Abstract
Periodontal disease is a bacterially mediated chronic inflammatory disease that results in destruction of the periodontal ligament (PDL) and alveolar bone that surround and support the dentition. While their precise roles are not well understood, periodontal pathogens, including Treponema denticola, are believed to initiate the destructive inflammatory responses and dysregulation of tissue homeostasis that characterize the disease. These responses are believed to result from both proinflammatory effects of acylated bacterial membrane components (lipopolysaccharides and lipoproteins) and degradative effects of secreted bacterial proteases. Host-derived matrix metalloproteinases (MMPs) are key enzymes both in tissue homeostasis and tissue destruction. MMP expression is modulated in part by specific proteolytic fragments of fibronectin (FN), which are associated with periodontal disease. FN is a predominant extracellular matrix component in the periodontium. We examined the ability of Treponema denticola and its acylated outer membrane PrtP protease complex to induce both activation of MMP-2 and generation of FN fragments in human PDL cell culture supernatants. T. denticola parent and isogenic mutant strains, as well as MMP-2 small interfering RNA and specific inhibitors of MMP-2 and PrtP activity, were used to examine protein expression, gelatinolytic activity, and FN fragmentation in culture supernatants. T. denticola and its purified protease induced both MMP-2 activation and FN fragmentation. Here, we demonstrate that PrtP proteolytic activity induces the activation of MMP-2 and that active MMP-2 is required for FN fragmentation. These results suggest a specific mechanism by which the T. denticola protease may disrupt homeostatic processes required for the maintenance of periodontal health.
Periodontal disease is a bacterially mediated chronic inflammatory disease that results in destruction of the gingival tissues, periodontal ligament (PDL), and alveolar bone that surround and support the dentition. Periodontal tissue destruction leads to tooth loss, speech and masticatory problems, and an overall reduced quality of life in a large proportion of the adult population in the United States and an even greater percentage worldwide. An important component of the pathogenesis of this disease is destruction of the extracellular matrix (ECM) that comprises the major structural support for these periodontal tissues. Dysregulation of normal tissue homeostatic processes in periodontal disease results in, among several other things, upregulation of host-derived matrix metalloproteinases (MMPs) that contribute directly to tissue destruction. This process is presumed to be initiated by bacterial components, such as lipopolysaccharides and secreted proteases, produced by several organisms identified as periodontal pathogens.
During the course of inflammatory periodontal disease, host-derived proteases cleave the ECM and release fragments of the ECM, including fibronectin (FN) fragments, into the inflammatory milieu. The presence of specific FN fragments (of 40, 68, and 120 kDa) in gingival crevicular fluid is a marker of periodontal disease status (18). Furthermore, FN fragments of 40 and 120 kDa produced by chymotrypsin cleavage induce apoptosis or suppress osteoblast differentiation of PDL cells (8, 21). Thus, it is of great interest to determine the mechanisms by which this proteolytic signature of FN fragments is generated in periodontal disease, including the relative contributions of bacterial and host-derived proteases.
Treponema denticola, an oral anaerobic spirochete, is among the oral pathogens that secrete ECM-degrading proteases (26, 29). T. denticola, a member of the “red complex” of subgingival plaque, is particularly associated with severe and refractory periodontal disease (36). Oral spirochetes are normally at or below detectable levels in healthy gingival plaque but are often the predominant organisms in severe periodontal disease (6, 10). T. denticola, the best-studied oral spirochete, produces an acylated serine protease complex (variously designated the PrtP complex [1], chymotrypsin-like protease [42], and dentilisin [20]) that degrades gelatin, laminin, various serum components, and bioactive peptides (15, 30, 42). The PrtP protease contributes to T. denticola's adherence to and cytotoxic effects on epithelial cells and fibroblasts (9, 12, 27) and penetration of epithelial tissue (4) and may play a role in complement-mediated bactericidal activity (31). While these behaviors are being studied by several groups, few direct links have been reported between the activity of T. denticola's protease and the cellular and tissue processes that drive periodontal tissue destruction.
MMPs comprise a large family of host-derived, zinc-dependent endopeptidases that are secreted in latent form and become activated in the pericellular environment. MMPs are involved in the tissue-destructive (2) and wound-healing (23) phases of human periodontal disease. In particular, MMP-2, -8, and -9 are implicated in tissue destruction in periodontal disease (3, 19, 24, 25, 28, 33-35, 38). Several groups have reported MMP-2 activation by cell surface components of periodontal pathogens. Treatment of gingival fibroblasts or PDL cells with Treponema lecithinolyticum resulted in MMP-2 activation, and the heat-sensitive activating component was proposed to be proteinaceous (5). Porphyromonas gingivalis culture supernatant activated MMP-2 in human PDL cells and was accompanied by an increase in the levels of MT1-MMP (32). MMP-2 activation was also induced in human PDL cells by the lipopolysaccharide of Aggregatibacter actinomycetemcomitans (40). Neither the mechanisms nor the consequences of MMP-2 activation were addressed in these studies.
