Abstract
Bacterial pathogens that colonize mucosal surfaces have acquired resistance to antimicrobials that are abundant at these sites. One of the main antimicrobials present on mucosal surfaces is lysozyme, a muramidase that hydrolyzes the peptidoglycan backbone of bacteria. Cleavage of the peptidoglycan backbone leads to bacterial cell death and lysis, which releases bacterial fragments, including peptidoglycan, at the site of infection. Peptidoglycan fragments can be recognized by host receptors and initiate an immune response that will aid in clearing infection. Many mucosal pathogens modify the peptidoglycan residues surrounding the cleavage site for lysozyme to avoid peptidoglycan degradation and the release of these proinflammatory fragments. This review will focus specifically on peptidoglycan modifications, their role in lysozyme resistance, and downstream effects on the host immune response to infection.
To establish infection, successful mucosal pathogens must be able to evade host antimicrobial enzymes, which are secreted at high levels by the epithelium and are also produced within professional phagocytes (10, 14, 87). Direct killing by antimicrobials may lead to the release of bacterial fragments, which can be sensed by host pattern recognition receptors (48, 69, 70). Sensing of microbial fragments initiates signaling cascades, which lead to the production of proinflammatory cytokines and chemokines and the influx of immune cells with additional antimicrobial substances to the site of infection. Infiltrating host cells may then impact the eventual clearance of the pathogen.
Many host antimicrobials target peptidoglycan, the primary structural component of the bacterial cell wall. Host hydrolytic enzymes, such as lysozyme, degrade peptidoglycan and cause bacterial cell lysis (34, 57). To prevent killing by antimicrobials, bacteria have acquired the ability to modify their peptidoglycan backbones. Modifications can prevent cell lysis, which allows bacteria to persist on the mucosal surface and may also affect the host immune response by limiting the release of bacterial fragments at the site of infection. This could specifically limit the release of peptidoglycan, which can play an important role in initiating the host immune response (6, 17, 20, 71, 80). The persistence of lysozyme-resistant organisms may also affect inflammation by prolonging host exposure to bacterial ligands (22, 23). This review will focus on the direct effects of peptidoglycan modifications on degradation by host enzymes and the effects of modifications on host sensing of bacterial infections.
HOST DEGRADATION AND RELEASE OF PEPTIDOGLYCAN
(i) Contribution of peptidoglycan modifications to lysozyme resistance.
To protect host mucosal surfaces from infection, the epithelium secretes a variety of antimicrobial enzymes. One of the most abundant proteins present on the mucosal surface is lysozyme (10), a host antimicrobial enzyme with muramidase and cationic antimicrobial peptide activities. Lysozyme is secreted by the epithelium and is also a major component within the granules of professional phagocytes, where it may help kill bacterial pathogens within phagolysosomes (10, 14, 23, 50, 87). The muramidase activity of lysozyme leads to hydrolysis of the β-1,4 glycosidic bond between the C-1 carbon of N-acetyl muramic acid (MurNAc) and the C-4 carbon of N-acetylglucosamine (GlcNAc) residues of the peptidoglycan backbone (Fig. 1A). This enzymatic activity breaks down the structural integrity of the cell wall, which leads to lysis of the bacterial cell. Evidence also suggests that lysozyme has nonmuramidase function; a 9-amino-acid cationic antimicrobial peptide within the protein is capable of disrupting bacterial membranes in the absence of enzymatic activity (34, 35, 57).
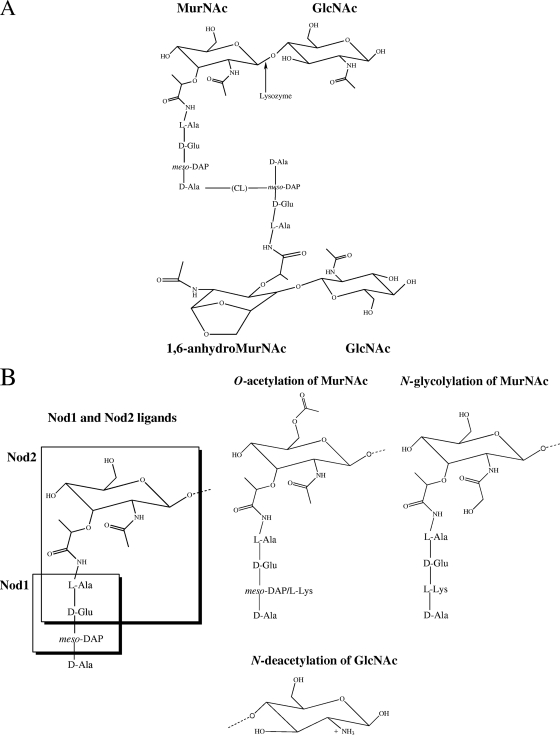
FIG. 1.
Peptidoglycan fragments and modifications. (A) Unmodified MurNAc-GlcNAc disaccharide residues are depicted with stem peptide structures, the cross-linking bridge (CL), and a disaccharide fragment from the adjacent glycan strand. The 1,6-anhydro-MurNAc residues are generated following cleavage by lytic transglycosylases, and this is depicted on one of the MurNAc residues. The structure depicted represents a fragment from a Gram-negative organism which has meso-DAP in the third residue of the stem peptide, while most Gram-positive organisms have an l-Lys at this position. (B) Nod1 and Nod2 ligands, shown with boxes around the minimal motifs sensed by these proteins, depicted on an unmodified, Gram-negative MurNAc residue. The three modifications to the glycan backbone discussed in the text are shown in the lower portion of the panel.
