Abstract
Chlamydiae are Gram-negative, obligate intracellular pathogens that replicate within a membrane-bounded compartment termed an inclusion. Throughout their development, they actively modify the eukaryotic environment. The type III secretion (TTS) system is the main process by which the bacteria translocate effector proteins into the inclusion membrane and the host cell cytoplasm. Here we describe a family of type III secreted effectors that are present in all pathogenic chlamydiae and absent in the environment-related species. It is defined by a common domain of unknown function, DUF582, that is present in four or five proteins in each Chlamydiaceae species. We show that the amino-terminal extremity of DUF582 proteins functions as a TTS signal. DUF582 proteins from C. trachomatis CT620, CT621, and CT711 are expressed at the middle and late phases of the infectious cycle. Immunolocalization further revealed that CT620 and CT621 are secreted into the host cell cytoplasm, as well as within the lumen of the inclusion, where they do not associate with bacterial markers. Finally, we show that DUF582 proteins are present in nuclei of infected cells, suggesting that members of the DUF582 family of effector proteins may target nuclear cell functions. The expansion of this family of proteins in pathogenic chlamydiae and their conservation among the different species suggest that they play important roles in the infectious cycle.
Chlamydiae are Gram-negative bacteria that constitute a distinct phylum. Long considered to comprise exclusively the family of Chlamydiaceae, which are obligate intracellular pathogens of vertebrates, chlamydiae now include a number of species described as symbionts of free-living amoebae and other eukaryotic hosts (16). The species that are pathogenic for humans include C. trachomatis, an agent of chronic genital and ocular infection, and C. pneumoniae, a prevalent cause of respiratory infections that is also possibly involved in atherosclerosis (30). Other species, which infect primarily animals, also have zoonotic potential.
All Chlamydia species share a unique, biphasic developmental cycle, which involves two distinct morphological and functional forms of the bacteria: the extracellular and invasive elementary body (EB) and the intracellular and replicative reticulate body (RB) (16, 23). Infection starts with the attachment of an EB to a host cell. Upon bacterial internalization, EBs gradually convert into RBs, which divide several times before differentiating back to the EB form at the end of the cycle. At 2 to 3 days after the initiation of infection, EBs are released in the extracellular space, ready to initiate a new cycle.
Importantly, throughout their developmental cycle, chlamydiae remain within a membrane-bounded compartment termed an inclusion (23). This localization restricts the interactions between the host and the bacteria. However, chlamydiae have acquired the ability to secrete a number of proteins into the host cell, including the inclusion membrane, presumably to create an environment favorable for survival and replication (reviewed in references 4 and 33). Most of these proteins, often called effectors, are secreted by a type III secretion (TTS) mechanism, which is also found in many Gram-negative pathogenic bacteria (11, 17, 18). Little is known about chlamydial effectors and how they manipulate host cellular processes (4, 33). In Chlamydia, efforts to identify effectors and their functions have been hampered by the absence of tools to genetically modify the bacteria and by their obligate intracellular lifestyle. The observation that TTS-dependent proteins of one bacterium can be secreted by a heterologous TTS apparatus of another bacterium opened the possibility of screening for chlamydial effectors. Using the heterologous TTS systems of Yersinia, Shigella, and Salmonella, numerous putative chlamydial effector proteins have been identified (10, 14, 31). TTS is active both during the intracellular multiplication phase of the cycle, as illustrated by the large family of proteins translocated into the inclusion membrane, the Inc proteins (27, 32), and during the entry step. Detection of TARP (translocated actin-recruiting phosphoprotein) in the host cytoplasm immediately after infection provided the first evidence that EBs are also capable of performing TTS across the plasma membrane (7).
In a directed screen to identify new chlamydial effector proteins we previously showed that the C. pneumoniae protein CPn0853 and its homologs in C. trachomatis and C. caviae (CT712 and CCA00914, respectively), possessed an amino-terminal signal recognized for TTS in Shigella flexneri (31). In this study, we show that these proteins belong to a large family of chlamydial proteins that share a domain of unknown function, referred to as DUF582. All pathogenic Chlamydia species sequenced so far possess four or five proteins belonging to the family. We provide evidence that members of this family are TTS substrates, which translocate into the host cell.
MATERIALS AND METHODS
Cell culture and bacterial culture conditions.
HeLa 229 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen). Chlamydia trachomatis L2 strain 434 (ATCC) was propagated in HeLa cells as previously described (6). For infection, semiconfluent monolayers of HeLa cells were inoculated with Chlamydia at a multiplicity of infection (MOI) of 0.5 to 1 for 1 h at 37°C. Infected cells were washed with phosphate-buffered saline (PBS) and incubated in fresh medium for the indicated times. Shigella flexneri mxiD and ipaB strains were grown as described previously (32). Escherichia coli strains DH5α and BL21 were cultivated at 37°C on Luria-Bertani (LB) plates or in LB liquid cultures.
Bioinformatics.
Protein sequences containing a DUF582 domain were obtained by a HMMER search (9) using the Pfam DUF582 model (12). The analysis was performed on eight Chlamydia genomes (C. trachomatis D/UW-3/CX, C. muridarum, C. abortus S26/3, C. caviae GPIC, C. felis Fe/C-56, C. pneumoniae CWL029, Protochlamydia amoebophila UWE25, and Waddlia chondrophila), resulting in a set of 26 sequences (no DUF582 domain was detected in the two environmental chlamydiae). Clustering of DUF582 protein families was performed by BLAST analysis after masking regions matching Pfam DUF582HMMs. DUF582 domain sequences were aligned using the multiple-sequence alignment program Promals (24). The plot of the sequence conservation of the multiple alignment was obtained by EMBOSS-Plotcon (26) with default parameters. Coiled-coil domains were detected by submitting the multiple alignment to Pcoil at the MPI Bioinformatics facilities (http://toolkit.tuebingen.mpg.de) (5).
Heterologous TTS assay.
