blaKPC-2 gene was expressed from pBR322-catI-blaKPC-2 in E. coli DH10B. The KPC-2 β-lactamase was isolated and purified (24). Anti-KPC-2 polyclonal antibodies were raised by Sigma-Genosys (The Woodlands, TX) and isolated from serum using protein G column purification (Sigma-Genosys) (24). KPC-producing K. pneumoniae isolates were grown in Luria-Bertani broth to an optical density at 600 nm of 0.8. Thirty microliters of each culture was mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, boiled, subjected to electrophoresis, and transferred to polyvinylidene difluoride membranes (Invitrogen, Carlsbad, CA) (14). After overnight incubation in blocking buffer (5% bovine serum albumin, 20 mM Tris HCl-buffered 150 mM saline, pH 7.5 [Sigma, St. Louis, MO]), the presence of KPC β-lactamase was detected by addition of 0.5 μg/ml anti-KPC antibody and incubation for 3 h at room temperature. Membranes were then washed four times, 10 min each, with Tris-buffered saline (pH 7.5) containing 0.05% Tween and further incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated protein G (Bio-Rad, Hercules, CA) in blocking buffer. After an additional four washes, the membranes were processed for film exposure using the Amersham ECL Plus Western blotting detection system (GE Healthcare, Buckinghamshire, United Kingdom) according to the protocol provided. For these experiments, 200 ng of purified KPC was used as a positive control.
Surveillance cultures for carbapenem-resistant Enterobacteriaceae were obtained during and following the outbreak. Rectal cultures were obtained and processed according to the CDC laboratory protocol published in 2009 (6).
In order to asses clonality, pulsed-field gel electrophoresis (PFGE) was conducted. Bacterial DNA was prepared and cleaved with 20 U SpeI endonuclease (New England Biolabs, Boston, MA). Electrophoresis was performed in a 1% agarose gel (BMA Products, Rockland, ME) prepared and run in 0.5× Tris-borate-EDTA buffer on a CHEF-DR III apparatus (Bio-Rad Laboratories, Richmond, CA). The initial switch time was 10 s, the final switch time was 45 s, and the run time was 23 h at 6 V/cm. Gels were then stained in ethidium bromide, destained in distilled water, and then photographed (GelDoc 2000; Bio-Rad) (16). GelCompar software was used to construct a dendrogram.
As an additional method to determine genetic relatedness among strains, repetitive extragenic palindromic PCR (Rep-PCR) using a semiautomated system (DiversiLab; bioMérieux, Marcy l'Etoile, France) was applied. Genomic DNA was extracted using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). PCR amplification of highly conserved, repetitive DNA sequences occupying distinct positions in the bacterial genome was performed using the DiversiLab Klebsiella kit. Rep-PCR products were separated by electrophoresis using a microfluidics chip. Analysis with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) yielded a profile of bands for each strain. Band patterns were compared with the modified Kullback-Leibler method in order to generate a dendrogram, using the DiversiLab software. As in previous studies, isolates with band patterns that were ≥95% similar were considered genetically related (10). Results of Rep-PCR typing were compared with those of previously characterized KPC-producing K. pneumoniae isolates (9, 10).
Statistical analysis.
All analyses were performed using SPSS software (SPSS, Chicago, IL, 2009). Fisher's exact test was used for the analysis of categorical variables, and the independent-sample t test or the Wilcoxon rank-sum test was used for the analysis of continuous variables. P values of ≤0.05 were considered significant. All P values were two sided.
RESULTS
The first culture of colistin-resistant, carbapenem-resistant K. pneumoniae was obtained on 27 July 2009, followed by four additional cases occurring between 16 and 22 August 2009. All isolations were from clinical cultures; none were from surveillance cultures. Table 1 depicts the patients' characteristics and outcomes. In general, patients were elderly, frail individuals with multiple complicated comorbid conditions who had numerous exposures to environments and settings where nosocomial infections can occur. Three of the patients reside permanently in nursing homes (different ones), and two reside in the LTAC facility attached to hospital A. In addition to the antimicrobials displayed in Table 1, all of the colistin-resistant, carbapenem-resistant K. pneumoniae strains were resistant to meropenem, ceftazidime, amikacin, and ciprofloxacin, while two out of five were susceptible to tigecycline (MIC = 2 μg/ml for both) and one was susceptible to tobramycin.
