Abstract
A PCR-sequencing assay was evaluated for direct detection of mutations in the quinolone resistance-determining region (QRDR) of gyrase A (gyrA) gene in fluoroquinolone-resistant Mycobacterium tuberculosis in respiratory specimens. As determined by gyrA QRDR analysis, complete concordance of genotypic and phenotypic fluoroquinolone resistance was demonstrated. Our results indicate that the assay is a rapid and reliable method for the diagnosis of fluoroquinolone-resistant tuberculosis, facilitating timely clinical management and public health control. Using the assay, we detected a novel gyrA Ala74Ser mutation in M. tuberculosis directly from sputum specimens. The functional effect of the Ala74Ser mutant was verified through the study of the DNA supercoiling inhibitory activity of fluoroquinolones against the recombinant gyrase. The drug-mediated gyrase-DNA cleavage complex model suggests perturbation of the gyrA-gyrA dimer interface caused by the Ala74Ser mutation probably disturbs the putative quinolone binding pocket and leads to the reduction of the drug binding affinity. A number of gyrA mutations (Glu21Gln, Ser95Thr, and Gly668Asp) were also characterized to be natural polymorphisms not associated with fluoroquinolone resistance.
Mycobacterium tuberculosis is the etiological agent of tuberculosis (TB), and it is estimated that about one-third of the global population is infected with this bacillus (51). Multidrug-resistant TB (MDR-TB), generally defined as disease with bacillary resistance to at least both isoniazid and rifampin, is a major threat to public health and the control of TB. It has been estimated that 400,000 new cases of MDR-TB emerged in 2006 (51). Extensively drug-resistant TB (XDR-TB) is caused by an MDR strain that is resistant to any fluoroquinolone (FQ) and at least one of the three second-line injectable drugs, namely, amikacin, capreomycin, or kanamycin (45). XDR-TB has also been reported worldwide (16, 50). Between 2000 and 2004, the global prevalence of XDR-TB among MDR-TB cases was 6.6% (16).
DNA gyrase is a unique type II topoisomerase present in M. tuberculosis, and the enzyme is thus the sole topoisomerase target for quinolones in M. tuberculosis (9). DNA gyrase is a tetrameric A2B2 protein. The A subunit carries the active site for double-stranded DNA breakage and reunion, whereas the B subunit promotes ATP hydrolysis (10). The A and B subunits are encoded by gyrase A (gyrA) and gyrase B (gyrB) genes, respectively. Mutations in the quinolone resistance-determining region (QRDR) of gyrA, mainly clustered in codons 90, 91, and 94, are the most important mechanism conferring FQ resistance (FQr) in M. tuberculosis (5, 31, 39, 43, 44). In the numbering system used for M. tuberculosis, the QRDR of gyrA extends from amino acid residues 74 to 113 (13, 14).
Detection of drug resistance in M. tuberculosis has traditionally been accomplished by time-consuming culture-based methods. Mycobacterial growth on culture requires 3 to 8 weeks, followed by an additional 2 to 3 weeks before anti-TB drug susceptibility testing result is available (8). Previous studies have demonstrated that molecular tests on resistance genes (e.g., rifampin resistance-determining region PCR-sequencing assay for rpoB gene and multiplex allele-specific [MAS]-PCRs for katG and mabA genes) can facilitate the rapid diagnosis of MDR-TB (21, 23, 24, 47).
FQs are generally regarded as having a pivotal role among the agents in multidrug regimens against MDR M. tuberculosis (48). The World Health Organization currently recommends inclusion of an FQ in standardized regimens for the treatment of MDR-TB (22). Resistance to FQs has been shown to portend an adverse treatment outcome in MDR-TB (22). Since the aminoglycosides or capreomycin also have potent anti-TB activity, the development of acquired resistance of M. tuberculosis to these second-line agents through their suboptimal use, in addition to bacillary resistance to the FQs, would result in XDR-TB with very poor prognosis (3, 28). Thus, early diagnosis of FQr MDR-TB and XDR-TB has predictive value on the management outcome of these complicated scenarios and could guide therapy accordingly (21).
In the present study, we developed a gyrA QRDR PCR-sequencing assay for rapid diagnosis of FQr TB. A total of 71 archived nonduplicate clinical isolates of M. tuberculosis and 535 respiratory specimens were used to validate the assay. A rare gyrA mutation at position 74 with a replacement of Ala by Ser was detected in sputum specimens collected from a patient suffering from pulmonary TB. Subsequent culture also revealed that the corresponding M. tuberculosis isolate harboring the gyrA Ala74Ser mutation was FQr. At present, no report has clearly defined the impact of the gyrA Ala74Ser mutation on drug binding affinity in the formation of bacteriostatic quinolone-gyrase-DNA complexes and the subsequent lethal activity of nongrowing M. tuberculosis cells. In the present study, the wild-type (WT) H37Rv gyrA, mutant gyrA bearing the Ala74Ser mutation, and WT H37Rv gyrB were overexpressed as His-tagged proteins in Escherichia coli. Using a DNA supercoiling assay, the concentrations of ofloxacin (OFX) and moxifloxacin (MXF) that inhibited DNA supercoiling by 50% (IC50s) were compared against the purified WT H37Rv gyrase complex and the purified gyrase complex reconstituted with gyrA Ala74Ser. A gyrA-MXF-DNA cleavage complex structure was presented accordingly, and the ternary complex structure of Ala74Ser conferring FQr is discussed in light of its drug-binding affinity.
