Abstract
Enterotoxigenic Escherichia coli (ETEC) is a major cause of diarrheal illness in children less than 5 years of age in low- and middle-income nations, whereas it is an emerging enteric pathogen in industrialized nations. Despite being an important cause of diarrhea, little is known about the genomic composition of ETEC. To address this, we sequenced the genomes of five ETEC isolates obtained from children in Guinea-Bissau with diarrhea. These five isolates represent distinct and globally dominant ETEC clonal groups. Comparative genomic analyses utilizing a gene-independent whole-genome alignment method demonstrated that sequenced ETEC strains share approximately 2.7 million bases of genomic sequence. Phylogenetic analysis of this “core genome” confirmed the diverse history of the ETEC pathovar and provides a finer resolution of the E. coli relationships than multilocus sequence typing. No identified genomic regions were conserved exclusively in all ETEC genomes; however, we identified more genomic content conserved among ETEC genomes than among non-ETEC E. coli genomes, suggesting that ETEC isolates share a genomic core. Comparisons of known virulence and of surface-exposed and colonization factor genes across all sequenced ETEC genomes not only identified variability but also indicated that some antigens are restricted to the ETEC pathovar. Overall, the generation of these five genome sequences, in addition to the two previously generated ETEC genomes, highlights the genomic diversity of ETEC. These studies increase our understanding of ETEC evolution, as well as provide insight into virulence factors and conserved proteins, which may be targets for vaccine development.
The enterotoxigenic Escherichia coli (ETEC) pathovar represents a significant global health problem. ETEC is the cause of nearly one billion cases of diarrheal disease annually, resulting in the deaths of 300,000 to 500,000 children under the age of five in low- and middle-income nations (79). ETEC is also the leading cause of diarrhea among travelers to these nations (79) and is estimated to cause ∼400,000 cases annually of diarrhea among individuals older than 15 years (79). In addition, there have been recent ETEC outbreaks in the United States (4, 17, 32) and the Centers for Disease Control and Prevention (CDC) has described ETEC as an “emerging” pathogen in recent years (8, 17).
ETEC was one of the first bacterial pathogens to be examined with molecular biology tools when the genes for the heat-labile toxin (LT) and the heat-stable toxin (ST) were cloned in the late 1970s (64, 65). In contrast, the first molecular characterization of another important E. coli pathovar, enteropathogenic E. coli (EPEC), was accomplished over a decade later, with the cloning and characterization of the eae gene required for the formation of attaching and effacing lesions on enterocytes (33). Although there have been significant and progressive advances in the molecular characterization of other E. coli pathovars, ETEC is still relatively poorly characterized. This lack of an identified repertoire of virulence-associated genes has hampered the identification and characterization of potential novel vaccine targets (21). Only recently has there been the description of novel virulence factors that are potential targets for vaccine development (24, 58, 59). Molecularly, ETEC is characterized by the presence and elaboration of LT and/or ST, which are both plasmid encoded (21, 34). LT is a classic AB5 toxin similar to cholera toxin in specificity and activity (74). The activity, ribosylation of an intracellular guanine nucleotide protein, results in the increase in intracellular cyclic AMP (cAMP) and phosphorylation of the cystic fibrosis transmembrane regulator (CFTR) by cAMP-dependent protein kinase. This phosphorylation event results in Cl− ion secretion and inhibition of Na+ and Cl− ion absorption, resulting in a net loss of water and observed diarrhea (62). ST is a proteolytically processed small cysteine-rich peptide that binds to the epithelial exposed extracellular domain of guanylyl cyclase C, activating the peptide to increase intracellular cGMP, which in turn activates cGMP-dependent protein kinase II to phosphorylate the CFTR in the same manner as LT, resulting in diarrhea (9, 31, 63). Epidemiological studies have identified multiple variants of the ST peptides in human and animal ETEC isolates (isolates 37, 40, 78, Moseley, and 1983 #427). Molecular studies have suggested that there is a difference in the virulence of ETEC strains producing two particular ST variants, STp and STh, with STh-producing strains seemingly being more pathogenic than their STp-producing counterparts (51, 71). However, STp-producing ETEC strains are capable of causing disease in humans (47).
In addition to the conserved enterotoxins, ETEC strains express multiple adhesins that play a distinct role in pathogenesis (66, 76). The majority of the studies on ETEC adhesins have been focused on the colonization factors (CFs) (reviewed in detail in reference 25). CFs are surface structures that facilitate binding of the bacterium to the epithelial cell surface and represent antigenically and structurally diverse targets; >25 types of CFs have been identified to date (25). Although many ETEC isolates express one or more CFs, there are many isolates that either do not produce any CFs or that produce as-yet-unidentified CFs (25, 28, 66). The paramount importance of CFs in inducing anti-colonizing immunity has recently been called into question as the examination of a cohort of West African children followed up to their second birthday indicates that factors other than CFs may contribute substantially to the naturally acquired protection against ETEC infections (70).
There have been a number of recent advances in understanding the mechanisms involved in ETEC attachment and colonization of the intestinal surface, including characterization of the toxigenic invasion loci A (tia) and B (tib), the etpBAC gene cluster, and the involvement of flagella. The tia and tib loci appear to have divergent functions (20). Tia is a 25-kDa outer membrane protein that mediates increased adherence to epithelial cells via surface proteoglycans (36). The TibA protein encodes a glycosylated autotransporter that mediates adhesion to surface epithelial cells, but the role in invasion is unclear (35). The etpBAC locus is responsible for the glycosylation (EtpC) and secretion (EtpB) of EtpA, a 170-kDa protein that appears to act as a bridge between the exposed regions of FliC at the flagellar tip and host surface structures (59, 61). In addition, vaccination with EtpA has shown promise as a protective antigen in an animal model (59, 60). Thus, the flagella have been shown to be essential in the adhesion and resulting toxin secretion required for the virulence of some ETEC strains (61); however, as with many other bacteria, there is significant genetic diversity among the flagellin subunits, and thus the requirements for interaction of all of these components are still being investigated.
One additional virulence factor not shown to play a role in adhesion is EatA, a protein of the serine protease autotransporters of the Enterobacteriaceae (SPATE) family (29). The eatA locus has been identified in a number of ETEC isolates by PCR, hybridization, and sequencing (49). Protein production has been demonstrated to increase ETEC virulence in an animal model; however, its exact function is currently unknown (49). A recent study using a mouse model of infection and human convalescent-phase sera has identified a set of ETEC peptides, including EatA, that are immunogenic. Further study is required to determine whether these are viable vaccine candidates (58).
The expression of virulence and colonization factors in ETEC has not yet been studied in a systematic genome-wide manner. The best-characterized regulon in ETEC is the Rns regulon (41, 43), which is characterized by the AraC-like regulator, Rns (also called CfaD in a CFA/I expression isolate) (50). In some strains, Rns controls toxin expression. This regulatory protein contains significant homology and functional similarity to AggR, of the AggR regulon in enteroaggregative E. coli (46). Rns has recently been characterized in the regulation of all class 5 fimbriae in ETEC (6).
