Abstract
Quinine is a standard drug for treating severe malaria in Africa, and it is also increasingly used to treat uncomplicated disease. However, failures of quinine therapy are common, and it is unknown if failures in Africa are due to drug resistance. Recent studies have identified associations between in vitro quinine sensitivity and polymorphisms in genes encoding putative transporters, including well-described polymorphisms in pfcrt and pfmdr1 and varied numbers of DNNND or DDNHNDNHNND repeats in microsatellite 4760 (ms4760) of the predicted sodium-hydrogen exchanger, pfnhe1. To better characterize mediators of quinine response, we assessed associations between genetic polymorphisms, in vitro quinine sensitivity, and quinine treatment responses in Kampala, Uganda. Among 172 fresh clinical isolates tested in vitro, decreasing sensitivity to quinine was associated with accumulation of pfmdr1 mutations at codons 86, 184, and 1246. Nearly all parasites had pfcrt 76T, preventing analysis of associations with this mutation. pfnhe1 ms4760 was highly polymorphic. Parasites with 2 copies of either ms4760 repeat showed modest decreases in quinine sensitivity compared to those with 1 or ≥3 repeats, but the differences were not statistically significant. None of the above polymorphisms predicted treatment failure among 66 subjects treated with quinine for uncomplicated malaria. Our data suggest that quinine sensitivity is a complex trait and that known polymorphisms in pfcrt, pfmdr1, and pfnhe1, while associated with quinine sensitivity, are not robust markers for quinine resistance.
Quinine was the first established antimalarial drug, and it has been used to treat malaria for centuries (26). Intravenous quinine is the standard therapy for severe Plasmodium falciparum malaria in Africa and many other areas, although intravenous artesunate recently showed superior efficacy in Asia (15). Quinine is also a second-line regimen used for the treatment of uncomplicated malaria in many countries after failure of initial therapy. Recent WHO guidelines suggest second-line use of quinine in combination with an antibiotic (55), but monotherapy is still commonly used (59). In addition, with failures of older therapies and limited availability of new artemisinin-based combination therapies, quinine is increasingly used as a first-line drug for treatment of uncomplicated malaria in Africa (1).
The use of quinine to treat uncomplicated malaria is problematic for a number of reasons. First, the efficacy of quinine for the treatment of uncomplicated malaria is uncertain. A number of studies and case reports have demonstrated apparent failures after quinine therapy for falciparum malaria in Asia (9, 41), South America (46, 60), and Africa (22, 31, 38). Recent studies incorporating detailed assessments of treatment efficacy or effectiveness have shown 28-day failure rates of >10% after 7-day courses of quinine for uncomplicated malaria in Sudan (3), Thailand (40), and Uganda (1). In Uganda, the 28-day genotype-corrected failure rate was 23%; some failures may have been due to poor treatment compliance, but no association between measures of compliance and treatment efficacy was identified (1). On the other hand, some studies have shown excellent efficacy for quinine, including over 95% success after 14 to 28 days for the treatment of falciparum malaria in Equatorial Guinea (45), in Venezuela (2), and in returned travelers in France (32). Second, quinine is poorly tolerated, especially later in the course of 7 days of treatment, suggesting that noncompliance with full treatment courses is common (19, 50). Noncompliance may limit treatment efficacy and help to select for drug-resistant parasites. Third, widespread quinine use engenders risks of serious toxicities, including cardiac effects, hypoglycemia, hemolysis, and thrombocytopenia (50). Fourth, quinine pharmacokinetics are variable, and metabolism can be altered by coadministration of a number of other drugs (42).
Diminished in vitro responsiveness of P. falciparum to quinine has been documented in parasites from Asia, South America, and Africa (57). Available studies have suggested that parasites from Africa generally remain sensitive to quinine (4, 21, 24, 37, 43, 51), but methodologies have varied, and strict cutoffs for in vitro drug resistance have not been established. Despite relatively low 50% inhibitory concentrations (IC50s), it is noteworthy that absolute sensitivities of African P. falciparum strains have varied greatly. For example, in a recent study in Uganda, the IC50s for quinine against freshly isolated parasites varied about 50-fold (29).
Mediators of P. falciparum resistance to quinine are poorly understood (58). The resistance phenotype is consistent with a complex genetic basis, as changes in sensitivity have appeared to evolve gradually. Quantitative trait locus analysis identified three genes predicted to play roles in the responsiveness of P. falciparum to quinine (18): pfcrt, which encodes a predicted transporter in which the 76T mutation is the principal mediator of resistance to chloroquine (12, 49); pfmdr1, which encodes a P-glycoprotein homolog for which different polymorphisms are associated with altered responses to a number of antimalarials (34, 44, 52); and pfnhe1, which encodes a putative sodium-hydrogen exchanger (18). Among known variations, an increased copy number of pfmdr1 is most clearly associated with the response to quinine, with its amplification being associated with 2- to 3-fold decreases in sensitivity (33, 47, 58). Increased pfmdr1 copy number has been very uncommon in Africa, although it has recently been reported in West Africa (56). Single nucleotide polymorphisms (SNPs) in pfcrt and pfmdr1 are also associated with alterations in quinine sensitivity (10, 27, 44, 49). In Ugandan field isolates, the pfmdr1 1246Y mutation was associated with diminished sensitivity to quinine (29). However, unlike analyses of chloroquine sensitivity, associations between quinine sensitivity and polymorphisms in pfcrt and pfmdr1 have been modest, suggesting important roles for additional polymorphisms, including pfnhe1 and other loci (18).
Considering variation in pfnhe1, reducing its expression by about 50% using allelic exchange led to a 30% increase in quinine sensitivity in some but not other parasite strains (28). Four studies have recently evaluated associations between polymorphisms at pfnhe1 microsatellite 4760 (ms4760) and in vitro sensitivity of parasites from individuals infected in countries where malaria is endemic. In 23 culture-adapted strains from multiple countries (20) and 60 freshly isolated strains from the China-Myanmar border (25), increased numbers of the DNNND repeat were associated with decreased quinine sensitivity, while increased numbers of the DDNHNDNHNND repeat within the same microsatellite were associated with increased quinine sensitivity (20). In 29 fresh isolates from the Kenyan coast, 2 DNNND repeats were associated with decreased sensitivity to quinine compared to the sensitivities of isolates with 1 or 3 copies, and there was no association between the number of DDNHNDNHNND repeats and quinine susceptibility (30). In 83 freshly isolated strains from individuals infected in Africa, multiple polymorphisms were seen in ms4760, but, in contrast to the above reports, significant associations were not seen between the numbers of the DNNND repeat and quinine sensitivity, and increased copies of the DDNHNDNHNND repeat were associated with decreased quinine sensitivity (5). Taken together, recent results suggest an uncertain role for pfnhe polymorphisms in quinine sensitivity.