MATERIALS AND METHODS
Clinical isolates and respiratory specimens.
Between September 2003 and March 2007, 71 clinical isolates of M. tuberculosis, 21 of which were FQr and 50 of which were FQ susceptible (FQs), were collected from patients in Queen Mary Hospital and Grantham Hospital in Hong Kong, China.
In addition, a total of 535 respiratory specimens (475 expectorated sputum, 34 bronchoalveolar lavage specimens, 16 bronchial aspirates, and 10 endotracheal aspirates) were collected from 407 in-patients with chest symptoms and/or chest radiographic infiltrates of undetermined origin between March 2008 and April 2009. After a direct smear for acid-fast bacilli (AFB), the respiratory specimens were decontaminated and concentrated as described previously (30). The digested sediments were divided equally for mycobacterial culture and subsequent PCR assays. Cultures positive for AFB were identified as described previously (46, 49).
Anti-TB drug susceptibility testing.
According to protocol M24-A (Clinical and Laboratory Standards Institute; formerly, the National Committee for Clinical and Laboratory Standards) guidelines, anti-TB drug susceptibility testing for clinical isolates were determined by 1% standard proportion method on Middlebrook 7H10 agar (Becton Dickinson) supplemented with 10% oleic acid-albumin-dextrose catalase (OADC enrichment; Becton Dickinson). MICs were defined as the lowest concentration of the antibiotics that inhibited 99% of the bacterial growth (8).
PCR for M. tuberculosis IS6110.
Genomic M. tuberculosis DNA in clinical isolates and respiratory specimens was extracted by using a Roche COBAS Amplicor extraction kit (Roche Diagnostics) as previously described (47). As an additional step, 90-μl portions of the DNA extracts of all direct respiratory specimens were cleaned up and concentrated by using a QIAquick PCR purification kit (Qiagen, Germany) according to the manufacturer's instructions, with 30 μl of eluted DNA (3-fold-concentrated extracts). Portions (5 μl) of DNA extract were used per 50-μl reaction mixtures of IS6110 one-tube nested PCR, gyrA QRDR PCR for direct sequencing, and PCRs for the construction of gyrA and gyrB protein expression vectors. The manual one-tube nested PCR for IS6110 was performed as described previously (4, 46, 47, 49).
Direct PCR and DNA sequencing for QRDR of gyrA.
The gyrA QRDR PCR procedure was essentially adapted from the work of Takiff et al. (40). The primers used to amplify the gyrA QRDR fragment were derived from nucleotide positions 78 to 97 (5′-CAGCTACATCGACTATGCGA-3′) and 397 to 379 (5′-GGGCTTCGGTGTACCTCAT-3′). With modification, the PCR mixture consisted of 1× PCR Buffer II (Applied Biosystems), 1.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate (dNTP), 15 pmol of each primer, and 1 U of AmpliTaq Gold polymerase (Applied Biosystems). AmpliTaq Gold polymerase in the master mix was first activated by incubation at 94°C for 12 min. The reaction mixture was then subjected to 45 cycles of amplification (denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min); this was followed by a final extension step for 10 min at 72°C. The amplicon size was 320 bp.
DNA sequencing (ABI Prism 3700 Genetic Analyzer; Applied Biosystems) was performed to investigatie point mutations in the QRDR of gyrA. The DNA sequences were assembled and edited by using BioEdit software version 7.0.9.0. The genetic polymorphisms of gyrA were compared to the complete nucleotide sequence of M. tuberculosis strain H37Rv in GenBank accession number NC_000962 (9).
DNA sequencing for gyrB-gyrA contig.
Complete sequence analysis of 2,517-bp gyrA and 2,145-bp gyrB genes of WT H37Rv strain, five randomly selected FQs clinical strains, and the FQr clinical strain harboring the gyrA Ala74Ser mutation were performed by DNA sequencing using the oligonucleotide primers shown in Table S1 in the supplemental material.
Construction of gyrA expression vectors.
The gyrA expression vectors of the WT H37Rv strain, the FQs clinical strain, and the Ala74Ser variant were constructed by using a Champion pET Directional TOPO expression kit (K101-01/D-TOPO; Invitrogen). Blunt-end PCR products, including the entire gyrA genes, were amplified with a forward primer (5′-CACCATGACAGACACGACGTTG-3′; the overhang sequence CACC before the ATG initiation codon of gyrA gene is underlined) and a reverse primer (5′-ATTGCCCGTCTGGTCTGCG-3′). The PCR mixture consisted of 1× PfuTurbo Cx reaction buffer (Stratagene), 0.2 mM concentrations of each dNTP, 100 ng of each primer, and 2.5 U of PfuTurbo Cx hotstart DNA polymerase (Stratagene). The gyrA genes were amplified for 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 1 min, with a final extension step for 10 min at 68°C. The blunt-end PCR products corresponding to 2.5-kb gyrA fragment were ligated into the linearized pET101/D-TOPO protein expression vector (Invitrogen) and then transformed into OneShot TOP10 E. coli (Invitrogen) by heat shock. Recombinant clones were selected from the resistant colonies growing on Luria-Bertani (LB) agar containing ampicillin (100 μg/ml).
Construction of gyrB expression vector.
The protocol for construction of the entire gyrB gene expression vector of WT H37Rv strain was essentially adapted from the work of Aubry et al. (1).