A phylogenetic analysis inferred from concatenated alignments from seven housekeeping genes used in multilocus sequence typing (MLST) analyses indicated that ETEC isolates have a diverse evolutionary history (68). Limited genomic comparisons, based on two genomes, suggests that ETEC isolates have relatively few pathovar-specific genes, as well as a greater number of isolate-specific genes than observed in other E. coli pathovars (54). However, from a genomic perspective, ETEC isolates are poorly characterized, with only two completed genomes (E24377A [54] and H10407 [12]) and one draft genome (B7A [54]) currently available in GenBank (54). The sequencing of additional ETEC genomes is therefore important not only from an evolutionary and phylogenetic perspective, but also as an important step in the identification of distribution profiles of virulence and colonization factors from a diverse set of ETEC isolates.
The purpose of the present study was to sequence and compare the genomes of isolates from important ETEC lineages to understand how these isolates are related genomically and phylogenetically. A gene-independent method utilized in the present study represents a valuable tool in the comparison of full genome sequences from a large number of isolates. In addition, this comparative genomic information will allow us to identify conserved regions that encode ETEC-specific genes that are potential targets for vaccine development. In addition, identification of the distribution of genes encoding virulence factors within ETEC genomes will help identify gene profiles that may help elucidate the mechanisms of ETEC virulence in humans.
MATERIALS AND METHODS
Strain selection.
The five ETEC strains selected for sequencing were isolated from children with diarrhea during a birth cohort study in Guinea-Bissau (GB-ETEC) and represent five distinct ETEC ancestral lineages and a range of different toxin and CF profiles (68); the details of each isolate are shown in Table 1 . Cultures were screened with multiplexed PCR for the identification of toxin and CF genes prior to sequencing as described by Rodas et al. (57). All predicted toxin and CF genes were confirmed to be present. The genome sequences for 44 E. coli isolates (see Table S1 in the supplemental material) were downloaded from GenBank and used for comparative analyses. Chromosome sequence only was used for comparative analyses between genomes.
TABLE 1.
Genomic characteristics of sequenced ETEC genomes
| Isolate | Clonal group | STa |
Presence (+) or absence (-) of toxinb |
Colonization factor(s) | Serotype(s)d | G+C (%) | ||
|---|---|---|---|---|---|---|---|---|
| EcMLST | PubMLST | ST | LT | |||||
| TW10598 | 1 | 171 | 4 | + | + | CS2, CS3, CS21 | O6:H16 | 50.7 |
| TW10722 | 5 | 706 | 443 | + | − | CS5, CS6 | O?:H5 | 50.7 |
| TW11681 | 8 | 713 | 728 | + | − | CFA/I, CS21 | O19:H45 | 50.7 |
| TW10828 | 3 | 127 | 173 | − | + | CS7 | O114:H49 | 50.7 |
| TW14425 | 2 | 88 | 23 | + | − | CS14 | O78:H9 | 50.5 |
| E24377Ac | 20 | 388 | 1308 | + | + | CS1, CS3 | CS1, CS3 | 50.6 |
| B7Ac | 4 | −52 | 94 | + | + | CS6 | CS6 | 50.7 |
DNA extraction.
Minimizing the number of passages of each strain, bacterial cultures were grown overnight from a population in 50 ml of Luria broth and genomic DNA was isolated according to standard methods (26). Briefly, bacterial cells were concentrated by centrifugation, washed and suspended in isolation buffer (0.15 M Tris, 0.1 M EDTA [pH 8.0]). Sodium dodecyl sulfate was then added to 1% (vol/vol) final concentration and allowed to incubate for 1 h at 55°C or until the solution cleared. Two volumes of phenol-chloroform-isoamyl alcohol (25:24:1) were added and briefly mixed by vortexing. The resulting solution was separated by centrifugation at 12,000 × g for 15 min at 4°C. The aqueous layer (top) was removed to a new tube and mixed with 2 volumes of chloroform. The mixture was separated by centrifugation at 12,000 × g for 15 min at 4°C, and the aqueous layer (top) was moved to a clean tube. The aqueous layer was then extracted with at least 10 volumes of ice-cold ethanol, and the precipitated DNA was spooled out of the mixture and suspended in ultrapure water. The purified mixture was further digested with RNase overnight at 37°C and reprecipitated the following day with 0.1 volumes of 3 M sodium acetate and 10 volumes of ice-cold ethanol. The pellet was allowed to air dry and then dissolved in a minimal volume of nuclease-free water (Ambion). The quantity and quality of genomic DNA were verified by gel electrophoresis, spectroscopy, and Picogreen assay.
Sequencing and assembly.
A sequencing approach using both Roche/454 Life Sciences and Solexa/Illumina sequencing technologies was used to generate draft genomes. We generated 8-kb, paired-end Roche/454 Life Sciences FLX and 300-bp paired-end Solexa/Illumina libraries for sequencing according to standard protocols suggested by the manufacturers and protocols used at the Institute for Genome Sciences Genome Resource Center at the University of Maryland School of Medicine. The 8-kb paired-end Roche/454 Life Sciences FLX sequence reads were assembled with Mira (10), Newbler, and the Celera Assembler (45). Solexa/Illumina reads were mapped to contigs from these assemblies with the Mosaik aligner (30). Assuming a chromosome length of 5 Mb, the expected coverage for each isolate was greater than 38× for each of the isolates. The best assembly was chosen by a combination of contig length, contig number, and the percentage of Solexa sequences that successfully mapped to 454 contigs. Based on these criteria, the Mira assembly was chosen for isolates TW10598, TW10722, TW14425, and TW10828, while the Celera assembly was chosen for isolate TW11681. The numbers of base pairs, reads, and contigs incorporated in the final assemblies are given in Table S2 in the supplemental material.
Sequence analysis and annotation.
Contigs from the assemblies were filtered for sequences of >500 nucleotides and with an average coverage of 9× to 11×, depending on the assembly. For annotation, the resulting contigs from assembly were first mapped to the E24377A chromosome with MUMmer (15). Contigs that failed to map to the reference were then compared by using BLAST at the nucleotide level to the GenBank nucleotide database using an E-value of 0.0001. Sequences that contained homology with previously annotated plasmids from enteric bacteria, including ETEC, were pooled together for annotation and further referred to as “plasmid-associated”; sequences that showed no homology to the ETEC chromosomal sequence or plasmids were annotated separately. Each binned group of contigs was then annotated with an annotation pipeline originally developed at the Institute for Genomic Research, which has previously been used for annotating other sequenced E. coli isolates (54).
BSR analysis.
BLAST score ratio (BSR) analysis (53) was conducted on genomes sequenced in the present study, as well as on the ETEC E24377A and B7A genomes. The BSR analysis divides the protein query BLAST score (2) by the reference blast score to determine relatedness between peptide sequences. Genes were considered to be conserved if they had a BSR > 0.8, unique if they had a BSR < 0.4, and divergent if they had a BSR in between these values.
MLST-based phylogenetic analyses.
