Abstract
Enhanced production of proinflammatory bradykinin-related peptides, the kinins, has been suggested to contribute to the pathogenesis of periodontitis, a common inflammatory disease of human gingival tissues. In this report, we describe a plausible mechanism of activation of the kinin-generating system, also known as the contact system or kininogen-kallikrein-kinin system, by the adsorption of its plasma-derived components such as high-molecular-mass kininogen (HK), prekallikrein (PK), and Hageman factor (FXII) to the cell surface of periodontal pathogen Porphyromonas gingivalis. The adsorption characteristics of mutant strains deficient in selected proteins of the cell envelope suggested that the surface-associated cysteine proteinases, gingipains, bearing hemagglutinin/adhesin domains (RgpA and Kgp) serve as the major platforms for HK and FXII adhesion. These interactions were confirmed by direct binding tests using microplate-immobilized gingipains and biotinylated contact factors. Other bacterial cell surface components such as fimbriae and lipopolysaccharide were also found to contribute to the binding of contact factors, particularly PK. Analysis of kinin release in plasma upon contact with P. gingivalis showed that the bacterial surface-dependent mechanism is complementary to the previously described kinin generation system dependent on HK and PK proteolytic activation by the gingipains. We also found that several P. gingivalis clinical isolates differed in the relative significance of these two mechanisms of kinin production. Taken together, these data show the importance of this specific type of bacterial surface-host homeostatic system interaction in periodontal infections.
Periodontitis is a common infectious disease in humans, characterized by gingival inflammation, followed by loss of connective tissue and bone around the root of the tooth, which may eventually lead to its exfoliation (59). The progression of periodontal disease depends on the interactions between different bacteria colonizing the gingival sulcus and the host immune system (46). Approximately 15% of the population is affected by severe forms of this disease (27), and, moreover, the occurrence of periodontal infection is often implicated as a risk factor for other pathological conditions, including cardiovascular diseases, adverse pregnancy outcomes, preterm low birth weight, diabetes, and rheumatoid arthritis (82, 84).
Porphyromonas gingivalis, an anaerobic black-pigmented Gram-negative bacterium, is found predominantly in the periodontal pocket, and its presence in large numbers is strongly correlated with periodontal destruction (80). Arguably, cysteine endopeptidases, referred to as gingipains, are the main virulence factors of P. gingivalis (20). They not only directly neutralize the antibacterial activity of the innate immune system by degrading defensins (14) and corrupting the complement system (40), but the gingipains also significantly contribute to the pathogenicity of P. gingivalis via (i) usurping control of local proteolysis (35), (ii) scavenging of iron and heme (44), (iii) stimulating expression of proinflammatory cytokines by host cells (18), and (iv) facilitating bacterial adherence to extracellular matrix proteins (63, 39) and host cells (3, 15) through their hemagglutinin/adhesin (HA) domains (HA1 to HA4) (67).
Apart from surface-bound gingipains, a variety of other bacterial surface proteins mediate the adhesion of P. gingivalis to host tissue constituents, with fimbriae and hemagglutinins being the major adhesins (17, 21). Fimbriae, the filamentous structures localized to the P. gingivalis cell surface, have been shown to interact with fibronectin (51) and to adhere to epithelial and endothelial cells and monocytes, subsequently triggering inflammation and expression of cell adhesion mediators by these host cells (2). The P. gingivalis hemagglutinins enable the bacterium to bind to host erythrocytes to access the heme/iron within (42, 58).
The host kinin-forming cascade is one of the major homeostatic systems involved in the regulation of many physiological processes. The cascade also plays an important role in the inflammatory process (for selected reviews, see references 16, 33, and 36). As a part of host defense against microbial infection, the system is often overactivated (25). Kinins, including bradykinin and a few related peptides, are produced in large amounts at local inflammatory foci to induce vasodilation, vascular permeability enhancement, cell migration, and pain (13). A major pathway for kinin production at these locations depends on the adsorption (the contact system) and activation of the Hageman factor (FXII) on the surfaces of endothelium and other host cells (5, 6, 23, 34, 56). Within a ternary complex assembled on the cell surface, the activated FXII converts plasma prekallikrein (PK) into the active enzyme which subsequently releases bradykinin from high-molecular-mass kininogen (HK).
The kinin-generating pathway can be hijacked by several bacterial and fungal pathogens which take advantage of the kinin-induced increase in vascular permeability for dissemination and/or delivery of plasma-derived nutrients to the site of infection (19). These organisms produce proteases that can either directly release kinins from kininogens (24, 31, 69) or activate FXII and/or prekallikrein directly (49, 50, 73). In the case of P. gingivalis, the arginine-specific gingipains (RgpA, RgpB) generate bradykinin via PK activation, while the lysine-specific gingipain (Kgp) releases kinin directly from its precursor (28, 30, 77). Although bacterial proteases have evolved different mechanisms to evoke the release of kinins (64), the assembly of contact factors at the microbial surface can also lead to activation of the contact system and the generation of kinins (7, 8, 9, 37, 48, 61, 70).
In the current work, we characterized a previously unexplored adsorption of contact system proteins on the P. gingivalis cell surface. Based on analysis of several P. gingivalis isogenic mutants and purified proteins, we have concluded that hemagglutinin/adhesin domain-bearing forms of gingipains (RgpA and Kgp) in the bacterial cell envelope constitute the major docking platform for this adsorption and kinin production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. gingivalis strains listed in Table 1 (except strain 381-derived strains) and clinical isolates (Table 2) were grown on an anaerobe blood agar plate (3% soy tryptone, 1.5% agar, 0.5% yeast extract, 0.05% l-cysteine, 0.01% dithiothreitol, 0.00005% vitamin K, 0.0005% hemin, 5% defibrinated sheep blood). The 381 strain and its Dpg3 mutant were grown on a brain heart infusion agar plate (1.85% brain heart infusion, 1.5% agar, 0.5% yeast extract, 0.00005% vitamin K, 0.0005% hemin, 5% defibrinated sheep blood). All strains were incubated for 7 days at 37°C in an anaerobic chamber (MACS500; Don Whitley Scientific Limited, Frederick, MD) in an atmosphere of 80% N2, 10% CO2, and 10% H2. Afterwards, bacteria were inoculated into 40 ml of appropriate broths and grown overnight under the same conditions. For growth selection of mutants on solid medium, 1 μg/ml tetracycline or 5 μg/ml erythromycin was used.
TABLE 1.