Verifying the fidelity of the gyrA and gyrB clones.
The fidelity of the gyrA and gyrB clones was verified by DNA sequencing using the T7 promoter primer (5′-TAATACGACTCACTATAGGG-3′), the T7 terminator primer (5′-GCTAGTTATTGCTCAGCGG-3′), and the oligonucleotide primers listed in Table S1 in the supplemental material.
Protein overexpression and purification of gyrA and gyrB subunits.
The pET101/D-TOPO plasmids carrying the gyrA gene and the pET-15b plasmid carrying the gyrB gene were separately transformed into BL21 Star (DE3) One Shot E. coli (Invitrogen) by heat shock. The recombinant His-tagged gyrA and gyrB proteins were separately overexpressed and purified by the same procedure. The BL21 clones were incubated with shaking in LB medium containing ampicillin (100 μg/ml) at 37°C until reaching log-phase growth. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to the culture at a final concentration of 1 mM to induce protein synthesis. After incubation at 30°C with shaking for 4 h, the cells were harvested by centrifugation at 5,000 × g at 4°C for 20 min and disrupted by sonication in prechilled binding buffer A (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 20 mM imidazole).
HisTrap HP 1-ml column (GE Healthcare/Amersham Biosciences) was connected to an ÄKTA Explorer 10 chromatography system (GE Healthcare/Amersham Biosciences) according to the manufacturer's instructions. Filtered and degassed protein sample in buffer A was pumped through the HisTrap HP column at 4°C for overnight. After protein binding, the column was washed with filtered and degassed buffer A at 4°C. A filtered, degassed, and prechilled elution buffer B (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 500 mM imidazole) gradient from 0 to 100% was used for the elution of protein. Elution peaks of gyrA and gyrB protein elutes were collected. The combined elution fractions was spun at 15,000 × g at 4°C for 30 min, and then the supernatant was dialyzed overnight at 4°C against 2.5 liters of 50 mM Tris-HCl (pH 7.9), 1 mM dithiothreitol, 1 mM EDTA, 50 mM l-arginine, 50 mM l-glutamic acid, and 30% glycerol. The protein concentrations were measured with a NanoDrop ND-1000 spectrophotometer. The proteins fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie brilliant blue staining.
DNA supercoiling assay.
DNA supercoiling assay was performed as described previously by Aubry et al. (1). The reaction mixture (30 μl) contained M. tuberculosis gyrase supercoiling assay buffer (40 mM Tris-HCl [pH 7.5], 25 mM KCl, 6 mM magnesium acetate, 2 mM spermidine, 4 mM dithiothreitol, 0.1 mg of tRNA/ml, 0.36 mg of bovine serum albumin/ml, 100 mM potassium glutamate, 1 mM ATP; pH 8.0 [Inspiralis, United Kingdom]), 0.5 μg of relaxed pBR322 plasmid DNA (Inspiralis) as the substrate, and purified recombinant M. tuberculosis gyrA and gyrB proteins. The reaction mixture was incubated at 37°C for 1 h. The reaction was terminated by the addition of 50% glycerol containing 0.25% bromophenol blue. The reaction mixture was resolved by 1% agarose gel electrophoresis for 6 h at 50 V, and then the agarose gel was stained with ethidium bromide (0.7 μg/ml). DNA supercoiling activity was tested with various ratios of purified gyrA and gyrB subunits. One unit of enzyme activity was defined as the amount of DNA gyrase that converted 0.5 μg of relaxed pBR322 plasmid DNA to supercoiled form in 1 h at 37°C. The inhibitory effect of OFX and MXF on DNA gyrase was assessed by determining the concentrations of drug that reduced the supercoiling activity of 2 U of gyrase complex by 50% (IC50). The intensity of DNA bands was quantified using ImageJ software version 1.42q.
RESULTS
Sensitivities and specificities of the IS6110 and the gyrA QRDR PCR assays for the detection of M. tuberculosis in respiratory specimens.
For the 535 respiratory specimens (Table 1), 140 (26.2%) specimens from 99 patients were culture positive for M. tuberculosis; among these, 124 (23.2%) were AFB smear positive. Of 395 specimens culture negative for M. tuberculosis, 12 were AFB smear negative but positive for both PCR assays. Retrospective investigation indicated that these 12 specimens were collected from 10 patients confirmed as culture-positive pulmonary TB within the last 2 months, and they were responding to anti-TB therapy. Both PCR assays exhibited a 100% specificity with no false-positive results toward 38 specimens growing nontuberculous mycobacteria (NTM) and 72 specimens growing other respiratory bacterial pathogens. Using culture as the gold standard, the overall sensitivities of IS6110 and gyrA PCR assays were 95.7% (134/140) and 82.9% (116/140), respectively. For AFB smear-positive specimens, IS6110 and gyrA PCR assays could attain higher sensitivities of 98.4% (122/124) and 87.1% (108/124), respectively.
TABLE 1.
Comparative sensitivities and specificities of IS6110 and gyrA PCR assays for direct detection of M. tuberculosis in respiratory specimens
| Respiratory specimens (no. of samples) | PCR for: |
|
|---|---|---|
| IS6110 | gyrA | |
| Total (535)a | ||
| M. tuberculosis culture positive (140) | ||
| AFB smear positive (124) | ||
| 108 | + | + |
| 14 | + | N |
| 2 | Ne | N |
| AFB smear negative (16) | ||
| 8 | + | + |
| 4 | + | N |
| 4 | N | N |
| NTM culture positive (38)b | N | N |
| Culture negative for Mycobacterium spp. (357) | ||
| AFB smear negative | ||
| 345c | N | N |
| 12d | + | + |
A total of 535 respiratory specimens were obtained from 407 patients. Overall, 140 specimens from 99 patients were M. tuberculosis culture positive.