We used the genomic sequences to extract the multilocus sequence typing (MLST) gene sequences for use in phylogenetic analyses. Two separate MLST systems were used, PubMLST (including the following genes: adk, fumC, gyrB, icd, mdh, purA, and recA) (80) and EcMLST (including the following genes: aspC, clpX, fadD, icdA, lysP, mdh, and uidA) (55). Alignments to each of these genomic regions were bioinformatically extracted from each genome sequence by using Mugsy (see below) and concatenated. To verify sequence typing, a BLAST search against a database of all possible sequence types was performed, and the typing for the top BLAST hit was extracted. MLST alignments for the isolates in the present study as well as from 44 genomes present in GenBank (see Table S1 in the supplemental material) were concatenated and aligned with Muscle (19) using default parameters. Columns that contained gaps, which represent missing data from draft genomes, were removed with Gblocks (72); this resulted in the analysis of 3,400 to 3,700 homologous gene sequence positions, depending on the system used. A phylogenetic tree for each MLST system was then inferred with the RAxML web server (67) and the general time-reversible model, with Escherichia fergusonii as the outgroup. A total of 1,000 bootstrap replicates were inferred, and the consensus topology was calculated.
Whole-genome phylogenetic analysis.
The sequence data for 44 E. coli/Shigella genomes were downloaded from GenBank and combined with sequence data from the five GB-ETEC isolates. Sequences were aligned with Mugsy (3), which incorporates MUMmer (15, 16) and SeqAn (18), to generate blocks of conserved, aligned sequence between species in the MAF file format. Blocks were then joined together and converted to a multifasta file with the bx-python toolkit (http://bitbucket.org/james_taylor/bx-python/wiki/Home). To validate the results of Mugsy, genomes were also aligned with progressiveMauve (13), and the alignment was converted to MAF and processed the same as for the Mugsy alignment. Columns with gaps in any one genome were removed with Gblocks (72) to create the core alignment, which consists of ∼2.7 Mb of genomic sequence. A phylogenetic tree was inferred by FastTree (52), with 1,000 bootstrap replicates and a general time-reversible model.
Shared genomic sequence.
The shared genomic sequence between GB-ETEC isolates, E24377A, and B7A was calculated using Mugsy (3). The amount of shared genomic sequence in the alignment of the seven ETEC genomes compared to the reference sequence length of the E24377A chromosome was calculated. We examined whether the GB-ETEC genomes share more genomic sequence with E24377A and B7A than with selected non-ETEC E. coli genomes. Five non-ETEC E. coli genomes from a pool of 42 were randomly selected with a custom Java script, and the absolute amount of genomic sequence they shared, irrespective of coding or noncoding status, with E24377A and B7A was calculated. This process was repeated so that each genome was represented at least once in the comparisons. In addition, the analysis was repeated and focused only to include groups of five genomes that were members of specific classical E. coli phylogenetic groups (55, 80). The lack of sufficient representatives from all of the pathovars prevented the examination in a pathovar-specific manner. We then used a two-tailed Student t test to determine whether the GB-ETEC strains had more genomic sequence in common with each other than with the five non-ETEC E. coli strains.
To compare the gene content between the E. coli core alignment and the ETEC alignment, conserved alignment blocks from the core alignment and the ETEC alignment were mapped to the genome of E24377A, and gene predictions were performed with Glimmer (14) using a minimum gene prediction length of 100 bp. Genes absent in the E. coli core alignment compared to the ETEC alignment were verified by a BLAST search of the genes against the E. coli core alignment; absent genes were then compared to the Kegg peptide database (http://www.genome.jp/kegg/) with BLASTX using a threshold of inclusion of E-value of <0.001. Gene annotations were inferred by comparison of the top BLAST hit to reference Kegg pathways.
Rns binding site identification.
A consensus sequence motif of known Rns binding sites, [TA][GTC]A[TA][TA][TA][TA][ATC][TA]TAT[CT][GAT] [GATC][ATC][TC]T (42), was identified in all GB-ETEC genomes with custom perl scripts. These scripts use genome coordinates and annotation to identify upstream and downstream genes from the identified motifs. Binding sites with the same motif and the same downstream gene annotations were identified.
Nucleotide sequence accession numbers.
The genomic data for these isolates have been submitted to GenBank under the following accession numbers: E. coli TW10598, AELA00000000; E. coli TW10722, AELB00000000; E. coli TW10828, AELC00000000; E. coli TW11681, AELD00000000; and E. coli TW14425, AELE00000000.
RESULTS
MLST validation.
The five isolates sequenced in the present study were selected based on MLST analyses of ETEC isolates from larger epidemiological studies (69). These isolates represent dominant and globally widespread ancestral ETEC lineages and virulence factor profiles (69). The comparison of the previously generated sequence for the MLST markers with the same marker sequence bioinformatically extracted from the genome sequences matched perfectly. This not only confirmed the quality of both analysis methodologies but also confirmed the initial sequence type classification of isolates using the MLST products.
Phylogenetic analysis.
Phylogenetic trees were inferred using sequence data both from MLST analyses and from a gene-independent whole-genome approach. From the 44 E. coli and Shigella genomes available in GenBank, 2.7 million bases of shared core genome were identified and extracted by using Mugsy. From this data set, over 290,000 variable positions that carry phylogenetic signal were identified. In contrast, the PubMLST scheme consists of ∼3,400 nucleotides with only 311 variable positions that carry phylogenetic signal in the E. coli alignment, and the EcMLST system consists of ∼3,700 nucleotides with only 256 variable positions identified. The whole-genome phylogenetic approach therefore provides 720- to 781-fold greater sequence data for analysis (depending on the system) and 935- to 1,130-fold greater phylogenetic resolution over traditional MLST analysis.
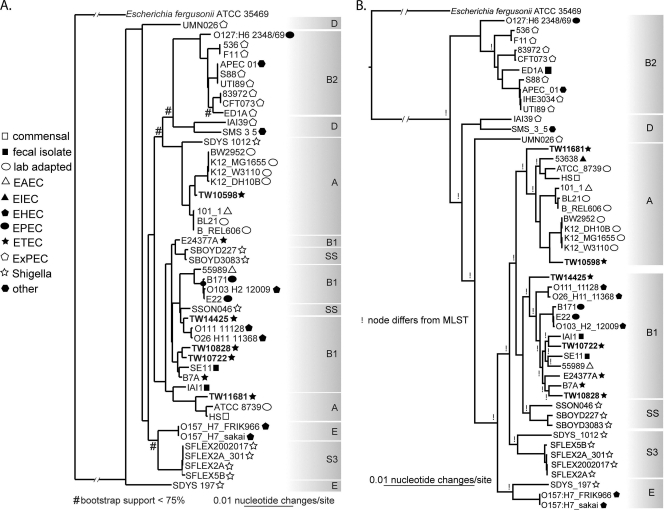
The topology of both seven-gene MLST trees (Fig. 1A, PubMLST tree shown) and the genomic core tree (Fig. 1B) differed at many nodes. For closely related species (e.g., K-12 isolates), the tip nodes were conserved. However, deeper branching nodes differed considerably between trees. The classical phylogenetic groups (A, B1, B2, D, and E) were applied to monophyletic groups in the genome core tree. Phylogenetic group D was found to be polyphyletic, as has been previously observed based on limited conserved genome data (73). When the phylogenetic groupings were then applied to the MLST tree for the same isolates, only phylogenetic group B2 was found to be monophyletic (Fig. 1A).
FIG. 1.