Descriptions of laboratory P. gingivalis strains used in this study
| P. gingivalis strain | Characteristics | Reference |
|---|---|---|
| W83 | Wild type | 52 |
| 381 | Wild type | 79 (culture collection of Forsyth Dental Center, Boston, MA) |
| HG66 | Wild type | Culture collection of Department of Microbiology, Jagiellonian University, Kraków, Poland |
| W83/PorT | porT Tcr | 54 |
| 381/Dpg3 | fimA Emr | 47 |
| W83/Sov | sov Tcr | 71 |
| W83/RgpA | rgpA Cmr | 53 |
| W83/KgpΔIg/HA | kgpΔ602 Emr | 81 |
| W83/K/RgpAB | kgpΔ598rgpArgpBΔ410 Tcr Cmr Emr | This work |
TABLE 2.
Descriptions of clinical P. gingivalis isolates used in this study
| Strain | % of cultivable flora | Patient |
||
|---|---|---|---|---|
| Diagnosis | Gender | Age (yr) | ||
| J-362-9 | 53 | Chronic periodontitis | Male | 47 |
| M-5-1-2 | 35 | Aggressive periodontitis | Female | 26 |
| J-426-1 | 48 | Chronic periodontitis | Female | 40 |
| J-424-1 | 25 | Aggressive periodontitis | Male | 21 |
| MaP4 | 52 | Chronic periodontitis | Male | 72 |
| MaRL | 36 | Chronic periodontitis | Male | 42 |
| D-2-4-3 | 40 | Chronic periodontitis | Male | 39 |
| J-384-1 | 11 | Chronic periodontitis | Female | 53 |
Creation of Sov and gingipain mutants.
Deletional inactivation of Sov was constructed using a technique similar to that described previously (53, 54). Briefly, two 1-kb regions flanking the sov gene were amplified by PCR using two sets of primers (set 1, CTGAGCTCGCCTACAATCTCAAGCGTATGC and TTAACCCGGGCGTGATCCTTCTTTGTTTGAGA; set 2, ACTCTAGATAGCCTAACGGCATTACCCAC and ATTAGTCGACCTTGTATGCAGGAAGCAGGATAG) and inserted into pUC19 at SacI-SmaI and XbaI-SalI sites (boldface nucleotides), respectively, with an intervening tetQ antibiotic resistance cassette (54). The resultant plasmid was electroporated into P. gingivalis W83 competent cells to facilitate homologous recombination into the genome, and transformants were selected on tetracycline-selective blood agar. The correct genotype was confirmed with DNA sequencing of the pertinent region, and the mutants displayed a phenotype that has been reported previously (71).
Creation of single gingipain mutants has been reported previously (53, 81). The RgpA-RgpB inactivation mutant was created by truncation of RgpB at amino acid position 410 to remove the Ig C-terminal domain (CTD) using deletional mutagenesis as previously described (53). To create the gingipain-null mutant W83/K/RgpAB, the ermF/AM cassette of the plasmid construct pREC-KgpΔIg/Cterm/HA (81) was swapped for a tetQ cassette (54) and transformed into the RgpA-RgpB mutant described above. The pertinent regions of all mutants were sequenced to confirm the correct construction, and gingipain biochemical assays were used to confirm the absence of gingipain activity.
Isolation of bacterial surface components.
Fimbriae from the P. gingivalis 381 strain were isolated by the method of Arai et al. (4), while the gingipains HRgpA, RgpB, and Kgp were purified from P. gingivalis strain HG66 as previously described (62, 65). Lipopolysaccharide (LPS) was obtained from the P. gingivalis W83 strain using phenol-water extraction according to the published procedure (83), and after dialysis against deionized water, the purity of the LPS preparation was checked by SDS-PAGE and zinc-imidazole staining (22).
Biotinylation of kinin-generating proteins.
Low-molecular-mass kininogen (LK), purified from human plasma according to the method of Johnson et al. (32), human HK, human plasma PK, and human FXII (all from Enzyme Research Laboratories, South Bend, IN) were incubated at 2 to 3 μM in 0.1 M carbonate buffer, pH 8.3 (total volume 500 μl), with biotin N-hydroxysuccinimide ester solution in dimethylformamide (40 μg/4 μl) for 4 h at 4°C in the presence of 2 μM aprotinin (for PK) or 0.1 mM phenylmethylsulfonyl fluoride (PMSF) (for FXII). After conjugation, the biotinylated (Bt) proteins (LK-Bt, HK-Bt, FXII-Bt, PK-Bt) were dialyzed overnight against phosphate-buffered saline (PBS) and stored frozen until use in binding experiments.
Binding of kinin-generating proteins to bacterial cell surface.
P. gingivalis cells from early-stationary-phase cultures (optical density, 0.6 to 0.8) were harvested by centrifugation (30 min, 3,500 × g), washed three times with PBS, and resuspended in PBS (108 cells per 200 μl) containing 1 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) for inactivation of the gingipains. Bacterial cell suspensions were incubated for 1 h at 37°C with biotinylated kinin-generating proteins at concentrations of 0 to 80 nM in Eppendorf tubes which had been pretreated with 1% bovine serum albumin (BSA) overnight at 4°C to minimize nonspecific binding of biotinylated proteins to the plastic surface during the binding experiments. After incubation, the cells were washed three times with PBS containing 0.1% BSA before bound biotinylated proteins were probed with horseradish peroxidase (HRP)-streptavidin conjugate (1:4,000 dilution, 1 h at room temperature). For HRP detection, 3,3′,5,5′-tetramethylbenzidine (TMB) was used as the substrate. After the reaction was stopped with 2 M HCl, the absorbance at 450 nm was determined in a Power Wave X Select microplate reader (BioTek Instruments, Winooski, VT) and the amounts of bound biotinylated proteins were estimated from the appropriate standard curves prepared for each of the analyzed proteins.
For the competition assay, cell suspensions (108 cells per 200 μl) were incubated with one of the biotinylated contact factors (3.8 pmol per tube) in the presence of increasing amount of selected, nonbiotinylated protein (0 to 38 pmol per tube). The incubation and detection conditions were the same as those described above, except that 1 μM aprotinin in the samples containing PK and 0.1 mM PMSF in the samples containing FXII were additionally applied to prevent the contact system activation.
Binding of purified bacterial surface components to contact system proteins.
To test the binding of contact system proteins to purified components of the P. gingivalis cell surface, a ligand binding microplate assay was applied. The surface components—FimA (4 μg per well), the gingipains (6 pmol per well), or LPS (4 μg per well)—were bound overnight at 4°C onto the wells of MaxiSorp microplates for enzyme-linked immunosorbent assays (ELISAs; Nunc, Roskilde, Denmark) in PBS or in 0.1 M bicarbonate buffer, pH 9.6 (for LPS only). Wells were washed three times with PBS and blocked for 2 h at 37°C with PBS containing 3% BSA, followed by three washes with 1% BSA in PBS. The high concentration of BSA was necessary to prevent the nonspecific binding of contact factors, especially of HK. Subsequently, solutions of biotinylated contact factors at concentrations between 0 and 160 nM were added and the microplates were incubated for 1.5 h at 37°C. For experiments on gingipain binding, all solutions additionally contained 1 mM TLCK. Bound proteins were detected with HRP-streptavidin conjugate (1:4,000 dilution, 1 h at room temperature) and TMB as the color-generating substrate.