Because of the abundance of lysozyme in both epithelial secretions and professional phagocytes (10, 14), many pathogens have evolved lysozyme resistance to prevent peptidoglycan hydrolysis. Lysozyme resistance is especially important for Gram-positive pathogens, which rely on a thick peptidoglycan layer to provide structural integrity for the cell. Gram-negative organisms have an outer membrane that can protect peptidoglycan from the direct effect of lytic enzymes. The primary mechanism for lysozyme resistance in both Gram-positive and Gram-negative organisms appears to be direct modification of peptidoglycan; however, modification of other cell wall-linked components, such as teichoic acid, may also contribute to resistance.
Modification of peptidoglycan structure was observed several decades ago in the naturally lysozyme-resistant organism Streptococcus pneumoniae. A significant proportion of the GlcNAc residues in wild-type (WT) S. pneumoniae peptidoglycan were N-deacetylated, and it was suggested that this may contribute to lysozyme resistance in this organism (43, 60). N-deacetylation was also observed in peptidoglycan from Bacillus anthracis, which is resistant to lysozyme. Treatment of B. anthracis cell walls with acetic anhydride, which N-acetylates peptidoglycan residues, led to lysozyme sensitivity, indicating that N-deacetylation may be important for lysozyme resistance (89). A distinct modification, O-acetylation, was observed in peptidoglycan from Neisseria gonorrhoeae, as well as in several other Gram-negative organisms (51, 66). Strains of N. gonorrhoeae with low levels of O-acetylation were sensitive to lysozyme, while strains with extensive O-acetylation were resistant to lysozyme (66, 74).
In the last 10 years the genes responsible for these modifications have been identified in several organisms. The first of these was oatA (O-acetyltransferase A), which acetylates MurNAc at the C-6 position (Fig. 1B); it was identified as the major determinant for lysozyme resistance in Staphylococcus aureus (3, 5). OatA is present only in lysozyme-resistant and pathogenic staphylococcal strains, and transformation of a lysozyme-sensitive, nonpathogenic staphylococcal strain with the S. aureus oatA gene was sufficient to confer lysozyme resistance (3). OatA was also found to provide protection against macrophage killing and limit host cytokine responses, both of which may be linked to its role in lysozyme resistance (72). O-acetylation has also been described in S. pneumoniae, and the gene responsible for this modification was identified based on its role in both lysozyme and penicillin resistance (15). This gene was termed adr (attenuator of drug resistance) and was identified as a peptidoglycan O-acetyltransferase with significant homology to oatA. Like OatA, Adr acetylates MurNAc at the C-6 position (Fig. 1B). Additional oatA homologs were identified in Enterococcus faecalis (30), in which OatA contributes to lysozyme resistance and evasion of macrophage killing, and Lactococcus lactis (79), in which OatA contributes to lysozyme resistance (Table 1). An unrelated peptidoglycan O-acetyltransferase protein was identified in the Gram-negative pathogens Neisseria meningitidis and Neisseria gonorrhoeae, and it was termed pacA (peptidoglycan acetylase A) (18). PacA requires a related protein called PacB for function, and it also modifies the C-6 residue of MurNAc, leading to lysozyme resistance. PacA/B proteins were also found to protect against a type of bacterial cell lysis called autolysis, which could be triggered following the initiation of peptidoglycan hydrolysis by lysozyme (Table 1).
TABLE 1.
Peptidoglycan modifications and their effects
| Peptidoglycan modification and genea | Species | Phenotype | Citation |
|---|---|---|---|
| O-acetylation of MurNAc in Gram-positive bacteriab | |||
| oatA | Staphylococcus aureus | Lysozyme resistance | Bera et al. (5) |
| Resistance to macrophage killing | Shimada et al. (72) | ||
| Limits host cytokine responses | Shimada et al. (72) | ||
| oatA (63) | Enterococcus faecalis | Lysozyme resistance | Hébert et al. (30) |
| Intracellular survival within macrophages | Hébert et al. (30) | ||
| oatA (58) | Lactococcus lactis | Lysozyme resistance | Veiga et al. (79) |
| Upregulated during envelope stress | Veiga et al. (79) | ||
| adr (52) | Streptococcus pneumoniae | Lysozyme resistance | Crisóstomo et al. (15) |
| Penicillin resistance | Crisóstomo et al. (15) | ||
| Virulence factor | Davis et al. (16) | ||
| O-acetylation of MurNAc in Gram-negative bacteriab | |||
| pacA/pacB | Neisseria gonorrhoeae | Lysozyme resistance | Dillard and Hackett (18) |
| Resistance to autolysis | Dillard and Hackett (18) | ||
| pacA/pacB (97) | Neisseria meningitidis | Lysozyme resistance | Dillard and Hackett (18) |
| Resistance to autolysis | Dillard and Hackett (18) | ||
| N-deacetylation of GlcNAc | |||
| pgdA | Streptococcus pneumoniae | Lysozyme resistance | Vollmer and Tomasz (83) |
| Virulence factor | Vollmer et al. (82) | ||
| pgdA (53) | Enterococcus faecalis | Lysozyme resistance | Hébert et al. (30) |
| pgdA (21) | Helicobacter pylori | Lysozyme resistance | Wang et al. (85) |
| Upregulated during oxidative stress | Wang et al. (85) | ||
| pgdA (60) | Listeria monocytogenes | Lysozyme resistance | Boneca et al. (6) |
| Limits host cytokine responses | Boneca et al. (6) | ||
| Intracellular survival within macrophages | Boneca et al. (6) | ||
| Upregulated during infection | Camejo et al. (7) | ||
| pgdA (70) | Streptococcus suis | Lysozyme resistance | Fittipaldi et al. (21) |
| Resistance to neutrophil killing | Fittipaldi et al. (21) | ||
| Promotes host cytokine responses | Fittipaldi et al. (21) | ||
| Upregulated during incubation with neutrophils | Fittipaldi et al. (21) | ||
| N-glycolylation of MurNAc | |||
| namH | Mycobacterium tuberculosis | Lysozyme resistance | Raymond et al. (65) |
| β-Lactam resistance | Raymond et al. (65) | ||
| Promotes host cytokine responses | Coulombe et al. (13) |
The first gene identified is listed first in each category, with the percent sequence similarity over the enzymatic domains listed next to the other genes.