Comparison of the phylogenetic trees using either the seven-gene PubMLST system (A) or a whole-genome alignment (B) of >2.7 Mb of sequence information as determined by Mugsy (3) to be the core genome of E. coli. The pathotype of each E. coli strain is depicted with a symbol described in the legend. The letters on the right of each tree indicate the phylogenetic group. Bootstrap values are greater than 75% (A) or 95% (B), unless stated otherwise. The PubMLST tree (A) was inferred by using the maximum-likelihood method, while the whole-genome tree (B) was inferred with a combination of neighbor-joining and maximum-likelihood methods. Nodes highlighted with an “!” are different in the whole-genome analysis (B) than they are in the PubMLST analysis (A). The whole-genome analysis consolidates a number of the other phylogenetic types that were previously separated on the MLST tree (groups A, B1, and E, as well as the Shigella group). EAEC, enteroaggregative E. coli; EHEC, enterohemorrhagic E. coli; EIEC, enteroinvasive E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; ExPEC, extraintestinal pathogenic E. coli.
The ETEC strains sequenced in the present study grouped with the classical E. coli phylogenetic groups A and B1. Strain TW10598 grouped near the lab-adapted K-12 strains, which has also been shown for ETEC H10407 based on MLST data (75). TW10828 shared a nearest common ancestor with the previously sequenced ETEC isolates, B7A and E24377A. The GB-ETEC strains are thought to be representative members of these distinct ETEC lineages (21), which to a large extent is supported by their diverse phylogenetic distribution identified by MLST and core genome analysis.
Shared genomic sequence between ETEC isolates.
No conserved segments of sequence longer than 100 nucleotides in the whole-genome alignment were found exclusively in all seven ETEC genomes, compared to the 42 non-ETEC E. coli and Shigella genomes. Gene-dependent views have failed to identify significant numbers of ETEC group-specific genes (54). However, to test whether ETEC isolates share a greater amount of genomic sequence than would be expected by chance, a Mugsy alignment was performed for E24377A (chromosome only), B7A, and the five GB-ETEC isolates. The results show that 80.2% (99% confidence range ± 1.45%) of the E24377A chromosome is conserved in sequence common to these seven ETEC genomes. In contrast, only 53.7% of the E24377A chromosome is present in all 44 publicly available E. coli genomes, based on homologous positions with >97% sequence identity. When any five non-ETEC E. coli genomes were randomly selected (10 iterations) from a pool of 42 present in GenBank and compared to E24377A and B7A, 67% (99% confidence range ± 2.8%) of the E24377A genome was conserved. These findings suggest that there is a conserved ETEC genomic core.
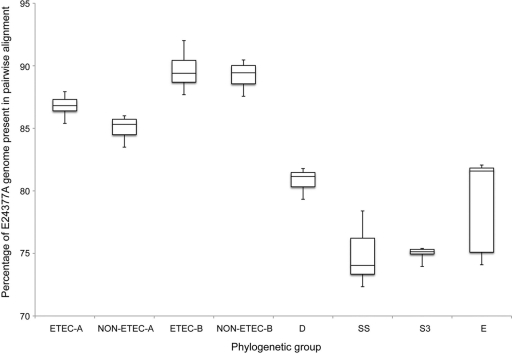
To test if this finding was simply a result of the fact that all ETEC genomes fall into phylogenetic groups A and B1 (Fig. 1), we examined 10 permutations of five randomly selected genomes from the 18 E. coli strains that are present in phylogenetic groups A and B1. Each permutation consisted of three genomes from group B1 and 2 from group A, which is the same distribution as for the GB-ETEC isolates. These results again demonstrate that although a higher percentage of the ETEC-core genome was present in this new alignment (77%; 99% confidence range ± 0.77%), the ETEC-only alignment contained a significantly higher percentage of shared genomic sequence (P = 0.009) (Fig. 2). The alignment using progressiveMauve (13) validated the results of the Mugsy alignment, with 75.9% (99% confidence range ± 1.6%) of E24377A in the A and B1 alignment and 79% (99% confidence range ± 0.95%) of E24377A in the ETEC-only alignment. The ETEC isolates from phylogenetic group A contain a significantly greater amount of sequence in common with other ETEC isolates than with other genomes in phylogenetic group A. This suggests that whereas ETEC strains possess very few group-specific genes compared to other E. coli isolates, ETEC isolates share a more common genomic sequence than would be expected by chance in relation to genomes with shared evolutionary history. A selected list of predicted genes present in the ETEC-only alignment and inconsistently present in the random alignment of closely related E. coli genomes is shown in Table S3 in the supplemental material.
FIG. 2.
Diversity of the E. coli genomes calculated on the conserved core in a gene-independent calculation. A box-and-whisker plot of the percent relatedness of genomes from defined phylogenetic groups to the chromosome of ETEC E24377A is shown. The percent relatedness was calculated by the amount of shared sequence in a whole-genome alignment divided by the genome sequence length of E24377A.
BSR analysis.
A BSR analysis was performed to search for conserved, divergent, and unique genes in the five draft genomes sequenced here compared to the publicly available ETEC genomes E24377A and B7A; the results of this analysis are presented in Table 2 . The results show that between 200 and 450 unique genes were identified in each genome (including plasmid sequence; B7A also contains plasmid sequence) compared to other ETEC genomes. A selected set of unique genes is listed in Table S4 in the supplemental material. The majority of these unique genes currently have no functional annotation.
TABLE 2.
Genetic characteristics of sequenced ETEC genomes based on BSR analysis
| Isolatea | No. of genesb |
|||
|---|---|---|---|---|
| Conserved genes | Divergent genes | Unique genes | Total genes | |
| TW10598 | 3,134 | 1,863 | 296 | 5,293 |
| TW10722 | 3,235 | 2,461 | 440 | 6,136 |
| TW10828 | 3,090 | 1,994 | 231 | 5,315 |
| TW11681 | 3,086 | 2,027 | 253 | 5,366 |
| TW14425 | 3,116 | 2,058 | 207 | 5,381 |
For each isolate listed, the plasmid sequence is included.
Conserved, blast score ratio (BSR) > 0.80; divergent, BSR between 0.4 and 0.8; unique, BSR < 0.40.
Adhesion and colonization factors, toxins, and virulence factors.
A number of genes have been linked with the virulence of ETEC isolates in humans (21). In addition to the ST, LT, and the CFs, there are a number of other putative adhesins and autotransporters thought to be produced during human infection by this pathovar. We bioinformatically examined each of the sequenced genomes using BLAST for known ETEC virulence factors (Table 3) . BLAST alignments longer than 50% of the query reference sequence and with >75% identity were considered to be conserved. As had been previously appreciated, the ETEC isolates sequenced to date do not have a consistent pattern of virulence gene presence (Table 3). Interestingly, the genes encoding LeoA (accessory protein for LT secretion) (23), TibA (autotransporter), Tia (surface protein) (22), ClyA (cytolysin), and EatA (serine protease autotransporter) have been associated with ETEC virulence (75), but they are not consistently present in all genomes.
TABLE 3.