Assay of gingipain activity on the surface of P. gingivalis W83 and clinical strains.
The proteolytic activities of Rgp and Kgp on the surface of P. gingivalis cells were determined by monitoring the hydrolysis of the chromogenic substrates benzoyl-l-arginine-p-nitroanilide (BApNA) and N-(p-tosyl)-Gly-Pro-Lys-p-nitroanilide (TGPKpNA), respectively, as previously described (65). Briefly, 108 cells were suspended in a Sarstedt microplate in 150 μl of assay buffer (200 mM Tris-HCl, 100 mM NaCl, 5 mM CaCl2, pH 7.6) supplemented with fresh 10 mM l-cysteine. The microplate was incubated at 37°C for 15 min prior to the addition of 50 μl 2 mM substrate in assay buffer. Formation of p-nitroanilide was measured as an increase in optical density at 405 nm over a 30-min period using the Power Wave X Select microplate reader. Each analysis was performed in triplicate.
Detection of kinin production from human plasma proteins adsorbed on P. gingivalis surface.
PBS-washed P. gingivalis W83 cells (109 cells per analyzed sample) were resuspended in 500 μl of 15 mM HEPES buffer, pH 7.0, containing 135 mM NaCl and 50 μM ZnCl2 with or without 2 mM TLCK. Bacteria were then mixed with 500 μl of human plasma and incubated at 37°C for 0 to 30 min. After incubation, the suspensions were centrifuged (2,000 × g) for 5 min at 4°C and the supernatants were diluted 2-fold with 1% trifluoroacetic acid (TFA) in water. After an additional centrifugation step, the supernatants were applied onto a C18 Sep column (Peninsula Laboratories) for extraction of the kinin peptides. Elution was performed with 1% TFA in 60% acetonitrile. After lyophilization of the samples, the amount of kinins liberated from human plasma was determined with an ELISA kit, according to the manufacturer's instructions (Peninsula Laboratories).
Data analysis.
All points in the figures represent the means ± standard deviations (SDs) from at least three independent experiments with duplicate measurements. For the analysis of both the saturable binding and the competition plots, a one-site model was fitted using GraphPad Prism software.
RESULTS
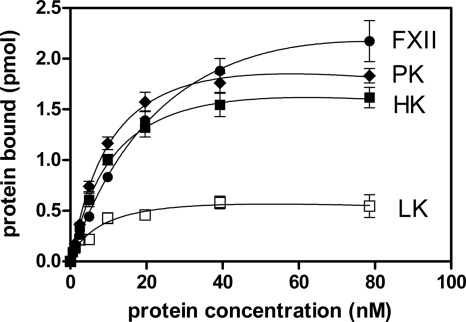
As shown in Fig. 1, biotinylated human kininogens (LK and HK) and the zymogens of the human plasma kinin-generating system (FXII and PK) were capable of tight binding to the cell surface of the P. gingivalis W83 strain. The saturable binding levels were the highest for FXII, reaching a value of 2 pmol bound protein per 108 P. gingivalis cells, followed by PK > HK > LK. Notably, LK binding reached saturation at a level as low as 0.5 pmol, which is much less than the levels for the three contact factors.
FIG. 1.
Binding of biotinylated contact factors and LK to the cell surface of P. gingivalis wild-type strain W83. FXII, PK, HK, and LK were incubated with 108 bacterial cells for 1 h at 37°C in PBS containing 1 mM TLCK. Bound proteins at the bacterial surface were detected with streptavidin-HRP conjugate. Data represent the means of three experiments; error bars denote the SDs.
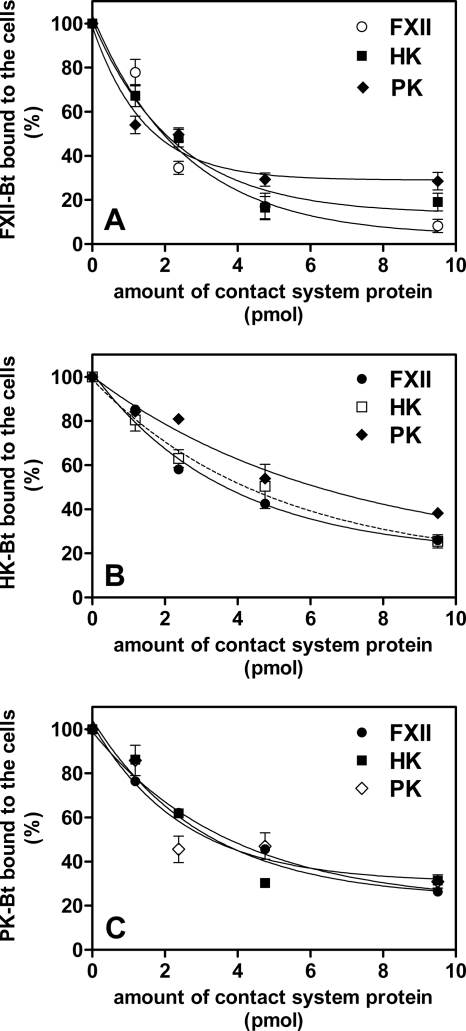
Unlabeled contact system proteins FXII, HK, and PK were able to compete with their corresponding biotinylated tracers at the cell surface of the W83 strain with 95%, 80%, and 70% efficiencies, respectively (Fig. 2). Moreover, all tested proteins could compete with each other to comparable extents, suggesting that they share most of the binding sites on the bacterial surface.
FIG. 2.
Competition of biotinylated contact factors with the native proteins at the bacterial surface. The suspension of P. gingivalis W83 (108 cells) was incubated with a mixture of biotinylated (3.8 pmol in a 200-μl total volume) FXII (A), HK (B), or PK (C) and increasing concentrations of nonbiotinylated competitors (HK, FXII, and PK). For the samples containing PK or FXII, the reaction mixture was supplemented with the selective inhibitors 1 μM aprotinin and 0.1 mM PMSF, respectively. The binding was detected with a streptavidin-HRP/TMB system.
Another P. gingivalis strain 381 bound contact factors to 2-fold lower capacity (Fig. 3A) than the W83 strain. The isogenic FimA-negative Dpg3 strain (47) was found to have a binding capacity for FXII comparable to that for wild-type strain 381 (Fig. 3B). However, there was a 2-fold increased binding for HK in Dpg3 compared with that in 381, and the level of PK binding was reduced by half.
