Abstract
Infections of the udder by Escherichia coli very often elicit acute inflammation, while Staphylococcus aureus infections tend to cause mild, subclinical inflammation and persistent infections. The molecular causes underlying the different disease patterns are poorly understood. We therefore profiled the kinetics and extents of global changes in the transcriptome of primary bovine mammary epithelial cells (MEC) after challenging them with heat-inactivated preparations of E. coli or S. aureus pathogens. E. coli swiftly and strongly induced an expression of cytokines and bactericidal factors. S. aureus elicited a retarded response and failed to quickly induce an expression of bactericidal factors. Both pathogens induced similar patterns of chemokines for cell recruitment into the udder, but E. coli stimulated their synthesis much faster and stronger. The genes that are exclusively and most strongly upregulated by E. coli may be clustered into a regulatory network with tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) in a central position. In contrast, the expression of these master cytokines is barely regulated by S. aureus. Both pathogens quickly trigger an enhanced expression of IL-6. This is still possible after completely abrogating MyD88-dependent Toll-like receptor (TLR) signaling in MEC. The E. coli-specific strong induction of TNF-α and IL-1 expression may be causative for the severe inflammatory symptoms of animals suffering from E. coli mastitis, while the avoidance to quickly induce the synthesis of bactericidal factors may support the persistent survival of S. aureus within the udder. We suggest that S. aureus subverts the MyD88-dependent activation of immune gene expression in MEC.
Inflammation of the udder (mastitis) is a frequent and costly disease in the dairy industry. Aside from the economic losses, mastitis impairs animal welfare, and food-borne diseases may affect human health (44). The resolution of mastitis may be classified as subclinical or clinical. The pathogenesis of Escherichia coli mastitis and that caused by other Gram-negative bacteria (3) is often characterized by an acute and severe inflammation, which, however, may eventually lead to pathogen clearance (54). Staphylococci are the bacteria most commonly isolated from cases of subclinical mastitis (51). Infection with these Gram-positive pathogens often causes mild signs of mastitis (2, 4). Ineffective pathogen clearance frequently leads to chronic infection. Recurrent and persistent mammary infections by Staphylococcus aureus pathogens are also a serious problem for women, owing to their eventual severity and difficulties in curing due to the antibiotic resistance of the pathogens (6). The molecular causes underpinning the pathogen-specific differences in the resolution of mammary gland infections are entirely unclear.
The mounting of an inflammation is a complex process involving many different regulatory steps. Conceivably, very different mechanisms are involved in establishing either acute or chronic inflammation. Knowledge about the molecular nature and time point of activation of those switches dictating acute or subclinical inflammation as an outcome of the infection is crucial for an understanding of the pathogen-specific reaction of the host and might be helpful for designing preventive measures against immune pathology or persistency of mastitis.
Many different mediators of inflammation are expressed at different times after pathogen or stimulus exposure. Cytokines are an important group of inflammatory mediators. Proinflammatory cytokines promote inflammation quickly after the perception of the pathogen anti-inflammatory cytokines suppress and confine the activity of proinflammatory cytokines. Interleukin-1 (IL-1) and tumor necrosis factor alpha (TNF-α) are major proinflammatory cytokines (10). They are locally produced by many cell types and are responsible for early responses. IL-1 and TNF-α trigger an inflammatory cascade and thereby eventually cause fever, inflammation, tissue damage, and, in some cases, toxic shock and death (10). IL-6 is the third master proinflammatory cytokine. It is one of the key mediators of the “acute-phase response” in inflammation (19). Chemokines recruit cellular factors of immune defense to the site of infection by facilitating the passage of leukocytes from the bloodstream into the tissues. The chronologically coordinated induction of their synthesis at the site of inflammation is decisive for an effective inflammatory response, including pathogen clearance, wound healing, and return to the normal state (9, 27). Disturbances in the well-balanced order and extent of the inflammatory response often lead to chronic inflammation and/or infection (27).
Pathogens evolved sophisticated molecular strategies to disturb and subvert host defenses (33). One principle is avoiding their recognition by impairing the activation of either pattern recognition receptors (PRR), notably the well-analyzed family of Toll-like receptors (TLRs) (25), or their downstream signaling. Lipopolysaccharides (LPSs) derived from Gram-negative bacteria such as E. coli activate the TLR4 receptor (37), while peptidoglycan, for example, from Gram-positive bacteria such as S. aureus, activates the inflammatory response via TLR2 (43). It was previously shown that heat-inactivated S. aureus strain 1027 particles block NF-κB activation in bovine mammary epithelial cells (MEC) (58), although they properly activate the bovine TLR2 receptor in the HEK293 reconstitution system of TLR signaling (7).
It proved to be difficult in the past to reliably analyze in vivo, in the udder, the time course of the pathogen-specific immune response (35). The E. coli-specific response of the udder was found previously to be massively confounded by systemic effects (32), while the response to S. aureus infection displayed the notorious animal individual manifestation of the subclinical phenotype (35). Furthermore, it is very difficult, if possible at all, to define the kinetics of bacterial growth in the udder with statistical significance. Multiple sampling of the udder tissue via biopsies would be necessary to profile the kinetics of the activation of immune response in the host, but this has severe drawbacks due to repeatedly wounding the udder. In addition, the individuality of the host response (8) combined with the necessarily limited number of animals hamper reaching statistical significance of the data. Thus, the usage of a relevant cell model to profile pathogen-specific immune mechanisms would obviously be advantageous.