Virulence factor presence or absence in sequenced genomes
| Virulence factor | Presence (+) or absence (-)a of virulence factor in isolate: |
||||||
|---|---|---|---|---|---|---|---|
| E24377A | B7A | TW10598 | TW10722 | TW10828 | TW11681 | TW14425 | |
| etpA | + | − | + | − | + | + | + |
| estA | + | + | + | + | − | + | + |
| eltA | + | + | + | − | + | − | − |
| eatA | + | − | + | + | + | + | − |
| ast | + | + | − | − | − | + | − |
| fimH | + | + | + | + | + | + | + |
| ecpA | + | − | + | + | + | − | + |
| tiab | − | − | − | − | − | − | − |
| tibA | + | − | − | − | − | − | + |
| leoAc | − | − | − | − | − | − | − |
| clyA | − | + | + | + | + | + | + |
| lngAd | − | − | + | − | − | + | − |
Genomic analysis of GB-ETEC genomes demonstrated that only isolate TW10598 contained both ST and LT genes (Table 3), as was previously predicted (Table 1). Both LT subunit genes (eltA and eltB) were present in two of the five genomes but absent in the other isolates. Conversely, the ST structural gene (estA) was verified to be present in four of the five GB-ETEC strains (Table 3).
The CFs demonstrate a wide distribution among the five GB-ETEC strains and the two ETEC strains for which whole genomic sequence data are publicly available (Table 1). Strains TW10598 and TW11681 shared the entire ∼14-kb operon that encodes a type IV pilus known as the longus pilus or CS21 (11, 27). Sequences from the longus operon were absent in all plasmid sequences from publicly available ETEC genomes. The structural subunit lngA, a colonization factor thought to contain high structural variability (27), exhibited an ∼99% nucleotide identity between TW10598, TW11681, and the published sequence available in GenBank (accession no. ABV57862), representing yet another adhesive mechanism of ETEC.
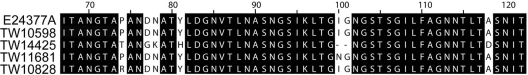
The etpA gene, which is part of the E. coli two-partner secretion locus (etpBAC) and is thought to act as a bridge between exposed regions of FliC of the ETEC flagella and host cell receptors (60, 61), was present in four of the five GB-ETEC strains (Table 3). EtpA may be a central peptide for ETEC adhesion, as well as a newly identified target for ETEC vaccine development (60, 61). A multiple sequence alignment of peptide sequences with the sequence from the homologous protein in E24377A identified several conserved motifs (Fig. 3).
FIG. 3.
Conservation and variation of the EtpA protein in ETEC. The visualization of a region of the EtpA global alignment performed by Muscle identifies conserved domains in the amino terminus. Numbers at the top indicate the relative peptide position of the alignment. Sequence blocks in black are identical among all five genomes.
In addition to ETEC colonization factors, several other genes associated with ETEC adherence were identified in the GB-ETEC genomes. The E. coli common pilus subunit gene (ecpA), shown to be present in 80% of ETEC strains (5) and in 96% of all major E. coli pathovars (56), was present in four of five of the GB-ETEC strains (absent only in TW11681), as well as the two previously sequenced ETEC genomes. Peptide sequences were highly conserved (>99%) across the four GB-ETEC isolates and E24377A and B7A. The proteins FimFGH, which are part of type I fimbriae, were successfully identified in all GB-ETEC strains. Although FimH has been associated with pathogenesis in other E. coli pathovars (21), its link to pathogenesis in ETEC strains is not currently understood.
Immunoreactive surface proteins.
A recent study by Roy et al. (58) identified 40 ETEC strain H10407 proteins that elicited an immune response in a mouse model and were reactive with human convalescent-phase sera. In addition to known antigens, such as colonization factors and enterotoxins, several hypothetical proteins and housekeeping genes were also found to be immunoreactive (Table 4). To identify the distribution of these genes across all sequenced ETEC genomes, a comparative approach of BSR values was used. Interestingly, the genes with the greatest ETEC specificity (i.e., common in ETEC strains and uncommon in non-ETEC strains) are EatA and EtpA. Both are identified exclusively in ETEC isolates and are ideal potential vaccine candidates. The results demonstrate that several of these genes are broadly conserved across sequenced ETEC genomes and largely absent in other E. coli/Shigella genomes (Table 4).
TABLE 4.
Distribution of immunoreactive genes in ETEC genomes
| Annotation | Accession no. | Presence (+) or absence (-)a in isolate: |
% Presentb in: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E24377A | B7A | TW10598 | TW10722 | TW10828 | TW11861 | TW14425 | HS | ETEC genomes | Non-ETEC genomes | ||
| FliC | YP_003229794 | − | − | − | − | − | − | − | − | 12.5 | 5.41 |
| EatA | AAO17297 | + | − | + | + | + | + | − | − | 75 | 0 |
| Antigen 43 | YP_001464349 | + | − | + | + | − | + | − | − | 62.5 | 35.1 |
| EtpA | AAX13509 | + | − | + | − | + | + | + | − | 75 | 0 |
| CfaA | AAC41414 | − | − | − | − | − | + | − | − | 25 | 0 |
| FimD2 | ABV18716 | + | + | + | + | + | + | + | + | 100 | 51.4 |
| CfaB | P02971 | − | − | − | − | − | + | − | − | 25 | 0 |
| LT-B | AAB02982 | − | − | + | − | + | − | − | − | 37.5 | 0 |
| CexE | ABM92275 | − | − | − | − | − | − | − | − | 12.5 | 0 |
| TibA | Q9XD84 | − | − | − | − | − | − | + | − | 37.5 | 0 |
| GroEL | AAR21890 | + | + | + | + | + | + | + | + | 100 | 100 |
| DnaK | NP_308041 | + | + | + | + | + | + | + | + | 100 | 100 |
| DegP/HtrA | NP_285857 | + | + | + | + | + | + | + | + | 100 | 100 |
| OmpA | NP_309068 | + | − | + | + | + | + | + | + | 87.5 | 100 |
| NmpC | AAB40749 | + | + | + | + | + | + | + | − | 100 | 62.2 |
| FecA | YP_405685 | + | + | − | + | + | + | + | − | 87.5 | 43.2 |
| OmpX | NP_752830 | + | + | + | + | + | + | + | + | 100 | 100 |
| Hypothetical | NP_288109 | + | + | + | + | + | + | + | + | 100 | 91.9 |
| Hypothetical | ZP_04533914 | − | − | + | − | − | − | − | − | 25 | 0 |
| Hypothetical | NP_755083 | − | − | + | + | + | + | + | − | 75 | 43.2 |
| Hypothetical | NP_311861 | − | + | + | + | + | + | + | + | 87.5 | 100 |
| Hypothetical | YP_853622 | − | − | − | + | − | − | + | − | 37.5 | 8.10 |
| Hypothetical | YP_002408614 | − | − | − | − | − | − | + | − | 25 | 5.41 |
| Hypothetical | NP_286070 | + | + | + | + | + | + | + | + | 100 | 75.7 |
| Lpp | NP_288111 | + | − | + | + | + | + | + | + | 87.5 | 97.3 |
| HYP9 | ZP_04533918 | − | − | + | − | − | − | − | − | 25 | 0 |
| YghJ | YP_002388453 | + | + | + | + | + | + | + | + | 100 | 59.5 |
| Hypothetical | NP_288218 | + | + | + | + | + | + | + | + | 100 | 100 |
| Hypothetical | YP_541993 | − | − | + | + | + | + | + | − | 75 | 43.2 |
| FusA | NP_312218 | + | − | + | + | + | + | + | + | 87.5 | 100 |
| H-NS | CAA47740 | + | + | + | + | + | + | + | + | 100 | 100 |
| aceE | ZP_03072175 | + | + | + | + | + | + | + | + | 100 | 100 |
| tpx | AAC43517 | + | + | + | + | + | + | + | + | 100 | 100 |
| trxA | AAA24696 | + | + | + | + | + | + | + | + | 100 | 97.3 |
| lpdA | NP_285812 | + | + | + | + | + | + | + | + | 100 | 97.3 |
+, Present with a BSR ≥ 0.80, which is similar to 80% peptide identity over 80% of the length of the peptide. −, Absent or demonstrating a BSR < 0.80.