FIG. 3.
Binding of biotinylated contact factors to the cell surface of P. gingivalis wild-type strain 381 (A) and isogenic FimA-deficient mutant (Dpg3) (B). The binding experiments were performed as described in Materials and Methods. Bound proteins were detected with streptavidin-HRP conjugate.
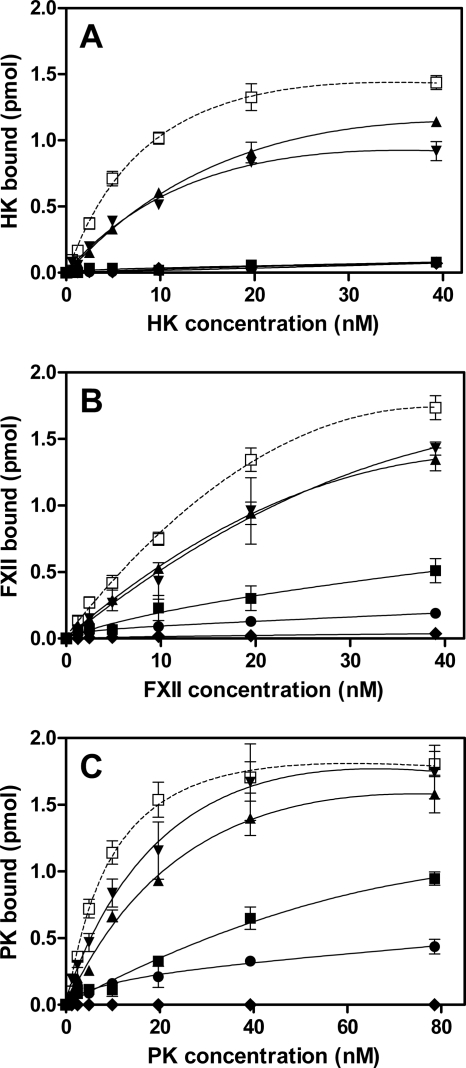
Our results show that FimA is one ligand for the contact factors, but the data do not exclude a possibility that other surface proteins might be involved. To address this experimentally, an array of isogenic W83-derived mutant strains lacking gingipains and other cell envelope proteins was employed (Table 1). These strains were tested in binding assays with biotinylated contact factors. Notably, the single rgpA and kgp mutant strains showed reduced binding capacities for HK and FXII in comparison to those of the parental strain, and the binding was almost entirely abolished by the elimination of all gingipain genes (Fig. 4A and B). The binding of HK was totally eliminated in the P. gingivalis mutants deficient in any of two essential proteins (PorT and Sov) of the gingipain secretion system (54, 71). In contrast to HK and FXII, the lack of a single gingipain, RgpA or Kgp, did not markedly affect the binding capacity for PK, and the triple-gingipain-knockout strain still exhibited residual PK binding (Fig. 4C). In contrast to the Sov-deficient strain, the PorT mutant strain showed reduced, but still significant, binding capacity for FXII and, to a lesser degree, for PK (Fig. 4B and C, respectively). Taken together, the analysis of gingipain-deficient strains clearly indicated that RgpA and Kgp are essential for adsorption of the contact system proteins on the P. gingivalis cell envelope.
FIG. 4.
Binding of biotinylated contact factors to P. gingivalis W83 wild-type strain and isogenic mutant strains. Biotinylated HK (A), FXII (B), and PK (C) were incubated with (108 per analytical point) P. gingivalis W83 wild-type strain (□) and isogenic mutant strains deficient in RgpA (▴), Kgp (▾), RgpA, RgpB, and Kgp (•), PorT (▪), and Sov (⧫) for 1 h at 37°C in PBS containing 1 mM TLCK. Bound proteins were detected with streptavidin-HRP conjugate.
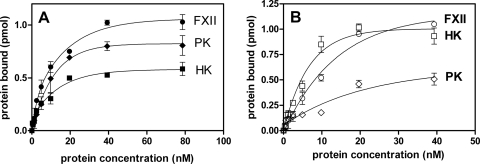
In another series of experiments, the binding of contact factors to FimA isolated from P. gingivalis 381 strain and immobilized to a microplate was tested (Fig. 5A). These results show that all contact factors have an affinity for FimA with decreasing binding affinity in the order of PK > FXII > HK. Microplate-immobilized LPS purified from P. gingivalis W83 was shown to also bind PK (Fig. 5B) but only at half of the level compared with that of immobilized FimA. Similarly, the binding of FXII and HK to LPS was weak, albeit detectable, suggesting that LPS does not significantly contribute to the adsorption of the last two contact factors to the P. gingivalis cell envelope.
FIG. 5.
Binding of biotinylated contact factors to immobilized FimA protein from strain 381 (A) and immobilized LPS from P. gingivalis W83 (B). Purified FimA and LPS were immobilized onto a MaxiSorb ELISA microplate at 4 μg per well in 200 μl PBS, and after the plate was blocked they were probed with various concentrations of biotinylated contact factors.
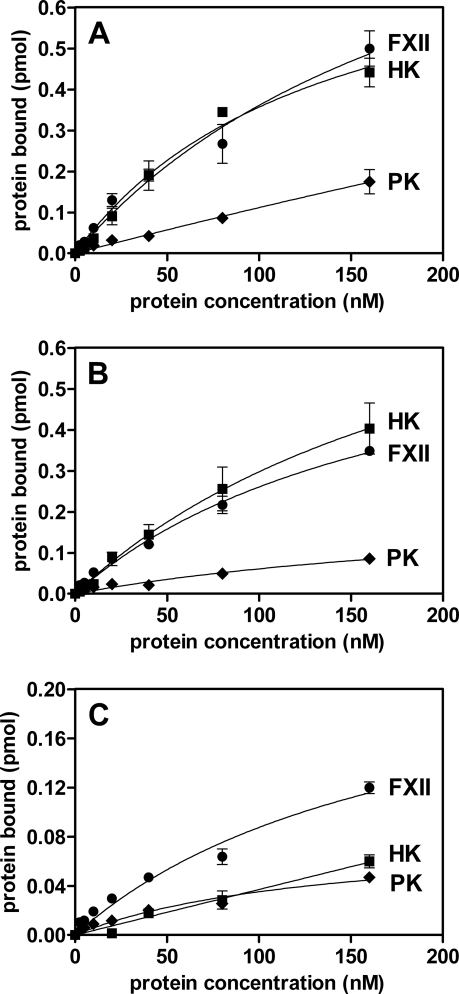
Similarly, to elucidate the role of the individual gingipains, we investigated the binding of HK, FXII, and PK to microplate-immobilized purified Kgp, HRgpA, and RgpB. RgpB is essentially identical to HRgpA, but it does not contain the hemagglutinin/adhesin domains (68). As presented in Fig. 6, immobilized Kgp and HRgpA bound FXII and HK to comparable levels which were 50- to 120-fold higher than the level of binding to immobilized FimA (recalculated according to an equimolar amount of immobilized 42-kDa FimA subunit). In comparison to FXII and HK, the level of PK binding to immobilized gingipains was lower (66 to 75%). Finally, in contrast to the high-molecular-mass gingipains, immobilized RgpB bound contact system proteins rather poorly (Fig. 6C), arguing that the major binding determinant(s) is located within the hemagglutinin/adhesin domain of the gingipains.