NTM, nontuberculous mycobacteria, including M. avium-M. intracellulare complex (n = 18), M. chelonei (n = 15), M. scrofulaceum (n = 1), and M. kansasii (n = 4).
Including 72 specimens culture positive for Streptococcus pneumoniae (n = 6), Haemophilus influenzae (n = 16), Pseudomonas aeruginosa (n = 33), Staphylococcus aureus (n = 12), Moraxella catarrhalis (n = 1), Nocardia spp. (n = 1), Aspergillus spp. (n = 2), and Penicillium spp. (n = 1).
Specimens were obtained from 10 patients confirmed within the last 2 months as having culture-positive pulmonary TB, and these patients were responding to anti-TB therapy.
N, negative.
Anti-TB drug susceptibilities and gyrA mutations.
In Table 1, the 140 M. tuberculosis isolates from respiratory specimens were collected from 99 patients. Further susceptibility testing of total 170 nonduplicate M. tuberculosis isolates (including 71 archived strains) revealed 28 FQr and 142 FQs M. tuberculosis strains (Table 2). For the 28 FQr M. tuberculosis strains, 14.3% (4/28) showed resistance to merely FQs, whereas 67.9% (19/28) were identified as MDR-TB strains (resistant to both isoniazid and rifampin). Of these 19 MDR-TB strains, 8 were also resistant to at least one of the injectable drugs amikacin, capreomycin, or kanamycin, fulfilling the definition of XDR-TB. Overall, 28.6% (8/28) of the FQr strains were identified as XDR-TB strains.
TABLE 2.
Comparative interpretation of gyrA PCR-sequencing assay and conventional FQ susceptibility testing of 170 nonduplicate M. tuberculosis isolatesa
| M. tuberculosis isolates (n) | No. of FQr strains according to CLSI guidance | MIC in μg/ml (relative MIC)b |
|
|---|---|---|---|
| OFX | MXF | ||
| gyrA mutation absent (142) | 0 | <2 | <1 |
| gyrA mutation present (28) | 25c | 8 (16) | 4 (16) |
| 1d | 16 (32) | 8 (32) | |
| 2e | 16 (32) | 4 (16) | |
Isolates include 71 archived strains and strains collected from respiratory specimens in 99 nonduplicate patients (i.e., Table 1).
The relative MIC (given in parentheses) is the MIC ratio against the WT H37Rv reference strain.
gyrA mutation patterns: Ala74Ser (n = 1), Gly88Cys (n = 1), Ala90Val (n = 5), Ser91Pro (n = 2), Asp94Gly (n = 13), and Asp94Ala (n = 3).
Asp94Asn (n = 1).
Asp94His (n = 1), Asp94Tyr (n = 1).
The 320-bp amplicons of gyrA PCR-positive respiratory specimens (Table 1), as well as 170 clinical isolates of M. tuberculosis (Table 2), were analyzed by DNA sequencing. Amplicons of respiratory specimens exhibited sequences identical to their corresponding M. tuberculosis culture isolates. The gyrA QRDR PCR-sequencing assay showed complete concordance with the phenotypic outcomes (Table 2). For the 12 gyrA PCR-positive and M. tuberculosis culture-negative respiratory samples (Table 1), gyrA mutations associated with FQr were not detected. Compared to the nucleotide sequence of gyrA of the laboratory reference strain, H37Rv, all clinical isolates of M. tuberculosis (regardless of susceptibility to FQs) harbored an amino acid modification at position 95, with a resulting Ser-to-Thr change. The Ser95Thr mutation was considered a nonfunctional polymorphism in our strains. Of 142 susceptible strains, no FQr-associated mutation was detected. In the present study, 92.9% (26/28) of the gyrA mutations clustered in hot spot codons 90, 91, and 94, among which codon 94 was the most frequently involved site (Table 2). Nineteen (67.9%) resistant strains had mutations at codon 94, exhibiting a total of five different types of amino acid changes. Five (17.9%) resistant strains bore the gyrA mutation Ala90Val, the second most predominant mutation site associated with FQr. Two (7.1%) resistant strains harbored the hot spot mutation Ser91Pro. Only one (3.6%) resistant strain harbored a Gly88Cys mutation. Of particular note, a rarely reported gyrA Ala74Ser mutation was detected in M. tuberculosis directly from sputum specimens and the corresponding culture isolate of a patient.
Characterization of complete gyrase sequences of the WT H37Rv strain, the FQs clinical strain, and the FQr gyrA Ala74Ser mutant strain.
As shown in Table 3, compared to WT H37Rv strain, one of the five FQs clinical strain harbored a novel combination of gyrA mutations composed of Glu21Gln, Ser95Thr, and Gly668Asp. The MICs for this FQs clinical strain were identical to the WT H37Rv strain for OFX and MXF (0.5 and 0.25 μg/ml, respectively). In addition to nucleotide alterations at positions 21, 95, and 668, one clinical strain harboring the Ala74Ser mutation exhibited high-level resistance to FQs (OFX MIC = 8 μg/ml and MXF MIC = 4 μg/ml). All three strains have an identical nucleotide sequence in gyrB.