The Shigella flexneri-based TTS assay was performed as previously described (32). Chimeras comprising the 5′ parts of different chlamydial genes upstream of the gene coding for the calmodulin-dependent adenylate cyclase of Bordetella pertussis were constructed by PCR as described previously (32). The constructs include about 30 nucleotides upstream from the proposed translation start sites and the first 24 to 30 codons of the chlamydial genes, using the following primers: CT619 (30 codons), AGTCAAGCTTGTAATAGTTTTGTTTTTATGTCTTCTTACTATTT and TCTCTAGAAAAATCTGAGCCAGGATTGG; CT621 (28 codons), AGTCAAGCTTGTAAATATTATTGGGATAGGTTCGC and AGTCTCTAGATGGTTTGGCAATCTTCTTTG; CPn0726 (24 codons), GTCAAGCTTGTAACTGTTCTTATCTAAGCAGACATTGA and AGTCTCTAGATGCAAAAGAATGCATTGAAGAC; and CPn0852 (26 codons), AGTCAAGCTTCTGATAAATACCGTGACGCTAC and AGTCTCTAGATGACGTATCAATCTTACCACCAG. The chimeric constructs were transformed in the S. flexneri strains SF401 and SF620, which are derivatives of M90T in which the mxiD and ipaB genes, respectively, have been inactivated (1, 22). Secretion in liquid cultures was assayed as described previously (32). Monoclonal antibodies against the adenylate cyclase (N. Guiso, Institut Pasteur) were used to detect the chimera, polyclonal antibodies against the Shigella type III effector IpaD (22) were used to control that TTS was not impaired by transformation of the various constructs, and antibodies against the cyclic AMP (cAMP) receptor protein (CRP) (A. Ullmann, Institut Pasteur) were used to control for bacterial lysis during fractionation.
Cloning, production of recombinant protein, and antibody production.
The open reading frames coding for the hypothetical proteins CT711, CT620, and CT621 from the C. trachomatis genome were amplified from C. trachomatis L2 DNA by PCR with Phusion high-fidelity DNA polymerase (Finnzyme, Espoo, Finland) according to the manufacturer's instructions and cloned into pET expression vector pET28 or pET30, providing an N-terminal six-histidine tag (Novagen, EMD Chemicals, Inc., Gibbstown, NJ).
For protein expression, constructs were transformed into E. coli BL21. The C. trachomatis proteins were expressed as 6×His-tagged N-terminal fusion proteins, and expression was induced in logarithmically growing cultures with isopropyl-β-d-thiogalactoside (IPTG) (Sigma). Bacterial culture pellets, resuspended in buffer containing 5 mM imidazole, 300 mM sodium chloride, and 20 mM Tris-HCl, were lysed with a French press. Lysates were spun to separate soluble and insoluble material. 6×His-tagged fusion proteins were purified from the soluble fraction by affinity chromatography using Ni-nitrilotriacetic acid (NTA) His Bind resin (Novagen, EMD Chemicals, Inc., Gibbstown, NJ) according to the manufacturer's guidelines. Purified proteins were used to immunize New Zealand White rabbits for production of polyclonal antisera (Agro-Bio, La Ferté Saint-Aubin, France).
Immunodetection.
For immunoblot analyses, samples of HeLa cells grown in six-well plates were infected with C. trachomatis L2 for the indicated time, washed twice in PBS, and detached in PBS supplemented with 0.5 mM EDTA. Where indicated, 100 μg/ml chloramphenicol dissolved in ethanol (34 mg/ml) was added 90 min prior to cell lysis; ethanol alone was added to the control cells. The cells were lysed in 1% sodium dodecyl sulfate (SDS), 6 M urea, 150 mM NaCl, and 30 mM Tris-HCl. The protein content of each lysate was quantified using the bicinchoninic acid (BCA) protein assay kit (Thermo) according to the manufacturer's instructions. An equal amount of total protein was loaded on 10 or 8% acrylamide gels, resolved by SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membranes. After incubation with primary antibodies to CT620, CT621, CT711, major outer membrane protein (MOMP) (mouse monoclonal antibody MyBioSource no. 310190), or actin (mouse monoclonal antibody clone AC-15; Sigma catalog no. 5441), membranes were probed with horseradish peroxidase-conjugated secondary antibodies and visualized with Amersham ECL Plus (GE Healthcare UK Limited).
C. trachomatis L2 EBs were purified on density gradients as described previously (29), lysed in the lysis buffer, and run on SDS-PAGE in parallel with the whole-cell lysates.
Immunofluorescence assay and microscopy.
HeLa cells grown on glass coverslips were infected with C. trachomatis L2 and fixed with 4% paraformaldehyde (PFA) at 31 h postinfection (p.i.). Cells were permeabilized in PBS containing 0.05% saponin (Sigma) and 1 mg/ml bovine serum albumin (BSA) for indirect immunofluorescence. Proteins were detected with antibodies against CT620 or CT621. Bacteria were labeled with mouse monoclonal anti-Hsp60 (Affinity BioReagents) or anti-MOMP (Argene no. 11-111) or with polyclonal rabbit antibodies to CT260 (a kind gift of R. Valdivia, Duke University). The respective antigens were visualized with Fluorolink Cy5-labeled goat anti-mouse antibody (Amersham) and Alexa 488-conjugated goat anti-rabbit antibody (Molecular Probes). DNA was stained with 0.5 μg/ml Hoechst 33342 (Molecular Probes) in the mounting medium. In antibody competition experiments, anti-CT620 or anti-CT621 antibodies were preincubated for 30 min with 4 μg purified His-CT620 or His-CT621 protein, respectively, or the irrelevant C. caviae protein CCA00037-His (31) in buffer used for immunolabelings. Images were acquired using an ApoTome microscope (Zeiss) equipped with a 63× objective and a Roper Scientific Coolsnap HQ camera, permitting optical sections of 0.7 μm.
Nuclear isolation.