TABLE 1.
Clinical characteristics of patients with colistin-resistant, carbapenem-resistant K. pneumoniae, Detroit, MI, 27 July to 22 August 2009
| Patient code | Age (yr) | Gender | Comorbid conditions | Admission source | Functional status at the time of admission | Days in hospital prior to isolation of CREm | Anatomic site of isolation | Primary clinical infectious syndrome | Associated bacteremia | MIC (μg/ml) |
LOSn | Discharge disposition | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Colistin | ||||||||||||
| 1 | 86 | Female | Colon cancer, HTN,a CRF,b dementia, stage IV pressure ulcers, recurrent UTIc | Nursing home | Bedridden | 9 | Urine | Colonization | No | >32 | 16 | 18 | Alive |
| 2 | 77 | Female | Parkinson's disease, HTN, DM,d dementia, dyslipidemia, stage IV pressure ulcers | LTAC | Partially dependent | 1 | Wound | Colonization | No | >32 | 8 | 39 | Dead |
| 3 | 76 | Male | CHF,e HTN, DM, COPD,f dementia, gastrostomy, stage IV pressure ulcers, CRF, pulmonary HTN | LTAC | Bedridden | 1 | Urine | UTI | No | >32 | 64 | 21 | Dead |
| 4 | 68 | Female | ESRF,g DM, AKA,h HTN, IHD,i CHF, DVT,j CVA,k hemiplegia, eye enucleation | Nursing home | Partially dependent | 11 | Tissue of legs' infected wound | Deep soft-tissue infection | Yes | >32 | 8 | 14 | Alive |
| 5 | 76 | Female | HTN, CHF, dyslipidemia, DM, neuropathy, morbid obesity, stage IV pressure ulcers, OA,l COPD, gastrostomy | Nursing home | Bedridden | 33 | Blood | Deep soft-tissue infection, UTI | Yes | >32 | 8 | 71 | Alive |
HTN, hypertension.
CRF, chronic renal failure.
UTI, urinary tract infection.
DM, diabetes mellitus.
CHF, congestive heart failure.
COPD, chronic obstructive pulmonary disease.
ESRF, end-stage renal failure.
AKA, above-knee amputation.
IHD, ischemic heart disease.
DVT, deep-vein thrombosis.
CVA, cerebrovascular attack.
OA, osteoarthritis.
CRE, carbapenem-resistant Enterobacteriaceae.
LOS, length of hospital stay.
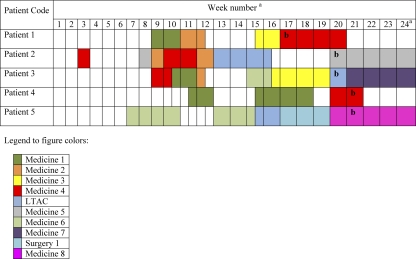
Isolates were from hospitals A and B and from one LTAC facility (Fig. 1). The index patient (patient 1, Fig. 1) was identified in hospital A on outbreak day 0 (week 17 in Fig. 1). Twenty days later (on outbreak day 20), patient 2 was identified at hospital B and patient 3 was identified at the LTAC facility. On day 25 and day 26, patients 4 and 5 were identified at hospital A. All of the patients, at some point, had stayed at hospital A, and patients 2, 3, and 5 had also stayed in the LTAC facility attached to hospital A. The mean number of opportunities for transmission between patients was 2.3 ± 0.5, and each patient had at least one opportunity for transmission involving one of the other patients. Figure 1 represents the time line and transmission opportunities for the patients. The order in which patients are presented does not necessarily reflect the true chronological sequence in which patients actually acquired the organism but is based on temporal isolation of the organism from a clinical culture.
FIG. 1.