To further characterize associations between polymorphisms in pfcrt, pfmdr1, and pfnhe1 and sensitivity to quinine, we evaluated key sequences in P. falciparum parasites from two studies in Kampala, Uganda. The first study provided access to a large number of clinical isolates for which in vitro sensitivity to quinine was determined (29). The second study provided samples from a recent assessment of the clinical effectiveness of quinine for the treatment of uncomplicated malaria (1). Our results are consistent with the conclusion that quinine responsiveness is a complex trait, with polymorphisms in multiple genes contributing to drug sensitivity.
MATERIALS AND METHODS
Clinical trials.
Plasmodium falciparum DNA was available from two clinical trials in Kampala. The first was a longitudinal comparison of the efficacies of three combination regimens against uncomplicated malaria in children aged 1 to 10 years at enrollment (601 enrolled from November 2004 to June 2005 and an additional 89 enrolled from January to May 2007) (11, 17). In this trial, patients were offered all medical care through the study clinic, so use of antimalarial drugs outside the study protocol was uncommon. Children were treated with quinine only for complicated malaria or after failure of a combination regimen, both uncommon events (17). Samples were collected for parasite culture between August 2006 and May 2008. The second trial was a randomized comparison of quinine and artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in children aged 6 to 59 months (1); 86 children were enrolled into the quinine treatment arm of the study. Responses over 28 days were graded on the basis of standard WHO criteria, with genotyping used to distinguish recurrences due to reinfection from those due to recrudescence (1). In both trials, when falciparum malaria was diagnosed on the basis of fever or a history of fever and parasites on Giemsa-stained blood smears, blood spots were collected on filter paper (Whatman 3MM) for subsequent molecular studies. All study subjects provided written informed consent. Both trials and the analysis of cultured parasites were approved by the Uganda National Council for Science and Technology, the Makerere University Research and Ethics Committee, and the University of California, San Francisco, Committee on Human Research.
Parasite culture and measurement of in vitro quinine sensitivity.
For the first trial described above, at the time of diagnosis of an episode of malaria and before the initiation of therapy, blood was collected in heparinized tubes and transported within 30 min to our laboratory. Giemsa-stained thin blood smears were examined, and if P. falciparum monoinfection was confirmed, culture was initiated. Blood was centrifuged, plasma and buffy coat were removed, and the erythrocyte pellet was washed twice with RPMI 1640 medium at 37°C. Parasites were diluted with 2% group O uninfected erythrocytes to obtain a density of 0.05%. Aliquots (200 μl) were then cultured in 10 ml RPMI 1640 medium supplemented with 25 mM HEPES, 0.2% NaHCO3, 0.1 mM hypoxanthine, 100 μg/ml gentamicin, and 0.5% Albumax II serum substitute to produce a packed cell volume of ∼2%. Sensitivities were measured for multiple drugs, including quinine, as previously described utilizing 96-well culture plates predosed with serial dilutions of drugs and a histidine-rich protein 2 (HRP-2)-based enzyme-linked immunosorbent assay (ELISA) (29). For this assay, optical density values were fitted to normal curves based on serial dilutions of HRP-2 standards, and IC50s were calculated on the basis of a nonlinear regression model.
Analysis of parasite genetic polymorphisms.
DNA was extracted from filter paper samples by extraction with Chelex, as previously described (35). We screened for polymorphisms at pfcrt K76T and pfmdr1 N86Y, Y184F, S1034C, N1042D, and D1246Y by PCR amplification of flanking sequences, sequence-specific restriction enzyme digestion, and evaluation of the digested fragments by agarose gel electrophoresis, all as previously reported (12, 16). Estimates of pfmdr1 gene copy number were carried out by quantitative PCR as previously reported (14, 39), using a 7500 real-time PCR system (Applied Biosystems).
The ms4760 locus of the pfnhe1 gene was amplified with primers NHE-A (5′-AGTCGAAGGCGAATCAGATG-3′) and NHE-B (5′-GATACTTACGAACATGTTCATG-3′) (53), using a Titanium PCR kit (Clontech). Each 20-μl PCR mixture included 14.8 μl H2O, 2 μl 10× buffer, 0.4 μl deoxynucleoside triphosphate mix (10 mM each), 0.2 μl of each 20 μM primer, 0.4 μl 50× Titanium Taq, and 2 μl of target DNA template. A touchdown PCR approach (23) used the following parameters: 94°C for 120 s; 10 cycles of 94°C for 15 s, 63°C for 30 s, and 68°C for 90 s; 25 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 90 s; and 1 cycle of 68°C for 7 min. The PCRs were performed on a Bio-Rad C1000 or S1000 thermocycler. PCR products were purified using ExoSAP-IT (USB). About 150 ng of each purified DNA was sequenced using 15 pmol primers NHE-C (5′-ATCCCTGTTGATATATCGAATG-3′) and NHE-D (5′-TTGTCATTAGTACCCTTAGTTG-3′) (53) at the University of California, San Francisco, Genomics Core Facility using ABI BigDye (version 3.1) Terminator sequencing chemistry and an ABI Prism 3730xl capillary DNA analyzer (Applied Biosystems).
For analysis of pfnhe1 ms4760, nucleotide traces were physically inspected using the SeqMan tool (DNAStar Lasergene 8 software). The sequences were then translated using the Edit Sequence tool and aligned using the MEGALIGN program (DNAStar). Microsatellite 4760 profiles that were not among the 35 previously reported profiles (5, 18, 25, 53), were confirmed by repeat template amplification and sequencing.
Statistical analysis.