The role of T. denticola's PrtP protease in ECM degradation and modulation of cellular responses in host-derived cells has not been described in detail. The purpose of the present study was to investigate the role of the T. denticola protease in the modulation of host cell-derived MMP activity and in the fragmentation of FN, a predominant ECM component of these cells. Here, we demonstrate that PrtP proteolytic activity induces MMP-2 activation and that active MMP-2 is required for FN fragmentation. These data suggest that the periodontal pathogen T. denticola is capable of perpetuating periodontal tissue destruction by activating MMP-2, which subsequently degrades the periodontal ECM. This ECM fragmentation, in turn, has deleterious effects on host cell survival and osteoblast differentiation, activities that are required for tissue restitution and homeostasis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
T. denticola ATCC 35405 and isogenic mutants P0760 and MHE (Table 1) were grown in new oral spirochete (NOS) broth medium under anaerobic conditions, as previously described (16). The culture medium was supplemented with erythromycin (40 μg ml−1) as appropriate. Strain P0760 carries an erm gene cassette (encoding erythromycin resistance) at the 5′ end of prcB, resulting in the loss of expression of the entire PrtP protease complex, as well as reduced expression of Msp, the major outer sheath protein (1, 14). Strain MHE, with an erm gene cassette inserted in the gene encoding Msp (13), produces no Msp but has native protease levels. The purity of the T. denticola cultures was monitored by dark-field microscopy prior to each experiment.
TABLE 1.
T. denticola strains used in this study
| T. denticola strain | Genotype | Relevant features | Reference or source |
|---|---|---|---|
| 35405 | Type strain | PrtP+ Msp+ | ATCCa |
| P0760 | 34505 prcB::ermF-ermB | PrtP− Msp− | 1 |
| MHE | 34505 msp::ermF-ermB | PrtP+ Msp− | 13 |
ATCC, American Type Culture Collection, Manassas, VA.
Primary culture of PDL cells.
PDL cells were obtained from extracted third molars or premolars of healthy patients. Cells were cultured as described previously (22) in alpha modification minimal essential medium (αMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin and used from passages 2 to 6. For use in experiments, culture medium was replaced with serum- and antibiotic-free αMEM. The use of human PDL cells for these studies was approved by the University of Michigan Health Sciences Institutional Review Board.
Treatment of PDL cells with T. denticola.
Four-day-old cultures of T. denticola parent and mutant strains were harvested by centrifugation at 10,000 × g (10 min, 4°C), washed twice in phosphate-buffered saline (PBS), and then suspended in serum- and antibiotic-free MEM to an optical density of 0.1 at 600 nm (2.4 × 108 cells ml−1). PDL cells were treated with T. denticola for 2 h at a multiplicity of infection (MOI) of 500 in serum- and antibiotic-free minimal essential medium. The bacteria were removed by washing 3 times with PBS, and fresh culture medium was added. Following incubation for an additional 7, 24, or 48 h, the conditioned medium was collected and stored at −80°C for further analysis by gelatin zymography and Western blotting.
For MMP-2 inhibition assays, PDL cells were challenged with T. denticola for 2 h and then washed and supplied with fresh medium as described above. The MMP-2-specific inhibitor ARP101 (0 to 40 μM in dimethyl sulfoxide; Sigma Chemical Co., St. Louis, MO) was added to the fresh medium following challenge. Conditioned medium was collected for assays after 24 h of incubation.
Purification of T. denticola protease.
Triton X-114 extraction and phase partitioning of treponemal outer membrane proteins were performed as described for Treponema pallidum (7), with slight modifications. T. denticola strain MHE (13) was harvested from 4-liter batch cultures by centrifugation at 4,000 × g (10 min at 4°C) and washed twice in PBS (pH 7.4), The cell pellet was suspended in 1/40 volume PBS containing 5 mM MgCl2 and 1% Triton X-114 and stirred gently overnight at 4°C. Detergent-extracted cells were centrifuged at 20,000 × g (10 min at 4°C). The supernatant, enriched for outer membrane components, was held at 37°C for 10 min and then partitioned into aqueous and detergent phases by centrifugation at 3,500 × g (10 min at 37°C). The detergent phase was diluted to a concentration of 1% Triton X-114 and subjected to two additional rounds of phase separation.
The PrtP protease complex was purified from the detergent phase of the Triton X-114 extracts of T. denticola MHE by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a model 491 Prep Cell (Bio-Rad Laboratories, Richmond, CA) as described previously (12), with slight modifications. The detergent phase of the extract was precipitated in acetone (17), resuspended in PBS plus standard SDS-PAGE sample buffer containing reducing agent, and then layered (without preheating) on the stacking gel matrix. Samples were electrophoresed at 60 mA at 4°C. The cathode buffer was 25 mM Tris (pH 8.3), 192 mM glycine, 0.1% SDS, and the anode and elution buffers consisted of 25 mM Tris (pH 8.3), 192 mM glycine. The eluate was collected in 5.0-ml fractions at a flow rate of 1 ml per min. Fractions were analyzed using silver-stained SDS-PAGE. Fractions containing proteins of interest were concentrated 10-fold in a stirred-cell ultrafiltration unit fitted with an Amicon XM50 filter (Millipore, Inc., Beverly, MA) and then subjected to buffer exchange by washing 3 times with 10 volumes of PBS containing 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. The final sample was then further concentrated 10-fold in an Amicon Ultracel 30 centrifugal concentration unit (Millipore). Following determination of protein concentration using a bicinchoninic acid (BCA) protein assay (Thermo Scientific, Rockford, IL), aliquots of the purified protease complex were stored at −80°C. The activity of purified protease samples was monitored periodically during storage by using the chromogenic substrate succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanine-p-nitroanilide (SAAPFNA) as described previously (15).