Plasmids.
The presence of enterotoxins and plasmid-encoded colonization factors demonstrates that each of the GB-ETEC strains most likely contains plasmids. ETEC plasmid-associated sequences were compared between all ETEC strains, largely due to the fact that the heat-stable and heat-labile toxins, and all but one known colonization factor, CS2, are plasmid encoded. A Mugsy alignment of plasmid sequences from E24377A, B7A, and the five ETEC isolates sequenced in the present study identified only ∼2,700 nucleotides of conserved sequence. A BLAST homology search showed that this conserved sequence fragment was largely from the pETEC74 plasmid isolated from E24377A. Conserved genes primarily included those encoding plasmid partition proteins; plasmid-encoded enterotoxins and colonization factors were not conserved across all comparable plasmid sequences, which was not expected based on their variable distribution within the ETEC pathovar (Table 3). This highlights the previously identified variability of the plasmids in ETEC (54).
Rns promoter region identification.
Based on a previously published genomic sequence motif (44), the Rns promoter region was bioinformatically identified in all GB-ETEC genomes. A selected list of gene annotations located downstream of these regions is shown in Table 5. Rns binding motifs were associated with colonization factors (Table 5), as reported previously (7). However, not all colonization factor genes present in GB-ETEC genomes were preceded by identifiable Rns binding sites in the promoter regions. Additional Rns binding motifs were consistently present in GB-ETEC genomes and were associated with a type VI secretion effector and an acetate operon repressor.
TABLE 5.
Distribution of Rns binding sites in ETEC isolates
| Rns binding motif | Downstream gene annotation | Presence (+) or absence (-) in isolate: |
||||
|---|---|---|---|---|---|---|
| TW10598 | TW10722 | TW10828 | TW11681 | TW14425 | ||
| ATATTTAATTATTTAACT | Inner membrane protein YafU | + | + | + | + | + |
| TTATTAATTTATTTATTT | Type VI secretion system effector | + | + | + | + | + |
| TCATTTAAATATCTCATT | Acetate operon repressor | + | + | + | + | + |
| AGATAATATTATTTCCCT | Putative kinase inhibitor | + | − | − | − | + |
| TTATTATTTTATCTCTTT | CFA/I fimbrial subunit D | − | − | − | + | − |
| TGATATATTTATTTTTTT | CS7 fimbrial major subunit | − | − | + | − | − |
| TGAAAAATATATCTTTCT | CFA/I fimbrial subunit D | − | − | − | − | + |
DISCUSSION
This study describes the sequencing and comparative genomic analysis of five ETEC isolates from children with diarrhea in Guinea-Bissau that were deliberately selected since they represent the five predominant and globally widespread ETEC ancestral lineages, as determined by MLST. This approach, made possible by lower sequencing costs, has allowed us to examine more than a single “reference” or “prototype” isolate and thereby describe dominant ETEC clonal lineages. This represents a new paradigm for genomic analysis. In addition, this methodology provides us with the opportunity to identify more subtle genomic features that would not be identified using only type strains.
Phylogenetic analysis.
The observed difference in tree topologies between the MLST (68) and the genomic core tree demonstrate that the limited information from MLST analysis alone is insufficient to resolve deep phylogenetic relationships between E. coli and Shigella species (Fig. 1). This genome core alignment provided by Mugsy and confirmed with the progressiveMauve aligner (13) represents a gene-independent view of evolution between closely related organisms. With the increased use of high-throughput sequencing methods, genomic data can more easily be obtained, the core alignment extracted, and phylogenies determined relatively cheaply. We have calculated that the time and effort spent on PCR amplification and Sanger sequencing of 7 to 10 MLST gene fragments will soon rival the cost of generating a draft genome sequence using “now-generation” and “next-generation” sequencing capabilities. These high-throughput methods can provide a framework for analyzing the relatedness of pathotypes, the passage of mobile elements between pathotypes, and the fate and emergence of virulence factors. Such a method is only now becoming possible as well as economically feasible.
The whole-genome alignment tree resolved the major phylogenetic groups within E. coli. Phylogenetic group D, however, was shown to be polyphyletic. Additional sequencing and placement of in-house draft genomes (data not shown) suggests that group D needs to be split into two separate, monophyletic groups. E. coli strain UMN026 appears to be divergent based on its deep branch with its closest common ancestor. However, additional sequencing is expected to expand and resolve this clade to form a new phylogenetic group within the E. coli.
ETEC shared sequence.
The whole-genome alignment phylogenetic tree demonstrates diversity within the ETEC pathovar. These data suggest that the genomic core of ETEC is not unique and that the acquisition of mobile elements encoding enterotoxins defines ETEC. However, the whole-genome alignment method demonstrates that the five GB-ETEC isolates sequenced in the present study generally contain more shared genomic sequence with previously sequenced ETEC genomes than five E. coli genomes chosen at random from similar phylogenetic distributions or diverse phylogenies. Inclusion of the newly published E. coli ETEC H10407 genome (12) in these analyses further supports these conclusions. Although none of the predicted gene sequences from this shared genomic core were explicitly linked with virulence or colonization, the presence of this shared sequence suggests a common evolutionary history between ETEC isolates that cannot simply be explained by the presence of mobile elements. The possibility exists that the increased relationship of five ETEC isolates from a limited geography, Guinea-Bissau, has some impact on the scope of the genomic conservation. However, the analysis of additional ETEC genomes isolated from diverse geographic locations suggests that isolation location has little effect on this phenomenon (J. W. Sahl and D. A. Rasko, unpublished). This is an important finding since it is the first time that the genomic core of an E. coli pathovar has been demonstrated to be conserved.
Enterotoxins and colonization factors.
The variable distribution of toxins and colonization factors within the ETEC isolates supports the lack of unique and totally conserved ETEC-specific genes. This also presents difficulties in designing traditional ETEC-specific vaccines. However, the addition of these five genomes allows for a more thorough sampling of the distribution of virulence and colonization factors within the ETEC pathovar.
One strategy for vaccine development has been to develop a killed whole-cell vaccine from strains of ETEC with a diverse set of colonization factors (1); such a strategy has been demonstrated to not be globally effective and may need to be improved (77). The sequencing of additional ETEC genomes will help identify all known colonization factors or surface-exposed antigens, and a vaccine could then be developed with a greater number of ETEC strains, or a selected set of region-selected strains, or with proteins derived from such a wider array of characterized ETEC strains.
Additional putative virulence factors.
In addition to the enterotoxins, colonization factors, and fimbriae, six additional virulence-associated genes (leoA, tibA, tia, clyA, eatA, and etpA) were screened for in the five GB-ETEC genomes sequenced. However, only eatA, etpA, and clyA were consistently identified in the GB-ETEC strains (Table 3). We have yet to determine whether these genes are absolutely required for ETEC virulence. These results confirm a previous study that showed that only ETEC H10407 contained all five of these genes in a collection of 116 human ETEC strains (75). Even though the presence of each of these genes is not necessary for ETEC pathogenesis, it is possible that a combination of these genes could increase ETEC virulence in humans.