FIG. 6.
Binding of biotinylated contact factors to isolated and microplate-immobilized gingipains. The MaxiSorp microplate was coated overnight at 4°C with solutions of purified gingipains (6 pmol per well) Kgp (A), HRgpA (B), and Rgp B (C) in PBS, and after the plate was blocked, it was probed with biotinylated HK, FXII, and PK. All experiments were performed in the presence of 1 mM TLCK.
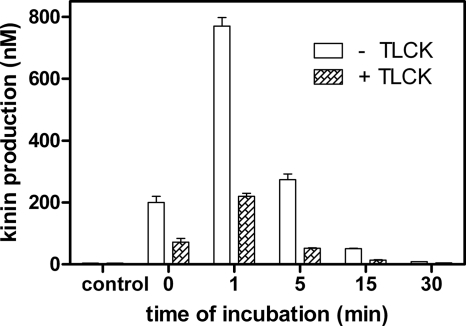
To test whether the interaction between contact factors and gingipains influences activation of the contact system, we compared P. gingivalis-induced kinin generation in human plasma in the presence or absence of a specific gingipain protease inhibitor (TLCK). When human plasma samples were mixed with a suspension of wild-type P. gingivalis W83 cells in the absence of TLCK, a momentary release of large amounts of kinins was observed within the first minute of incubation (Fig. 7). Subsequently, the kinin level dropped quickly, probably owing to the actions of numerous kinin-degrading enzymes (kininases) present in plasma (10). Bacterial pretreatment with TLCK markedly lowered the levels of kinin production. This residual gingipain activity-independent kinin generation was most likely to be from the endogenous contact system activation on the bacterial cell envelope. This result further corroborates our hypothesis that P. gingivalis uses both gingipain activity and cell surface binding of contact system components to generate kinins.
FIG. 7.
Kinin generation by P. gingivalis W83 strain cells upon contact with human plasma. A mixture of human plasma and P. gingivalis W83 strain cells (109 cells) in 7.5 mM HEPES buffer, pH 7.0, containing 75 mM NaCl, 25 μM ZnCl2, and 1 mM TLCK was incubated at 37°C. After the specified times of incubation, the supernatant was applied on a C18 Sep column. Eluted kinins were quantified with a Peninsula Laboratories ELISA kit (specific to the kinin 5-amino-acid C-terminal sequence), according to the manufacturer's instructions. For control samples, plasma was incubated for 30 min without any gingipain added.
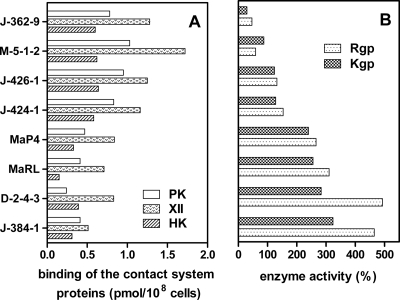
From previously published data and results presented here, it is clear that gingipains contribute to increased levels of kinins at sites of infections by a dual mechanism, either through PK activation and direct bradykinin release from soluble HK (29) or through adsorption of contact system proteins on the bacterial surface. To evaluate which mechanism of the kinin generation is used by clinical isolates of P. gingivalis (Table 2), gingipain activities and the ability to bind contact factors were compared. This analysis revealed a general trend showing that strains with lower total enzymatic activities of Kgp and Rgp were more active in the contact system attraction, and vice versa (Fig. 8). Hence, the two pathways of the activation of the kinin release at the surface of P. gingivalis cells seemed to be complementary.
FIG. 8.
Relation between binding of the contact factors (A) and the gingipain proteolytic activity (B) in clinical isolates of P. gingivalis. The binding experiments were performed as described in Materials and Methods with a 100 nM concentration of the contact factors. Gingipain activity was determined for 108 cells of P. gingivalis clinical strains in the assay buffer (200 mM Tris-HCl, pH 7.6, 100 mM NaCl, 5 mM CaCl2) supplemented with cysteine (10 mM) in the presence of chromogenic substrate BApNA for Rgp and the TGPKpNA substrate for Kgp. The release of p-nitroanilide was quantified at 405 nm. The proteolytic activity of the clinical strains was expressed relative to the activity of the wild-type W83 strain.
DISCUSSION
Kinins are powerful proinflammatory mediators, and they have been implicated in the pathogenesis of periodontitis due to their potential to strongly upregulate bone resorption (11, 12, 43, 45). By facilitating P. gingivalis dissemination across the vascular barrier (26), kinins may also contribute to the pathogenesis of systemic diseases that associate with periodontitis. Although the role of the contact system in the pathology of P. gingivalis infections has been known for some decades, the pathways of kinin production in the inflamed gingival tissue are poorly characterized. For instance, only the proteolytic action of P. gingivalis gingipains on host kininogens and on the kallikrein zymogen has been reported so far (28, 30, 77). In this study, we aimed to investigate a more specific mechanism of periodontitis-associated enhancement of kinin production which depends on the adsorption of the kinin-forming system of the host on the pathogen cell surface.
Our data show that all investigated strains of P. gingivalis, including reference strains W83 and 381 as well as several clinical isolates, have remarkable affinities for contact factors, with the binding capacities being in the order FXII > PK > HK. Under the experimental conditions used, 30% and 13% of the total amount of added HK was adsorbed by the W83 and 381 strains, respectively. Comparable yields of HK immobilization were reported for Staphylococcus aureus, Escherichia coli, and Salmonella spp. (9). The low level of binding of LK (Fig. 1) suggests that the specific surface-binding domain of the HK light chain is critically involved in this protein adsorption, which is in agreement with similar results reported for other microbial pathogens (8).
The competitive binding of all three contact system proteins onto the P. gingivalis cell surface (Fig. 2) suggests that they largely share affinity to the same structures on the bacterial cell envelope with some minor preferences. In this respect, PK seems to partially occupy unique sites (such as LPS; Fig. 5B) which do not interact with HK and FXII.