MMP-2 activation in cell-free PDL culture supernatants.
PDL cells were cultured in serum- and antibiotic-free αMEM for 24 h to allow release of pro-MMP-2 into the medium (22). Conditioned medium containing pro-MMP-2 was collected, and then 2-ml samples were incubated with 2 μl of purified T. denticola PrtP protease complex at various concentrations for 2 h at 37°C. Samples were concentrated approximately 10-fold in Amicon centrifugal concentrators (10,000-molecular-weight cutoff) prior to gelatin zymography and Western immunoassays. In a control experiment, the activity of the T. denticola PrtP protease was inhibited by treatment with 2 mM phenylmethylsulfonyl fluoride (PMSF) prior to incubation with cell-free PDL culture supernatant.
Transfection of PDL cultures with MMP-2 siRNA.
Small interfering RNA (siRNA) specific for MMP-2 (Santa Cruz Biotechnology, Santa Cruz, CA) was used to inhibit MMP-2 expression in PDL cells, according to the manufacturer's instructions. Briefly, PDL cells were washed with siRNA transfection medium and treated for 6 h with MMP-2 siRNA or an irrelevant siRNA (control) in transfection reagent. The transfection reagent was then removed and replaced with normal growth medium, and the cells were incubated for an additional 18 h, after which they were challenged with T. denticola for 2 h at an MOI of 500, as described above.
Immunoblotting.
Conditioned medium from PDL cell cultures was concentrated 10-fold as described above. Equal volumes of concentrated samples were loaded into each well, resolved by 4 to 12% SDS-PAGE, and electroblotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA) by a semidry transfer blot method (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline and Tween 20 (TBST) for 1 h at room temperature, incubated with primary antibodies, incubated with appropriate horseradish peroxidase-conjugated secondary antibodies in blocking buffer for 2 h at room temperature, washed with TBST, and developed using a West-Pico ECL kit (Pierce Chemical Co., Rockford, IL). The following specific primary antibodies were used in individual experiments: anti-MMP-2 catalytic domain polyclonal antibody (Chemicon International, MA) and antifibronectin rabbit polyclonal antibody (Santa Cruz Biotechnology, CA).
Gelatin zymography.
Equal volumes of concentrated PDL cell-conditioned medium were mixed with 4× sample buffer (0.25 M Tris base, 0.8% SDS, 40% glycerol, and 0.05% bromophenol blue) and subjected to SDS-PAGE at 4°C on 12.5% gels containing 2 mg ml−1 gelatin. After electrophoresis, SDS was removed by washing the gels in renaturing buffer (2.5% Triton X-100, 50 mM Tris base) for 30 min with one change of washing buffer. The gels were incubated at 37°C for 20 h in developing buffer (50 mM Tris base, pH 8, 10 mM CaCl2, and 0.02% NaN3), stained with 0.05% Coomassie blue, and destained in 10% acetic acid and 40% methanol until clear bands in a blue background were visible.
RESULTS
T. denticola induces PrtP-dependent fibronectin degradation in PDL cell culture supernatants.
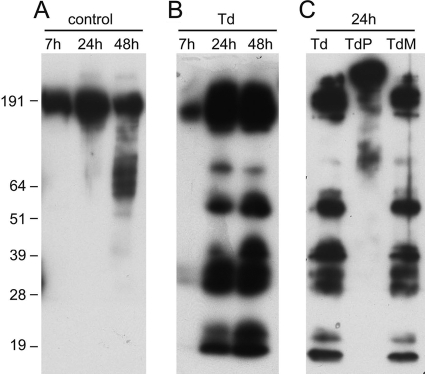
Over the course of 48 h in serum- and antibiotic-free medium, PDL cell culture supernatants accumulate a high-molecular-weight form of FN that undergoes limited fragmentation by 48 h (Fig. 1A). However, challenge with T. denticola for 24 h or more results in extensive fragmentation of FN (Fig. 1B) that persists after T. denticola cells nonadherent to PDL cells have been removed. Consistent with the reported FN-degrading activity of the purified T. denticola protease (15), no FN degradation above baseline was observed in PDL cell culture supernatants challenged with protease-deficient mutant T. denticola P0760 (Fig. 1C). Interestingly, the FN-binding T. denticola Msp protein is not required for T. denticola-mediated FN degradation in PDL cell culture supernatants (Fig. 1C).
FIG. 1.