MEC are highly relevant sentinel and effector cells of udder immunity (16, 26, 49). These cells line the alveoli of the milk parenchyma. They form the early line of defense against invading pathogens due to their exposed position and their abundance in the healthy mammary gland. In response to a pathogenic stimulus, they may secrete large amounts of cytokines and chemokines for leukocyte recruitment and activation (38). However, they can also express factors contributing directly to fighting off pathogens, including the bactericidal β-defensins (LAP and BNBD5), some complement factors, and acute-phase proteins (14, 16, 50, 58).
Primary bovine mammary epithelial cells (pbMEC) are an established model for the molecular analysis of immune defense mechanisms in the udder (17, 49, 58). We recently characterized their substantial potential to express sentinel as well as effector genes of immune defense subsequent to stimulation with heat-killed particles of E. coli (16). By comparing these data to those from fully inflamed udders after infection with live E. coli pathogens, this study also validated the potential and some limitations of this cell model for monitoring aspects of immune regulation in the udder. We now used this cell model to compare the time courses of the pathogen-specific reprogramming of the transcriptome after challenge with heat-killed particles of E. coli and S. aureus and used such strains that were previously shown to cause in vivo acute and subclinical mastitis, respectively (35).
MATERIALS AND METHODS
Cell culture and challenge with pathogens.
The preparation of pbMEC, their culturing in RPMI 1640 medium on collagen-coated dishes, and general conditions for challenging them were previously described (16). We challenged these cells with heat-inactivated particles of the mastitis-causing pathogens E. coli strain 1303 and S. aureus strain 1027 (35). Details regarding the culturing of the E. coli pathogens and their use to challenge pbMEC were described previously (16). S. aureus strain 1027 was grown (37°C) in brain heart infusion broth (Sigma-Aldrich, Munich, Germany) to the logarithmic phase of culture growth (optical density [OD] at 600 nm [OD600] of 0.5; ∼5 × 107 cells/ml). Plating of dilution series was used to calibrate cell counts from the OD readings. Incubation of the cells for 1 h at 80°C killed all cells, as verified by control plating. Subsequently, cells were spun down, washed twice with RPMI 1640 medium, and resuspended therein at a density of 5 × 108 cells/ml. Aliquots were stored frozen at −20°C. pbMEC were challenged for different times with 107 particles/ml of either S. aureus 1027 or E. coli 1303 just as described previously (16). We validated for both pathogen species that heat inactivation and inactivation with UV light resulted in preparations of particles displaying similar biological activities (see Fig. S2 in the supplemental material).
RNA extraction and qRT-PCR.
We used Trizol (Invitrogen, Karlsruhe, Germany) for RNA extraction from cultured pbMEC. The quality of the RNA was assessed by agarose gel electrophoresis (2.2 M formaldehyde) and ethidium bromide staining. The quantification of relative mRNA concentrations by quantitative real-time PCR (qRT-PCR) was done with a LightCycler instrument and the SYBR green Plus reagent kit (both from Roche, Basel, Switzerland), essentially as described previously (14, 16). Sequences of the oligonucleotide primers used are given in Table S1 in the supplemental material.
Microarray hybridization.
We followed the recommendations given by Affymetrix precisely throughout. Three biological replicates were performed for each bacterial strain and experimental condition (1 h, 3 h, 6 h, 24 h, and control). Ten micrograms of total RNA was first reverse transcribed with a T7 oligo(dT) primer. The subsequent synthesis of double-stranded cDNA exploited the One-Cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA). The cDNA was linearly amplified with T7 RNA polymerase in an in vitro transcription reaction and labeled with biotinylated ribonucleotides (GeneChip IVT labeling kit). Ten micrograms of biotin-labeled, purified, and fragmented cRNA was then hybridized (16 h at 45°C) onto GeneChip bovine genome arrays (Affymetrix). Posthybridization washing and staining were performed by using an automated protocol of the Fluidics Station 450 instrument (Affymetrix). Finally, the arrays were scanned with a GeneChip 3000 7G scanner.
Microarray data analysis.
The evaluation of the microarray data was done by using the R statistical language. In a first step all arrays were admitted after having passed a quality control. Background correction, normalization, and summation were then performed by using guanine cytosine robust multiarray analysis (GCRMA) (56). Only such genes exhibiting at least three “present calls” were further processed. The gene expression values for the various points in time were estimated with a linear model. In order to account for multiple testing, a false discovery rate approach described previously (48) was used, and we selected only the genes with a q value of ≤0.05. In a further filtering step, only genes with a fold change of ≥2 in the time profile were kept. Finally, the genes exhibiting a clear maximum or minimum in their expression over time were detected and classified accordingly.
The identification of human orthologs of regulated bovine genes was based on data described previously (20a). Gene ontology information for regulated genes was obtained by using Ingenuity Pathways Analysis (IPA 7.0) software (Ingenuity Systems, Redwood City, CA). Heat maps of gene expression were established with Expander (EXPression ANalyzer and DisplayER) software (52).