TABLE 3.
Inhibitory activities of OFX and MXF against gyrase complexes reconstituted with WT H37Rv, the FQs clinical strain, and the gyrA Ala74Ser variant
| Strain | Amino acid change in DNA gyrasea |
MIC (μg/ml) |
IC50 (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
gyrA |
gyrB | OFX | MXF | OFX | MXF | |||||
| E21Q | S95T | G668D | A74S | Other | ||||||
| WT H37Rv | ND | ND | ND | ND | ND | ND | 0.5 | 0.25 | 1.92 | 0.98 |
| FQs clinical strain | + | + | + | ND | ND | ND | 0.5 | 0.25 | 3.30 | 1.62 |
| Clinical strain harboring the Ala74Ser mutation in gyrA | + | + | + | + | ND | ND | 8.0 | 4.0 | 15.56 | 14.37 |
+, Detected; ND, not detected.
Purification and DNA supercoiling activities of recombinant M. tuberculosis gyrA and gyrB subunit proteins.
The entire amplified gyrA and gyrB genes were inserted separately in frame downstream of a T7 promoter in pET101/D-TOPO and pET-15b protein expression vectors, respectively. The recombinant gyrA (97-kDa) and gyrB (82-kDa) proteins carried hexahistidine tags at the C-terminal and N-terminal ends, respectively. The soluble gyrA subunits of the WT H37Rv strain, the FQs clinical strain, and the Ala74Ser mutant, as well as the soluble gyrB subunit of WT H37Rv strain, were purified by immobilized metal ion affinity chromatography and visualized by SDS-PAGE (see Fig. S1 in the supplemental material). The four gyrase subunit proteins were obtained at high purity (>95%) in milligram amounts. Neither the gyrA nor the gyrB subunit alone induced DNA supercoiling activity. E. coli topoisomerase activity contaminated from the BL21 host was undetectable. As shown in Fig. S2 in the supplemental material, the combination of recombinant gyrA and gyrB proteins catalyzed the supercoiling of relaxed pBR322 plasmid, demonstrating that they reconstituted functional DNA gyrase activities.
Inhibitory activities of FQs against gyrase complexes reconstituted with WT H37Rv and gyrA Ala74Ser.
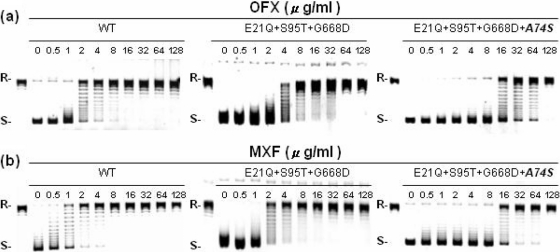
As shown in Fig. 1 and Table 3, the recombinant gyrase complex harboring the gyrA Glu21Gln, Ser95Thr, and Gly668Asp mutations exhibited IC50s of OFX and MXF similar to the WT H37Rv gyrase complex (<2-fold difference). The results indicated that the mutation combination at positions 21, 95, and 668 of gyrA is not associated with FQr, and thus these are regarded as nonfunctional or natural polymorphisms. In marked contrast, the IC50s of OFX and MXF against the recombinant gyrase complex bearing the mutation combination plus Ala74Ser mutation in gyrA were 8- and 14-fold higher than the WT H37Rv gyrase complex (Fig. 1 and Table 3). The results clearly demonstrate that the gyrA Ala74Ser mutation confers FQr in M. tuberculosis.
FIG. 1.
Comparative inhibitory effects of OFX (a) and MXF (b) on the DNA supercoiling activities of M. tuberculosis DNA gyrase complexes reconstituted with WT H37Rv, FQs clinical strain gyrA harboring E21Q, S95T, and G668D mutations, as well as variant gyrA harboring E21Q, S95T, G668D, and A74S mutations. R and S, relaxed and supercoiled DNA, respectively.
DISCUSSION
Corroborating previous reports (7, 12, 36, 40), the gyrA Ser95Thr mutation is not associated with FQr in M. tuberculosis and thus considered a nonfunctional polymorphism. In the present study, we report two novel polymorphisms, including Glu21Gln and Gly668Asp, which are located outside the QRDR of gyrA and occur in combination with the gyrA Ser95Thr mutation in the clinical strains of M. tuberculosis in our geographic region. As shown in Table 2, all 28 nonduplicate M. tuberculosis strains that harbored an additional mutation in QRDR of gyrA exhibited cross-resistance to OFX and MXF with high MICs (OFX MIC ≥ 8 μg/ml; MXF MIC ≥ 4 μg/ml). Our findings suggest that high-level phenotypic resistance to FQs among clinical isolates of M. tuberculosis is predominantly due to mutations in the QRDR of gyrA and mainly clustered in codons 90, 91, and 94, corroborating other reports (5, 31, 39, 43, 44). Compared to OFX, MXF (C-8-methoxy derivative) is more active with lower MICs for all isolates. These findings also concur with the results of other studies (7, 17, 33, 37, 38).
Heteroresistance refers to the phenomenon that concomitant presence of susceptible and resistant organisms in the same patient (32). It might result from infection by two different strains or one strain splitting into two types of organisms (15). Previous studies have reported that heteroresistance of M. tuberculosis to isoniazid, rifampin, and FQs could be detected by sequencing analysis of the rpoB, katG, and gyrA genes (15, 18, 42). Our previous report also demonstrated the current gyrA PCR-sequencing assay was able to identify mixed resistant genotypes in direct samples and indicated that two FQr variants coexisted in a patient (21). However, coexistence of susceptible and resistant populations was not detected among our samples in the present study.