Quinine IC50s were calculated using a polynomial regression model and HN-NonLin software (http://malaria.farch.net) and were considered continuous variables. Differences in quinine IC50s were examined using the Mann-Whitney U test for comparisons of two groups or the Kruskal-Wallis test with Dunn's multiple-comparison posttest for comparisons of more than two groups. Infections with mixed mutant/wild-type genotypes at the pfcrt and pfmdr1 loci were analyzed as mutant, in view of the expected phenotype of the infection. Associations between alleles and treatment outcomes were assessed using Fisher's exact two-tailed test. Tests were carried out using GraphPad Prism software. In all cases the statistical significance level was set at a P value of <0.05.
Nucleotide sequence accession numbers.
Nucleotide sequences for the Ugandan isolates were deposited in the GenBank database under accession numbers HQ412347 to HQ412386.
RESULTS
Samples available for study.
We evaluated polymorphisms in two sets of samples from patients infected with P. falciparum in Kampala. First, we evaluated samples from a cohort of children who were followed in a drug efficacy trial comparing three combination regimens for uncomplicated falciparum malaria (17). Blood samples collected upon presentation with uncomplicated falciparum malaria and before the initiation of therapy were used to inoculate cultures, and in vitro sensitivities to quinine were determined immediately after sample collection (29). Second, we evaluated 79 samples from the quinine treatment arm of a trial with children randomized to treatment of uncomplicated falciparum malaria with either artemether-lumefantrine or quinine (1). Patients were followed over 28 days after presentation, and treatment outcomes were classified on the basis of WHO criteria (54); in vitro sensitivities were not assessed for samples from this trial.
Associations between polymorphisms and in vitro quinine sensitivity.
The in vitro sensitivities to quinine of isolates collected in Kampala varied widely, with a geometric mean IC50 of 94.4 nM and an IC50 range of from 15 to 761 nM (29). We first assessed known polymorphisms in pfcrt and pfmdr1 and searched for associations with the sensitivity of cultured parasites to quinine. The pfcrt 76T mutation was seen in all except 1 of 169 tested samples. Thus, many parasites were highly sensitive to quinine, despite the presence of pfcrt 76T, and it was not possible to assess the role of this polymorphism in quinine sensitivity. For pfmdr1, the N86Y and 1246Y mutations were also very common, although they were not fixed; the 184F mutation was less common; and mutations at positions 1034 and 1042, which have principally been described in samples from regions other than Africa, were not seen (Table 1). For the 86Y, 184F, and 1246Y polymorphisms, mutant parasites had diminished sensitivity to quinine (Table 1), but none of these differences attained statistical significance. Considering combinations of polymorphisms, there was a trend toward decreased quinine sensitivity with increased numbers of mutations (Table 2). The difference was statistically significant, after adjustment for multiple comparisons, between isolates having any 1 of the pfmdr1 mutations and isolates having all 3 pfmdr1 mutations (P = 0.02). Increased copy number of pfmdr1 has been associated with diminished quinine sensitivity in Asia but has been seen uncommonly in African isolates. To consider a potential contribution of pfmdr1 copy number on results from Uganda, we analyzed 58 samples with a range of quinine sensitivities (26 samples with IC50s of from 15 to 22 nM, 10 with IC50s of from 41 to 298 nM, 22 with IC50s of from 308 to 755 nM). All of these parasites contained only a single copy of the pfmdr1 gene.
TABLE 1.
pfmdr1 polymorphisms seen in Kampala and in vitro quinine sensitivities of isolates
| Parameter | Result for isolates with the following polymorphisms: |
|||||
|---|---|---|---|---|---|---|
| N86Y |
Y184F |
D1246Y |
||||
| WTa | Mutant | WT | Mutant | WT | Mutant | |
| No. of isolates | 24 | 143 | 143 | 21 | 37 | 133 |
| Median IC50 (nM) | 84 | 123 | 112 | 132 | 84 | 125 |
| 25th-75th percentile IC50 (nM) | 24-223 | 38-259 | 34-255 | 33-242 | 23-180 | 38-263 |
| Geometric mean IC50 (nM) | 74 | 101 | 95 | 108 | 71 | 103 |
| 95% CIb (nM) | 45-121 | 84-121 | 79-114 | 64-181 | 49-102 | 85-125 |
WT, wild type.
CI, confidence interval.
TABLE 2.
Impact of combinations of pfmdr1 polymorphisms on in vitro quinine sensitivitya
| Parameter | Result for isolates with the following polymorphisms: |
|||
|---|---|---|---|---|
| All WT | 1 mutation | 2 mutations | 3 mutations | |
| No. of isolates | 12 | 23 | 125 | 5 |
| Median IC50 (nM) | 64 | 84 | 123 | 278 |
| 25th-75th percentile IC50 (nM) | 20-244 | 22-170 | 37-259 | 179-601 |
| Geometric mean IC50 (nM) | 65 | 65 | 101 | 312 |
| 95% CIb (nM) | 30-140 | 40-105 | 83-123 | 142-687 |
In vitro IC50s are shown for parasites with all wild-type (WT) sequences at the five pfmdr1 alleles studied or with one, two, or three mutations at pfmdr1 codon 86, 184, or 1246.
CI, confidence interval.