HeLa cells (in two 10-cm dishes) infected for 40 h were washed once with PBS, pelleted, and resuspended in 0.4 ml buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, and 1 mM EGTA plus protease inhibitor cocktail [Sigma, P8340]) for 10 min before addition of 0.2% NP-40 and four passages through a 26-gauge syringe. Nuclei were pelleted at 800 × g for 5 min, while the supernatant was saved as the cytoplasmic fraction. Nuclei were washed two times with 1 ml buffer A, resuspended in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% SDS plus protease inhibitor cocktail) for 30 min, and centrifuged at 16,000 × g for 10 min to obtain the nuclear soluble fraction in the supernatant. Protein concentrations were determined using the Bradford assay (Bio-Rad protein assay), and equal quantities of the cytosolic and nuclear fractions (5 μg) were loaded for analysis by Western blotting. In addition to the antibodies already described, antibodies to poly(ADP ribose) polymerase (PARP), EF-Tu, and Rab-GDP dissociation inhibitor β (GDIβ) were used. Monoclonal anti-PARP antibody was purchased from Trevigen, monoclonal antibody against chlamydial EF-Tu was a kind gift from Y.-X. Zhang (Boston), and polyclonal rabbit antibody against GDIβ was a kind gift from B. Goud (Institut Curie, France).
Ectopically expressed GFP fusion proteins.
The open reading frames coding for the hypothetical proteins CT711, CT620, and CT621 from the C. trachomatis genome were amplified from C. trachomatis L2 DNA by PCR with Phusion high-fidelity DNA polymerase (Finnzyme, Espoo, Finland) according to the manufacturer's instructions and cloned into a plasmid pEGFP-derived destination vector providing a N-terminal green fluorescent protein (GFP) tag using the Gateway technology. HeLa cells were transiently transfected with these plasmids for 24 h using Fugene reagent (Roche), and the cells were fixed, briefly permeabilized with 0.05% saponin, mounted in Mowiol supplemented with 0.5 μg/ml Hoechst, and observed with an ApoTome microscope as described above.
RESULTS
Identification of the chlamydial DUF582 family of proteins.
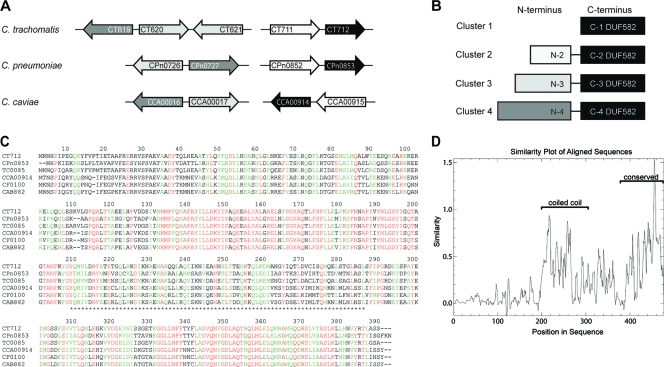
We have previously reported that the C. trachomatis protein CT712 and its homologs in C. pneumoniae and C. caviae are putative TTS substrates because they were secreted by the heterologous secretion system of S. flexneri (31). CT712 is predicted to encode a 390-amino-acid protein, consisting essentially of a domain of unknown function, annotated as DUF582 in the PFAM database. Bioinformatics analysis showed that this domain is also found at the carboxyl-terminal extremities of four other C. trachomatis proteins, CT711, CT619, CT620, and CT621. Moreover, DUF582 proteins are present in all pathogenic chlamydiae sequenced so far, and only in these bacteria. Each genome codes for four different DUF582 proteins, with the exceptions of C. trachomatis and C. muridarum, which have five DUF582 proteins (Table 1). Multiple-alignment analysis of all DUF582 domains showed that they have 17 to 88% sequence identity. Most DUF582 domains are associated with a second domain located in the N-terminal parts of the proteins. These domains were analyzed for their sequence similarity relationships and found to define four different clusters, with each cluster containing one (or two, for CT620/CT621 and TC0910/TC0911) proteins from each genome (Table 1 and Fig. 1 A and B). Interestingly, DUF582 domains are more conserved between orthologs (i.e., within the same cluster) than between paralogs (i.e., within the same species). For example, DUF582 proteins of cluster 1 show 45 to 88% identity, while identity between DUF582 domains within the C. trachomatis species ranges between 18% (CT711-CT712) and 39% (CT620-CT621).
TABLE 1.
Proteins with DUF582 domains in different Chlamydia species
| Chlamydia species | Protein (aa) in cluster: |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| C. trachomatis | CT712 (390) | CT711 (767) | CT620 (838), CT621 (832) | CT619 (877) |
| C. pneumoniae | CPn0853 (389) | CPn0852 (766) | CPn0726 (831) | CPn0727 (872) |
| C. caviae | CCA00914 (388) | CCA00915 (747) | CCA00017 (838) | CCA00016 (870) |
| C. muridarum | TC0085 (390) | TC0084 (769) | TC0910 (828), TC0911 (831) | TC0909 (875) |
| C. felis | CF0100 (388) | CF0099 (746) | CF0989 (849) | CF0990 (864) |
| C. abortus | CAB882 (388) | CAB833 (746) | CAB017 (885) | CAB016 (866) |
FIG. 1.
Conservation of the DUF582 proteins in Chlamydiaceae. (A) Schematic overview of the genes encoding DUF582-containing proteins in the C. trachomatis, C. pneumoniae, and C. caviae genomes. (B) Schematic representation of the Chlamydia DUF582 proteins. The proteins are grouped in four clusters based on sequence similarity. In all of them, DUF582 is located at the carboxy terminus. Proteins belonging to clusters 2, 3, and 4 contain additional N-terminal domains. The gray-level code corresponds to the DUF582 genes depicted in panel A. (C) Alignment of cluster 1 DUF582 proteins in six chlamydial genomes: C. trachomatis (CT), C. pneumoniae (CPn), C. muridarum (TC), C. caviae (CCA), C. felis (CF), and C. abortus (CAB). Identical residues are shown in red. Amino acids with high similarity are highlighted in green. Asterisks delimit the segment predicted to adopt a coiled-coil conformation. (D) Sequence conservation in the multiple alignment of all DUF582 sequences.
Within one cluster, the amino-terminal domains show between 25 and 75% identity within their primary sequence, while no similarity was found when the amino-terminal domains of proteins from different clusters were compared. Database iterative searches using the N-terminal multiple alignments retrieved no homolog sequences, meaning that these N-terminal domains are also Chlamydia specific.