Correlations were evaluated with Spearman's rank or Pearson moment correlation analysis programs using SigmaStat 3.5 software (Systat Software Inc., San Jose, CA).
Generation of stable transfectants.
Bovine MAC-T cells were stably transfected (Lipofectamine 2000; Invitrogen) with pSV2-neo (47) and plasmids with expression under the control of the cytomegalovirus (CMV) promoter trans-dominant negative (DN) variants of the bovine factor myeloid differentiation primary response 88 (MyD88) (DN-MyD88) or TIR-domain-containing adapter-inducing IFN-β (TRIF). The latter variant was expressed together with the DN-MyD88 variant on a single polypeptide chain. Briefly, 1 μg of the respective DN plasmid and 0.1 μg of the pSV2-neo vector were cotransfected into a single well of a 6-well plate (35 mm in diameter). The cells were grown overnight in Dulbecco's modified Eagle's medium (DMEM) (Lonza, Basel, Switzerland) supplemented with 5% fetal calf serum (FCS) (PAN-Biotech, Aidenbach, Germany), 4 mM l-glutamine (Biochrom, Berlin, Germany), 100 μg/ml kanamycin (Serva, Heidelberg, Germany), and 100 μg/ml penicillin-streptomycin (Sigma-Aldrich). From the next day on, Geneticin G418 (200 μg/ml; Lonza) was added to the medium, and pools of stable transfectants were selected for the next 2 weeks. All pools consisted of more than 500 individual clones.
Western blotting.
Total protein was extracted from unstimulated pbMEC and from cells challenged for 2 or 4 h with 107 cells/ml of either heat-inactivated S. aureus 1027 or E. coli 1303 particles. Cell lysates were prepared as described previously (17). Twenty micrograms of total lysate protein was separated on 12% polyacrylamide gels (Mini-Protean TGX; Bio-Rad, Munich, Germany) and transferred onto 0.2 μM nitrocellulose membranes (Pall Corporation, Port Washington, NY) by semidry electrophoretic transfer (Trans-Blot SD; Bio-Rad). Nonspecific antibody binding sites were blocked for 1 h with 2.5% bovine serum albumin (Serva) in phosphate-buffered saline (PBS) containing 0.1% Tween 20. IL-1A was detected with a goat polyclonal antibody against the C terminus of IL-1A of human origin (sc-1253; Santa Cruz Biotechnology, Heidelberg, Germany). Therefore, the blots were incubated with a 1:200 (vol/vol) dilution of the primary antibody in blocking buffer overnight at 4°C with gentle agitation. Immune complexes were detected with horseradish peroxidase-conjugated donkey anti-goat immunoglobulin (sc-2020; Santa Cruz Biotechnology). The secondary antibody was diluted 1:5,000 (vol/vol) in blocking buffer and applied for 1 h at room temperature. Immunoreactive proteins were visualized with an enhanced chemiluminescence detection system (ECL-plus; GE Healthcare, Chalfont St. Giles, United Kingdom) essentially according to the recommendations provided by the manufacturer.
Microarray data accession number.
The complete microarray data sets can be accessed via the GEO database under accession no. GSE25413.
RESULTS
We first attempted to analyze the pathogen-specific kinetics of the mounting of the mammary immune defense in an animal model of clinical mastitis (35). This model was based on sequential infections of three quarters of healthy first-lactation cows with pathogenic E. coli strain 1303 or S. aureus strain 1027. However, we were unable to define with statistical significance a clear time course in the modulated expression of infection-regulated genes in E. coli-infected animals, due to variation between individuals. The individual-specific influence of immune reactivity was even more pronounced in the S. aureus-infected animals. An obvious alternative is the use of primary cultures of mammary epithelial cells from cows (pbMEC) for monitoring the kinetics of the activation of immune mechanisms. Pilot experiments had shown that pbMEC cultures reflect in part the kinetic aspects of immune response regulation as they are observed in mammary tissue, eventually displaying less variance in different biological replicas than encountered in samplings with live cows after different times of infection (see Fig. S1 and Table S2 in the supplemental material). Hence, we used pbMEC cultures to globally profile pathogen-specific transcriptome alterations.
Gene expression profiling of the pbMEC response after challenge with E. coli or S. aureus.
We challenged pbMEC cultures with identical concentrations (107 particles/ml) of either heat-inactivated E. coli 1303 or S. aureus 1027 for 1, 3, 6, and 24 h and compared their transcriptomes to those of untreated control cells. The experiment included three biological replicas for each pathogen stimulus, each with pbMEC derived from a different cow and stimulating the cultures in parallel with both pathogens.
We used the Affymetrix bovine genome array for global transcription analysis. Transcripts were considered differentially expressed if their q value was ≤0.05 and their fold change compared to the control was ≥2. qRT-PCR was used to validate the quality of the data derived from microarray hybridizations. The mRNA concentrations were measured for 10 different genes in all three individual pbMEC culture experiments across all five time points (control and inductions for 1, 3, 6, and 24 h). A comparison of these data to the corresponding values determined from the microarrays revealed a high degree of correlation (r = 0.86 to 1.00) (see Table S3 in the supplemental material).