Fibronectin degradation in culture supernatants of PDL cells. PDL cells were challenged with T. denticola for 2 h at an MOI of 500 and then washed and incubated in fresh medium for the indicated times. Culture supernatants were then analyzed in Western blots probed with an anti-FN antibody. (A) Unchallenged control PDL cell supernatants following 7, 24, or 48 h of incubation in fresh medium. (B) PDL cell supernatants following 7, 24, or 48 h of postchallenge incubation in fresh medium. (C) PDL cell supernatants following 24 h of postchallenge incubation in fresh medium. T. denticola strains are 35405 (Td), P0760 (TdP; protease mutant), and MHE (TdM; Msp mutant). Molecular weight standards (in thousands) are indicated.
T. denticola induces PrtP-dependent activation of MMP-2 in PDL cell culture supernatants.
Pro-MMP-2 is constitutively expressed by PDL cells and secreted into the culture medium. Challenge with T. denticola results in the activation of MMP-2 from the 72-kDa pro-MMP-2 form to the active 64-kDa form of MMP-2 in culture supernatants after removal of nonadherent T. denticola (Fig. 2). Challenge with protease-deficient T. denticola P0760 did not induce activation of MMP-2, whereas Msp-deficient T. denticola MHE induced MMP-2 activation at equal or higher levels than wild-type T. denticola 35405. The T. denticola protease complex is visible as a gelatinolytic band at approximately 100 kDa present in supernatants from cultures challenged with T. denticola 35405 and MHE. This band is absent in supernatants from unchallenged PDL cell cultures and in supernatants from PDL cells challenged with protease-deficient T. denticola P0760.
FIG. 2.
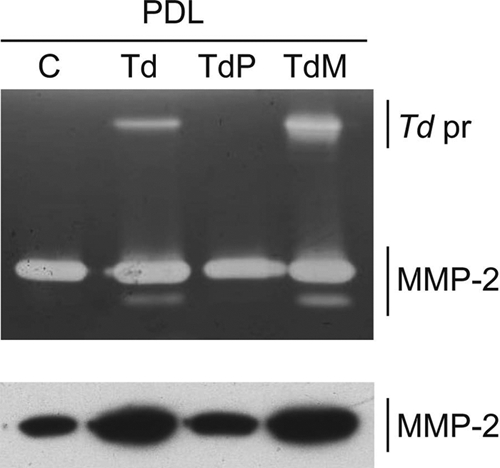
MMP-2 activity in 24-h supernatants of PDL cell cultures (PDL) following a 2-h challenge with T. denticola. Gelatin zymogram (top) and Western immunoblot (bottom) probed with an anti-MMP-2 catalytic domain antibody. Lanes: C, PDL no-challenge control; Td, PDL challenged with T. denticola 35405; TdP, PDL challenged with T. denticola P0760; TdM, PDL challenged with T. denticola MHE. The locations of T. denticola protease (Td pr) and MMP-2 are indicated.
The T. denticola protease induces both MMP-2 activation and FN fragmentation.
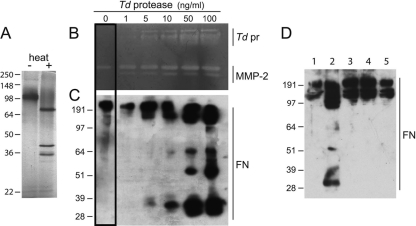
To confirm the role of the PrtP protease complex in inducing MMP-2-dependent FN degradation, PDL cell culture supernatants were collected and incubated with a range of concentrations of purified T. denticola protease complex. The purity of the T. denticola PrtP protease complex was confirmed (Fig. 3A) by silver-stained SDS-PAGE of unheated (active protease complex, approximately 100 kDa) and heated (denatured PrtP, PrcA1, and PrcA2 at approximately 70, 40, and 30 kDA, respectively) samples. The purified protease complex also contains detectable levels of PrcB, a 22-kDa protease complex-associated protein (J. C. Fenno, unpublished data). PDL cell culture supernatants contain 72-kDa pro-MMP-2, which is constitutively expressed in PDL cells. As shown in Fig. 3B, both the gelatinase activity of the T. denticola protease and the activation of MMP-2 to the 64-kDa form were detectable at protease concentrations of 5 ng ml−1. FN fragmentation was detected (Fig. 3C) at a slightly higher concentration of the T. denticola protease complex (10 ng ml−1). To confirm the dependence of FN fragmentation on the activity of T. denticola protease, PDL cell culture supernatants were incubated with purified PrtP protease complex (final concentration, 100 ng ml−1) or with PrtP protease complex that had been inactivated either by boiling for 5 min or by treatment with 2 mM PMSF. As shown in Fig. 3D, FN fragmentation was detected only in PDL cell culture supernatants treated with native PrtP protease complex.
FIG. 3.
Treatment of cell-free PDL culture supernatants with T. denticola PrtP protease complex. Supernatants of PDL cultures were incubated with purified T. denticola PrtP protease complex (Td pr) at the indicated final concentrations for 2 h at 37°C. (A) Silver-stained SDS-PAGE of purified T. denticola PrtP protease complex (0.4 μg per lane). Lanes: −, unheated sample showing native PrtP complex; +, heated sample showing protease complex components PrtP, PrcA2, and PrcA1. (B) Gelatin zymogram of protease-treated PDL cell culture supernatants. Bands showing gelatinolytic activity of T. denticola PrtP protease complex and MMP-2 are indicated. (C) Western immunoblot of PDL cell culture supernatants probed with an anti-FN antibody. (D) Western immunoblot of PDL cell culture supernatants treated with T. denticola PrtP protease complex at 100 ng ml−1. Lanes: 1, PDL without Td protease (control); 2, PDL with Td protease; 3, PDL with Td protease pretreated with 2 mM PMSF; 4, PDL with Td protease pretreated by boiling for 5 min; 5, PDL with 2 mM PMSF and without Td protease. The blot was probed with an anti-FN antibody. Numbers on the left of gels indicate molecular weight standards (in thousands).