In the present study, all eight bacillary strains from XDR-TB patients harbored a gyrA hot spot mutation, including Ala90Val (n = 1), Asp94Gly (n = 5), Asp94Ala (n = 1), and Asp94Tyr (n = 1). Among the 28 nonduplicate strains harboring FQr gyrA mutations, 28.6% (8 of 28) were regarded as XDR-TB strains. These data suggest that successful detection of FQr M. tuberculosis by genotypic means would enable early diagnosis of FQr MDR-TB or XDR-TB.
Molecular methods other than PCR-DNA sequencing, including MAS-PCR (11), locked nucleic acid probe real-time PCR (42), and line probe (12), have been evaluated for screening of FQr-associated gyrA mutations in M. tuberculosis. These rapid genotypic assays were demonstrated to detect FQr in purified clinical isolates (6). This is the first report to validate PCR and nucleotide sequencing for amplifying the gyrA QRDR of M. tuberculosis in respiratory specimens and for the successful detection of FQr-associated hot spot and novel mutations with high specificity. The average turnaround time for this assay is 3 days, with a net time gain of 2 to 3 months over conventional mycobacterial culture plus anti-TB susceptibility testing. Early diagnosis facilitates effective clinical management and public health control (21). In the present study, the sensitivity of the IS6110 PCR assay was 12.8% higher than the gyrA QRDR method for the detection of M. tuberculosis in clinical samples. The nested IS6110 PCR assay enhances the sensitivity for the detection of M. tuberculosis complex in direct samples over gyrA QRDR PCR, which is only a single PCR (49). Due to the high cost of the DNA-sequencing assay and the superior sensitivity of the IS6110 PCR over the gyrA QRDR PCR (Table 1), the latter should be performed for TB-PCR positive cases at high risk for MDR- or XDR-TB, such as patients originating from high-incidence areas or suffering from recurrent TB, treatment failure, and irregular prior treatment.
The gyrA Ala74Ser mutation was previously reported in combination with a hot spot gyrA mutation Asp94Gly in clinical isolates (35, 39). The association of the gyrA Ala94Gly mutation alone with high-level FQr in M. tuberculosis has been functionally characterized and reported (2); however, the functional effect of the gyrA Ala74Ser mutation on FQ susceptibility and affinity in the formation of ternary drug-mediated gyrase-DNA complex in M. tuberculosis remains unclear. We identified one FQr M. tuberculosis isolate bearing the gyrA Ala74Ser mutation independently with no other FQr-associated gyrA and gyrB mutations (Table 3). The effect of the gyrA Ala74Ser mutation on gyrase function was further investigated by purifying the recombinant mutant gyrase complex and reconstituting the enzymatic DNA supercoiling activity. In the DNA supercoiling assay, the recombinant mutant gyrase complex reconstituted with gyrA Ala74Ser was 8- and 14-fold more resistant than the WT H37Rv gyrase complex, as well as 4- and 8-fold more resistant than the FQs clinical strain gyrase complex, to inhibition by OFX and MXF, respectively (Fig. 1 and Table 3). These findings correlated well with the results of phenotypic susceptibilities. This is the first detailed enzymatic analysis of the association of gyrA Ala74Ser mutation with FQr in M. tuberculosis.
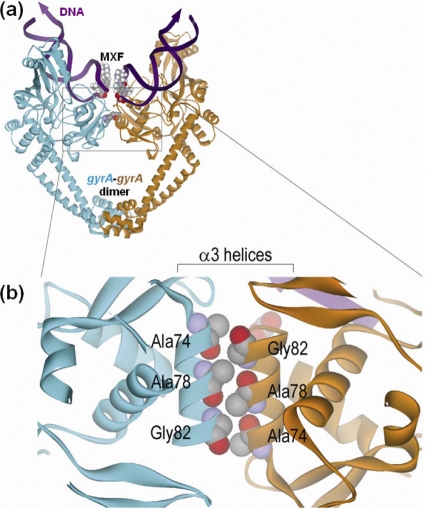
The 59-kDa N-terminal breakage-reunion domain of gyrA protein is the principal drug-binding region of the DNA gyrase (41). X-ray crystallography has demonstrated the DNA-cleavage complex is stabilized by the action of FQs and a novel antibacterial dione (19, 20). Alanine at position 74 in gyrA of M. tuberculosis, which corresponds to the alanine at position 67 in gyrA of E. coli, is an amino acid lying in the α3 helix domain which forms a large hydrophobic interface between gyrA-gyrA dimer (Fig. 2) and is located just beneath the α4 helix domain (the DNA recognition site). It is believed that the gyrA Ala74Ser mutation perturbs a gyrA-gyrA dimer interface by interrupting the hydrophobic interaction between two α3 helix domains (25, 27). Recently, structural studies of type IIA topoisomerases (19, 20, 34) revealed a new molecular connection by which DNA breakage is coordinated with the association and dissociation of intersubunit interfaces during strand scission. Intriguingly, one of the inhibitor-trapped gyrA structures shows that the α3 helices slide ∼6 Å past each other on the formation of the complex, suggesting the conformational flexibility of α3 helices at the intersubunit interface during the enzymatic reaction. Therefore, perturbation of the intersubunit interface caused by the Ala74Ser mutation probably disturbs the putative binding pocket of the drug due to the restriction of the movement of α3 helix domain, and the distance effects lead to the reduction of the drug binding affinity.