Finally, DUF582 proteins were analyzed with structure prediction tools. DUF582 is predicted to be mainly α-helical (80% as estimated by the Gor IV secondary-structure prediction method [13] and confirmed in the Promals alignment) and to contain a segment adopting a coiled-coil conformation (Fig. 1C). When all DUF582 sequences are compared, the coiled-coil domain and the carboxy-terminal part stand as the best conserved segments of this domain of unknown function (Fig. 1D). The amino-terminal parts of the proteins of clusters 2 to 4 are also predicted to be rich in alpha helices, possibly adopting coiled-coil structures.
DUF582-containing proteins are TTS substrates.
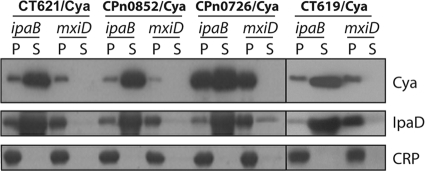
We have previously described a secretion assay in Shigella that allowed us to identify potential chlamydial TTS substrates (31). Three proteins of cluster 1 of the DUF582 protein family were shown to possess TTS signals (CT712, CPn0853, and CCA00914). To investigate whether other members of the family possessed TTS signals, we tested proteins representative of each cluster in the Shigella secretion assay. Because the CT620/Cya and CT711/Cya chimeras were not well expressed (data not shown), we constructed chimeras with the C. pneumoniae gene of the same cluster. Fusion constructs were made between the N-terminal part of the respective chlamydial gene and a reporter molecule, the calmodulin-dependent adenylate cyclase of Bordetella pertussis (Cya). Each construct was transformed into the TTS-competent Shigella ipaB strain and into the mxiD strain, which is deficient for TTS (1, 22). Shigella organisms expressing the chlamydial constructs were grown to exponential phase and harvested. Cultures were fractionated into cell culture supernatant and cell pellet and analyzed by Western blotting (Fig. 2). CPn0852/Cya (cluster 2), CT621/Cya and CPn0726/Cya (both from cluster 3), and CT619/Cya (cluster 4) were all detected in the supernatant of the transformed ipaB strain, indicating that they were secreted by the bacteria. Importantly, CRP, a cytosolic Shigella protein, was detected only in the pellet fractions, showing that detection of the chimera in the supernatant did not result from bacterial lysis. An endogenous TTS substrate of Shigella, IpaD, was also detected in the supernatant, indicating that expression of the chimera did not prevent TTS of Shigella effectors. In contrast, the chimeras were not detected in the supernatant of transformed mxiD cultures, which are deficient for type III secretion. This result implies that secretion of the four chimeras observed in the ipaB background was type III dependent. In conclusion, this experiment demonstrates that the amino-terminal regions of all the DUF582 family members tested contain TTS signals recognized by S. flexneri. This result strongly supports the hypothesis that all the members of the DUF582 family are Chlamydia TTS effectors.
FIG. 2.
DUF582 proteins CT621, Cpn0852, CPn0726, and CT619 have TTS signals. The amino-terminal segments of the indicated proteins (24 to 30 amino acids, depending on the constructs) were fused to the Cya reporter protein and expressed in a Shigella flexneri ipaD (constitutive TTS) or mxiD (defective TTS) strain. Exponential-phase cultures expressing the reporter fusion protein were fractionated into supernatants (S) and pellets (P). Samples were resolved by SDS-PAGE, transferred to a PVDF membrane, and probed with anti-Cya (to detect chlamydial fusion proteins), anti-IpaD (Shigella secreted protein), or anti-CRP (Shigella nonsecreted protein) antibodies. All results shown are representative of at least two separate experiments.
C. trachomatis DUF582 proteins are expressed in the middle and late phases of the infectious cycle.
For the remainder of our study, we concentrated on the C. trachomatis DUF582 proteins: CT712 (cluster 1; 390 amino acids [aa]; expected molecular mass, 44 kDa), CT711 (cluster 2; 767 aa; expected molecular mass, 86 kDa), CT620 (cluster 3; 838 aa; expected molecular mass, 93kDa), CT621 (cluster 3; 832 aa; expected molecular mass, 93 kDa), and CT619 (cluster 4; 877 aa; expected molecular mass, 97 kDa).
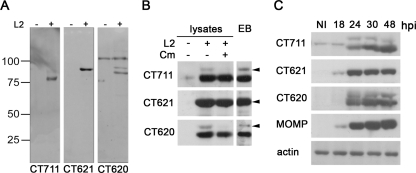
All proteins were expressed as His-tagged recombinant proteins in E. coli. CT620, CT621, and CT711 were purified by affinity chromatography and used to immunize rabbits to obtain antiserum. CT619 was expressed at a very low level, and CT712 was insoluble; both were excluded from further analysis. The antisera obtained for CT620, CT621, and CT711 were tested for their specificity on HeLa cell lysates infected or not for 30 h with C. trachomatis L2 (Fig. 3A). Antibodies against CT621 reacted with a one protein product of the expected molecular mass which was present only in the infected samples. In infected-cell lysates, antibodies against CT620 reacted mainly with a 83-kDa product and only faintly with a 93-kDa product, which is the expected molecular mass of this protein. In addition, the serum reacted with a higher-molecular-mass product that was also present in noninfected cells and which we therefore considered nonspecific. Strikingly, the same pattern of migration was observed with antibodies against CT711: the full-length protein was hardly visible in total cell extracts, and the antibodies reacted mainly with a lower-molecular-mass product that could represent a truncation of 10 kDa (Fig. 3A).
FIG. 3.