Fibronectin fragmentation in T. denticola-challenged PDL cell cultures is due to MMP-2 activity.
To further determine whether the observed FN fragmentation was directly due to T. denticola's proteolytic activity, PDL cultures that had been challenged with T. denticola 35405 for 2 h were incubated in fresh medium containing ARP101, a specific inhibitor of MMP-2 activity. As shown in Fig. 4 A, the residual gelatinolytic activity of T. denticola PrtP in culture supernatants was not inhibited by ARP101, while MMP-2 gelatinase activity decreased with increased ARP101. MMP-2 cleavage is not blocked by ARP101, but dose-dependent binding of ARP101 to the MMP-2 catalytic domain is indicated by decreasing detection of MMP-2 by catalytic domain-specific anti-MMP-2 antibody (Fig. 4B). Furthermore, as shown in Fig. 4C, treatment of PDL cell cultures with ARP101 resulted in dose-dependent decreases in FN fragmentation in T. denticola-challenged PDL culture supernatants, indicating that T. denticola perpetuates the fragmentation of PDL cell-derived FN by activation of secreted MMP-2.
FIG. 4.
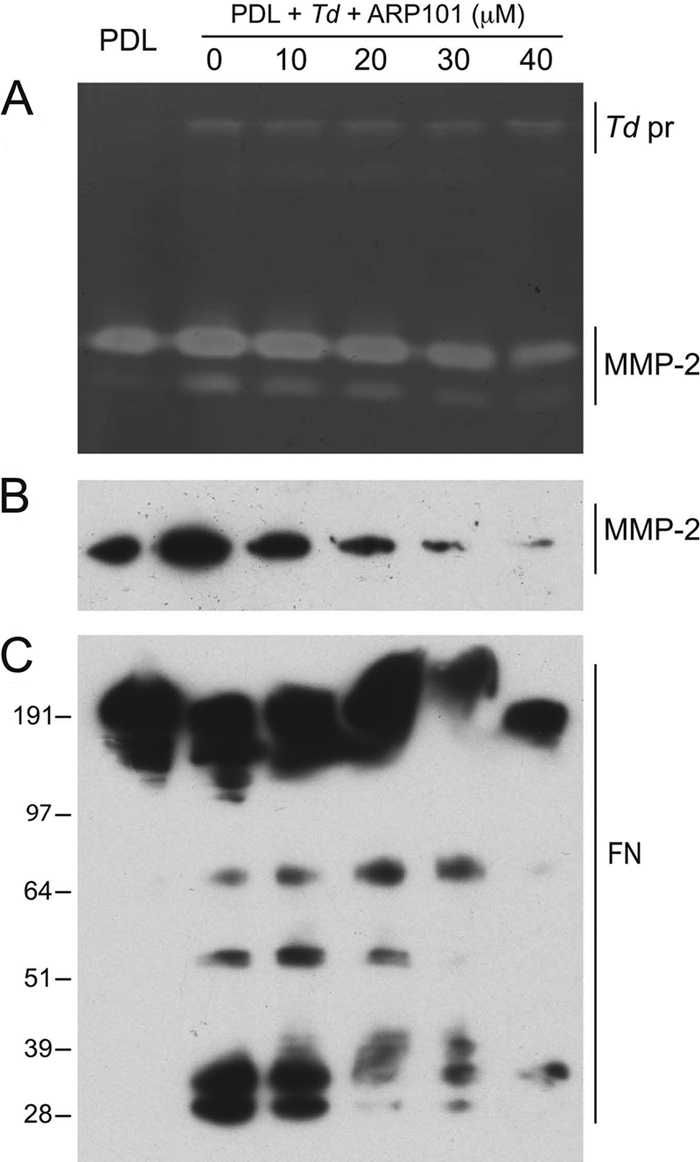
MMP-2-specific inhibitor blocks MMP-2 activation and inhibits FN fragmentation. Lanes contain PDL culture supernatant alone (PDL) or after 2 h of treatment with T. denticola 35405, washing, and addition of fresh medium (PDL + Td). ARP101 was added to the fresh medium at the indicated concentrations. (A) Gelatin zymogram with positions of T. denticola protease complex (Td pr) and MMP-2 bands indicated. (B) Western immunoblot probed with anti-MMP-2 catalytic domain antibody. (C) Western immunoblot probed with an anti-FN antibody. Molecular weight standards (in thousands) are indicated.