Three groups of major P. gingivalis cell surface components (fimbriae, LPS, and gingipains) were tested for their ability to bind the contact system proteins. To assess the role of fimbriae in binding to these components, we compared two reference strains, strains W83 and 381 (see Fig. 1 versus Fig. 3A), which have been reported to differ in the level of fimbriation. The former strain is considered to be sparsely fimbriated, with the minor fimbriae not being expressed, while the latter strain has been reported to be heavily decorated with both major and minor fimbriae (2, 55). Since markedly higher levels of FXII, HK, and PK were bound by W83 than by 381, this suggested that fimbriae are not the main surface structure interacting with the contact system proteins on the P. gingivalis surface. This conclusion was supported by the fact that fimA inactivation did not affect FXII binding but, instead, enhanced 2-fold the HK binding capacity of the mutant Dpg3 (Fig. 3B). The increased HK binding capacity of the fimA mutant could arise from exposure of additional HK binding sites in the absence of fimbrial structures. In contrast, binding of PK to the fimA mutant was markedly impaired in comparison to that to the wild-type strain, indicating that among contact system proteins, mainly PK has affinity for FimA. These findings were further supported by the binding of PK to immobilized, purified FimA (Fig. 5A), which adds PK to a list of proteins, including laminin, fibronectin, fibrinogen, and vitronectin, that have been reported to interact with P. gingivalis fimbriae (1).
Tight binding of HK to E. coli LPS has been reported recently (60). In this study, however, we have found that LPS isolated from P. gingivalis did not show any remarkable affinity to HK or FXII (Fig. 5B). The lack of interaction is most likely due to significant differences in the structures of LPSs derived from P. gingivalis and E. coli (41, 57, 76). Again, unequivocal binding was noted mainly for PK, but only at half the capacity observed for FimA.
In most P. gingivalis strains, the gingipains RgpA and Kgp are tightly associated with the bacterial cell surface (66) and their hemagglutinin/adhesin domains mediate the adhesion of bacteria to the cells of host gingival tissue (3, 15); extracellular matrix proteins such as fibrinogen, fibronectin, and laminin (63); and hemoglobin (58). Gingipain translocation across the outer membrane to the cell surface is dependent on a novel secretion system composed of several proteins (74), including integral outer membrane β-barrel proteins PorT and Sov (54, 72). Inactivation of any of these proteins resulted in the failure in gingipain maturation and the accumulation of inactive progingipains in the periplasm (71, 75). Here, we find that the sov mutant is not able to bind any of the contact system proteins, while deletion of the porT gene eliminated HK binding and lowered FXII and PK adsorption by 70% and 50%, respectively (Fig. 4). These results argue that HK and FXII interact with bacterial proteins transported to the cell surface through this secretion system but that other surface components, like FimA, LPS, and possibly Sov itself, additionally contribute to the PK adherence.
Although several P. gingivalis proteins bearing a conservative C-terminal domain (54, 78) are apparently secreted by the PorT/Sov-dependent pathway (38), only the surface exposure of RgpA and Kgp was shown to be essential for HK and FXII binding by P. gingivalis. Interestingly, deficiency of a single gingipain, either RgpA or Kgp, exerted only a modest effect, suggesting that neither gingipain alone is absolutely essential for binding of the contact system proteins but that together, RgpA, RgpB, and Kgp seem to cooperate for maximal sorption of the contact system components (Fig. 4). Similarly, the relatively modest reduction in PK immobilization in the gingipain mutants clearly indicates the presence of alternative binding sites for this protein on the P. gingivalis cell envelope. Results obtained using whole P. gingivalis cells, including single- and triple-gingipain-knockout mutants, were fully corroborated by the finding that both purified Kgp and RgpA, but not RgpB, strongly bind HK and FXII (Fig. 6). Further, the affinities of FXII and HK to immobilized gingipains were found to be much higher than those to immobilized FimA. Taken together, it can be concluded that hemagglutinin/adhesin domains of RgpA and Kgp provide the major binding sites for HK and FXII, whereas PK binding is less specific and may also include FimA and LPS.
Our analysis of kinin release from human plasma on contact with P. gingivalis clearly demonstrates that the activation of the contact system after adsorption of its components on the bacterial surface complement kinin generation by the proteolytic action of the gingipains (28, 30, 77) (Fig. 7). We also found that in the case of P. gingivalis clinical isolates with low gingipain activity compared to that of reference strain W83, the contact activation pathway on the bacterial surface was a significant source of kinins (Fig. 8). These data suggest that these two systems operate concurrently to produce kinins under pathophysiological conditions.
In conclusion, the current work describes the ability of P. gingivalis to concentrate the host plasma-derived kinin-forming system on its cell surface to trigger kinin production. The adhesive contact system components HK (the kinin precursor) and FXII (the PK activator) were found to dock primarily to the cell-bound gingipains, with their hemagglutinin/adhesin domains being the most likely binding determinants.
Acknowledgments
This work was supported in part by the Ministry of Science and Higher Education, Poland (grant nos. 2P04C 035 29 and 1642/B/P01/2008/35 to A.K. and J.P., respectively), the Jagiellonian University (statutory funds DS/15/WBBiB and DS/9/WBBiB), the National Institutes of Health (grant DE 09761), and the Foundation for Polish Science (Team/2009-4/8). H.H. was supported by a grant from the Swedish Research Council (project 7480). The Faculty of Biochemistry, Biophysics, and Biotechnology of Jagiellonian University is a beneficiary of structural funds from the European Union (grant no. POIG.02.01.00-12-064/08, Molecular biotechnology for health).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 22 November 2010.