E. coli causes faster and stronger changes in mRNA abundance than does S. aureus.
A total of 104, 382, 325, and 306 genes were significantly regulated at 1, 3, 6, and 24 h after challenge with E. coli particles. In contrast, we found only 17, 27, 33, and 111 genes to be differentially expressed at those time points after challenge with S. aureus (Table 1). For subsequent analyses, we will only consider genes with known human orthologs, and we use that time after challenge at which the mRNA concentration of the genes was maximally altered as a principal criterion to cluster genes according to their regulation kinetics (Table 1). The complete lists of genes regulated by the E. coli and S. aureus challenges are compiled in Tables S4A and S4B, respectively, in the supplemental material. Genes are ranked in those lists according to the extents of their mRNA concentration alteration. The annotated gene functions are indicated.
TABLE 1.
Significantly differentially expressed genes at each time pointa
| Gene sorting method | No. of genes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
E. coli challenge |
S. aureus challenge |
|||||||||
| 1 h | 3 h | 6 h | 24 h | Total | 1 h | 3 h | 6 h | 24 h | Total | |
| Total significantly differentially expressedb | ||||||||||
| All | 104 | 382 | 325 | 306 | 17 | 27 | 33 | 111 | ||
| Upregulated | 82 | 287 | 272 | 231 | 15 | 27 | 28 | 81 | ||
| Downregulated | 22 | 95 | 53 | 75 | 2 | 0 | 5 | 30 | ||
| According to time to reach max altered mRNA concnc | ||||||||||
| All | 61 | 243 | 157 | 164 | 625 | 13 | 14 | 10 | 101 | 138 |
| Concn increased | 42 | 160 | 130 | 100 | 432 | 11 | 14 | 8 | 71 | 104 |
| Concn decreased | 19 | 83 | 27 | 64 | 193 | 2 | 0 | 2 | 30 | 34 |
| Exceeding 10-fold regulation | 14 | 25 | 17 | 13 | 69 | 1 | 3 | 0 | 7 | 11 |
Considered are only genes with known IPA-eligible human orthologs.
Total numbers of significantly differentially expressed genes at each time point.
Number of genes sorted according to the time (p.i.) of reaching their maximally altered mRNA concentration.
The global characteristics of pathogen-specific transcriptome profiling show that both the number and extent of differentially expressed genes were much higher after challenge with E. coli than after challenge with S. aureus (Table 1). The E. coli challenge significantly altered the expression of 625 annotated genes, while the S. aureus challenge altered the expression of only 138 genes. Moreover, the E. coli challenge increased the mRNA concentration of 69 annotated genes to more than 10-fold compared to the unstimulated control, while those from only 11 genes were found to exceed this strong level of regulation after the S. aureus challenge. The E. coli challenge provoked a much faster response than did the S. aureus challenge, although those particles from both pathogen species had been applied at the same time and at identical concentrations. The E. coli challenge maximally altered the mRNA concentration of 243 genes as early as 3 h after the challenge, while it took 24 h after the S. aureus challenge to achieve the maximum impact on the mRNA levels of 101 genes altogether (Table 1). We found only 34 genes to be exclusively but very weakly (2- to 3-fold) regulated by the S. aureus challenge, while the expressions of as many as 521 genes were regulated exclusively by the E. coli stimulus.
Time course of the regulation of inflammatory response genes in E. coli- and S. aureus-challenged pbMEC.
For the analysis of functional pathways governing the immune response in these cells, we first considered the time course of the expression of “inflammatory response” genes defined by use of IPA software and subdivided the time pattern according to the response elicited by E. coli.
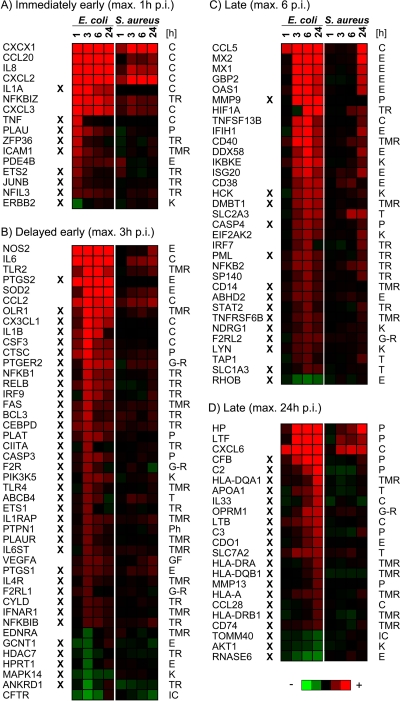
Immediately early primary response (maximum alteration at 1 h postinduction [p.i.]).
The E. coli stimulation swiftly increased the mRNA concentrations of cytokine-encoding genes (such as CXCL1, CXCL2, CCL20, and IL-8) and transcription regulators (e.g., NFKBIZ and ZFP36). Also, the entire group of inflammatory genes (20 from a total of 61 genes) (see Table S4A in the supplemental material) contributes prominently to the very early and strongly regulated genes of this cluster (Fig. 1A). The mRNA concentrations of all these chemokines and of NFKBIZ were also significantly affected by S. aureus but at later times after the stimulus.
FIG. 1.