Only clyA, a cytolysin, was present and conserved in the genomes of all available ETEC genomes. However, even laboratory-adapted E. coli strains, such as K-12, have been shown to express a cytolytic phenotype (48). Research has shown that clyA is regulated by a complex system of regulation, including HN-S, cyclic AMP, and slyA (38, 48), which are present in all sequenced ETEC genomes.
In addition, we are examining the peptides in a global analysis. It is possible that conserved domains within some of these peptides are important but do not meet the threshold for identification in our studies. Detailed functional genomics analysis will be required to determine these capabilities.
Vaccine targets.
A comparison of the two previously sequenced ETEC strains with the five GB-ETEC strains failed to identify genes or genomic regions shared comprehensively and exclusively by the ETEC strains. The more common pattern observed is highlighted by the virulence factors etpA and lngA that were conserved among some, but not all, of the ETEC isolates (Table 3). This suggests that a strategy targeting multiple surface antigens may be the most effective approach when developing vaccines to protect against as diverse a phylogenetic set of ETEC strains as possible.
A previous study suggested that the E. coli common pilus (ECP), due to its widespread distribution within ETEC, also be considered as an ETEC vaccine target (5). Eighty percent of the strains sequenced in the present study contained ECP, and the peptide sequences were >99% identical to the two previously published ETEC genomes (54). These results verify that ecpA is widely distributed and well conserved across phylogenetically diverse ETEC isolates. However, the role of ECP in ETEC colonization should be determined before research into a vaccine is conducted.
Previous studies with ETEC strains H10407 and E24377A have demonstrated that the EtpA glycoprotein can provide protection against ETEC colonization in mice (59, 61). Genomic sequence analysis performed here identified common peptide motifs across ETEC strains (Fig. 3), which suggests that a vaccine with EtpA glycoprotein could protect across a wide phylogenetic range of ETEC. Interestingly, Fleckenstein et al. identified EtpA protein secretion in E24377A and H10407 (24). Additionally, these authors demonstrated that 58% of the ETEC isolates examined contained the etpA gene, but only 44% secreted the EtpA protein in vitro (24). Of the five strains sequenced here, four had similar EtpA peptide sequences (Fig. 3), despite a diverse phylogenetic distribution. Protein secretion, however, has not yet been examined. Furthermore, etpA is present and immunoreactive in H10407 (58), whereas it is absent in all non-ETEC E. coli genomes (Table 4). Additional sequencing of ETEC genomes will help identify the conservation of etpA, and further analysis of protein secretion will help determine whether EtpA vaccination is a viable approach for designing and testing global ETEC vaccinations.
Another potential vaccine target is the longus pilus, or CS21, structural subunit (lngA), thought to be present in only 10 to 38% of ETEC strains (11, 27, 39). Despite the fact that lngA is thought to be under strong selection for structural diversification (27), the highly conserved nature of the sequence in two of the GB-ETEC strains suggests that it could be used, perhaps in combination with other surface proteins, as a potential vaccine target. Recent work has demonstrated reduced adherence of ETEC due to the presence of serum longus antibodies (39).
In addition to previously identified antigens in ETEC, a recent study identified genes that elicit an immune response in a mouse model infected with ETEC H10407 (58). In addition to genes associated with ETEC virulence, hypothetical proteins not associated with virulence were broadly conserved across ETEC genomes and less commonly conserved or absent in non-ETEC genomes. The broad conservation of these immunoreactive proteins suggests that these peptides could be potential candidates to include in a vaccine targeting a diverse set of ETEC strains.
Overall, the sequencing and comparison of these five new ETEC genomes has resulted in a number of important findings, the most important of which is that while the ETEC pathovar virulence factors are varied, a pathovar-specific genomic core exists. This is the first time that such a conserved genomic pathovar core has been identified. We must also note that the inclusion of these five diverse ETEC genomes represents the largest group of diverse strains of any one pathovar analyzed to date. There are more EHEC O157:H7 genomes completed, but they represent clonal expansion and the sequencing of multiple isolates from outbreaks. We have also confirmed the variability of virulence and antigenically dominant genes in the ETEC pathovar and indicate that the variability extends beyond just the virulence genes to the genome core. This genomic study suggests that any ETEC vaccine will need to be multivalent to address a wide range of clonal lineages. Taken together, these findings shed genomic light on one of the most important diarrheal pathogens known to mankind and provide a paradigm for the examination of additional E. coli and Shigella pathovars.
Supplementary Material
Acknowledgments
We thank the Guinea-Bissau Childhood Diarrhea Research Team, in particular Palle Valentiner-Branth and Kåre Mølbak, for providing ETEC strains and the members of the Institute for Genome Sciences Genomics Resource Center and Informatics Resource Center for their contributions to these studies. In addition, we thank David Lacher for the in silico serotyping analysis.
This study was supported in part by the Global Health and Vaccination (GLOBVAC) Research Programme under the Research Council of Norway (www.rcn.no) contract 185872/S50 and startup funds from the State of Maryland to the Rasko laboratory.
Editor: S. M. Payne
Footnotes
Published ahead of print on 15 November 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Ahren, C., M. Jertborn, and A. M. Svennerholm. 1998. Intestinal immune responses to an inactivated oral enterotoxigenic Escherichia coli vaccine and associated immunoglobulin A responses in blood. Infect. Immun. 66:3311-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Angioli, S., and S. L. Salzberg. 9 December 2010. MUGSY: rapid and accurate whole genome reference independent alignment. Bioinformatics doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed]
- 4.Beatty, M. E., et al. 2006. Epidemic diarrhea due to enterotoxigenic Escherichia coli. Clin. Infect. Dis. 42:329-334. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, D., et al. 2009. Distribution of the Escherichia coli common pilus among diverse strains of human enterotoxigenic E. coli. J. Clin. Microbiol. 47:1781-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodero, M. D., E. A. Harden, and G. P. Munson. 2008. Transcriptional regulation of subclass 5b fimbriae. BMC Microbiol. 8:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2005. Diarrheagenic Escherichia coli (non-Shiga toxin-producing E. coli). CDC, Atlanta, GA. http://www.cdc.gov/ncidod/dbmd/diseaseinfo/diarrecoli_t.htm.