O-acetylation genes were separated based on Gram strain reaction, because Gram-negative organisms use two proteins to acetylate MurNAc wherease Gram-positive organisms use a single protein.
The gene responsible for a second modification, deacetylation of peptidoglycan, was first identified in S. pneumoniae and was termed pgdA (peptidoglycan deacetylase A) (83). PgdA deacetylates GlcNAc at the C-2 residue (Fig. 1B), contributing to lysozyme resistance. PgdA was identified as a virulence factor based on the in vivo attenuation of a pgdA mutant (82, 83). A similar phenotype was also demonstrated in the animal pathogen Streptococcus suis, where a mutant pgdA strain was sensitive to lysozyme and was attenuated in vivo (21). Infection with the pgdA-deficient strain indicated that PgdA may also play a role in the host response to this organism; the mutant strain was more sensitive to neutrophil killing and elicited lower levels of cytokines in plasma compared to the WT strain. Additional pgdA functional homologs were identified in E. faecalis (30), Helicobacter pylori (85), and Listeria monocytogenes (6), based on the contributions of these genes to lysozyme resistance (Table 1). In L. monocytogenes, PgdA also promoted intracellular survival within macrophages and limited host cytokine responses (6). The occurrence of these peptidoglycan modifications across bacteria, their enzymatic activities, and their biological relevance have recently been reviewed elsewhere (81).
In some organisms, both deacetylation at the C-2 residue of GlcNAc and acetylation at the C-6 residue of MurNAc contribute to lysozyme resistance. A mutant strain of S. pneumoniae lacking both modifications was hypersensitive to the effects of lysozyme and was attenuated during colonization of the nasopharynx in the presence, but not the absence, of lysozyme (16). An Enterococcus faecalis mutant lacking both an O-acetyltransferase, oatA, and an N-acetylglucosamine, pgdA, was also hypersensitive to the effects of lysozyme (30). This mutant strain had impaired survival following phagocytosis, a phenotype described for other lysozyme-sensitive pathogens in both macrophages (6) and neutrophils (21). However, for both pneumococcal and enterococcal infection models, it seems that one of the two modifications played a predominant role in lysozyme resistance. This may be due in part to differences in expression of the peptidoglycan-modifying enzymes.
Several studies have focused on the transcriptional regulation of peptidoglycan-modifying enzymes. The oatA O-acetyltransferase gene of L. lactis was found to be upregulated in response to envelope stress sensed by the two-component system CesSR (79). Many other Gram-positive organisms possess a similar two-component system that functions as an envelope stress sensor, which is termed LiaSR in Bacillus subtilis and VraSR in S. aureus (38). In the case of L. lactis, CesSR responded to cell wall damage by lysozyme (79), which led to the upregulation of oatA. Because of the abundance of lysozyme within phagocytes and on the mucosal surface, this response could be especially relevant during infection, and indicates that O-acetylation of peptidoglycan could be increased in vivo. In vivo upregulation of the pgdA gene has been shown in L. monocytogenes and could also occur in other organisms during infection (7). Additionally, the H. pylori pgdA gene was upregulated during oxidative stress (85), which would be encountered following uptake by phagocytes or interaction with antimicrobials on the mucosal surface. The S. suis pgdA gene was upregulated during interactions with neutrophils, which could also be a result of oxidative stress (21). These studies indicate that peptidoglycan modifications may be differentially regulated to confer lysozyme resistance during infection and that, in different infection models, upregulation of one modification may be sufficient to provide resistance, whereas in other models more than one modification may be necessary. The proportion of modified residues may also vary under different conditions or between organisms and should be considered when determining the importance of a particular modification.
(ii) Additional peptidoglycan-degrading enzymes.
Peptidoglycan recognition proteins (PGRPs) are host proteins that interact with peptidoglycan. Insects have many PGRPs, and sensing of peptidoglycan by these proteins can initiate a signaling cascade that results in the production of antimicrobial peptides (67). Mammals have four PGRPs, termed PGLYRP-1, -2, -3, and -4; however, none of these proteins has been directly linked to the sensing of peptidoglycan. PGLYRP-1, -3, and -4 all have direct bactericidal activities that seem to occur through the binding and disruption of peptidoglycan synthesis, in a manner similar to penicillin (45). PGLYRP-2 has amidase activity (24, 86) and can cleave between the N-acetyl muramic acid residue and the stem peptide to release adjacent peptidoglycan polymers, leading to cell wall instability and cell lysis. PGLYRP-2 can contribute to inflammation, and it was required in a model of arthritis induced by peptidoglycan (68). The antimicrobial activities of PGLYRPs could release peptidoglycan and other bacterial fragments during infection, which would be sensed by the host and activate the immune response.
Bacteria also have peptidoglycan-degrading enzymes that cleave at the same sites as host antimicrobial enzymes, but they aid in normal growth and turnover of the cell wall. This could explain why bacteria remain sensitive to many of the host degradative enzymes, such as amidases. Bacterial peptidoglycan-degrading enzymes are tightly regulated to maintain the structural integrity of the cell wall during growth. However, these enzymes can be activated following cell damage, leading to lysis of the bacterial cell. This can occur following interaction with β-lactam antibiotics, such as penicillin, which target penicillin-binding proteins (PBPs) that cross-link stem peptides from adjacent strands of peptidoglycan. In pneumococci, covalent binding of β-lactams to PBPs prevents peptidoglycan cross-linking and leads to lysis of bacterial cells by a process that activates LytA (75, 76). LytA is the major autolysin in this organism, and like PGLYRP-2 it has amidase activity, which releases adjacent peptidoglycan polymers and is important in peptidoglycan remodeling during growth (32). Autolysins are present in both Gram-negative and Gram-positive bacteria and could be activated in a similar way following antibiotic treatment of other bacterial species. Evidence suggests that killing by penicillin can also occur by a nonlytic mechanism that is independent of LytA (55), indicating that lysis may not be necessary for killing.