Time line and transmission opportunities among patients during the outbreak. Each row represents a patient, and each column represents a week. The colors represent the different units/wards as presented in the legend at the bottom. A transmission opportunity was deemed to have occurred if two patients were in the same ward during the same time period. For example, patients 1 and 2 had a transmission opportunity during week 12 in the Medicine 2 ward. Superscript a: week1, 29 March to 4 April 2009; week 24, 5 to 11 September 2009. Superscript b: date of clinical culture.
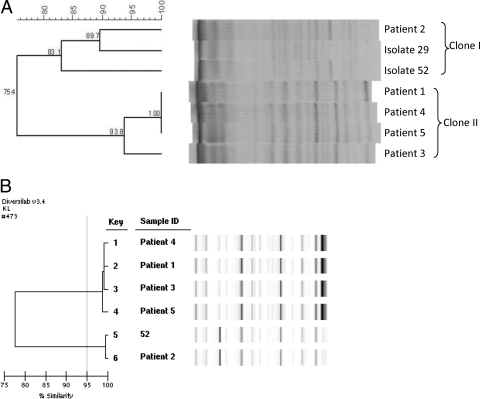
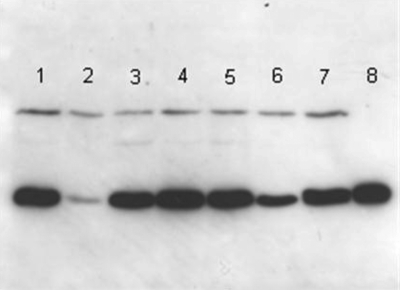
The PFGE and Rep-PCR analysis results are displayed in Fig. 2 A and B, respectively. Both methods reveal that there are two clones (I and II). Four of the five colistin-resistant, carbapenem-resistant K. pneumoniae isolates belonged to a single clone designated clone II (Fig. 2A). This clone consisted of isolates 1, 3, 4, and 5 from Table 1 and Fig. 1. There was a second clone of carbapenem-resistant K. pneumoniae at DMC, designated clone I (Fig. 2A), which included the fifth colistin-resistant case (case 2 in Table 1 and Fig. 1), and “representative” colistin-susceptible, carbapenem-resistant K. pneumoniae from DMC (isolates 29 and 52, Fig. 2A). Immunoblotting revealed that there is increased expression of KPC β-lactamase in clone II compared to that in clone I (Fig. 3).
FIG. 2.
Dendrograms of carbapenem-resistant Klebsiella species strains, DMC, 2009. (A) Dendrogram showing the relatedness of colistin-resistant KPC-producing K. pneumoniae based on PFGE patterns after digestion with XbaI. Isolates showing 80% similarity are considered related. (B) Dendrogram showing the relatedness of colistin-resistant isolates based on Rep-PCR. Isolates 95% similar are considered related. According to both methods, isolates from patients 1, 3, 4, and 5 are related and represent a single clone. Isolates 29 and 52 are representative colistin-susceptible, KPC-producing K. pneumoniae strains isolated at DMC during the outbreak period.
FIG. 3.
Immunoblot analysis of KPC production by colistin- and carbapenem-resistant K. pneumoniae isolates. Lanes 1 to 5 are the colistin-resistant isolates (MIC, >32 μg/ml for patients 1 through 5). Lanes 6 and 7 are two representative strains of colistin-susceptible, carbapenem-resistant (MIC = 2 μg/ml for both) K. pneumonia (isolates 29 and 52) isolated at DMC during the outbreak period. Lane 8 represents the activity of 200 ng of purified KPC beta-lactamase. Comparison of the blots gives an indication of the relative levels of KPC production.