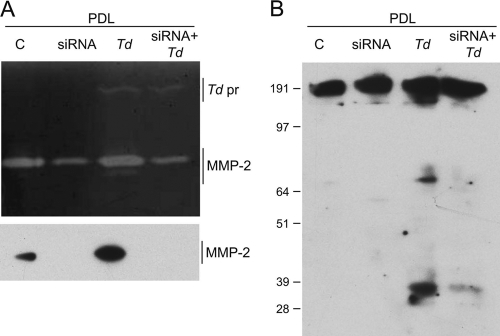
MMP-2 siRNA blocks FN fragmentation in T. denticola-challenged PDL cells.
To further explore the mechanism of T. denticola-dependent FN degradation, PDL cells were transfected with MMP-2-specific siRNA. As shown in Fig. 5 A, MMP-2 pro and active forms were reduced in PDL cell cultures transfected with MMP-2 siRNA, as examined by zymography and immunoblotting. Challenge with T. denticola induced MMP-2 activation in PDL cultures transfected with scrambled-sequence siRNA, but MMP-2 activation was not detected in MMP-2 siRNA-transfected cultures with or without T. denticola challenge. T. denticola's gelatinolytic activity in culture supernatants was not affected by the transfection. As shown in Fig. 5B, transfection with MMP-2 siRNA blocked FN fragmentation in PDL cultures treated with T. denticola, providing further confirmation of the key role of activated MMP-2 in T. denticola-induced FN fragmentation.
FIG. 5.
FN degradation is blocked in supernatants of MMP-2 siRNA-transfected PDL cell cultures. Cell-free supernatants from PDL cultures transfected with MMP-2-specific siRNA or scrambled siRNA control were challenged or not challenged with T. denticola 35405 (Td). (A) Top, Gelatin zymogram with positions of T. denticola protease complex (Td pr) and MMP-2 bands indicated. Bottom, Western immunoblot probed with anti-MMP-2 catalytic domain antibody. (B) Western immunoblot probed with anti-FN antibody. Molecular weight standards (in thousands) are indicated.
DISCUSSION
Defining the specific roles of the microbes identified as periodontal pathogens in the induction of cellular and tissue responses seen in periodontal disease continues to be an elusive goal. While it is highly unlikely that a single organism “causes” the disease, it is well established that periodontal disease is initiated by a characteristic microbial population shift in which a particular group of proteolytic Gram-negative anaerobes become a significant (and often predominant) segment of the total subgingival flora. These microbes, including T. denticola and other members of the “red complex,” exhibit a range of proteolytic and immunostimulatory behaviors that is consistent with identification as pathogens.
Proteolytic enzymes of periodontopathogens have been identified as likely virulence determinants in periodontal diseases. Previous in vitro studies of the T. denticola outer membrane acylated PrtP protease complex demonstrated its ability to degrade numerous host cell and tissue proteins. Cell challenge studies support a major role for the T. denticola protease in binding to and penetration of tissue (reviewed in reference 10). However, there is limited evidence to date of a direct role of bacterial proteases in periodontal tissue destruction and bone loss. In the present study, we focused on the role of the T. denticola protease in inducing certain key cell and tissue responses that are characteristic of periodontal diseases.
A specific pattern of FN fragments in gingival crevicular fluid is an indicator of periodontal disease status (18), and at least one of these fragments (of 120 kDa, containing the central cell-binding domain of FN) induces the expression of PDL cell collagenase, stromelysin, and urokinase plasminogen activator. Furthermore, other specific FN fragments induce apoptosis and suppress osteoblast differentiation of PDL cells (7, 17). Our experiments, designed to mimic in vivo interactions between cells of the PDL and oral spirochetes, demonstrated that T. denticola induces FN fragmentation in the supernatant of PDL cultures. While Ellen et al. (11) showed degradation of cell-associated FN in gingival fibroblasts, the present study is, to our knowledge, the first to examine FN released from cultured PDL cells following T. denticola challenge. Following the removal of nonadherent spirochetes and two washes in PBS, we assayed FN fragmentation in supernatants of PDL cultures grown in fresh medium for various periods. T. denticola protease activity was still detectable in the supernatants, most likely due to protease expressed and secreted by remaining cell-adherent spirochetes.
MMP-2, one of several MMPs involved in tissue homeostasis and remodeling, is constitutively expressed in PDL cells as pro-MMP-2. Thus, control of the process or rate of activation of MMP-2 can be viewed as a key regulatory step. Because FN fragments are reported to induce activation of MMPs in PDL cultures, our initial experiments examining the activation of the constitutively expressed MMP-2 in T. denticola-challenged PDL cultures were undertaken with the hypothesis that T. denticola-dependent FN fragmentation might result in the activation of MMPs. We readily demonstrated that MMP-2 in PDL supernatants was activated in a PrtP protease-dependent manner, as shown by the absence of FN fragmentation following challenge with an isogenic protease mutant and following challenge with PMSF-treated purified PrtP protease complex. The activity of T. denticola PrtP is highly sensitive to the serine protease inhibitor PMSF (42), while MMP-2 activity is not affected by PMSF (39). Experiments using both PMSF and a specific inhibitor of MMP-2 activity showed that active T. denticola protease was not sufficient to induce FN fragmentation when MMP-2 activity was inhibited. T. denticola protease's activity was not inhibited by MMP-2 inhibitor ARP101 (Fig. 4A), which blocks MMP-2 activation by binding the zinc-dependent catalytic site of MMP-2 (41). These results indicate that FN fragmentation in PDL culture supernatants was not due to direct effects of T. denticola protease but was dependent on MMP-2 activity and that activation of pro-MMP-2 was dependent on T. denticola protease activity. The dependence of FN fragmentation on MMP-2 activation was further confirmed by siRNA knockdown of MMP-2 expression in PDL cultures in the presence of T. denticola protease activity. These unexpected results suggest a general mechanism by which T. denticola (or other MMP-2-activating periodontal pathogens, such as T. lecithinolyticum, P. gingivalis, or A. actinomycetemcomitans) may disrupt the regulation of MMP-2 activation, leading to the generation of FN fragments that contribute to disregulation of periodontal tissue turnover processes, including osteoblast function and other homeostatic mechanisms.