FIG. 2.
(a) Front view of the gyrA-DNA complex of M. tuberculosis stabilized by MXF. The diagram was adapted with permission from the crystal structure of M. tuberculosis GyrA59 (41). (b) Close-up top view of the gyrA α3 helix domain.
Information on the lethality activity of quinolone in M. tuberculosis cells beyond the quinolone-gyrase-DNA complex formation is very limited. Previous work has already shown that chloramphenicol, an inhibitor of protein synthesis, affects the lethal activity of all C-8-methoxy group FQs tested, except MXF against M. tuberculosis (26). It appears that the presence of the C-8-methoxy group and the diazabicyclo C-7 ring system in the MXF compound are both important for rapid killing of nongrowing M. tuberculosis cells. The gyrA amino acid modification with amino acid change from Ala to Ser at position 67 in E. coli is expected to perturb the gyrA-gyrA dimer interface near the DNA gate (27, 29), which allowed nalidixic acid to fragment chromosomes and kill nongrowing cells in the absence of protein synthesis. Simultaneously, the Ala67Ser mutation not only made a noninducible lexA mutant hypersusceptible to nalidixic acid but also facilitated immunoprecipitation of DNA fragments by gyrA antiserum following nalidixic acid treatment of cells (27). These results proved that the gyrA Ala67Ser mutation in E. coli destabilize the ternary drug- gyrase-DNA complex and increased the rate of killing. Since the modes of quinolone action appear to be similar in many organisms, further study of the effect of the gyrA Ala74Ser mutation on quinolone lethality and chromosome fragmentation (27) may provide elucidate the lethal pathway(s) of nongrowing M. tuberculosis cells.
Supplementary Material
Acknowledgments
We are grateful to Bayer HealthCare Pharmaceuticals for providing the MXF powder.
This study was supported by grants from the National Chinese Grant for Infectious Diseases (grant 2008ZX10003-012) and the Research Fund for the Control of Infectious Diseases of the Food and Health Bureau of the Hong Kong SAR Government.
We also have no conflict of interest to declare.
Footnotes
Published ahead of print on 18 October 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 48:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, A., et al. 2006. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob. Agents Chemother. 50:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caminero, J. A. 2008. Likelihood of generating MDR-TB and XDR-TB under adequate National Tuberculosis Control Programme implementation. Int. J. Tuberc. Lung Dis. 12:869-877. [PubMed] [Google Scholar]
- 4.Chan, C. M., et al. 1996. Single-tube nested PCR in the diagnosis of tuberculosis. J. Clin. Pathol. 49:290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, R. C., et al. 2007. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J. Antimicrob. Chemother. 59:866-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, K. C., W. W. Yew, and R. C. Chan. 2010. Rapid assays for fluoroquinolone resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. J. Antimicrob. Chemother. 65:1551-1561. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, A. F., et al. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standards, vol. 23, no. 18. M24-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed]
- 9.Cole, S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Cozzarelli, N. R. 1980. DNA gyrase and the supercoiling of DNA. Science 207:953-960. [DOI] [PubMed] [Google Scholar]
- 11.Evans, J., and H. Segal. 2010. Novel multiplex allele-specific PCR assays for the detection of resistance to second-line drugs in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65:897-900. [DOI] [PubMed] [Google Scholar]
- 12.Giannoni, F., et al. 2005. Evaluation of a new line probe assay for rapid identification of gyrA mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillemin, I., E. Cambau, and V. Jarlier. 1995. Sequences of conserved region in the A subunit of DNA gyrase from nine species of the genus Mycobacterium: phylogenetic analysis and implication for intrinsic susceptibility to quinolones. Antimicrob. Agents Chemother. 39:2145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillemin, I., V. Jarlier, and E. Cambau. 1998. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42:2084-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann-Thiel, S., et al. 2009. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur. Respir. J. 33:368-374. [DOI] [PubMed] [Google Scholar]
- 16.Jain, A., and P. Dixit. 2008. Multidrug-resistant to extensively drug resistant tuberculosis: what is next? J. Biosci. 33:605-616. [DOI] [PubMed] [Google Scholar]
- 17.Kam, K. M., et al. 2006. Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb. Drug Resist. 12:7-11. [DOI] [PubMed] [Google Scholar]
- 18.Karahan, Z. C., and N. Akar. 2005. Restriction endonuclease analysis as a solution for determining rifampin resistance mutations by automated DNA sequencing in heteroresistant Mycobacterium tuberculosis strains. Microb. Drug Resist. 11:137-140. [DOI] [PubMed] [Google Scholar]
- 19.Laponogov, I., et al. 2010. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5:e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laponogov, I., et al. 2009. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16:667-669. [DOI] [PubMed] [Google Scholar]
- 21.Lau, R. W., et al. 2010. Rapid diagnosis of multidrug-resistant smear-positive pulmonary tuberculosis. Int. J. Antimicrob. Agents 35:202-203. [DOI] [PubMed] [Google Scholar]
- 22.Leimane, V., et al. 2005. Clinical outcome of individualized treatment of multidrug-resistant tuberculosis in Latvia: a retrospective cohort study. Lancet 365:318-326. [DOI] [PubMed] [Google Scholar]
- 23.Leung, E. T., et al. 2006. Molecular characterization of isoniazid resistance in Mycobacterium tuberculosis: identification of a novel mutation in inhA. Antimicrob. Agents Chemother. 50:1075-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung, E. T., et al. 2003. Detection of katG Ser315Thr substitution in respiratory specimens from patients with isoniazid-resistant Mycobacterium tuberculosis using PCR-RFLP. J. Clin. Microbiol. 52:999-1003. [DOI] [PubMed] [Google Scholar]
- 25.Lu, T., X. Zhao, and K. Drlica. 1999. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob. Agents Chemother. 43:2969-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik, M., and K. Drlica. 2006. Moxifloxacin lethality against Mycobacterium tuberculosis in the presence and absence of chloramphenicol. Antimicrob. Agents Chemother. 50:2842-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810-825. [DOI] [PubMed] [Google Scholar]
- 28.Migliori, G. B., et al. 2008. Resistance to second-line injectables and treatment outcomes in multidrug-resistant and extensively drug-resistant tuberculosis cases. Eur. Respir. J. 31:1155-1159. [DOI] [PubMed] [Google Scholar]
- 29.Morais Cabral, J. H., et al. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 30.Nolte, F. S., and B. Metchock. 1995. Mycobacterium, p. 400-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, DC.