Expression of the C. trachomatis DUF582 proteins CT711, CT620, and CT621 during chlamydial infection. (A) Polyclonal antiserum obtained after immunization of rabbits with recombinant CT711, CT620, and CT621 was tested for its specificity on whole-cell lysates. HeLa cells were mock infected (−) or infected with C. trachomatis L2 (+) for 30 h. Total cell lysates were resolved in polyacrylamide gels and probed by immunoblotting with antibodies to CT711, CT620, and CT621. (B) HeLa cells were infected or not with C. trachomatis L2 for 40 h at 37°C, prior to addition of 100 μg/ml chloramphenicol (Cm) in the indicated sample. Ninety minutes later, the cells were collected and whole-cell lysates were probed with the indicated antibodies. Density gradient-purified EBs were lysed and run in parallel with the whole-cell lysates. Arrowheads point to the expected molecular size for each DUF582 protein. (C) HeLa cell lysates infected with C. trachomatis L2 for the indicated time were run on SDS-PAGE and transferred to membranes. Membranes were probed with antibodies to CT711, CT621, CT620, and MOMP. Antibodies against actin were used as a loading control.
This similarity in patterns, using two different antibodies, suggests that the two proteins undergo similar processing. We suspected that the small amount of full-length CT620 or CT711 detected by Western blotting was due to a rapid processing into the truncated forms. To test this hypothesis, we blocked bacterial protein synthesis by incubating infected cells for 90 min in 100 μg/ml chloramphenicol before cell lysis. Following this treatment, the amounts of full-length CT620 and CT711 detected in cell lysates strongly decreased compared to those in lysates of untreated cells, demonstrating that these two forms have a short half-life (Fig. 3B). In comparison, inhibition of protein synthesis for 90 min only slightly decreased the amounts of CT621 and of the truncated forms of CT620 and CT711, showing that these products are stable. This difference in stability between the full-length and truncated forms can account for their respective abundances in total cell lysates. Importantly, bacterial lysates made from EBs purified on a density gradient showed the same double-band migration profile for CT620 and CT711 as previously observed on whole infected-cell lysates, and in the same proportion of full-length versus truncated protein products, suggesting that processing occurs in the bacteria (Fig. 3B). However, it is also possible that the shorter forms of CT620 and CT711 result from spurious postlysis degradation of these proteins rather than specific processing.
Although these three proteins share a DUF582 domain, we did not expect the sera to cross-react with the different members of the family because the domain is not very conserved between paralogs. Indeed, we did not observe cross-reaction when testing our sera against the different purified DUF582 proteins (data not shown). Altogether, we concluded that our sera were specific for the proteins against which they were raised and could be used for detection by immunoblotting.
Next, we analyzed when the C. trachomatis DUF582 containing proteins are expressed during the course of infection. We found that all three proteins were expressed by 24 h postinfection, with a sharp increase between the 18-h and 24-h time points, which corresponds to an increase in bacterial load observed with the antibody against MOMP (Fig. 3C). By microarray analysis, transcription of the genes coding for C. trachomatis DUF582 proteins was first detected at 8 h postinfection (p.i.), when RB-to-EB conversion is completed (3). These data are consistent with our finding that the DUF582 proteins are detected coincidently with an increase in bacterial number and remain present throughout the remainder of the cycle.
Localization of C. trachomatis DUF582-containing proteins in the host cell cytoplasm and in the lumen of the chlamydial inclusion.
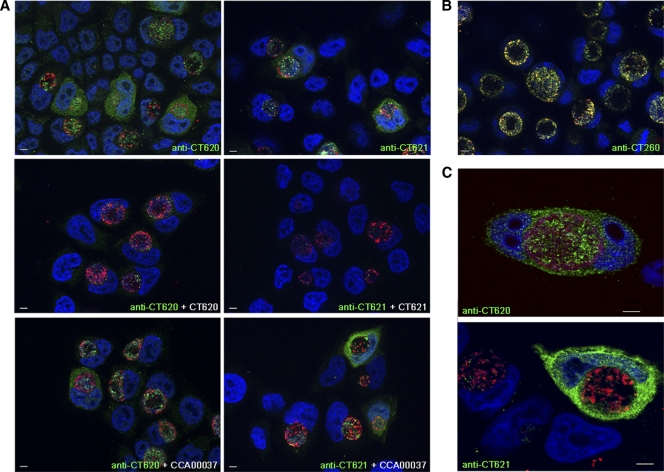
We next analyzed the expression and localization of DUF582-containing proteins by indirect immunofluorescence microscopy. Our antibody for CT711 failed to detect a specific signal, probably because the protein is expressed at a level below the detection threshold with this technique, and was therefore excluded from our immunolocalization studies. HeLa cell monolayers were infected with C. trachomatis for 31 h, fixed, and labeled for CT620 or CT621. Both proteins were observed in association with the inclusion and with the cytoplasm of infected HeLa cells (Fig. 4A). To control for the specificity of these stainings, we verified that they disappeared when the sera were used in the presence of an excess of the purified proteins against which they were raised but not in the presence of an irrelevant His-tagged purified protein, CCA00037-His (Fig. 4A). When we performed a time course of infection, the earliest detectable signal was obtained at 18 to 20 h p.i., and at these early time points the labeling was restricted to the inclusion (data not shown). Cells with cytoplasmic staining for CT620 and CT621 were frequently observed at 31 h p.i., and by later time points, these proteins were found in the cytoplasm of almost all infected cells.
FIG. 4.