Comparison of differentially expressed genes in response to challenges with E. coli or S. aureus particles. Considered are genes contributing to the group of “inflammatory response” genes, as defined by IPA software. Heat maps (red, increase; green, decrease) visualize changes in mRNA concentrations relative to the unstimulated controls at different times after challenge (p.i.). Genes are grouped according to the time to reach their maximally altered mRNA concentrations after an E. coli challenge. The X symbol indicates genes regulated solely by E. coli. Gene functions according to the IPA annotation are given to the right (C, cytokine; E, enzyme; G-R, G-protein-coupled receptor; IC, ion channel; K, kinase; P, peptidase; Ph, phosphatase; T, transporter; TMR, transmembrane receptor; TR, transporter).
Delayed early primary response (maximum alteration at 3 h p.i.).
Most of the genes that were significantly regulated by the challenge with E. coli particles belong to the group of delayed early primary response genes (n = 243) (Table 1 and see Table S4A in the supplemental material), comprising transcripts of numerous cytokine receptors (e.g., FAS, IL1RAP, and IL6ST), pattern recognition receptors (TLR2 and TLR4), and transcriptional regulators. The latter include the NF-κB and C/EBP families of factors (e.g., RELB, NFKB1, BCL3, and CEBPD) (Fig. 1B). The regulation of this battery of transcription factors highlights that E. coli provoked significant changes in the signal transduction machinery of the host cell. In addition, the concentrations of mRNAs encoding factors important for host defense (e.g., NOS2, SOD2, and PDGS1/2) were also maximally increased at that time after the stimulus. Only few of those genes were significantly regulated by S. aureus, including NOS2, IL-6, TLR2, SOD, and CCL2, but they all reached their maximally altered mRNA concentration at later times and with lower levels of modulation.
Late (secondary) response (maximum alteration at 6 to 24 h p.i.).
Genes featuring maximal altered mRNA concentrations at 6 h and 24 h after E. coli challenge are considered together, since their separate analyses did not very obviously indicate functional differences.
The late inflammatory response after E. coli challenge regulated the transcript concentrations of many antigen-presenting receptors of the major histocompatibility complex (MHC) (e.g., HLA-DQA1, -DRA, -DQB1, -A, and -DRB1 and CD74), of enzymes known to be expressed in response to type I interferon (IFN) (e.g., MX1/2, GBP2, and OAS1), and of peptidases contributing to the acute-phase response (e.g., HP, LTF, the matrix metallopeptidases [MMPs] MMP9 and MMP13, and complement factors) (Fig. 1C and D). Some cytokines and transcription regulators are also part of the late response elicited by E. coli.
The secondary response of pbMEC caused by the S. aureus challenge was characterized by the expression of IFN-induced enzymes. Transcription factors regulated late after the S. aureus challenge (HIF1A, NFKB2, SP140, and IFN regulatory factor 7 [IRF7]) were also found to be regulated late after E. coli stimulation. However, the S. aureus challenge, unlike E. coli, did not cause any alteration in the levels of mRNAs encoding MHC receptors as well as of matrix metallopeptidases and complement peptidases (Fig. 1C and D).
Downregulation.
Overall, we found that the mRNA abundance of many more genes was increased rather than decreased subsequent to challenge with both E. coli and S. aureus (Table 1). Furthermore, the extent of the downregulation was relatively weak (see Tables S4A and S4B in the supplemental material). Only a few inflammatory response genes were significantly downregulated. Among them, the E. coli challenge significantly but weakly (−2-fold) decreased the level of expression of components from relevant signaling cascades, including MAPK1 (synonym, extracellular signal-regulated kinase 2 [ERK2]), MAPK14 (synonym, p38), v-akt murine thymoma viral oncogene homolog 1 (AKT1), and transforming growth factor beta receptor 1 (TGFBR1; this factor is not comprised in the IPA category “inflammatory response”) (Fig. 1 and Table S4A). These genes were not regulated by the S. aureus challenge (Table S4B). Endothelin receptor type A (EDNRA) was 2-fold downregulated by both the E. coli and S. aureus challenges (Fig. 1B). This represents a rare example of a factor whose expression is similarly diminished by both pathogen species. Interestingly, two downstream targets of this receptor signaling, the histone deacetylase HDAC7 and the growth factor receptor tyrosine kinase ERBB2, were both found to be downregulated by E. coli only (both −2-fold). Thus, E. coli but not S. aureus significantly reduces the signaling of endothelin growth factor.
Aside from those genes relevant to inflammation, we note that E. coli but not S. aureus caused a quite strong downregulation of genes encoding structural molecules (e.g., keratin 15 was −7-fold and −4-fold regulated at 1 h and 3 h p.i., respectively) and members of the tight-junction protein complex (e.g., the claudins CLDN23 and CLDN3, vinculin, and tight-junction protein 3, with a range of changes of expressional regulation of −6-fold to −3-fold) (see Tables S4A and S4B in the supplemental material). Moreover, the E. coli challenge reduced the level of expression of the well-known tumor suppressor breast cancer 1, early onset (BRCA1) (−2 and −3-fold at 6 h and 24 h p.i., respectively).
Pathogen-specific cytokine expression profile.