Antibiotic-induced cell lysis of S. pneumoniae leads to the release of bacterial fragments that can stimulate host receptors (54). This effect was reported to be specific to β-lactams and was not seen with antibiotics of other classes, such as moxifloxacin or erythromycin, which do not inhibit cell wall synthesis (53, 54). Treatment with the glycopeptide cell wall synthesis inhibitor vancomycin did not release proinflammatory fragments, possibly because it binds directly to the stem peptide of peptidoglycan instead of binding PBPs. For S. pneumoniae, release of bacterial fragments appeared to be LytA dependent; however, treatment with β-lactams that act through nonlytic mechanisms, such as cefotaxime and faropenem, also led to the release of stimulatory fragments (53). This indicates that interaction with PBPs, instead of lysis, may be the key step in releasing fragments. These studies indicate that treatment with β-lactams could initiate an inflammatory response in the host, and this needs to be considered when treating patients with antibiotics.
Additional structures on the cell surface may also be important in preventing peptidoglycan degradation. In S. aureus, cell wall-linked teichoic acid interferes with the interaction between the cell surface and lysostaphin, an enzyme that specifically degrades the S. aureus cell wall (28). Lysostaphin has three distinct enzymatic activities, amidase, endo-glucosaminidase, and glycylglycine endopeptidase activities, the latter of which is specific to the structure of S. aureus peptidoglycan. All these activities contribute to the breakdown of S. aureus peptidoglycan, leading to lysis and release of bacterial products. The addition of cell wall-linked teichoic acid, which is attached to peptidoglycan at the C-2 position of MurNAc, seems to interfere with lysostaphin activity and has also been linked with lysozyme resistance (4). In this case, the addition of cell wall teichoic acid, instead of the direct modification of the peptidoglycan backbone, may help prevent cell lysis.
HOST SENSING OF PEPTIDOGLYCAN AND MODIFIED PEPTIDOGLYCAN
Peptidoglycan modifications can prevent direct killing of bacteria by providing resistance to host enzymes during infection. Modifications may also prevent elimination of bacteria by altering the host immune response. Bacteria with unmodified peptidoglycan are lysed following interaction with lysozyme, which releases peptidoglycan fragments that can be sensed by host proteins called pattern recognition receptors (PRRs) (Fig. 2) (8, 25, 26, 37). Sensing by these proteins activates host signaling cascades and initiates an immune response that aids in clearing infection. Bacteria with modified peptidoglycan are resistant to hydrolysis by lysozyme and remain intact, which may limit inflammation (6, 21, 72). Alternatively, infection with lysozyme-resistant organisms may lead to the persistence of bacterial ligands at the site of infection, which could also be proinflammatory (22, 23).
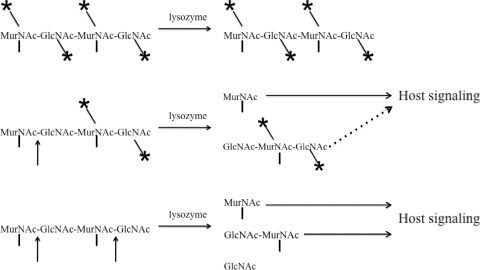
FIG. 2.
Model for sensing of peptidoglycan fragments by host receptors. Intact peptidoglycan is depicted by the tetrasaccharide MurNAc-GlcNAc-MurNAc-GlcNAc. Modifications to individual residues are represented by asterisks, and lysozyme cleavage sites are depicted with arrows between tetrasaccharide residues. Vertical lines below MurNAc residues represent the stem peptide. Fully modified peptidoglycan is resistant to hydrolysis by lysozyme and does not release peptidoglycan fragments following interaction with the enzyme. Partially modified peptidoglycan has some cleavage sites for lysozyme and releases some fragments that can be sensed by the host, which is depicted by an arrow with a solid line. Larger released fragments may or may not contribute to host sensing, which is depicted by an arrow with a dotted line. Unmodified peptidoglycan is hydrolyzed by lysozyme, which releases bacterial fragments that initiate host signaling cascades.
Bacteria contain numerous ligands that are sensed by host PRRs. The major families of host receptors that sense bacterial products include the Toll-like receptors (TLRs) and the Nod-like receptors (NLRs). The TLRs are predominantly surface-associated host proteins, while NLRs are cytoplasmic proteins. Toll-like receptor-2 (TLR-2) senses lipoteichoic acid (LTA) and lipoproteins, which are associated with bacterial membranes (69, 77), and may also sense peptidoglycan, although this remains controversial (1, 70). Peptidoglycan is sensed primarily through two host receptors in the NLR family. Nucleotide-binding oligomerization domain-containing protein 1 (Nod1) is a host intracellular protein that is expressed in virtually all cell types (36). Nod1 senses meso-diaminopimelic acid (meso-DAP) containing peptidoglycan, which is found primarily in Gram-negative bacteria and a few Gram-positive organisms, such as Bacillus species and L. monocytogenes (8, 25, 36) (Fig. 1B). Nod1 senses fragments of the stem peptide terminating in meso-DAP, with or without MurNAc attached, and may also sense stem peptide fragments terminating in d-Ala in some hosts (Fig. 1B) (46). Nod2 is a host intracellular protein that is expressed in myelomonocytic cells, such as monocytes, macrophages, and neutrophils, and is also expressed by stimulated epithelial cells in culture (29, 59). Nod2 senses the muramyl dipeptide (MDP) residue of bacteria, which is common to all bacterial peptidoglycans (26, 37, 59) (Fig. 1B). Both Nod1 and Nod2 contain nucleotide oligomerization binding (Nod) domains that define the receptor family, in addition to C-terminal leucine-rich repeat (LRR) domains which sense peptidoglycan and N-terminal caspase recruitment domains (CARD) which interact with adaptor molecules to initiate signaling cascades following receptor activation (36, 59). Signaling downstream of Nod receptors involves Rip2 and the IKK complex and results in NF-κB activation and the production of proinflammatory cytokines, chemokines, and antimicrobials such as defensins (41, 52, 63, 84). Together, these cytokines aid in the recruitment of immune cells into the site of infection to initiate the immune response and also help activate phagocytes to efficiently eliminate pathogens.