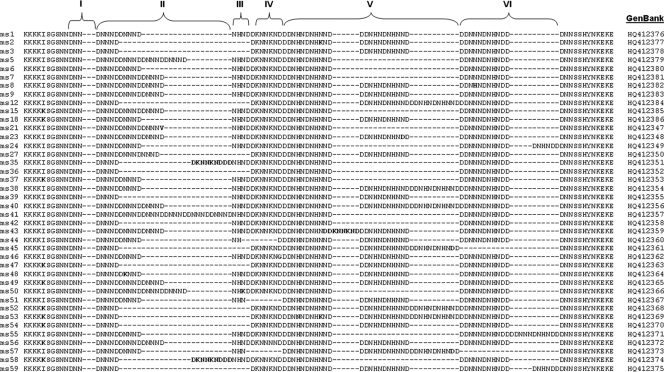
We next evaluated associations between polymorphisms in pfnhe1 ms4760 and quinine sensitivity. We sequenced 240 isolates, and in vitro quinine sensitivity data were obtained for 172 isolates. We found a high degree of polymorphism at this locus. We found 46% (16/35) of the previously described ms4760 haplotypes. In addition, we identified 24 previously undescribed ms4760 haplotypes, now designated ms4760-36 to ms4760-59 (Table 3 and Fig. 1). Nucleotide sequences for the Ugandan isolates were deposited in the GenBank database under the accession numbers indicated above and in Fig. 1. The number of DNNND repeats ranged from 1 to 6 (mean ± standard deviation [SD], 2.12 ± 0.86), and that of DDNHNDNHNND repeats ranged from 1 to 3 (mean ± SD, 1.83 ± 0.54). The quinine sensitivities of isolates with different numbers of DNNND and DDNHNDHNND repeats were compared. Sensitivities varied greatly within each category, but parasites with 2 copies of either repeat were somewhat less sensitive to quinine than those with 1 or 3 or more repeats (Table 4), although the associations between repeat number and quinine IC50 were not significant (P = 0.20 and 0.15 for DNNND and DDNHNDHNND, respectively). Considered together, the impacts of known polymorphisms in pfcrt, pfmdr1, and pfnhe1 did not lead to identification of significant associations. Considering the combinations leading to the least and the most sensitive parasites, those with the wild-type sequence at pfmdr1 positions 86 and 1246 and 3 DNNND repeats were more sensitive to quinine (IC50, 55 nM) than parasites with mutant pfmdr1 sequences and 2 DNNND repeats (IC50, 151 nM), but the differences were not statistically significant (P = 0.13; Table 5).
TABLE 3.
pfnhe1 microsatellite 4760 profiles detected among Ugandan P. falciparum isolates
| ms4670 profilea | No. of repeats |
Quinine IC50 (nM) | No. of isolates | |
|---|---|---|---|---|
| DNNND | DDNHNDNHNND | |||
| 35, 36, 42, 58 | 1 | 1 | 22.3-298.0 | 3 |
| 2, 3, 39, 47, 48, 53, 54, 59 | 1 | 2 | 15.4-760.9 | 39 |
| 12, 45, 52 | 1 | 3 | 63.5-96.8 | 2 |
| 6, 24, 37, 55 | 2 | 1 | 16.6-527.1 | 11 |
| 1, 18, 27, 44, 46, 51 | 2 | 2 | 15.4-754.9 | 58 |
| 38, 57 | 2 | 3 | 15.4-193.6 | 2 |
| 7, 15, 21, 43 | 3 | 1 | 15.4-447.5 | 24 |
| 9, 23, 49, 56 | 3 | 2 | 15.4-483.0 | 19 |
| 8, 40 | 3 | 3 | 15.4-275.2 | 9 |
| 5, 50 | 4 | 1 | 15.4-292.3 | 4 |
| 41 | 6 | 1 | 278.4 | 1 |
FIG. 1.
Alignment of 40 sequences of pfnhe1 microsatellite 4760 identified in 240 Ugandan P. falciparum isolates. Blocks I to VI have been described previously (53). The DNNND repeats are in block II, and the DDNNNDNHNDD repeats are in block V. Profiles ms4760-1 to ms4760-35 have been described previously (5, 18, 25, 53); ms4760-36 to ms4760-59 are described in this study. Profiles 4, 10, 11, 13, 14, 16, 17, 19, 20, 22, 25, 26, 28, 29, 30, 31, 32, 33, and 34 were not observed in the present study. Other differences among sequences are indicated in boldface. Gaps were created for optimal alignment.
TABLE 4.
In vitro quinine sensitivities of parasites with different numbers of repeats in pfnhe1 ms4760
| Parameter | Result for the indicated no. of repeats of: |
|||||
|---|---|---|---|---|---|---|
| DNNND |
DDNHNDNHNND |
|||||
| 1 | 2 | ≥3 | 1 | 2 | 3 | |
| No. of isolates | 44 | 71 | 57 | 43 | 116 | 13 |
| Median IC50 (nM) | 105 | 124 | 72 | 93 | 124 | 55 |
| 25th-75th percentile IC50 (nM) | 32-233 | 55-273 | 25-217 | 25-271 | 36-253 | 16-166 |
| Geometric mean IC50 (nM) | 91 | 113 | 76 | 87 | 103 | 55 |
| 95% CIa (nM) | 64-130 | 87-147 | 56-104 | 61-123 | 84-127 | 28-108 |
CI, confidence interval.
TABLE 5.
In vitro quinine sensitivities of isolates with various combinations of polymorphismsa
| Parameter | Result for isolates with polymorphisms at codons 86 and 1246 and the indicated no. of DNNND repeats: |
|||||
|---|---|---|---|---|---|---|
| Mutant |
WT |
|||||
| 1 | 2 | 3 | 1 | 2 | 3 | |
| No. of isolates | 33 | 49 | 42 | 4 | 10 | 5 |
| Median IC50 (nM) | 115 | 151 | 124 | 111 | 111 | 55 |
| 25th-75th percentile IC50 (nM) | 33-275 | 60-292 | 30-249 | 42-200 | 18-238 | 27-182 |
| Geometric mean IC50 (nM) | 99 | 129 | 90 | 90 | 75 | 61 |
| 95% CIb (nM) | 65-153 | 96-175 | 64-128 | 20-410 | 29-194 | 18-206 |
In vitro IC50s are shown for parasites with mutant or wild-type (WT) sequences at pfmdr1 codons 86 and 1246 and with the indicated number of pfnhe1 repeats.
CI, confidence interval.
Associations between polymorphisms and quinine treatment responses.
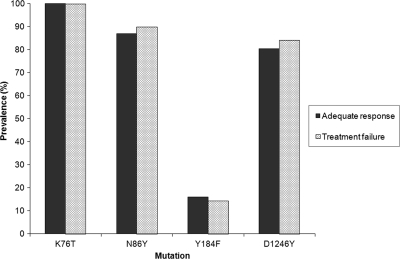
Of 79 samples available from the quinine treatment arm of a recent drug effectiveness trial in Kampala (1), we excluded 12 samples from patients classified by genotyping as clinical failures due to new infection during follow-up and 1 due to failed amplification, leaving 66 samples for analysis, including 46 (70%) from patients who experienced adequate clinical and parasitological responses and 20 (30%) from treatment failures due to recrudescent parasites (2 early treatment failures, 11 late clinical failures, and 7 late parasitological failures). All evaluable isolates (2 failed to amplify) had the pfcrt 76T mutation. For pfmdr1, the prevalence rates of the 86Y, 184F, and 1246Y mutations were very similar between isolates that led to adequate response or treatment failure (Fig. 2). Combinations of any 2 or 3 pfmdr1 mutations were also not associated with treatment response. As with isolates collected for in vitro analyses, mutations at codons 1034 and 1042 (tested in all 66 isolates) or increases in copy number (tested in 55 samples) of pfmdr1 were not seen in any of the clinical isolates.