Once the cluster was recognized, infection control practices were enforced at all three study institutions. Patients were subjected to strict contact precautions, in single-occupancy rooms with dedicated staff, and infection control preventionists educated health care workers regarding the importance of hand hygiene and strict adherence to precautions. Infection control interventions were implemented, including active surveillance culturing (rectal) of all patients who were exposed to case patients and/or were in the same patient care unit as case patients. Overall, 112 surveillance rectal cultures were obtained and none revealed colistin-resistant, carbapenem-resistant K. pneumoniae. All patients admitted from LTAC facilities to a DMC facility were empirically subjected to contact precautions, and surveillance cultures for carbapenem-resistant Enterobacteriaceae were obtained. At the LTAC facility, infection control interventions included active surveillance of all new admissions to the LTAC facility, cohorting of patients who were culture positive for carbapenem-resistant Enterobacteriaceae, and initiation of daily baths with chlorhexidine (2%) for all LTAC facility patients. Additional colistin-resistant, carbapenem-resistant Enterobacteriaceae cases were not reported from DMC hospitals in the subsequent months.
When the 5 colistin-resistant, carbapenem-resistant K. pneumoniae case patients were compared to 60 patients with colistin-susceptible, carbapenem-resistant K. pneumoniae, the MIC to imipenem was significantly higher for case patients (a median of 64 μg/ml versus a median of 2 μg/ml; P < 0.001 between groups). The mean age of case patients was greater than the mean age of patients with colistin-susceptible, carbapenem-resistant K. pneumoniae (77 ± 6 years and 62 ± 8 years, respectively, P = 0.05). The mortality rate and length of hospital stay were higher among case patients than among controls, though these differences did not reach statistical significance (mortality rates were 40% and 26% for case patients and controls, respectively, and the mean lengths of hospital stay were 33 ± 23 days and 30 ± 23 days, respectively; P > 0.05 for both comparisons). All five case patients had cocolonization with a nonfermenter; three were cocolonized with P. aeruginosa, and two were cocolonized with A. baumannii. In contrast, only 23 (38%) of 60 controls were cocolonized with a nonfermenter (odds ratio, 2.44; 95% confidence interval, 1.8 to 3.3; P = 0.01). The two cocolonizing strains of A. baumannii were not resistant to colistin, and the three P. aeruginosa isolates were not tested for colistin susceptibility. All five nonfermenters had MICs of >32 μg/ml to imipenem.
Colistin use during the 3-month period prior to the outbreak (1 April to 30 June 2009) did not differ from that in the previous quarter (1 January to 31 March 2009) (mean DDD, 20.7 ± 3.4/1,000 patient days for the entire 18 months studied). None of the five patients received colistin in the 3 months preceding their colistin-resistant, carbapenem-resistant K. pneumoniae isolation. Polymyxin B had rarely been used at DMC in the past 4 years.
DISCUSSION
This report of a colistin-resistant, carbapenem-resistant K. pneumoniae outbreak is notable for four reasons. First, as far as we know, it is the first report from the United States that describes an outbreak involving colistin-resistant, carbapenem-resistant K. pneumoniae. There have been previous case reports of infections of carbapenem-resistant K. pneumoniae resistant to polymyxin B in New York (4, 8, 17), but these cases occurred sporadically and not as part of an outbreak. Second, three different institutions are involved. Previous reports of outbreaks from other parts of the world involved only single institutions (2, 28, 37). Third, patient-to-patient transmission seems like the major mechanism instead of emergence of resistance. Finally, the MICs to imipenem were particularly high among these isolates (all were >32 μg/ml). Most carbapenem-resistant K. pneumoniae strains within our health system and elsewhere usually display an MIC of 1 to 4 μg/ml to imipenem according to automated systems. Although it is unclear whether the high level of resistance present in this outbreak is associated with increased resistance to other categories of antimicrobials, this issue is plausible and troubling. Based on pharmacokinetic synergy studies, infections caused by isolates that are nonsusceptible to carbapenems (MICs ranging between 4 and 8 μg/ml) can be treated with higher doses of carbapenem (21, 25), but a high level of resistance might not, therefore, be a major therapeutic issue.