Because NLRs are cytoplasmic receptors, it was originally thought that Nod proteins recognized primarily intracellular pathogens (20, 71). However, several recent papers have shown that Nod1 and Nod2 can also sense extracellular pathogens and that these receptors play a key role in the immune response to extracellular pathogens as well (17, 61, 64, 90). Because extracellular pathogens are not actively dividing and shedding peptidoglycan within host cells like intracellular pathogens, the exact mechanism by which their peptidoglycan accesses Nod receptors remains somewhat unclear. Several hypotheses exist, such as release from phagolysosomes into host cytoplasm following uptake of whole bacteria, or uptake of peptidoglycan by phagocytosis or clathrin-dependent endocytosis (40, 44, 49, 72). Gram-negative organisms shed peptidoglycan fragments during normal growth, which can stimulate Nod1-dependent responses (9, 12, 58). Outer membrane vesicles (OMVs) may contain peptidoglycan fragments and help deliver these ligands to nonphagocytic epithelial cells (39). It has also been reported that bacterial pore-forming toxins, which insert into host membranes, may aid in Nod1 ligand delivery into the host cytosol (64). Additionally it has been shown that host cells express peptide transporters that can transport both Nod1 and Nod2 ligands directly into the host cytosol (2, 78).
The role of Nod receptors in controlling early bacterial infection has been shown in the context of several different infections (17, 20, 33, 61, 71, 90), which suggests that it would be beneficial for pathogens to be able to evade Nod sensing. To prevent sensing of Nod ligands, bacteria can modify peptidoglycan to prevent the release of fragments. Peptidoglycan that is fully modified is resistant to lysozyme and does not generate fragments that can be sensed by the host following contact with lysozyme, whereas unmodified peptidoglycan is easily broken down by lysozyme and releases proinflammatory fragments (Fig. 2). However, many organisms have a proportion of residues that are unmodified (81). Partially modified peptidoglycan could be broken down at unmodified residues and lead to the release of larger peptidoglycan fragments that may be sensed by the host (Fig. 2). However, the effect of fragment size on the efficiency of Nod-mediated signaling remains unclear. Modification to the MurNAc residues present within these larger fragments may also directly interfere with Nod2 sensing of these sugars. In addition, there are several stem peptide modifications that alter Nod1 sensing, such as amidation of meso-DAP or glutamic acid, or the presence of l-ornithine instead of meso-DAP in the stem peptide (27, 88). Although these stem peptide modifications do not affect hydrolysis of the peptidoglycan backbone, they indicate there may be selective pressure within pathogens to avoid host Nod sensing.
Several studies have confirmed that peptidoglycan modifications could alter Nod signaling by preventing lysozyme degradation and the release of Nod ligands. A lysozyme-sensitive mutant, lacking the cell wall modification by PgdA, was shown to elicit more cytokine production by macrophages, in a Nod1-dependent manner (6). Lysozyme digestion was also found to contribute to the release of ligands sensed by another NLR, Nalp3, leading to interleukin-1β production. Digestion of an S. aureus oatA mutant strain, which lacks O-acetylation of MurNAc, led to the production of heightened levels of proinflammatory cytokines (72), supporting the hypothesis that peptidoglycan modifications can prevent the release of ligands and prevent sensing.
Peptidoglycan modifications may also alter the immune response to pathogens by affecting the sensing of peptidoglycan by Nod receptors. Gram-negative bacteria shed peptidoglycan during growth, releasing ligands that can be sensed by Nod receptors. Stem peptide modifications that affect Nod sensing may have evolved to allow bacteria to replicate and minimize the host response (27, 88). Alternatively, increased sensing could also be beneficial to bacteria if they require an immune response to persist in the host, such as the recruitment of immune cells that form the replicative niche for some intracellular pathogens. One example of this is Mycobacterium tuberculosis, which modifies its peptidoglycan to stimulate an immune response and also replicates within host macrophages. The N-glycolylated peptidoglycan of Mycobacterium species was shown to elicit significantly more Nod2-dependent immune activation than the N-acetylated peptidoglycan that is common across bacteria (13, 62). This modification was performed with NamH, which acts on the C-2 carbon of MurNAc of mycobacteria (Fig. 1B). Peptidoglycan modification by NamH was also associated with both lysozyme resistance and resistance to β-lactam antibiotics (65), which may indicate an additional role in preventing peptidoglycan digestion (Table 1). Because N-glycolylation of MurNAc leads to an increased immune response, it may indicate that host inflammation could be beneficial for mycobacteria during infection. It will be interesting to determine if other pathogens that replicate within host immune cells may have also evolved a peptidoglycan that promotes inflammation to recruit these cells to the site of infection.
Evidence suggests that other NLR proteins, specifically Nalp1, may also play a role in the sensing of MDP residues (33). Nalp1 contains the characteristic Nod domain but has a pyrin domain instead of a CARD. This pyrin domain recruits different adaptor proteins following ligand binding, to help form an inflammasome complex and eventual caspase-1 activation. Nalp1 has been shown to form a complex with Nod2 following stimulation by MDP, which may mean that this signaling pathway could also be affected by peptidoglycan modifications, by altering stimulation of Nod2.