Further studies are required to determine the mechanism by which T. denticola PrtP protease induces the activation of pro-MMP-2. One possibility is that T. denticola protease degrades one or more of the tissue inhibitors of metalloproteinases (TIMPs), resulting in increased MMP-2 activation. However, preliminary experiments showed no decrease in TIMP-1 or TIMP-2 levels in PDL supernatants challenged with T. denticola (data not shown). This is consistent with the observations of Sorsa et al., who reported that while purified T. denticola protease could activate purified pro-MMP-1 in vitro, it could not reduce the capacity of TIMP-1 to inhibit collagenase activity (37). However, this mechanism of MMP-2 activation cannot yet be ruled out. In addition, we are particularly interested in characterizing the activities of specific FN fragments resulting from T. denticola protease challenge of PDL cells and comparing them with those of the FN fragments found in gingival crevicular fluid from periodontal lesions.
Acknowledgments
This work was supported by Public Health Service grants DE018221 (J.C.F.) and DE013725 (Y.L.K.) from the National Institute of Dental and Craniofacial Research and by support from the China Scholarship Council (D.M.).
Editor: S. R. Blanke
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Bian, X.-L., H.-T. Wang, Y. Ning, S. Y. Lee, and J. C. Fenno. 2005. Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect. Immun. 73:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkedal-Hansen, H. 1993. Role of matrix metalloproteinases in human periodontal diseases. J. Periodontol. 64:474-484. [DOI] [PubMed] [Google Scholar]
- 3.Cesar Neto, J. B., et al. 2004. Matrix metalloproteinase-2 may be involved with increased bone loss associated with experimental periodontitis and smoking: a study in rats. J. Periodontol. 75:995-1000. [DOI] [PubMed] [Google Scholar]
- 4.Chi, B., M. Qi, and H. K. Kuramitsu. 2003. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 154:637-643. [DOI] [PubMed] [Google Scholar]
- 5.Choi, B. K., et al. 2001. Activation of matrix metalloproteinase-2 by a novel oral spirochetal species Treponema lecithinolyticum. J. Periodontol. 72:1594-1600. [DOI] [PubMed] [Google Scholar]
- 6.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, T. M., E. M. Walker, J. N. Miller, and M. A. Lovett. 1988. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, R., A. Iwama, S. Wang, and Y. L. Kapila. 2005. Disease-associated fibronectin matrix fragments trigger anoikis of human primary ligament cells: p53 and c-myc are suppressed. Apoptosis 10:503-512. [DOI] [PubMed] [Google Scholar]
- 9.Ellen, R. P., J. R. Dawson, and P. F. Yang. 1994. Treponema denticola as a model for polar adhesion and cytopathogenicity of spirochetes. Trends Microbiol. 2:114-119. [DOI] [PubMed] [Google Scholar]
- 10.Ellen, R. P., and V. B. Galimanas. 2005. Spirochetes at the forefront of periodontal infections. Periodontol. 2000 38:13-32. [DOI] [PubMed] [Google Scholar]
- 11.Ellen, R. P., M. Song, and C. A. McCulloch. 1994. Degradation of endogenous plasma membrane fibronectin concomitant with Treponema denticola 35405 adhesion to gingival fibroblasts. Infect. Immun. 62:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenno, J. C., et al. 1998. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect. Immun. 66:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenno, J. C., G. W. K. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 14.Godovikova, V., et al. 2010. Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (dentilisin) complex. J. Bacteriol. 192:3337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grenier, D., V.-J. Uitto, and B. C. McBride. 1990. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haapasalo, M., U. Singh, B. C. McBride, and V.-J. Uitto. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hager, D. A., and R. R. Burgess. 1980. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal. Biochem. 109:76-86. [DOI] [PubMed] [Google Scholar]
- 18.Huynh, Q. N., et al. 2002. Specific fibronectin fragments as markers of periodontal disease status. J. Periodontol. 73:1101-1110. [DOI] [PubMed] [Google Scholar]
- 19.Ingman, T., et al. 1996. Matrix metalloproteinases and their inhibitors in gingival crevicular fluid and saliva of periodontitis patients. J. Clin. Periodontol. 23:1127-1132. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joseph, J., Y. L. Kapila, T. Hayami, and S. Kapila. 2010. Disease-associated extracellular matrix suppresses osteoblastic differentiation of human periodontal ligament cells via MMP-1. Calcif. Tissue Int. 86:154-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapila, Y. L., S. Kapila, and P. W. Johnson. 1996. Fibronectin and fibronectin fragments modulate the expression of proteinases and proteinase inhibitors in human periodontal ligament cells. Matrix Biol. 15:251-261. [DOI] [PubMed] [Google Scholar]