- 9.Chao, A. C., et al. 1994. Activation of intestinal CFTR Cl− channel by heat-stable enterotoxin and guanylin via cAMP-dependent protein kinase. EMBO J. 13:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevreux, B., T. Wetter, and S. Suhai. 1999. Computer science and biology. Proc. German Conf. Bioinformatics 99:45-56. [Google Scholar]
- 11.Clavijo, A. P., J. Bai, and O. G. Gomez-Duarte. 2010. The longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents. Microb. Pathog. 48:230-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crossman, L. C., et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darling, A. E., B. Mau, and N. T. Perna. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher, A. L., A. Phillippy, J. Carlton, and S. L. Salzberg. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delcher, A. L., S. L. Salzberg, and A. M. Phillippy. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics Chapter 10:Unit 10-13. [DOI] [PubMed] [Google Scholar]
- 17.Devasia, R. A., et al. 2006. Endemically acquired food-borne outbreak of enterotoxin-producing Escherichia coli serotype O169:H41. Am. J. Med. 119:168 e167-e110. [DOI] [PubMed] [Google Scholar]
- 18.Doring, A., D. Weese, T. Rausch, and K. Reinert. 2008. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment with reduced time and space complexity. BMC Bioinformatics 5:113-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsinghorst, E. A., and D. J. Kopecko. 1992. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 60:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleckenstein, J. M., et al. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleckenstein, J. M., L. E. Lindler, E. A. Elsinghorst, and J. B. Dale. 2000. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 26.Ge, Z., and D. E. Taylor. 1992. H. pylori DNA transformation by natural competence and electroporation, p. 145-152. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed]
- 27.Gomez-Duarte, O. G., et al. 2007. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J. Bacteriol. 189:9145-9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harel, J., et al. 1991. Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J. Clin. Microbiol. 29:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 30.Hillier, L. W., et al. 2008. Whole-genome sequencing and variant discovery in Caenorhabditis elegans. Nat. Methods 5:183-188. [DOI] [PubMed] [Google Scholar]
- 31.Hughes, J. M., F. Murad, B. Chang, and R. L. Guerrant. 1978. Role of cyclic GMP in the action of heat-stable enterotoxin of Escherichia coli. Nature 271:755-756. [DOI] [PubMed] [Google Scholar]
- 32.Jain, S., et al. 2008. An outbreak of enterotoxigenic Escherichia coli associated with sushi restaurants in Nevada, 2004. Clin. Infect. Dis. 47:1-7. [DOI] [PubMed] [Google Scholar]
- 33.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lasaro, M. A., et al. 2008. Genetic diversity of heat-labile toxin expressed by enterotoxigenic Escherichia coli strains isolated from humans. J. Bacteriol. 190:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lortie, L. A., J. D. Dubreuil, and J. Harel. 1991. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J. Clin. Microbiol. 29:656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig, A., et al. 2010. Mutations affecting export and activity of cytolysin A from Escherichia coli. J. Bacteriol. 192:4001-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazariego-Espinosa, K., A. Cruz, M. A. Ledesma, S. A. Ochoa, and J. Xicohtencatl-Cortes. 2010. Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192:2791-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moseley, S. L., M. Samadpour-Motalebi, and S. Falkow. 1983. Plasmid association and nucleotide sequence relationships of two genes encoding heat-stable enterotoxin production in Escherichia coli H-10407. J. Bacteriol. 156:441-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munson, G. P., L. G. Holcomb, H. L. Alexander, and J. R. Scott. 2002. In vitro identification of Rns-regulated genes. J. Bacteriol. 184:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 45.Myers, E. W., et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196-2204. [DOI] [PubMed] [Google Scholar]
- 46.Nataro, J. P., D. Yikang, D. Yingkang, and K. Walker. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishikawa, Y., et al. 1998. Epidemiology and properties of heat-stable enterotoxin-producing Escherichia coli serotype O169:H41. Epidemiol. Infect. 121:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of hemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 49.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pilonieta, M. C., M. D. Bodero, and G. P. Munson. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 189:5060-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porat, N., A. Levy, D. Fraser, R. J. Deckelbaum, and R. Dagan. 1998. Prevalence of intestinal infections caused by diarrheagenic Escherichia coli in Bedouin infants and young children in Southern Israel. Pediatr. Infect. Dis. J. 17:482-488. [DOI] [PubMed] [Google Scholar]
- 52.Price, M. N., P. S. Dehal, and A. P. Arkin. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasko, D. A., G. S. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasko, D. A., et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 56.Rendon, M. A., et al. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodas, C., et al. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J. Clin. Microbiol. 47:1218-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, K., S. Bartels, F. Qadri, and J. M. Fleckenstein. 2010. Enterotoxigenic Escherichia coli elicit immune responses to multiple surface proteins. Infect. Immun. 78:3027-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy, K., D. Hamilton, K. P. Allen, M. P. Randolph, and J. M. Fleckenstein. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy, K., D. Hamilton, M. M. Ostmann, and J. M. Fleckenstein. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601-4608. [DOI] [PubMed] [Google Scholar]
- 61.Roy, K., et al. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sack, R. B., et al. 1971. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J. Infect. Dis. 123:378-385. [DOI] [PubMed] [Google Scholar]
- 63.Schulz, S., C. K. Green, P. S. Yuen, and D. L. Garbers. 1990. Guanylyl cyclase is a heat-stable enterotoxin receptor. Cell 63:941-948. [DOI] [PubMed] [Google Scholar]
- 64.So, M., H. W. Boyer, M. Betlach, and S. Falkow. 1976. Molecular cloning of an Escherichia coli plasmid determinant than encodes for the production of heat-stable enterotoxin. J. Bacteriol. 128:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.So, M., W. S. Dallas, and S. Falkow. 1978. Characterization of an Escherichia coli plasmid encoding for synthesis of heat-labile toxin: molecular cloning of the toxin determinant. Infect. Immun. 21:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sommerfelt, H., et al. 1996. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J. Infect. Dis. 174:768-776. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis, A., P. Hoover, and J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57:758-771. [DOI] [PubMed] [Google Scholar]
- 68.Steinsland, H., D. W. Lacher, H. Sommerfelt, and T. S. Whittam. 2010. Ancestral lineages of human enterotoxigenic Escherichia coli. J. Clin. Microbiol. 48:2916-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steinsland, H., P. Valentiner-Branth, P. Aaby, K. Molbak, and H. Sommerfelt. 2004. Clonal relatedness of enterotoxigenic Escherichia coli strains isolated from a cohort of young children in Guinea-Bissau. J. Clin. Microbiol. 42:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinsland, H., et al. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286-291. [DOI] [PubMed] [Google Scholar]
- 71.Steinsland, H., et al. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740-1747. [DOI] [PubMed] [Google Scholar]
- 72.Talavera, G., and J. Castresana. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564-577. [DOI] [PubMed] [Google Scholar]
- 73.Touchon, M., et al. 2009. Organized genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsai, S. C., M. Noda, R. Adamik, J. Moss, and M. Vaughan. 1987. Enhancement of choleragen ADP-ribosyltransferase activities by guanyl nucleotides and a 19-kDa membrane protein. Proc. Natl. Acad. Sci. U. S. A. 84:5139-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner, S. M., et al. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44:4528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valvatne, H., H. Steinsland, and H. Sommerfelt. 2002. Clonal clustering and colonization factors among thermolabile and porcine thermostable enterotoxin-producing Escherichia coli. APMIS 110:665-672. [DOI] [PubMed] [Google Scholar]
- 77.Walker, R. I., D. Steele, and T. Aguado. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic Escherichia coli (ETEC) disease. Vaccine 25:2545-2566. [DOI] [PubMed] [Google Scholar]
- 78.Weikel, C. S., K. M. Tiemens, S. L. Moseley, I. M. Huq, and R. L. Guerrant. 1986. Species specificity and lack of production of STb enterotoxin by Escherichia coli strains isolated from humans with diarrheal illness. Infect. Immun. 52:323-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.WHO. 2006. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly. Epidemiol. Record 81:97-104. [PubMed] [Google Scholar]
- 80.Wirth, T., et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.