ADDITIONAL EFFECTS OF PEPTIDOGLYCAN MODIFICATIONS
Peptidoglycan modifications prevent the release of bacterial fragments, which includes peptidoglycan, and also other cell wall components that can be sensed by host receptors. Following digestion, cell walls from S. pneumoniae have been shown to elicit tumor necrosis factor alpha production from peripheral blood mononuclear cells (48). The fragments carrying proinflammatory activity were identified as branched stem peptide residues, which did not contain known Nod ligands, indicating that additional receptors can respond to cell wall fragments following digestion. One of these additional receptors may be TLR-2, since it was shown that S. pneumoniae cell walls could be sensed in a TLR-2-dependent manner (56). Lipid-modified cell components are associated with cell membranes and are also released following peptidoglycan hydrolysis of L. monocytogenes (6), leading to the TLR-2-dependent sensing of bacterial fragments.
Modifications to peptidoglycan may have additional effects on structures covalently attached to peptidoglycan, such as cell wall-linked teichoic acid (11, 42), proteins, or capsular polysaccharide (capsule) (73). The capsule may not directly affect host signaling, but it can prevent opsonophagocytosis and clearance by neutrophils (31, 47). Increased expression of capsule was observed in pgdA mutants, which have neutrally charged GlcNAc residues that may alter attachment of the negatively charged capsule (16). Increased capsule expression has been linked to decreased sensitivity to both penicillin and vancomycin through prevention of autolysis, which could alter the release of peptidoglycan following antibiotic treatment (19).
CONCLUSIONS
Peptidoglycan modifications are a common mechanism used by pathogens to avoid degradation and bacterial lysis following exposure to host antimicrobials. One of the most abundant host antimicrobials on mucosal surfaces is lysozyme, and pathogens that colonize these sites have evolved lysozyme resistance by modifying their peptidoglycan. In addition to preventing bacterial lysis during early interactions with the host, these modifications may also indirectly allow bacteria to establish infection, by preventing release of bacterial products that can initiate the host immune response. The persistence of lysozyme-resistant organisms at the site of infection can also increase inflammation (22, 23), which may indicate that peptidoglycan modifications are anti-inflammatory early in infection but actually indirectly contribute to inflammation later in infection.
Although lysozyme resistance can prevent early release of bacterial products within the host, pathogens can also release fragments during infection as part of normal growth. This shedding of fragments occurs in Gram-negative organisms, which can release peptidoglycan fragments and also OMVs that may deliver bacterial fragments to host cells (9, 12, 39, 58). Intracellular pathogens that are actively dividing within host cells may also deliver fragments in a more direct manner, by shedding peptidoglycan and other bacterial ligands into the host cytosol (58). Additionally, bacterial products could be delivered by type IV secretion systems along with bacterial effectors (80). All of these mechanisms would allow the host to recognize bacteria in a lysozyme-independent manner and initiate the host response to pathogens that have evolved high levels of lysozyme resistance.
The occurrence of lysozyme resistance and corresponding peptidoglycan modifications across both Gram-positive and Gram-negative bacterial pathogens indicate that the host imparts a significant selective pressure on the cell wall of bacteria that reside on mucosal surfaces (81). Lysozyme resistance may provide a selective advantage by allowing bacteria to persist on the mucosal surface, where they can remain initially undetected. Resistance to lysozyme will also prevent the release of bacterial fragments early in infection and in this way can impact the initiation of the host immune response. Peptidoglycan modifications may also directly impact the host immune response by interfering with sensing of peptidoglycan fragments. Peptidoglycan modifications can impact all of these aspects of the host immune response, which may help explain the abundance of peptidoglycan-modifying enzymes in bacterial pathogens.
Acknowledgments
We thank T. B. Clarke for critical reading of the manuscript.
This work was supported by U.S. Public Health Service grants AI44231, AI78538, and AI38446 to J.N.W.
Editor: A. T. Maurelli
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Asong, J., M. Wolfert, K. Maiti, D. Miller, and G. Boons. 2009. Binding and cellular activation studies reveal that toll-like receptor 2 can differentially recognize peptidoglycan from Gram-positive and Gram-negative bacteria. J. Biol. Chem. 284:8643-8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahadduri, P., et al. 2005. Functional characterization of the peptide transporter PEPT2 in primary cultures of human upper airway epithelium. Am. J. Respir. Cell Mol. Biol. 32:319-325. [DOI] [PubMed] [Google Scholar]
- 3.Bera, A., R. Biswas, S. Herbert, and F. Götz. 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74:4598-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bera, A., et al. 2007. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189:280-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bera, A., S. Herbert, A. Jakob, W. Vollmer, and F. Götz. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778-787. [DOI] [PubMed] [Google Scholar]
- 6.Boneca, I., et al. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camejo, A., et al. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamaillard, M., M. Hashimoto, Y. Horie, J. Masumoto, and S. Qiu. 2003. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4:702-707. [DOI] [PubMed] [Google Scholar]
- 9.Cloud-Hansen, K., K. Hackett, D. Garcia, and J. Dillard. 2008. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J. Bacteriol. 190:5989-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, A., et al. 2002. Cationic polypeptides are required for antibacterial activity of human airway fluid. J. Immunol. 169:6985-6991. [DOI] [PubMed] [Google Scholar]
- 11.Coley, J., E. Tarelli, A. Archibald, and J. Baddiley. 1978. The linkage between teichoic acid and peptidoglycan in bacterial cell walls. FEBS Lett. 88:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Cookson, B., A. Tyler, and W. Goldman. 1989. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry 28:1744-1749. [DOI] [PubMed] [Google Scholar]