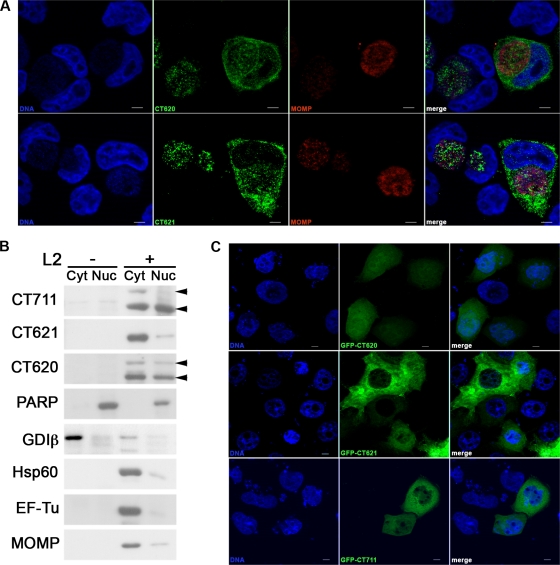
The C. trachomatis DUF582 proteins CT620 and CT621 are detected in the host cytoplasm during chlamydial infection. HeLa cells were infected with C. trachomatis L2 and fixed 31 h later. (A) C. trachomatis DUF582 proteins were immunolabeled with anti-CT620 and anti-CT621 as indicated, and bacteria were detected with a mouse anti-Hsp60 antibody. Coverslips were subsequently incubated with Alexa 488-conjugated anti-rabbit antibody to detect the DUF582 proteins (green) and with Cy5-conjugated anti-mouse antibody for Hsp60 (red). Host cell nuclei and bacterial DNA were stained with Hoechst stain (blue). For antibody competition, anti-CT620 and anti-CT621 were incubated in the presence of an excess of His-CT620, His-CT621, or CCA00037-His (an irrelevant His-tagged protein), as indicated. Arrows point to cells in which cytoplasmic staining for CT620 and CT621 is observed. Note that in addition to the cytoplasmic staining, CT620 and CT621 also localize to the lumen of the chlamydial inclusion with only little colocalization with the bacterial protein Hsp60. (B) Infected cells were labeled with antibodies to the chlamydial chaperones CT260 (green) and Hsp60 (red), which colocalize. (C) C. trachomatis DUF582 proteins were immunolabeled with anti-CT620 and anti-CT621 as indicated, and bacteria were detected with a mouse anti-MOMP antibody. Coverslips were subsequently incubated with Alexa 488-conjugated anti-rabbit antibody to detect the DUF582 proteins (green) and with Cy5-conjugated anti-mouse antibody for MOMP (red). Host cell nuclei and bacterial DNA were stained with Hoechst stain (blue). Images were acquired using an ApoTome fluorescence microscope. Bars, 5 μm.
In addition to their cytosolic localization, CT620 and CT621 were detected in the inclusion. The inclusion staining for CT620 and CT621 did not fully overlap with the bacterial marker Hsp60 and was found in areas devoid of bacteria (Fig. 4A). In comparison, rabbit serum against the bacterial chaperone CT260 showed a perfect colocalization with Hsp60 (Fig. 4B). Similar results were observed after methanol fixation (data not shown), indicating that it is not an artifact of fixation. As an additional control, we used anti-MOMP antibodies to stain the outer membrane of bacteria. Again, a large proportion of the luminal signal for CT620 or CT621 did not colocalize with MOMP (Fig. 4C). These observations indicate that in addition to being secreted in the host cytoplasm, a portion of CT620 and CT621 is secreted from the bacteria within the lumen of the inclusion.
DUF582 proteins are detected in the nuclei of infected cells.
During the course of this study, CT621 was reported to be detected in the nuclei of infected cells (15). Our anti-CT620 and anti-CT621 sera also detected proteins within the nuclei of infected cells (Fig. 5A). Importantly, for both markers, nuclear staining was observed in about 80% (n = 20 for each antibody) of the infected cells in which the proteins were also observed in the cytoplasm. In noninfected cells, or in cells in which the DUF582 proteins were observed only in the inclusion, no nuclear staining was visible with either anti-CT620 or anti-CT621 antibodies, indicating that the staining is specific. To confirm the nuclear localization of DUF582 proteins by another approach, we subjected infected cells to a subcellular fractionation protocol to separate the cytoplasmic material (including the bacteria) from the nuclei. To control for the purity of our fractions, we probed the membrane for GDIβ (found only in the cytoplasmic fractions) and PARP (found only in the nuclear fractions). In addition, antibodies against three different nonsecreted chlamydial proteins (Hsp60, EF-Tu, and MOMP) were used to determine the level of contamination of the nuclear fractions with dense bacteria. In addition to a cytoplasmic distribution, CT620 and CT711 were found to be enriched in the nuclear fraction of infected cells compared to the three bacterial markers (Fig. 5B). Interestingly, although there was less full-length than truncated CT620 in the nucleus, it was still significantly enriched in this fraction relative to its total expression level. The low level of expression of full-length CT711 does not allow us to ascertain its presence in the nuclear fraction, while the truncated form was clearly visible. In contrast, CT621 was not clearly enriched in the nuclear fraction compared to other nonsecreted chlamydial proteins. This result is in slight contradiction with the detection of CT621 in the nuclei of infected cells by microscopy. It could be accounted for by the observation that although CT621 is more abundant than CT620 and CT711 (as assessed by immunofluorescence and Western blotting), the nuclear staining for CT621 was not stronger than that for CT620 (Fig. 5A). Therefore, the proportion of nuclear CT621 is probably lower than that of CT620 and just below the detection level by the subcellular fractionation technique. It is important to note that equal protein amounts were loaded in each lane. This represents about a 5-fold concentration of the nuclear fractions relative to the cytoplasmic fractions. Therefore, even though DUF582 proteins are present in the nuclear fractions, they distribute primarily in the cytoplasmic fractions, in agreement with our microscopy observations.
FIG. 5.
DUF582 proteins are detected in the host nucleus during infection. (A) HeLa cells were infected with C. trachomatis L2 and fixed 31 h later. C. trachomatis DUF582 proteins were immunolabeled with anti-CT620 and anti-CT621 as indicated, and bacteria were detected with a mouse anti-MOMP antibody. Coverslips were subsequently incubated with Alexa 488-conjugated anti-rabbit antibody to detect the DUF582 proteins (green) and with Cy5-conjugated anti-mouse antibody for MOMP (red). Host cell nuclei and bacterial DNA were stained with Hoechst stain (blue). Both CT620 and CT621 were observed in the cell nuclei. (B) HeLa cells were infected with C. trachomatis for 40 h. Nuclei were isolated as described in Materials and Methods, and both cytosolic and nuclear fractions were analyzed by Western blotting. The quality of the fractionation was controlled using GDIβ as a cytosolic marker and PARP as a nuclear marker. In addition, three nonsecreted chlamydial proteins, EF-Tu, Hsp60, and MOMP, were used to assess the degree of contamination of the nuclear fraction with bacteria. Note that the proportion of CT621 detected in the nuclear fraction is similar to what is observed for the nonsecreted proteins. In contrast, both CT620 and CT711 (arrowheads) are enriched in the nuclear fractions, above the contamination level. Blots are representative of three separate experiments. (C) HeLa cells were transfected for 24 h with the indicated GFP fusion proteins before fixation. Nuclei were stained with Hoechst stain (blue). While GFP-CT620 and GFP-CT711 are observed in the nuclei of all transfected cells, nuclear staining was not detected in all GFP-CT621-transfected cells. Bars, 5 μm.