REFERENCES
- 1.Amano, A. 2007. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front. Biosci. 12:3965-3974. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., I. Nakagawa, N. Okahashi, and N. Hamada. 2004. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 39:136-142. [DOI] [PubMed] [Google Scholar]
- 3.Andrian, E., D. Grenier, and M. Rouabhia. 2006. Porphyromonas gingivalis-epithelial cell interactions in periodontitis. J. Dent. Res. 85:392-403. [DOI] [PubMed] [Google Scholar]
- 4.Arai, M., N. Hamada, and T. Umemoto. 2000. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol. Lett. 193:75-81. [DOI] [PubMed] [Google Scholar]
- 5.Barbasz, A., I. Guevara-Lora, M. Rapala-Kozik, and A. Kozik. 2008. Kininogen binding to the surfaces of macrophages. Int. Immunopharmacol. 8:211-216. [DOI] [PubMed] [Google Scholar]
- 6.Barbasz, A., and A. Kozik. 2009. The assembly and activation of kinin-forming systems on the surface of human U-937 macrophage-like cells. Biol. Chem. 390:269-275. [DOI] [PubMed] [Google Scholar]
- 7.Bengtson, S. H., et al. 2009. Activation of TAFI on the surface of Streptococcus pyogenes evokes inflammatory reactions by modulating the kallikrein/kinin system. J. Innate Immun. 1:18-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Nasr, A., et al. 1997. Absorption of kininogen from human plasma by Streptococcus pyogenes is followed by the release of bradykinin. Biochem. J. 326:657-660. [PMC free article] [PubMed] [Google Scholar]
- 9.Ben Nasr, A., A. Olsén, U. Sjöbring, W. Muller-Esterl, and L. Björck. 1996. Assembly of human contact phase protein and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20:927-935. [DOI] [PubMed] [Google Scholar]
- 10.Bhoola, K. D., C. D. Figueroa, and K. Worthy. 1992. Bioregulation of kinins: kallikreins, kininogens, and kininases. Pharmacol. Rev. 44:1-80. [PubMed] [Google Scholar]
- 11.Brechter, A. B., and U. H. Lerner. 2007. Bradykinin potentiates cytokine-induced prostaglandin biosynthesis in osteoblasts by enhanced expression of cyclooxygenase 2, resulting in increased RANKL expression. Arthritis Rheum. 56:910-923. [DOI] [PubMed] [Google Scholar]
- 12.Brechter, A. B., E. Persson, I. Lundgren, and U. H. Lerner. 2008. Kinin B1 and B2 receptor expression in osteoblasts and fibroblasts is enhanced by interleukin-1 and tumour necrosis factor-alpha. Effects dependent on activation of NF-kappaB and MAP kinases. Bone 43:72-83. [DOI] [PubMed] [Google Scholar]
- 13.Calixto, J. B., A. Cabrini, J. Ferreira, and M. M. Campos. 2000. Kinins in pain and inflammation. Pain 87:1-5. [DOI] [PubMed] [Google Scholar]
- 14.Carlisle, M. D., R. N. Srikantha, and K. A. Brogden. 2009. Degradation of human α- and β-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J. Innate Immun. 1:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, T., and M. J. Duncan. 2004. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb. Pathog. 36:205-209. [DOI] [PubMed] [Google Scholar]
- 16.Colman, R. W., and A. H. Schmaier. 1997. Contact system: a vascular biology modulator with anticoagulant, profibrinolytic, antiadhesive, and proinflammatory attributes. Blood 90:3819-3843. [PubMed] [Google Scholar]
- 17.Cutler, C. W., J. R. Kalmar, and C. A. Genco. 1995. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3:45-51. [DOI] [PubMed] [Google Scholar]
- 18.Fitzpatrick, R. E., et al. 2009. High molecular weight gingipains from Porphyromonas gingivalis induce cytokine responses from human macrophage-like cells via a nonproteolytic mechanism. J. Innate Immun. 1:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frick, I. M., L. Björck, and H. Herwald. 2007. The dual role of the contact system in bacterial infectious disease. Thromb. Haemostasis 98:497-502. [PubMed] [Google Scholar]
- 20.Guo, Y., K. A. Nguyen, and J. Potempa. 2010. Dichotomy of gingipain action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins. Periodontol. 2000 54:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamada, S., et al. 1998. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol. Immunol. 13:129-138. [DOI] [PubMed] [Google Scholar]
- 22.Hardy, E., E. Pupo, L. Castellanos-Serra, J. Reyes, and C. Fernández-Patrón. 1997. Sensitive reverse staining of bacterial lipopolysaccharides on polyacrylamide gels by using zinc and imidazole salts. Anal. Biochem. 244:28-32. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, L., C. D. Figueroa, W. Muller-Esterl, and K. D. Bhoola. 1994. Assembly of contact-phase factors on the surface of the human neutrophil membrane. Blood 84:474-482. [PubMed] [Google Scholar]
- 24.Herwald, H., M. Collin, W. Müller-Esterl, and L. Björck. 1996. Streptococcal cysteine proteinase releases kinins: a virulence mechanism. J. Exp. Med. 84:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herwald, H., M. Morgelin, and L. Bjorck. 2003. Contact activation by pathogenic bacteria: a virulence mechanism contributing to the pathophysiology of sepsis. Scand. J. Infect. Dis. 36:604-607. [DOI] [PubMed] [Google Scholar]
- 26.Hu, S. W., C. H. Huang, H. C. Huang, Y. Y. Lai, and Y. Y. Lin. 2006. Transvascular dissemination of Porphyromonas gingivalis from a sequestered site is dependent upon activation of the kallikrein/kinin pathway. J. Periodontal Res. 41:200-207. [DOI] [PubMed] [Google Scholar]
- 27.Hugoson, A., and O. Norderyd. 2008. Has the prevalence of periodontitis changed during the last 30 years? J. Clin. Periodontol. 35:338-345. [DOI] [PubMed] [Google Scholar]
- 28.Imamura, T., J. Potempa, R. N. Pike, and J. Travis. 1995. Dependence of vascular permeability enhancement on cysteine proteinases in vesicles of Porphyromonas gingivalis. Infect. Immun. 63:1999-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamura, T., J. Travis, and J. Potempa. 2003. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr. Protein Pept. Sci. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 30.Imamura, T., R. N. Pike, J. Potempa, and J. Travis. 1994. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalis induces vascular permeability enhancement through activation of the kallikrein/kinin pathway. J. Clin. Invest. 94:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura, T., et al. 2005. Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J. Exp. Med. 201:1669-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, D. A., G. Salvesen, M. A. Brown, and A. J. Barrett. 1987. Rapid isolation of human kininogens. Thromb. Res. 48:187-193. [DOI] [PubMed] [Google Scholar]
- 33.Joseph, K., and A. P. Kaplan. 2005. Formation of bradykinin: a major contributor to the innate inflammatory response. Adv. Immunol. 86:159-208. [DOI] [PubMed] [Google Scholar]
- 34.Joseph, K., B. Ghebrehiwet, and A. P. Kaplan. 2001. Activation of the kinin-forming cascade on the surface of endothelial cells. Biol. Chem. 382:71-75. [DOI] [PubMed] [Google Scholar]
- 35.Kantyka, T., et al. 2009. Elafin is specifically inactivated by RgpB from Porphyromonas gingivalis by distinct proteolytic cleavage. Biol. Chem. 390:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan, A. P., K. Joseph, and M. Silverberg. 2002. Pathways for bradykinin formation and inflammatory disease. J. Allergy Clin. Immunol. 109:195-209. [DOI] [PubMed] [Google Scholar]