FIG. 2.
Prevalence of pfcrt and pfmdr1 mutations among children who experienced adequate response or treatment failure after therapy with quinine.
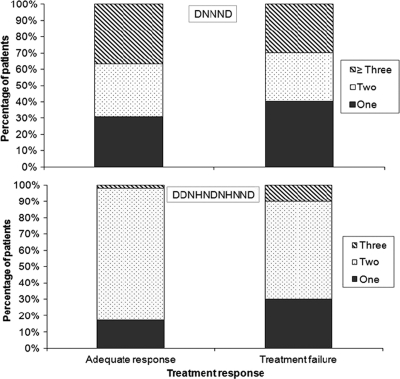
We next considered the impact of polymorphisms in pfnhe1 on treatment responses. The numbers of DNNND and DDNHNDHNND repeats varied somewhat between groups, but there was no significant association between the number of repeats and clinical response (Fig. 3). In particular, there was no significant difference in quinine treatment response between individuals harboring parasites with one versus two or more DNNND or DDNHNDNHNND repeats.
FIG. 3.
Percentage of isolates from patients treated with quinine with the indicated numbers of pfnhe1 ms4760 DNNND and DDNHNDNHNND repeats.
DISCUSSION
Polymorphisms in three P. falciparum genes have been linked to varied responses of malaria parasites to quinine. We considered associations between these polymorphisms and in vitro quinine sensitivity in samples from children with uncomplicated malaria in Uganda. We found modest associations between known polymorphisms in pfmdr1 and quinine sensitivity, including a significant decrease in sensitivity with an increased prevalence of combinations of the 86Y, 184F, and 1246Y mutations. Considering newly described polymorphisms in pfnhe1, we found a modest and statistically insignificant association between two copies and one or three or more copies of two adjacent repeats and decreased quinine sensitivity. We also assessed associations between P. falciparum polymorphisms and clinical outcomes using samples from a clinical trial of quinine for the treatment of uncomplicated malaria in Uganda. None of the tested polymorphisms was predictive of quinine treatment failure. Taken together, our results confirm prior findings showing modest associations between polymorphisms in pfmdr1 and quinine sensitivity, suggest that associations between pfnhe1 repeat numbers and quinine sensitivity are not as great as suggested in a recent paper (20), and indicate that the known polymorphisms evaluated in the present study are unlikely to serve as reliable markers of quinine treatment outcomes.
Although quinine has retained good antimalarial efficacy in most areas, its clinical efficacy has decreased in some regions, especially in Asia (51). Further, some reports have identified worrisome limitations in the efficacy of quinine in Africa, including our recent identification of a 23% failure rate for the treatment of uncomplicated falciparum malaria in Uganda (1). The recent Ugandan study did not include directly observed therapy, methodologies have varied among other studies, and the true incidence of quinine treatment failure in Africa is uncertain. Further, the relative contributions of different factors to treatment failure, including parasite resistance to quinine, poor compliance with therapeutic regimens, varied drug absorption and pharmacokinetics, and other factors, are unknown.
Of note, even in areas where quinine efficacy has appeared to remain good, such as sub-Saharan Africa, the sensitivity of individual P. falciparum isolates to the drug has varied widely. Thus, in older studies the IC50s for isolates collected in Senegal were 31 to 765 nM (7), and those for isolates collected in Cameroon were 23 to 780 nM (8). Many other studies did not report full ranges of sensitivity, but wide variations have commonly been seen. In the samples from Uganda utilized in this study, quinine IC50s ranged from 15 to 761 nM (29). Methodologies for in vitro assessments have varied, and associations between in vitro values and clinical outcomes are uncertain. Nonetheless, the wide range in quinine sensitivity seen across Africa and recent evidence for quinine treatment failure in Africa suggest that the evolution of parasites with decreased drug sensitivity may be contributing to diminished clinical responses to the drug.
The mediators of decreased quinine sensitivity of P. falciparum are uncertain. The well-described K76T polymorphism in the gene encoding the putative transporter pfcrt, which is the principal mediator of resistance to chloroquine (13, 49), also appears to play a role in mediating quinine sensitivity, on the basis of the results of laboratory studies (49, 58). The pfcrt 76T mutation was nearly universal in Ugandan isolates, so associations between this polymorphism and in vitro or clinical outcomes could not be studied. Polymorphisms in the gene encoding the multidrug resistance homolog pfmdr1 also play roles in resistance to multiple drugs (52, 58). Amplification of pfmdr1 decreases sensitivity to quinine (39, 58). The single nucleotide polymorphisms in pfmdr1 that are common in Africa, N86Y, Y184F, and D1246Y, have had less marked impacts on quinine sensitivity in laboratory studies (44, 48). In evaluations of clinical isolates, associations between pfmdr1 polymorphisms and in vitro quinine sensitivity have been uncertain, but evaluations considering the common polymorphisms seen in Africa have been limited (6). We recently evaluated pfmdr1 polymorphisms in the most and least sensitive of 196 isolates for which quinine sensitivity was measured in Uganda. The most resistant parasites were significantly more likely to harbor the pfmdr1 1246Y mutation, but not 86Y or 184F (29). Considering overall results, parasites with any of the three pfmdr1 polymorphisms seen in our isolates were somewhat more resistant to quinine than those without the polymorphism (Table 1), but the differences were not statistically significant. Considering the mutations together, an increasing number of mutations was associated with progressively greater quinine IC50s (Table 2).