Finally, to determine whether DUF582 proteins were able to translocate to the cell nucleus even in the absence of infection, we expressed N-terminal GFP fusion proteins by transfection in HeLa cells. GFP-CT620, GFP-CT621, and GFP-CT711 were all detected at least to some extent in the nuclei of transfected cells, with differences between constructs (Fig. 5C). Nuclear staining was easily detected for GFP-CT620, while it was visible in only some of the GFP-CT621-transfected cells. GFP-CT711 was very poorly expressed and could be seen in the nuclei of transfected cells. Altogether, GFP fusion proteins reproduced what was observed in infection in terms of level of expression (with GFP-CT621 being better expressed than GFP-CT620 and GFP-CT711 being very poorly expressed) and in terms of nuclear localization (with GFP-CT620 and GFP-CT711 being more enriched in the nucleus than GFP-CT621).
DISCUSSION
Chlamydiae, like other Gram-negative pathogens, use a TTS system to translocate effector proteins into their host to modulate cellular functions. In this study, we have identified a new family of chlamydial effector proteins, called the DUF582 proteins, that are secreted by a TTS mechanism during infection. This conclusion is based on (i) the presence of a TTS signal in the seven DUF582 proteins from three different species that we tested (this report and reference 31), (ii) the demonstration of the cytoplasmic localization of two DUF582 proteins in infected cells, and (iii) the detection of three DUF582 proteins in the nuclei of infected cells. The family consists of 26 members in the six annotated genomes that we analyzed.
We have previously shown that the DUF582 proteins CPn0853, CT712, and CCA00914 are recognized by the heterologous TTS system of Shigella flexneri (31). These proteins share a domain of unknown function, DUF582, and we hypothesized that other DUF582 proteins might be recognized for secretion by the TTS system. Indeed, we showed here that four other members of the family possessed a TTS signal recognized by the Shigella TTS system. Altogether, we now have demonstrated the presence of a TTS signal in at least one member of each of the four different DUF582 subfamilies that can be distinguished based on sequence analysis, suggesting that the whole family serves as substrates for TTS. So far, no well-conserved sequence motif has been identified in TTS substrates. However, two independent research groups presented a sequence-based computational approach to predict TTS signals (2, 28). Both groups applied this approach to chlamydial genomes and identified a number of putative TTS effectors. Interestingly, the C. trachomatis DUF582 proteins CT620 and CT711 were among the top 10% of predicted chlamydial effectors by one of the predictors (28). However, CT712, CT619, and CT621 were not predicted to be TTS substrates, showing the limitations of this approach. Of the seven DUF582 proteins for which we demonstrated the presence of a TTS signal, the second predictor (2) successfully classified CCA00914, CPn0726, CPn0852, and CT621 as potential TTS substrates but not CPn0853, CT619, or CT712, indicating again that the experimental approach using heterologous secretion remains to date the best method to identify potential TTS effectors.
Most soluble TTS effectors studied so far are produced in small amounts, hampering their detection by immunofluorescence. CT621 and, to a lesser extent, CT620 were abundantly expressed, allowing for detection by Western blotting and immunofluorescence. Both proteins were observed in the cytoplasm of infected cells, demonstrating that they are secreted during infection. CT711 expression was lower, and the protein was not detected by immunofluorescence. In subcellular fractionation experiments analyzed by Western blotting, CT711 was observed in the nuclear fraction of infected cells, bringing evidence that this protein is also secreted out of the inclusion during infection. CT620 was also detected in the nuclear fraction and observed in the nucleus by microscopy techniques. While we were able to confirm earlier CT621 detection in the nuclei of infected cells (15) by microscopy, only a small proportion of the total pool of CT621 is nuclear, making it difficult to detect over bacterial contamination by subcellular fractionation techniques.
The detection of all three DUF582 proteins tested in the nuclei of infected cells is intriguing. These proteins are too large to diffuse passively into the nucleus, suggesting that active import occurs, although no obvious nuclear localization signal was identified in their sequence. Interestingly the proteins were also observed in the nucleus when ectopically expressed by transfection, indicating that other bacterial factors are not needed for their translocation in the nucleus in infection. The only other nuclear effector of Chlamydia identified so far, NUE, is found predominantly in the nucleus (25). This is not the case for the DUF582 proteins, which are more abundant in the cytosol than in the nucleus and might shuttle between these locations. NUE has histone methyltransferase activity, suggesting that chlamydial proteins may target host gene expression (25). Whether DUF582 proteins function in the cytoplasm and/or in the nucleus during infection requires further investigation.
In addition to their cytoplasmic localization, CT620 and CT621 were also observed in the lumen of the inclusion. Proteins stored in the bacteria prior to secretion do not account for the totality of the luminal staining, because it does not fully overlap with the distribution of two bacterial markers, Hsp60 and MOMP. More likely, most of the luminal signal corresponds to proteins secreted out of the bacteria inside the inclusion lumen. This observation suggests that TTS can occur independently of the formation of a translocation pore across the inclusion membrane. In Shigella, it has been reported that there is always 4 to 5% secretion into the extracellular medium even in the absence of host cells (21). In addition, it now appears that several membranous compartments from the host might make their way to the inside of the inclusion (reference 8 and our own observations), which could provide the necessary trigger for TTS within the inclusion. The luminal staining pattern for CT620 and CT621 appears to be more punctate than the diffuse staining pattern we observed in the cytosol. One could speculate that once DUF582 proteins reach the lumen of the inclusion they tend to aggregate because they are not in the right cytosolic environment. This would be consistent with Shigella proteins that also aggregate if they are secreted by “leakage” into the culture medium (21). Aggregation would concentrate the signal and might explain why when looking at CT620 and CT621 distribution over time, we always observed the luminal staining, while cytoplasmic staining was visible only at later times of infection. Whether luminal DUF582 proteins have activity or represent leakage remains to be determined. Two other chlamydial proteins of unknown function, Pls1 and Pls2, were shown to localize to globular structures within the inclusion lumen and at the inclusion membrane (19).