- 37.Karkowska-Kuleta, J., A. Kozik, and M. Rapala-Kozik. 2010. Binding and activation of human plasma kinin-forming system on the cell walls of Candida albicans and Candida tropicalis. Biol. Chem. 391:97-103. [DOI] [PubMed] [Google Scholar]
- 38.Kondo, Y., et al. 2010. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 78:2846-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kontani, M., et al. 1996. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblast and matrix proteins. Infect. Immun. 64:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krauss, J. L., J. Potempa, J. D. Lambris, and G. Hajishengallis. 2010. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol. 2000 52:141-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumada, H., Y. Haishima, T. Umemoto, and K. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lépine, G., R. P. Ellemn, and A. Progulske-Fox. 1996. Construction and preliminary characterization of three hemagglutinin mutants of Porphyromonas gingivalis. Infect. Immun. 64:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lerner, U. H. 1995. Stimulation of bone resorption by the kallikrein-kinin system and the coagulation cascade. Acta Orthop. Scand. Suppl. 266:45-50. [PubMed] [Google Scholar]
- 44.Lewis, J. P. 2010. Metal uptake in host-pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 52:94-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ljunggren, O., and U. H. Lerner. 1988. Stimulation of bone resorption in cultured mouse calvaria by Met-Lys-bradykinin. J. Periodontal Res. 23:75-77. [DOI] [PubMed] [Google Scholar]
- 46.Madianos, P. N., Y. A. Bobetsis, and D. F. Kinane. 2005. Generation of inflammatory stimuli: how bacteria set up inflammatory responses in the gingiva. J. Clin. Periodontol. 32(Suppl. 6):57-71. [DOI] [PubMed] [Google Scholar]
- 47.Malek, R., et al. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattson, E., et al. 2001. Staphylococcus aureus induces release of bradykinin in human plasma. Infect. Immun. 69:3877-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyoshi, S., H. Watanabe, T. Kawas, H. Yamada, and S. Shinoda. 2004. Generation of active fragments from human zymogens in the bradykinin-generating cascade by extracellular proteases from Vibrio vulnificus and V. parahaemolyticus. Toxicon 44:887-893. [DOI] [PubMed] [Google Scholar]
- 50.Molla, A., T. Yamamoto, T. Akaike, S. Miyoshi, and H. Maeda. 1989. Activation of Hageman factor and prekallikrein and generation of kinin by various microbial proteinases. J. Biol. Chem. 264:10589-10594. [PubMed] [Google Scholar]
- 51.Murakami, Y., et al. 1996. Porphyromonas gingivalis fimbrillin is one of the fibronectin-binding proteins. Infect. Immun. 64:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson, K. E., et al. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen, K. A., J. Travis, and J. Potempa. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J. Bacteriol. 189:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen, K. A., et al. 2009. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishiyama, S., et al. 2007. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153:1916-1925. [DOI] [PubMed] [Google Scholar]
- 56.Oehmcke, S., M. Mörgelin, and H. Herwald. 2009. Activation of the human contact system on neutrophil extracellular traps. J. Innate Immun. 1:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa, T. 1993. Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolisaccharide. FEBS J. 332:197-201. [DOI] [PubMed] [Google Scholar]
- 58.Olczak, T., W. Simpson, X. Liu, and C. A. Genco. 2005. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol. Rev. 29:119-144. [DOI] [PubMed] [Google Scholar]
- 59.Page, R. C. 2002. The etiology and pathogenesis of periodontitis. Compend. Contin. Educ. Dent. 23(Suppl. 5):11-14. [PubMed] [Google Scholar]
- 60.Perkins, R., M. D. Ngo, F. Mahdi, and Z. Shariat-Madar. 2008. Identification of lipopolysaccharide binding site on high molecular weight kininogen. Biochem. Biophys. Res. Commun. 366:938-943. [DOI] [PubMed] [Google Scholar]
- 61.Persson, K., et al. 2000. Severe lung lesions caused by Salmonella are prevented by inhibition of contact system. J. Exp. Med. 192:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pike, R., W. McGraw, J. Potempa, and J. Travis. 1994. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 269:406-411. [PubMed] [Google Scholar]
- 63.Pike, R. N., J. Potempa, W. McGraw, T. H. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complex from Porphyromonas gingivalis. J. Bacteriol. 178:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potempa, J., and R. N. Pike. 2009. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 1:70-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Potempa, J., et al. 1998. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273:21648-21657. [DOI] [PubMed] [Google Scholar]
- 66.Potempa, J., R. N. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potempa, J., A. Sroka, T. Imamura, and J. Travis. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 68.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24:153-192. [DOI] [PubMed] [Google Scholar]
- 69.Rapala-Kozik, M., et al. 2010. Degradation of human kininogens with the release of kinin peptides by extracellular proteinases of Candida spp. Biol. Chem. 391:823-830. [DOI] [PubMed] [Google Scholar]
- 70.Rapala-Kozik, M., et al. 2008. Kininogen adsorption to the cell surface of Candida spp. Int. Immunopharmacol. 8:237-241. [DOI] [PubMed] [Google Scholar]
- 71.Saiki, K., and K. Konishi. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51:483-491. [DOI] [PubMed] [Google Scholar]
- 72.Saiki, K., and K. Konishi. 2010. The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis. FEMS Microbiol. Lett. 302:166-174. [DOI] [PubMed] [Google Scholar]
- 73.Sakata, Y., et al. 1996. Bradykinin generation triggered by Pseudomonas proteases facilitates invasion of the systemic circulation by Pseudomonas aeruginosa. Microbiol. Immunol. 40:415-425. [DOI] [PubMed] [Google Scholar]
- 74.Sato, K., et al. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato, K., et al. 2005. Identification of a new membrane-associated protein that influences transport/maturation gingipains and adhesion of Porphyromonas gingivalis. J. Biol. Chem. 280:8668-8677. [DOI] [PubMed] [Google Scholar]
- 76.Sawada, N., T. Ogawa, Y. Asai, Y. Makimura, and A. Sugiyama. 2007. Toll-like receptor 4-dependent recognition of structurally different forms of chemically synthesized lipid as of Porphyromonas gingivalis. Clin. Exp. Immunol. 148:529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott, C. F., E. J. Whitaker, B. F. Hammond, and R. W. Colman. 1993. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J. Biol. Chem. 268:7935-7942. [PubMed] [Google Scholar]
- 78.Seers, C. A., et al. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135-187. [DOI] [PubMed] [Google Scholar]
- 81.Sztukowska, M., et al. 2004. The C-terminal domains of the gingipain K polyprotein are necessary for assembly of the active enzyme and expression of associated activities. Mol. Microbiol. 54:1393-1408. [DOI] [PubMed] [Google Scholar]
- 82.Wegner, N., et al. 2010. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-91. [Google Scholar]
- 84.Wiliams, R. C., and S. Offenbacher. 2000. Periodontal medicine: the emergence of a new branch of periodontology. Periodontol. 2000 23:9-12. [DOI] [PubMed] [Google Scholar]