Recently, quantitative trait locus analysis identified variations in the pfnhe1 gene, in addition to pfcrt and pfmdr1, as potentially mediating quinine sensitivity (18). Specifically, varied numbers of repeats in the ms4760 microsatellite were associated with quinine sensitivity. Following up on this report, four groups evaluated associations between ms4760 repeat numbers and in vitro quinine sensitivity. In 23 culture-adapted strains from multiple regions (20) and in 60 fresh isolates from the China-Myanmar border (25), increasing numbers of the DNNND repeat and decreasing numbers of the DDNHNDNHNND repeat were associated with decreased quinine sensitivity. In contrast, in 83 freshly isolated strains from Africa, a significant association was not seen between quinine sensitivity and the number of DNNND repeats, and an increased number of DDNHNDNHNND repeats was associated with decreased quinine sensitivity (5). Lastly, in 29 fresh isolates from Kenya, two DNNND repeats were associated with decreased sensitivity to quinine compared to the sensitivity of parasites with one or three copies, and there was no association between the number of DDNHNDNHNND repeats and quinine sensitivity (30). Our study adds the analysis of a larger number of isolates from Africa. We found only a modest trend toward decreased quinine sensitivity in isolates with two copies of either repeat. We also offer the first consideration of pfnhe1 polymorphisms and clinical outcomes. The number of either repeat was not associated with response to quinine therapy. Our results add to uncertainty regarding the importance of pfnhe1 polymorphisms in mediating quinine sensitivity and treatment response. More broadly, our finding of only modest associations between pfmdr1 and pfnhe1 polymorphisms and quinine sensitivity and treatment response are consistent with the conclusion that quinine sensitivity is a complex trait, with polymorphisms in multiple genes probably contributing to the phenotype (18, 28).
Simple molecular markers for resistance of malaria parasites to available drugs are valuable, as they allow the ready characterization of the sensitivity of parasites without the complex infrastructure required for clinical trials or in vitro analysis of parasites (36). Such data can help to guide treatment policy. For quinine, our results and prior studies suggest contributions of polymorphisms in multiple genes to drug sensitivity, but no single mutation or set of polymorphisms has consistently been a robust marker for in vitro quinine sensitivity. Consistent with this result, we saw, at best, only modest associations between previously identified polymorphisms and in vitro quinine sensitivity or quinine treatment outcomes. Thus, at present, straightforward markers for the responsiveness of P. falciparum to quinine are not available. Our analysis was limited to known markers of interest, and it is likely that additional polymorphisms contribute to quinine sensitivity. Further studies, including whole-genome sequencing strategies, may be needed to better characterize the molecular basis of quinine sensitivity and allow a better appreciation of the extent to which alterations in quinine sensitivity are limiting the antimalarial efficacy of this important drug.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AI52142, AI075045, and TW01506), the Department for International Development (DFID) through the Malaria Consortium (CNTR 04 5432), and the Doris Duke Charitable Foundation, with which P.J.R. is a distinguished clinical scientist. F.N.B. was the recipient of a Ruth L. Kirschstein National Research Service Award (5T32AI060537).
We thank the participants in the clinical trials from which samples were collected, their parents and guardians, and our clinical study and laboratory teams. We thank Harald Noedl, Medical University of Vienna, for valuable advice regarding drug sensitivity assays; Elizabeth Krow-Lucal, Edwin Ochong, and Christian Dokomajilar for helpful discussions; and the three anonymous reviewers for their helpful comments on the manuscript. Control P. falciparum strains used with in vitro assays were from the Malaria Research and Reference Reagent Resource Center.
Footnotes
Published ahead of print on 15 November 2010.
REFERENCES
- 1.Achan, J., J. K. Tibenderana, D. Kyabayinze, F. Wabwire Mangen, M. R. Kamya, G. Dorsey, U. D'Alessandro, P. J. Rosenthal, and A. O. Talisuna. 2009. Effectiveness of quinine versus artemether-lumefantrine for treating uncomplicated falciparum malaria in Ugandan children: randomised trial. BMJ 339:b2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ache, A., M. Escorihuela, E. Vivas, E. Paez, L. Miranda, A. Matos, W. Perez, O. Diaz, and E. Izarra. 2002. In vivo drug resistance of falciparum malaria in mining areas of Venezuela. Trop. Med. Int. Health 7:737-743. [DOI] [PubMed] [Google Scholar]
- 3.Adam, I., I. Salih, and M. I. Elbashir. 2005. Quinine for the treatment of uncomplicated Plasmodium falciparum malaria in eastern Sudan. Trans. R. Soc. Trop. Med. Hyg. 99:736-738. [DOI] [PubMed] [Google Scholar]