Colistin is the mainstay of therapy for carbapenem-resistant Enterobacteriaceae and is often the last available active antimicrobial agent (12). Although we were unable to demonstrate an association between colistin use and colistin resistance, other investigators have demonstrated an association between the use of colistin and the emergence of resistance in carbapenem-resistant K. pneumoniae, though results are conflicting (2, 17, 37). The pharmacologic characteristics of colistin are still poorly understood. CLSI has not published susceptibility breakpoints for this agent against Enterobacteriaceae, and pharmacokinetic characteristics are controversial and not clearly defined (13, 26). The emergence of resistance to colistin is extremely troubling, particularly since colistin resistance might be associated with poor outcomes (37). Since optimal dosing regimens have not been determined for colistin, there is a great risk that inappropriate dosing might lead to continued spread of colistin resistance among carbapenem-resistant K. pneumoniae strains and other pathogens (36).
Patient-to-patient transmission appeared to play an important role in this outbreak. Hospital A, where all of the patients resided during the study period, was the location where the most transmission opportunities occurred. The outbreak terminated when strict infection control practices were implemented at all three health care institutions, including active surveillance and chlorhexidine bathing of residents at the LTAC facility. LTAC facilities care for patients with severe illnesses, admit patients from a variety of different hospitals, and are an important reservoir of resistant organisms (9, 22, 23). LTAC facilities have been implicated in outbreaks of multidrug-resistant (MDR) Gram-negative bacilli (9). Interventions to limit the emergence and spread of carbapenem-resistant Enterobacteriaceae in hospitals should include referring LTAC facilities.
Another intriguing characteristic of this outbreak was that all five case patients were cocolonized with A. baumannii or P. aeruginosa, both Gram-negative bacilli which are non-lactose fermenters. High-level resistance to carbapenems and colistin is more common in these pathogens than in Enterobacteriaceae. The mechanisms of colistin resistance are not well described in nonfermenters or Enterobacteriaceae. While it is possible that genetic resistance determinants might have passed from nonfermenters to the outbreak strains, in vitro evidence to date does not support such a mechanism of transfer (1). A stronger hypothesis is that the association between cocolonization with nonfermenters and colistin-resistant, carbapenem-resistant K. pneumoniae is that the patient populations colonized or infected by nonfermenters share the same risk factors as patients colonized by or infected with MDR Enterobacteriaceae. Unfortunately, the nonfermenter strains were unavailable for molecular analysis. It seems advisable to separate patients colonized by or infected with carbapenem-resistant Enterobacteriaceae from patients with Gram-negative non-lactose fermenters. Further analysis of the impact of infection control interventions on the spread of genetic resistance determinants is warranted.
Ongoing studies further elucidating the mechanisms involved in colistin resistance and high-level carbapenem resistance among carbapenem-resistant K. pneumoniae strains are in progress. Infection control and antimicrobial stewardship interventions geared toward preventing the dissemination of these essentially untreatable pathogens and their associated resistance determinants are needed.
Acknowledgments
We have no conflicts of interest to declare relating to the content of this paper.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Adams, M. D., et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniadou, A., et al. 2007. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J. Antimicrob. Chemother. 59:786-790. [DOI] [PubMed] [Google Scholar]
- 3.Bergen, P. J., et al. 2008. Comparison of once-, twice- and thrice-daily dosing of colistin on antibacterial effect and emergence of resistance: studies with Pseudomonas aeruginosa in an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 61:636-642. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., et al. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128-132. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb. Mortal. Wkly. Rep. 58:256-260. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2009. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing, [sic] Klebsiella spp. and E. coli from rectal swabs. Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement. Approved standard M100-S19 Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Elemam, A., J. Rahimian, and W. Mandell. 2009. Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin. Infect. Dis. 49:271-274. [DOI] [PubMed] [Google Scholar]
- 9.Endimiani, A., et al. 2009. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J. Antimicrob. Chemother. 64:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endimiani, A., et al. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern U. S. A. J. Antimicrob. Chemother. 63:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing. 2009. Clinical breakpoints. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden.