CT620 and CT711 share an intriguing migration profile. We have shown that both proteins are present in whole-cell lysates as a high-molecular-mass product, which corresponds to the expected size of the full-length protein, and a truncated product with a molecular mass 10 kDa lower. The high-molecular-mass product is unstable, disappearing within 90 min of inhibition of protein synthesis, while the truncated product is stable. Both forms were detected in density gradient-purified EBs in the same proportions as in the whole-cell lysates, suggesting that cleavage occurs before translocation out of the bacteria. These observations suggest that the full-length CT620 and CT711 are rapidly processed into truncated forms. However, we cannot at this stage rule out the hypothesis that the shorter forms result from postlysis degradation of the proteins.
DUF582 proteins are present in all pathogenic chlamydiae sequenced so far and are absent from the two environmental chlamydiae sequenced, Protochlamydia amoebophila and Waddlia chondrophila (this study and reference 15). Their conservation in pathogenic bacteria, together with our finding that they probably all represent TTS effectors, suggests that they may likely play an important role in pathogenesis. CT620, CT621, and CT711 were first detected only about 18 to 24 h after infection, which corresponds to the middle to late phase of the C. trachomatis serovar L2 developmental cycle. More broadly, in transcriptomic analyses, all genes coding for DUF582 proteins came up as midcycle/tardy genes (3, 20), suggesting that our results can be extrapolated to all DUF582 proteins. Sequence analysis failed to provide clues to the potential function of these proteins, which show no similarity with known proteins. Importantly, while they share the DUF582 domain, they diverge largely regarding the N-terminal part. This domain is absent from members of cluster 1, or very different in sequence in members from clusters 2, 3, and 4, suggesting that each cluster might represent addition of a different module to the same functional DUF582 domain. Their expression profile suggests that they could be involved in the middle and/or late steps of development, including exit from the host cell. The CT620 and CT621 distributions suggest that these DUF582 proteins, and maybe other members of the family, are abundantly present in the host cytoplasm during infection. This property, together with the observation that they are found in all pathogenic chlamydiae and not in other bacteria, makes them interesting candidates for major histocompatibility complex (MHC) class I presentation of bacterial antigens. Our present efforts are concentrating on the identification of interacting proteins for several members of the family, which might give clues to the putative function(s) of the conserved DUF582 domain.
Acknowledgments
We thank Valérie Malardé for excellent technical help and Scot Ouellette for donation of density gradient-purified EBs and critical reading of the manuscript.
This work was supported by the European Marie Curie program European Initiative for Basic Research in Microbiology and Infectious Diseases and by the Agence Nationale pour la Recherche (ANR-06-JCJC-0105).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 15 November 2010.
REFERENCES
- 1.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol. Microbiol. 7:59-68. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, R., S. Brandmaier, F. Kleine, P. Tischler, E. Heinz, S. Behrens, A. Niinikoski, H. W. Mewes, M. Horn, and T. Rattei. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., G. M. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, H. J., K. Wolf, and K. A. Fields. 2009. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12:81-87. [DOI] [PubMed] [Google Scholar]
- 5.Biegert, A., C. Mayer, M. Remmert, J. Soding, and A. N. Lupas. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34:W335-W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boleti, H., A. Benmerah, D. Ojcius, N. Cerf-Bensussan, and A. Dautry-Varsat. 1999. Chlamydia infection of epithelial cells expressing dynamin and Eps15 mutants: clathrin-independent entry into cells and dynamin-dependent productive growth. J. Cell Sci. 112:1487-1496. [DOI] [PubMed] [Google Scholar]
- 7.Clifton, D. R., K. A. Fields, N. A. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchiaro, J. L., Y. Kumar, E. R. Fischer, T. Hackstadt, and R. H. Valdivia. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379-9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 10.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048-1060. [DOI] [PubMed] [Google Scholar]
- 11.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671-683. [DOI] [PubMed] [Google Scholar]
- 12.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 14.Ho, T. D., and M. N. Starnbach. 2005. The Salmonella enterica serovar Typhimurium-encoded type III secretion systems can translocate Chlamydia trachomatis proteins into the cytosol of host cells. Infect. Immun. 73:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobolt-Pedersen, A.-S., G. Christiansen, E. Timmerman, K. Gevaert, and S. Birkelund. 2009. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn, M. 2008. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62:113-131. [DOI] [PubMed] [Google Scholar]
- 17.Hsia, R.-C., Y. Pannekoek, E. Ingerowski, and P. Bavoil. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25:351-359. [DOI] [PubMed] [Google Scholar]
- 18.Hueck, C. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen, I., and R. H. Valdivia. 2008. Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect. Immun. 76:3940-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer, A. P., A. Mehlitz, H. J. Mollenkopf, and T. F. Meyer. 2007. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 3:752-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ménard, R., P. Sansonetti, and C. Parsot. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei, J., B. H. Kim, M. Tang, and N. V. Grishin. 2007. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 35:W649-W652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennini, M. E., S. p. Perrinet, A. Dautry-Varsat, and A. Subtil. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 6:e1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 27.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 28.Samudrala, R., F. Heffron, and J. E. McDermott. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scidmore, M. A. 2005. Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr. Protoc. Microbiol. 11:A11.11-A11.25. [DOI] [PubMed] [Google Scholar]
- 30.Stephens, R. S. (ed.) 1999. Chlamydia. Intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC.
- 31.Subtil, A., C. Delevoye, M. E. Balañá, L. Tastevin, S. Perrinet, and A. Dautry-Varsat. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 56:1636-1647. [DOI] [PubMed] [Google Scholar]
- 32.Subtil, A., C. Parsot, and A. Dautry-Varsat. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39:792-800. [DOI] [PubMed] [Google Scholar]
- 33.Valdivia, R. H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53-59. [DOI] [PubMed] [Google Scholar]