- 4.Agnamey, P., P. Brasseur, P. E. de Pecoulas, M. Vaillant, and P. Olliaro. 2006. Plasmodium falciparum in vitro susceptibility to antimalarial drugs in Casamance (southwestern Senegal) during the first 5 years of routine use of artesunate-amodiaquine. Antimicrob. Agents Chemother. 50:1531-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andriantsoanirina, V., D. Menard, S. Rabearimanana, V. Hubert, C. Bouchier, M. Tichit, J. L. Bras, and R. Durand. 2010. Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am. J. Trop. Med. Hyg. 82:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basco, L. K., and P. Ringwald. 2002. Molecular epidemiology of malaria in Cameroon. X. Evaluation of PFMDR1 mutations as genetic markers for resistance to amino alcohols and artemisinin derivatives. Am. J. Trop. Med. Hyg. 66:667-671. [DOI] [PubMed] [Google Scholar]
- 7.Brandicourt, O., P. Druilhe, F. Diouf, P. Brasseur, P. Turk, and M. Danis. 1986. Decreased sensitivity to chloroquine and quinine of some Plasmodium falciparum strains from Senegal in September 1984. Am. J. Trop. Med. Hyg. 35:717-721. [DOI] [PubMed] [Google Scholar]
- 8.Brasseur, P., J. Kouamouo, R. Moyou-Somo, and P. Druilhe. 1992. Multi-drug resistant falciparum malaria in Cameroon in 1987-1988. I. Stable figures of prevalence of chloroquine- and quinine-resistant isolates in the original foci. Am. J. Trop. Med. Hyg. 46:1-7. [DOI] [PubMed] [Google Scholar]
- 9.Chongsuphajaisiddhi, T., A. Sabchareon, and P. Attanath. 1983. Treatment of quinine resistant falciparum malaria in Thai children. Southeast Asian J. Trop. Med. Public Health 14:357-362. [PubMed] [Google Scholar]
- 10.Cooper, R. A., M. T. Ferdig, X. Z. Su, L. M. Ursos, J. Mu, T. Nomura, H. Fujioka, D. A. Fidock, P. D. Roepe, and T. E. Wellems. 2002. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol. Pharmacol. 61:35-42. [DOI] [PubMed] [Google Scholar]
- 11.Davis, J. C., T. D. Clark, S. K. Kemble, N. Talemwa, D. Njama-Meya, S. G. Staedke, and G. Dorsey. 2006. Longitudinal study of urban malaria in a cohort of Ugandan children: description of study site, census and recruitment. Malar. J. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourte, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, C. V. Plowe, and D. Coulibaly. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257-263. [DOI] [PubMed] [Google Scholar]
- 13.Djimde, A., O. K. Doumbo, R. W. Steketee, and C. V. Plowe. 2001. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet 358:890-891. [DOI] [PubMed] [Google Scholar]
- 14.Dokomajilar, C., S. L. Nsobya, B. Greenhouse, P. J. Rosenthal, and G. Dorsey. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondorp, A., F. Nosten, K. Stepniewska, N. Day, and N. White. 2005. Artesunate versus quinine for treatment of severe falciparum malaria: a randomised trial. Lancet 366:717-725. [DOI] [PubMed] [Google Scholar]
- 16.Dorsey, G., M. R. Kamya, A. Singh, and P. J. Rosenthal. 2001. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J. Infect. Dis. 183:1417-1420. [DOI] [PubMed] [Google Scholar]
- 17.Dorsey, G., S. Staedke, T. D. Clark, D. Njama-Meya, B. Nzarubara, C. Maiteki-Sebuguzi, C. Dokomajilar, M. R. Kamya, and P. J. Rosenthal. 2007. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. JAMA 297:2210-2219. [DOI] [PubMed] [Google Scholar]
- 18.Ferdig, M. T., R. A. Cooper, J. Mu, B. Deng, D. A. Joy, X. Z. Su, and T. E. Wellems. 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 52:985-997. [DOI] [PubMed] [Google Scholar]
- 19.Fungladda, W., E. R. Honrado, K. Thimasarn, D. Kitayaporn, J. Karbwang, P. Kamolratanakul, and R. Masngammueng. 1998. Compliance with artesunate and quinine + tetracycline treatment of uncomplicated falciparum malaria in Thailand. Bull. World Health Organ. 76(Suppl. 1):59-66. [PMC free article] [PubMed] [Google Scholar]
- 20.Henry, M., S. Briolant, A. Zettor, S. Pelleau, M. Baragatti, E. Baret, J. Mosnier, R. Amalvict, T. Fusai, C. Rogier, and B. Pradines. 2009. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob. Agents Chemother. 53:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry, M., I. Diallo, J. Bordes, S. Ka, B. Pradines, B. Diatta, P. S. M'Baye, M. Sane, M. Thiam, P. M. Gueye, B. Wade, J. E. Touze, J. M. Debonne, C. Rogier, and T. Fusai. 2006. Urban malaria in Dakar, Senegal: chemosusceptibility and genetic diversity of Plasmodium falciparum isolates. Am. J. Trop. Med. Hyg. 75:146-151. [PubMed] [Google Scholar]
- 22.Jelinek, T., P. Schelbert, T. Loscher, and D. Eichenlaub. 1995. Quinine resistant falciparum malaria acquired in east Africa. Trop. Med. Parasitol. 46:38-40. [PubMed] [Google Scholar]
- 23.Korbie, D. J., and J. S. Mattick. 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 3:1452-1456. [DOI] [PubMed] [Google Scholar]
- 24.Menard, D., F. Yapou, A. Manirakiza, D. Djalle, M. D. Matsika-Claquin, and A. Talarmin. 2006. Polymorphisms in pfcrt, pfmdr1, dhfr genes and in vitro responses to antimalarials in Plasmodium falciparum isolates from Bangui, Central African Republic. Am. J. Trop. Med. Hyg. 75:381-387. [PubMed] [Google Scholar]
- 25.Meng, H., R. Zhang, H. Yang, Q. Fan, X. Su, J. Miao, L. Cui, and Z. Yang. 2010. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob. Agents Chemother. 54:4306-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meshnick, S. R., and M. J. Dobson. 2001. The history of antimalarial drugs, p. 15-25. In P. J. Rosenthal (ed.), Antimalarial chemotherapy: mechanisms of action, resistance, and new directions in drug discovery. Humana Press Inc., Totowa, NJ.