The induction of cytokines/chemokines is crucial for establishing the cellular branch of immune defense in the udder. We directly compared the kinetics of their regulation after the E. coli and S. aureus challenges (Table 2).
TABLE 2.
Alteration of mRNA levels of cytokine/chemokine-encoding genes after stimulation with E. coli or S. aureus
| Chemokine or cytokinea | Fold changeb |
|||||||
|---|---|---|---|---|---|---|---|---|
|
E. coli |
S. aureus |
|||||||
| 1 h | 3 h | 6 h | 24 h | 1 h | 3 h | 6 h | 24 h | |
| CXCL1 | 779 | 142 | 78 | 43 | 3 | 17 | 7 | 13 |
| CCL20 | 529 | 479 | 137 | 69 | 2 | 2 | 3 | |
| IL-8 | 384 | 144 | 103 | 71 | 5 | 4 | 4 | |
| CXCL2 | 348 | 223 | 144 | 98 | 4 | 24 | 13 | 16 |
| IL-1A | 123 | 24 | 5 | 4 | ||||
| CXCL3 | 17 | 7 | 8 | 6 | 3 | 2 | 2 | |
| TNF | 7 | |||||||
| IL-6 | 32 | 39 | 13 | 11 | 3 | 3 | 4 | |
| CCL2 | 6 | 13 | 7 | 8 | 3 | 3 | 4 | |
| CX3CL1 | 12 | 7 | 4 | |||||
| IL-1B | 3 | 11 | 6 | |||||
| CSF3 | 3 | 11 | 3 | 3 | ||||
| CCL5 | 7 | 134 | 175 | 106 | 31 | |||
| TNFSF13B | 6 | 12 | 3 | 2 | ||||
| IL-15 | 3 | |||||||
| CXCL6 | 8 | 19 | 18 | 20 | 5 | 7 | 6 | |
| IL-33 | 2 | 5 | ||||||
| LTB | 2 | 3 | 5 | |||||
| CCL28 | 2 | |||||||
Boldface type indicates master cytokines; chemokines are underlined.
Values in boldface type indicate maximal alteration compared to the control blanks, not significantly regulated.
We found 19 cytokines/chemokines to be significantly regulated by E. coli. Ten of them were also significantly induced by S. aureus. However, E. coli always elicited a much stronger alteration of the mRNA levels quantitatively than did S. aureus.
E. coli significantly induced the expression of 10 chemokines. Eight of those were significantly regulated as early as 1 h poststimulation. The concentration of mRNAs encoding CXCL1, CXCL2, CXCL3, and IL-8 peaked at this early time point. Subsequently, they were quickly and strongly downregulated. These chemokines interact with the CXCR2 receptor, known to be predominantly expressed on neutrophils. Hence, the MEC signals to recruit those cells into the infected udder and to activate them.
The mRNAs encoding chemokines important for the recruitment of monocytes and lymphocytes are known to be induced later than those factors recruiting neutrophils. The mRNA concentrations of the monocyte-recruiting factor CCL2 and of the adhesion mediator CX3CL1 reached their maximum 3 h after E. coli stimulation, while the mRNAs encoding the predominantly T-cell-recruiting chemokines CCL5 and CCL28 reached their maximal concentrations 6 and 24 h after E. coli challenge (maximally induced 175- and 2-fold, respectively). The induction kinetics of CCL20 differ from those of the other lymphocyte-recruiting factors. This chemokine was massively (529-fold) and more swiftly (1 h p.i.) induced after E. coli stimulation.
S. aureus significantly regulated the expression of eight chemokines in pbMEC. Their induction was retarded and weak compared to those found with the E. coli challenge. Nevertheless, CXCL1, CXCL2, CXCL3, and IL-8 were again the earliest maximally induced chemokines after pathogen contact. CXCL2 and CXCL1 are among the immune factors most strongly induced by S. aureus (maximum inductions of 24- and 17-fold, respectively, at 3 h p.i.). CCL2 and CCL5 were again found to be induced later than the CXCL factors. The induction of CCL20 occurred later and was much weaker (only 3-fold at 24 h p.i.) than that of the E. coli challenge.
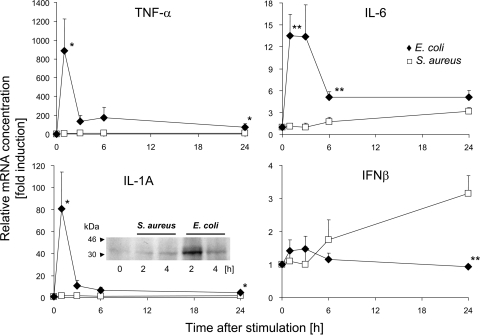
The expression of three key proinflammatory cytokines (IL-1A and IL-1B, TNF-α, and IL-6) was quickly and strongly induced by the E. coli challenge (Table 2). S. aureus, in contrast, did not significantly induce the expression of IL-1 and TNF-α. We reevaluated these observations by qRT-PCR (Fig. 2). These measurements revealed a statistically significant induction of TNF-α 24 h after S. aureus challenge. However, the maximum expression level after S. aureus challenge was only 1% of that recorded after the E. coli challenge. IL-1A was not found to be significantly regulated by the S. aureus challenge.