- 31.Pitaksajjakul, P., et al. 2005. Mutations in the gyrA and gyrB genes of fluoroquinolone-resistant Mycobacterium tuberculosis from TB patients in Thailand. Southeast Asian J. Trop. Med. Public Health 36:228-237. [PubMed] [Google Scholar]
- 32.Rinder, H., K. T. Mieskes, and T. Loscher. 2001. Heteroresistance in Mycobacterium tuberculosis. Int. J. Tuber. Lung Dis. 5:339-345. [PubMed] [Google Scholar]
- 33.Rodriguez, J. C., M. Ruiz, A. Climent, and G. Royo. 2001. In vitro activity of four fluoroquinolones against Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 17:229-231. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt, B. H., A. B. Burgin, J. E. Deweese, N. Osheroff, and J. M. Berger. 2010. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465:641-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, R., J. Zhang, C. Li, Y. Kazumi, and I. Sugawara. 2006. Emergence of ofloxacin resistance in Mycobacterium tuberculosis clinical isolates from China as determined by gyrA mutation analysis using denaturing high-pressure liquid chromatography and DNA sequencing. J. Clin. Microbiol. 44:4566-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqi, N., et al. 2002. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob. Agents Chemother. 46:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somasundaram, S., and N. C. Paramasivan. 2006. Susceptibility of Mycobacterium tuberculosis strains to gatifloxacin and moxifloxacin by different methods. Chemotherapy 52:190-195. [DOI] [PubMed] [Google Scholar]
- 38.Sulochana, S., F. Rahman, and C. N. Paramasivan. 2005. In vitro activity of fluoroquinolones against Mycobacterium tuberculosis. J. Chemother. 17:169-173. [DOI] [PubMed] [Google Scholar]
- 39.Sun, Z., J. Zhang, X. Zhang, S. Wang, Y. Zhang, and C. Li. 2008. Comparison of gyrA gene mutations between laboratory-selected ofloxacin-resistant Mycobacterium tuberculosis strains and clinical isolates. Int. J. Antimicrob. Agents 31:115-121. [DOI] [PubMed] [Google Scholar]
- 40.Takiff, H. E., et al. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tretter, E. M., A. J. Schoeffler, S. R. Weisfield, and J. M. Berger. 2010. Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis. Proteins 78:492-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doorn, H. R., et al. 2008. Fluoroquinolone resistance detection in Mycobacterium tuberculosis with locked nucleic acid probe real-time PCR. Int. J. Tuberc. Lung Dis. 12:736-742. [PubMed] [Google Scholar]
- 43.Von Groll, A., et al. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob. Agents Chemother. 53:4498-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, J. Y., et al. 2007. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates: associated genetic mutations and relationship to antimicrobial exposure. J. Antimicrob. Chemother. 59:860-865. [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization. 2006. Extensively drug-resistant tuberculosis (XDR-TB): recommendations for prevention and control. Wkly. Epidemiol. Rec. 81:430-432. [PubMed] [Google Scholar]
- 46.Yam, W. C., K. Y. Yuen, and W. H. Seto. 1998. Direct detection of Mycobacterium tuberculosis in respiratory specimens using an automated DNA amplification assay and a single tube nested polymerase chain reaction (PCR). Clin. Chem. Lab. Med. 36:597-599. [DOI] [PubMed] [Google Scholar]
- 47.Yam, W. C., et al. 2004. Direct detection of rifampin-resistant Mycobacterium tuberculosis in respiratory specimens by PCR-DNA sequencing. J. Clin. Microbiol. 42:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yew, W. W., et al. 2000. Outcomes of patients with multidrug-resistant pulmonary tuberculosis treated with ofloxacin/levofloxacin-containing regimens. Chest 117:744-751. [DOI] [PubMed] [Google Scholar]
- 49.Yuen, K. Y., W. C. Yam, L. P. Wong, and W. H. Seto. 1997. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 35:1385-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Y., and W. W. Yew. 2009. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 13:1320-1330. [PubMed] [Google Scholar]
- 51.Zignol, M., et al. 2006. Global incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 194:479-485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.