- 12.Giamarellou, H., and G. Poulakou. 2009. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs 69:1879-1901. [DOI] [PubMed] [Google Scholar]
- 13.Gilad, J., and Y. Carmeli. 2008. Treatment options for multidrug-resistant Acinetobacter species. Drugs 68:165-189. [DOI] [PubMed] [Google Scholar]
- 14.Hujer, A. M., C. R. Bethel, and R. A. Bonomo. 2004. Antibody mapping of the linear epitopes of CMY-2 and SHV-1 beta-lactamases. Antimicrob. Agents Chemother. 48:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitchel, B., et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leavitt, A., S. Navon-Venezia, I. Chmelnitsky, M. J. Schwaber, and Y. Carmeli. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J., G. Patel, S. Huprikar, D. P. Calfee, and S. G. Jenkins. 2009. Decreased susceptibility to polymyxin B during treatment for carbapenem-resistant Klebsiella pneumoniae infection. J. Clin. Microbiol. 47:1611-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, J., et al. 2007. Antibiograms of multidrug-resistant clinical Acinetobacter baumannii: promising therapeutic options for treatment of infection with colistin-resistant strains. Clin. Infect. Dis. 45:594-598. [DOI] [PubMed] [Google Scholar]
- 19.Marchaim, D., et al. 2007. Molecular and epidemiologic study of polyclonal outbreaks of multidrug-resistant Acinetobacter baumannii infection in an Israeli hospital. Infect. Control Hosp. Epidemiol. 28:945-950. [DOI] [PubMed] [Google Scholar]
- 20.Markou, N., et al. 2008. Colistin serum concentrations after intravenous administration in critically ill patients with serious multidrug-resistant, Gram-negative bacilli [sic] infections: a prospective, open-label, uncontrolled study. Clin. Ther. 30:143-151. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, S. J., and J. W. Lancaster. 2009. Doripenem monohydrate, a broad-spectrum carbapenem antibiotic. Clin. Ther. 31:42-63. [DOI] [PubMed] [Google Scholar]
- 22.Munoz-Price, L. S. 2009. Long-term acute care hospitals. Clin. Infect. Dis. 49:438-443. [DOI] [PubMed] [Google Scholar]
- 23.Munoz-Price, L. S., et al. 2010. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae at a long-term acute care hospital. Infect. Control Hosp. Epidemiol. 31:341-347. [DOI] [PubMed] [Google Scholar]
- 24.Papp-Wallace, K. M., et al. 2010. Inhibitor resistance in the KPC-2 beta-lactamase, a preeminent property of this class A beta-lactamase. Antimicrob. Agents Chemother. 54:890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasteran, F., T. Mendez, L. Guerriero, M. Rapoport, and A. Corso. 2009. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J. Clin. Microbiol. 47:1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plachouras, D., et al. 2009. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by gram-negative bacteria. Antimicrob. Agents Chemother. 53:3430-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samra, Z., O. Ofir, Y. Lishtzinsky, L. Madar-Shapiro, and J. Bishara. 2007. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int. J. Antimicrob. Agents 30:525-529. [DOI] [PubMed] [Google Scholar]
- 29.Schwaber, M. J., and Y. Carmeli. 2008. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA 300:2911-2913. [DOI] [PubMed] [Google Scholar]
- 30.Schwaber, M. J., et al. 2008. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob. Agents Chemother. 52:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souli, M., et al. 2010. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin. Infect. Dis. 50:364-373. [DOI] [PubMed] [Google Scholar]
- 32.Suh, J. Y., et al. 2010. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob. Agents Chemother. 54:560-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, T. Y., and S. Y. Ng. 2006. The in-vitro activity of colistin in gram-negative bacteria. Singapore Med. J. 47:621-624. [PubMed] [Google Scholar]
- 34.Wallace, S. J., et al. 2008. Subacute toxicity of colistin methanesulfonate in rats: comparison of various intravenous dosage regimens. Antimicrob. Agents Chemother. 52:1159-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodford, N., et al. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62:1261-1264. [DOI] [PubMed] [Google Scholar]
- 36.Yau, W., et al. 2009. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the western Pacific region in the SENTRY antimicrobial surveillance programme. J. Infect. 58:138-144. [DOI] [PubMed] [Google Scholar]
- 37.Zarkotou, O., et al. 2010. Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. J. Clin. Microbiol. 48:2271-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]