- 13.Coulombe, F., et al. 2009. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 206:1709-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cramer, E., and J. Breton-Gorius. 1987. Ultrastructural localization of lysozyme in human neutrophils by immunogold. J. Leukoc. Biol. 41:242-247. [DOI] [PubMed] [Google Scholar]
- 15.Crisóstomo, M., et al. 2006. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol. Microbiol. 61:1497-1509. [DOI] [PubMed] [Google Scholar]
- 16.Davis, K., H. Akinbi, A. Standish, and J. Weiser. 2008. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 4:e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshmukh, H., et al. 2009. Critical role of Nod2 in regulating the immune response to Staphylococcus aureus. Infect. Immun. 77:1376-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillard, J., and K. Hackett. 2005. Mutations affecting peptidoglycan acetylation in Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 73:5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernebro, J., et al. 2004. Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J. Infect. Dis. 189:328-338. [DOI] [PubMed] [Google Scholar]
- 20.Ferwerda, G., et al. 2005. NOD2 and Toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 1:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fittipaldi, N., et al. 2008. Significant contribution of the pgdA gene to the virulence of Streptococcus suis. Mol. Microbiol. 70:1120-1135. [DOI] [PubMed] [Google Scholar]
- 22.Fleming, T., D. Wallsmith, and R. Rosenthal. 1986. Arthropathic properties of gonococcal peptidoglycan fragments: implications for the pathogenesis of disseminated gonococcal disease. Infect. Immun. 52:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganz, T., et al. 2003. Increased inflammation in lysozyme-M deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood 101:2388-2392. [DOI] [PubMed] [Google Scholar]
- 24.Gelius, E., C. Persson, J. Karlsson, and H. Steiner. 2003. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem. Biophys. Res. Commun. 306:988-994. [DOI] [PubMed] [Google Scholar]
- 25.Girardin, S., I. Boneca, L. Carneiro, A. Antignac, and M. J.éhanno. 2003. NOD1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- 26.Girardin, S., et al. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869-8872. [DOI] [PubMed] [Google Scholar]
- 27.Girardin, S., et al. 2003. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278:41702-41708. [DOI] [PubMed] [Google Scholar]
- 28.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. 188:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez, O., et al. 2002. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-κB activation. J. Biol. Chem. 277:41701-41705. [DOI] [PubMed] [Google Scholar]
- 30.Hébert, L., P. Courtin, R. Torelli, M. Sanguinetti, and M. Chapot-Chartier. 2007. Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75:5390-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hostetter, M. 1986. Serotypic variations among virulent pneumococci in deposition and degradation of covalently bound C3b: implications for phagocytosis and antibody production. J. Infect. Dis. 153:682-693. [DOI] [PubMed] [Google Scholar]
- 32.Howard, L., and H. Gooder. 1974. Specificity of the autolysin of Streptococcus (Diplococcus) pneumoniae. J. Bacteriol. 117:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu, L., et al. 2008. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1b secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc. Natl. Acad. Sci. U. S. A. 105:7803-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim, H., T. Matsuzaki, and T. Aoki. 2001. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 506:27-32. [DOI] [PubMed] [Google Scholar]
- 35.Ibrahim, H., U. Thomas, and A. Pellegrini. 2001. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J. Biol. Chem. 276:43767-43774. [DOI] [PubMed] [Google Scholar]
- 36.Inohara, N., et al. 1999. Nod1, an Apaf-like activator of caspase-9 and nuclear factor-κB. J. Biol. Chem. 274:14560-14567. [DOI] [PubMed] [Google Scholar]
- 37.Inohara, N., et al. 2003. Host recognition of bacterial muramyl dipeptide mediated through Nod2: implications for Crohn's disease. J. Biol. Chem. 278:5509-5512. [DOI] [PubMed] [Google Scholar]
- 38.Jordan, S., M. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 39.Kaparakis, M., et al. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell. Microbiol. 12:372-385. [DOI] [PubMed] [Google Scholar]
- 40.Kapetanovic, R., et al. 2007. Contribution of phagocytosis and intracellular sensing for cytokine production by Staphylococcus aureus-activated macrophages. Infect. Immun. 75:830-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi, K., et al. 2002. RICK/Rip2/CARDIAK mediates signaling for receptors of the innate and adaptive immune systems. Nature 416:194-199. [DOI] [PubMed] [Google Scholar]
- 42.Kojima, N., Y. Araki, and E. Ito. 1985. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J. Bacteriol. 161:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacks, S., and M. Neuberger. 1975. Membrane location of a deoxyribonuclease implicated in the genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 124:1321-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, J., et al. 2009. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J. Biol. Chem. 284:23818-23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, X., et al. 2006. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 281:5895-5907. [DOI] [PubMed] [Google Scholar]
- 46.Magalhaes, J., et al. 2005. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 6:1201-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magee, A., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majcherczyk, P., et al. 1999. Digestion of Streptococcus pneumoniae cell walls with its major peptidoglycan hydrolase releases branched stem peptides carrying proinflammatory activity. J. Biol. Chem. 274:12537-12543. [DOI] [PubMed] [Google Scholar]
- 49.Marina-García, N., et al. 2009. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J. Immunol. 182:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markart, P., T. Korfhagen, T. Weaver, and H. Akinbi. 2004. Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am. J. Respir. Crit. Care Med. 169:454-458. [DOI] [PubMed] [Google Scholar]
- 51.Martin, J., J. Fleck, M. Mock, and J. Ghuysen. 1973. The wall peptidoglycans of Neisseria perflava, Moraxella glucidolytica, Pseudomonas alcaligenes and Proteus vulgaris strain P18. Eur. J. Biochem. 38:301-306. [DOI] [PubMed] [Google Scholar]