FIG. 2.
Alteration in the mRNA concentrations of TNF-α, IL-1A, IL-6, and IFN-β (IFN-β1 and IFN-β2) in pbMEC after challenge with E. coli (⧫) and S. aureus (□) particles. Shown are values for mean fold induction (ordinate, ± standard error of the mean [SEM]; n = 3) at times after challenge (abscissa) relative to the mRNA concentration measured for unstimulated cells (set as 1). Asterisks indicate a statistical significance (*, P ≤ 0.05; **, P ≤ 0.01 [by t test]) of the difference between the E. coli and S. aureus values at the time indicated. The IL-1A insert shows a Western blot analysis of IL-1A in lysates (20 μg/slot) from unchallenged pbMEC (0 h) and from those challenged for 2 h or 4 h with S. aureus or E. coli particles (107 particles/ml). The band shows the 31-kDa IL-1A precursor protein.
The microarray analysis revealed IL-6 to be the only master cytokine to be significantly induced by S. aureus. Its mRNA concentration was increased by about 3-fold, as measured by qRT-PCR (Fig. 2). While this was quantitatively less of an induction than that seen after the E. coli challenge (13-fold), it nevertheless amounted to approximately 20% of the E. coli-induced enhancement in concentrations.
We realized that, confusingly, two different genes are annotated for the bovine genome (Bovine Genome Assembly 4.2) as encoding the factor IL-6, while for humans, only one IL-6-encoding gene is known. This human gene is synonymous with IFN-β2. Its bovine ortholog is represented on the bovine Affymetrix array and corresponds to the gene localized on Bos taurus autosome 4 (BTA4) (IL-6 [IFN-β2]; Entrez GeneID 280826). The other bovine gene, IL-6 (IFN-β2), is localized on BTA8 within an IFN gene cluster (Entrez GeneID 517016). This gene is an ortholog of the human IFN-β1 gene and has no sequence similarity to the former bovine IL-6 gene but shares 84% DNA sequence similarity to the bovine IFN-β1-encoding gene. We refer throughout to the IL-6 gene (GeneID 280826) as encoding IL-6 and to that gene denoted by GeneID 517016 as encoding IFN-β2.
In the microarray experiment the mRNA concentration of IFN-β1 was below the level of detection, and the array contained no probe set for IFN-β2. Given that IFN-βs are known as relevant immune factors, we measured the alteration of their mRNA abundances in response to E. coli and S. aureus by qRT-PCR. We applied a pair of primers amplifying both IFN-β1- and IFN-β2-encoding messages. Surprisingly, we observed a significant and late induction of these factors only in S. aureus- and not in E. coli-stimulated cells (Fig. 2). Thus, we identified the group of IFN-βs as immune factors exclusively regulated by S. aureus.
It was reported previously that increased mRNA concentrations of proinflammatory cytokines eventually poorly correlate with the respective protein synthesis or abundance (40, 41). Taking IL-1A as an example for one of our key factors, we therefore examined changes in the intracellular abundance of the 31-kDa IL-1A precursor protein subsequent to our pathogen challenges by Western blotting. We found that E. coli stimulation strongly induced its synthesis, while the S. aureus challenge barely changed the abundance (Fig. 2, insert in IL-1A graph). Thus, the protein abundance of this key factor reflects the respective mRNA concentration in pbMEC. It is known that the abundance of the IL-1A precursor exceeds that of the cleaved 17-kDa IL-1A factor itself and that both factors are bioactive (20).
Differential regulation of functional networks.
We next examined gene interactions revealed by their known contribution to regulatory networks in order to identify key regulatory principles of the pathogen-specific immune response in the MEC.
E. coli.
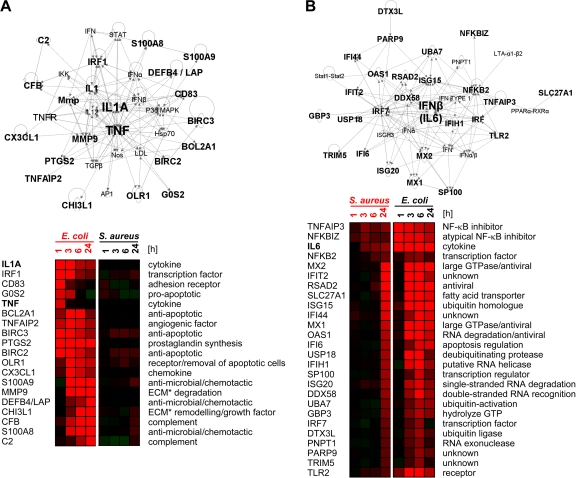
The large number of E. coli-regulated genes necessitated the introduction of constraints in order to reduce the complexity of the network analysis. Therefore, we focused our analysis on the most strongly regulated genes. We selected the 20 most strongly induced genes from each of the four time groups (maximum at 1, 3, 6, and 24 h p.i.). Twenty-seven genes from among these 80 top regulated genes were found to be regulated exclusively by E. coli. The latter genes were subjected to an IPA-Network analysis. Nineteen of them were identified as part of the top-ranked network (network score of 49). This network features the key proinflammatory cytokines IL-1A and TNF-α in the central position (Fig. 3A).