- 23.Kinane, D. F., et al. 2003. Changes in gingival crevicular fluid matrix metalloproteinase-8 levels during periodontal treatment and maintenance. J. Periodontal Res. 38:400-404. [DOI] [PubMed] [Google Scholar]
- 24.Korostoff, J. M., et al. 2000. Analysis of in situ protease activity in chronic adult periodontitis patients: expression of activated MMP-2 and a 40 kDa serine protease. J. Periodontol. 71:353-360. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, M. S., G. Vamsi, R. Sripriya, and P. K. Sehgal. 2006. Expression of matrix metalloproteinases (MMP-8 and -9) in chronic periodontitis patients with and without diabetes mellitus. J. Periodontol. 77:1803-1808. [DOI] [PubMed] [Google Scholar]
- 26.Kuramitsu, H. K. 1998. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol. Immunol. 13:263-270. [DOI] [PubMed] [Google Scholar]
- 27.Leung, W. K., M. Haapasalo, V.-J. Uitto, P. M. Hannam, and B. C. McBride. 1996. The surface proteinase of Treponema denticola may mediate attachment of the bacteria to epithelial cells. Anaerobe 2:39-46. [Google Scholar]
- 28.Makela, M., T. Salo, V. J. Uitto, and H. Larjava. 1994. Matrix metalloproteinases (MMP-2 and MMP-9) of the oral cavity: cellular origin and relationship to periodontal status. J. Dent. Res. 73:1397-1406. [DOI] [PubMed] [Google Scholar]
- 29.Mäkinen, K. K., and P. L. Mäkinen. 1996. The peptidolytic capacity of the spirochete system. Med. Microbiol. Immunol. 185:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Mäkinen, P. L., K. K. Mäkinen, and S. A. Syed. 1995. Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect. Immun. 63:3567-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDowell, J. V., B. Huang, J. C. Fenno, and R. T. Marconi. 2009. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect. Immun. 77:1417-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattamapun, K., S. Tiranathanagul, T. Yongchaitrakul, J. Kuwatanasuchat, and P. Pavasant. 2003. Activation of MMP-2 by Porphyromonas gingivalis in human periodontal ligament cells. J. Periodontal Res. 38:115-121. [DOI] [PubMed] [Google Scholar]
- 33.Rodini, C. O., et al. 2008. Morphologic evaluation and expression of matrix metalloproteinases-2 and 9 and nitric oxide during experimental periodontal disease in rat. J. Mol. Histol. 39:275-282. [DOI] [PubMed] [Google Scholar]
- 34.Seguier, S., B. Gogly, A. Bodineau, G. Godeau, and N. Brousse. 2001. Is collagen breakdown during periodontitis linked to inflammatory cells and expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human gingival tissue? J. Periodontol. 72:1398-1406. [DOI] [PubMed] [Google Scholar]
- 35.Silva, J. A., et al. 2008. The influence of type I diabetes mellitus on the expression and activity of gelatinases (matrix metalloproteinases-2 and -9) in induced periodontal disease. J. Periodontal Res. 43:48-54. [DOI] [PubMed] [Google Scholar]
- 36.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 37.Sorsa, T., et al. 1992. Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect. Immun. 60:4491-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tervahartiala, T., et al. 2000. The in vivo expression of the collagenolytic matrix metalloproteinases (MMP-2, -8, -13, and -14) and matrilysin (MMP-7) in adult and localized juvenile periodontitis. J. Dent. Res. 79:1969-1977. [DOI] [PubMed] [Google Scholar]
- 39.Theret, N., K. Lehti, O. Musso, and B. Clement. 1999. MMP2 activation by collagen I and concanavalin A in cultured human hepatic stellate cells. Hepatology 30:462-468. [DOI] [PubMed] [Google Scholar]
- 40.Tiranathanagul, S., T. Yongchaitrakul, K. Pattamapun, and P. Pavasant. 2004. Actinobacillus actinomycetemcomitans lipopolysaccharide activates matrix metalloproteinase-2 and increases receptor activator of nuclear factor-kappaB ligand expression in human periodontal ligament cells. J. Periodontol. 75:1647-1654. [DOI] [PubMed] [Google Scholar]
- 41.Tuccinardi, T., et al. 2006. Amber force field implementation, molecular modelling study, synthesis and MMP-1/MMP-2 inhibition profile of (R)- and (S)-N-hydroxy-2-(N-isopropoxybiphenyl-4-ylsulfonamido)-3-methylbutanamides. Bioorg. Med. Chem. 14:4260-4276. [DOI] [PubMed] [Google Scholar]
- 42.Uitto, V.-J., D. Grenier, E. C. Chan, and B. C. McBride. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]