- 27.Mu, J., M. T. Ferdig, X. Feng, D. A. Joy, J. Duan, T. Furuya, G. Subramanian, L. Aravind, R. A. Cooper, J. C. Wootton, M. Xiong, and X. Z. Su. 2003. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 49:977-989. [DOI] [PubMed] [Google Scholar]
- 28.Nkrumah, L. J., P. M. Riegelhaupt, P. Moura, D. J. Johnson, J. Patel, K. Hayton, M. T. Ferdig, T. E. Wellems, M. H. Akabas, and D. A. Fidock. 2009. Probing the multifactorial basis of Plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium-proton exchanger PfNHE. Mol. Biochem. Parasitol. 165:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nsobya, S. L., M. Kiggundu, S. Nanyunja, M. Joloba, B. Greenhouse, and P. J. Rosenthal. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob. Agents Chemother. 54:1200-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okombo, J., S. M. Kiara, J. Rono, L. Mwai, L. Pole, E. Ohuma, S. Borrmann, L. I. Ochola, and A. Nzila. 2010. In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob. Agents Chemother. 54:3302-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmieri, F., N. Petrosillo, M. G. Paglia, A. Conte, D. Goletti, L. P. Pucillo, M. Menegon, A. Sannella, C. Severini, and G. Majori. 2004. Genetic confirmation of quinine-resistant Plasmodium falciparum malaria followed by postmalaria neurological syndrome in a traveler from Mozambique. J. Clin. Microbiol. 42:5424-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parola, P., S. Ranque, S. Badiaga, M. Niang, O. Blin, J. J. Charbit, J. Delmont, and P. Brouqui. 2001. Controlled trial of 3-day quinine-clindamycin treatment versus 7-day quinine treatment for adult travelers with uncomplicated falciparum malaria imported from the tropics. Antimicrob. Agents Chemother. 45:932-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peel, S. A., P. Bright, B. Yount, J. Handy, and R. S. Baric. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648-658. [DOI] [PubMed] [Google Scholar]
- 34.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. S. Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plowe, C. V., A. Djimde, M. Bouare, O. Doumbo, and T. E. Wellems. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 52:565-568. [DOI] [PubMed] [Google Scholar]
- 36.Plowe, C. V., C. Roper, J. W. Barnwell, C. T. Happi, H. H. Joshi, W. Mbacham, S. R. Meshnick, K. Mugittu, I. Naidoo, R. N. Price, R. W. Shafer, C. H. Sibley, C. J. Sutherland, P. A. Zimmerman, and P. J. Rosenthal. 2007. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar. J. 6:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pradines, B., P. Hovette, T. Fusai, H. L. Atanda, E. Baret, P. Cheval, J. Mosnier, A. Callec, J. Cren, R. Amalvict, J. P. Gardair, and C. Rogier. 2006. Prevalence of in vitro resistance to eleven standard or new antimalarial drugs among Plasmodium falciparum isolates from Pointe-Noire, Republic of the Congo. J. Clin. Microbiol. 44:2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pradines, B., T. Pistone, K. Ezzedine, S. Briolant, L. Bertaux, M. C. Receveur, D. Parzy, P. Millet, C. Rogier, and D. Malvy. 2010. Quinine-resistant malaria in traveler returning from Senegal, 2007. Emerg. Infect. Dis. 16:546-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pukrittayakamee, S., A. Chantra, S. Vanijanonta, R. Clemens, S. Looareesuwan, and N. J. White. 2000. Therapeutic responses to quinine and clindamycin in multidrug-resistant falciparum malaria. Antimicrob. Agents Chemother. 44:2395-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pukrittayakamee, S., W. Supanaranond, S. Looareesuwan, S. Vanijanonta, and N. J. White. 1994. Quinine in severe falciparum malaria: evidence of declining efficacy in Thailand. Trans. R. Soc. Trop. Med. Hyg. 88:324-327. [DOI] [PubMed] [Google Scholar]
- 42.Pukrittayakamee, S., S. Wanwimolruk, K. Stepniewska, A. Jantra, S. Huyakorn, S. Looareesuwan, and N. J. White. 2003. Quinine pharmacokinetic-pharmacodynamic relationships in uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 47:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quashie, N. B., N. O. Duah, B. Abuaku, and K. A. Koram. 2007. The in-vitro susceptibilities of Ghanaian Plasmodium falciparum to antimalarial drugs. Ann. Trop. Med. Parasitol. 101:391-398. [DOI] [PubMed] [Google Scholar]
- 44.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 45.Roche, J., A. Guerra-Neira, J. Raso, and A. Benito. 2003. Surveillance of in vivo resistance of Plasmodium falciparum to antimalarial drugs from 1992 to 1999 in Malabo (Equatorial Guinea). Am. J. Trop. Med. Hyg. 68:598-601. [DOI] [PubMed] [Google Scholar]
- 46.Segurado, A. A., S. M. di Santi, and M. Shiroma. 1997. In vivo and in vitro Plasmodium falciparum resistance to chloroquine, amodiaquine and quinine in the Brazilian Amazon. Rev. Inst. Med. Trop. Sao Paulo 39:85-90. [DOI] [PubMed] [Google Scholar]
- 47.Sidhu, A. B., A. C. Uhlemann, S. G. Valderramos, J. C. Valderramos, S. Krishna, and D. A. Fidock. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidhu, A. B., S. G. Valderramos, and D. A. Fidock. 2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913-926. [DOI] [PubMed] [Google Scholar]
- 49.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, W. R., and N. J. White. 2004. Antimalarial drug toxicity: a review. Drug Saf. 27:25-61. [DOI] [PubMed] [Google Scholar]
- 51.Tinto, H., C. Rwagacondo, C. Karema, D. Mupfasoni, W. Vandoren, E. Rusanganwa, A. Erhart, C. Van Overmeir, E. Van Marck, and U. D'Alessandro. 2006. In-vitro susceptibility of Plasmodium falciparum to monodesethylamodiaquine, dihydroartemisinin and quinine in an area of high chloroquine resistance in Rwanda. Trans. R. Soc. Trop. Med. Hyg. 100:509-514. [DOI] [PubMed] [Google Scholar]
- 52.Valderramos, S. G., and D. A. Fidock. 2006. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 27:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinayak, S., M. T. Alam, M. Upadhyay, M. K. Das, V. Dev, N. Singh, A. P. Dash, and Y. D. Sharma. 2007. Extensive genetic diversity in the Plasmodium falciparum Na+/H+ exchanger 1 transporter protein implicated in quinine resistance. Antimicrob. Agents Chemother. 51:4508-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.WHO. 2003. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria, vol. WHO/HTM/RBM/2003.5. World Health Organization, Geneva, Switzerland.
- 55.WHO. 2010. Guidelines for the treatment of malaria, 2nd ed. World Health Organization, Geneva, Switzerland.
- 56.Witkowski, B., X. Iriart, P. N. Soh, S. Menard, M. Alvarez, V. Naneix-Laroche, B. Marchou, J. F. Magnaval, F. Benoit-Vical, and A. Berry. 2010. pfmdr1 amplification associated with clinical resistance to mefloquine in West Africa: implications for efficacy of artemisinin combination therapies. J. Clin. Microbiol. 48:3797-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 58.Woodrow, C. J., and S. Krishna. 2006. Antimalarial drugs: recent advances in molecular determinants of resistance and their clinical significance. Cell. Mol. Life Sci. 63:1586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeka, A., J. Achan, U. D'Alessandro, and A. O. Talisuna. 2009. Quinine monotherapy for treating uncomplicated malaria in the era of artemisinin-based combination therapy: an appropriate public health policy? Lancet Infect. Dis. 9:448-452. [DOI] [PubMed] [Google Scholar]
- 60.Zalis, M. G., L. Pang, M. S. Silveira, W. K. Milhous, and D. F. Wirth. 1998. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am. J. Trop. Med. Hyg. 58:630-637. [DOI] [PubMed] [Google Scholar]