- 52.McCarthy, J., J. Ni, and V. Dixit. 1998. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J. Biol. Chem. 273:16968-16975. [DOI] [PubMed] [Google Scholar]
- 53.Moore, L., A. Pridmore, S. Dower, and R. Read. 2004. The glycopeptide vancomycin does not enhance toll-like receptor 2 (TLR2) activation by Streptococcus pneumoniae. J. Antimicrob. Chemother. 54:76-78. [DOI] [PubMed] [Google Scholar]
- 54.Moore, L., A. Pridmore, S. Dower, and R. Read. 2003. Penicillin enhances the toll-like receptor 2-mediated proinflammatory activity of Streptococcus pneumoniae. J. Infect. Dis. 188:1040-1047. [DOI] [PubMed] [Google Scholar]
- 55.Moreillon, P., Z. Markiewicz, S. Nachman, and A. Tomasz. 1990. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob. Agents Chemother. 34:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreira, L., et al. 2008. The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory cytokine response to Gram-positive cell walls. Cell. Microbiol. 10:2067-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nash, J., T. Ballard, T. Weaver, and H. Akinbi. 2006. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J. Immunol. 177:519-526. [DOI] [PubMed] [Google Scholar]
- 58.Nigro, G., et al. 2008. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell. Microbiol. 10:682-695. [DOI] [PubMed] [Google Scholar]
- 59.Ogura, Y., et al. 2001. Nod2, a Nod1/Apaf family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276:4812-4818. [DOI] [PubMed] [Google Scholar]
- 60.Ohno, N., T. Yadomae, and T. Miyazaki. 1982. Identification of 2-amino-2-deoxyglucose residues in the peptidoglucan of Streptococcus pneumoniae. Carbohydr. Res. 107:152-155. [DOI] [PubMed] [Google Scholar]
- 61.Opitz, B., et al. 2004. Nucleotide binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J. Biol. Chem. 279:36426-36432. [DOI] [PubMed] [Google Scholar]
- 62.Pandey, A., et al. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park, J., et al. 2007. RICK/Rip2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 178:2380-2386. [DOI] [PubMed] [Google Scholar]
- 64.Ratner, A., J. Aguilar, M. Shchepetov, E. Lysenko, and J. Weiser. 2007. Nod1 mediates cytoplasmic sensing of combinations of extracellular bacteria. Cell. Microbiol. 9:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raymond, J., S. Mahapatra, D. Crick, and M. J. Pavelka. 2005. Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280:326-333. [DOI] [PubMed] [Google Scholar]
- 66.Rosenthal, R., J. Blundell, and H. Perkins. 1982. Strain-related differences in lysozyme sensitivity and extent of O-acetylation of gonococcal peptidoglycan. Infect. Immun. 37:826-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Royet, J., and R. Dziarski. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defenses. Nat. Rev. Microbiol. 5:264-277. [DOI] [PubMed] [Google Scholar]
- 68.Saha, S., et al. 2009. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe 5:137-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schröder, N., et al. 2003. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 278:15587-15594. [DOI] [PubMed] [Google Scholar]
- 70.Schwandner, R., R. Dziarski, R. Wesche, M. Rothe, and C. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 71.Shimada, K., et al. 2009. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 5:e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimada, T., et al. 2010. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1β secretion. Cell Host Microbe 7:38-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sørensen, U., J. Henrichsen, H. Chen, and S. Szu. 1990. Covalent linkage between the capsular polysaccharide and the cell wall peptidoglycan of Streptococcus pneumoniae revealed by immunochemical methods. Microb. Pathog. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 74.Swim, S., M. Gfell, C. Wilde III, and R. Rosenthal. 1983. Strain distribution in extents of lysozyme resistance and O-acetylation of gonococcal peptidoglycan determined by high-performance liquid chromatography. Infect. Immun. 42:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 76.Tomasz, A., and S. Waks. 1975. Mechanism of action of penicillin: triggering of pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc. Natl. Acad. Sci. U. S. A. 72:4162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Travassos, L., et al. 2004. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 5:1000-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vavricka, S., et al. 2004. hPepT1 transports muramyl dipeptide, activating NF-κB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology 127:1401-1409. [DOI] [PubMed] [Google Scholar]
- 79.Veiga, P., et al. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342-19354. [DOI] [PubMed] [Google Scholar]
- 80.Viala, J., et al. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166-1174. [DOI] [PubMed] [Google Scholar]
- 81.Vollmer, W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287-306. [DOI] [PubMed] [Google Scholar]
- 82.Vollmer, W., and A. Tomasz. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 70:7176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 84.Wada, A., et al. 2001. Helicobacter pylori-mediated transcriptional regulation of the human beta-defensin 2 gene requires NF-κB. Cell. Microbiol. 3:115-123. [DOI] [PubMed] [Google Scholar]
- 85.Wang, G., A. Olczak, L. Forsberg, and R. Maier. 2009. Oxidative stress-induced peptidoglycan deacetylase in Helicobacter pylori. J. Biol. Chem. 284:6790-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang, Z., et al. 2003. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 278:49044-49052. [DOI] [PubMed] [Google Scholar]
- 87.Welsh, I., and J. Spitznagel. 1971. Distribution of lysosomal enzymes, cationic proteins, and bactericidal substances in subcellular fractions of human polymorphonuclear leukocytes. Infect. Immun. 4:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolfert, M., A. Roychowdhury, and G. Boons. 2007. Modification of the structure of peptidoglycan is a strategy to avoid detection by nucleotide-binding oligomerization domain protein 1. Infect. Immun. 75:706-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zipperle, G. J., J. J. Ezzell, and R. Doyle. 1984. Glucosamine substitution and muramidase susceptibility in Bacillus anthracis. Can. J. Microbiol. 30:553-559. [DOI] [PubMed] [Google Scholar]
- 90.Zola, T., E. Lysenko, and J. Weiser. 2008. Mucosal clearance of capsule-expressing bacteria requires both TLR and nucleotide-binding oligomerization domain 1 signaling. J. Immunol. 181:7909-7916. [DOI] [PubMed] [Google Scholar]