FIG. 3.
Network analysis of regulatory relationships. (A) E. coli network. IPA analysis reveals an interaction network dominated by IL-1A and TNF-α as having the highest statistical probability (network score of 49) of the 27 top-ranked genes regulated exclusively by E. coli (top). Heat maps (bottom) (red, increase; green, decrease) compare the regulation intensities of those genes after E. coli (right) or S. aureus (left) challenge. (B) Consideration of all S. aureus-regulated gene (n = 138) yields with the highest statistical significance (network score of 48) for a network dominated by IL-6. Heat maps (bottom) show the regulation intensities of those genes after E. coli (right) or S. aureus (left) challenge. ECM, extracellular matrix.
S. aureus.
All 138 annotated genes regulated by S. aureus were included in this analysis. The top-ranked network (network score of 48) comprised 26 genes with IFN-β in a central position (Fig. 3B). The Ingenuity software subsumes under IFN-β both factors IFN-β1 and IL-6. Only three genes of this S. aureus-regulated network were induced relatively early, from 3 h on (Fig. 1B and see Table S4B in the supplemental material). These genes are IL-6 and two inhibitors of NF-κB signaling (NFKBIZ and TNFAIP3). NFKBIZ is known as an essential inducer of IL-6 expression.
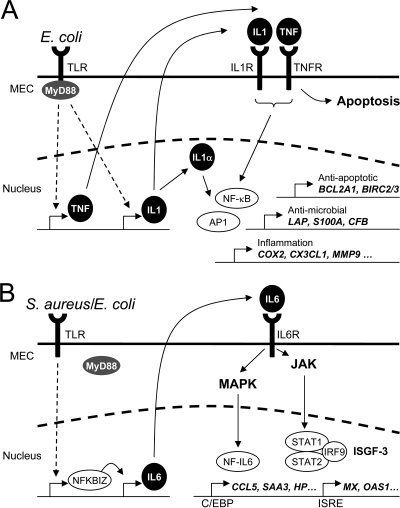
Considering the orchestration of the immune response in MEC, our data together indicate that the coordinated gene regulation governed by the activation of IL-1A and TNF-α signaling is an E. coli-specific response feature. The S. aureus response, in contrast, induces a functional network dominated by IFN-β/IL-6.
Aside from considering genes relevant to the immune response and inflammation, we note that S. aureus induced quickly and very strongly the expression of genes involved in the detoxification of xenobiotic substances (see Table S4B in the supplemental material). These genes include the monooxygenases CYP1B1 (25-fold induced 1 h after challenge) and CYP1A1 (maximum induction 3 h after challenge; 391-fold) as well as the gene encoding the 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TIPARP) (4-fold induced 1 h after challenge). These three genes are key factors for the oxidative metabolism of various toxic substances and showed similar patterns of regulation after the E. coli challenge as well.
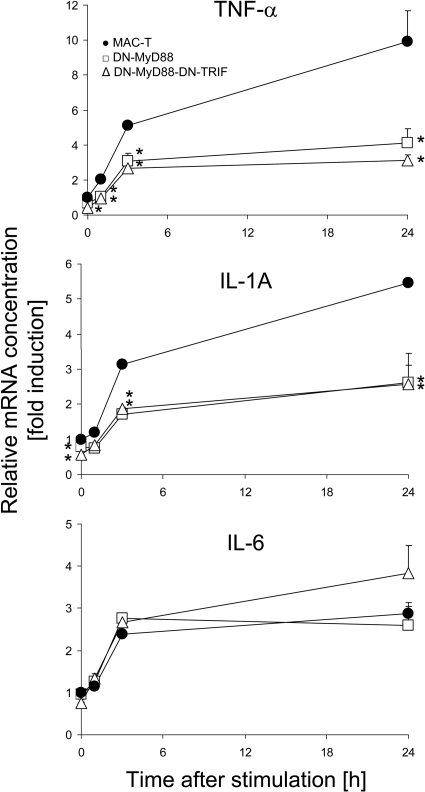
Functional knockdown of MyD88-dependent signaling reduces the induction of TNF-α and IL-1A but does not influence the induction of IL-6 expression in E. coli-stimulated MEC.
We wanted to examine if the blockage of TLR-mediated signaling in MEC would still allow an induction of IL-6 expression in this cell type upon stimulation with E. coli particles. Central to TLR signaling are receptor-proximal adaptors. Among these molecules, myeloid differentiation primary response 88 (MyD88) is widely used from almost all TLRs, while TIR-domain-containing adapter-inducing IFN-β (TRIF) is specific for TLR4 and TLR3 signaling. The functional knockdown of TLR signaling can be achieved by stably overexpressing trans-dominant negative (DN) mutants of the factors MyD88 and TRIF in these cells (34, 58). Therefore, we stably transfected vectors expressing these DN factors into cells of the transformed and permanent bovine MEC cell line MAC-T. Wild-type (WT) MAC-T cells and DN mutants expressing pools of clones were challenged with E. coli for 1, 3, and 24 h. The change in the abundance of mRNAs encoding TNF-α, IL-1A, and IL-6 was measured over time (Fig. 4 ). While the extent of TNF-α and IL-1A expression was significantly reduced in transfectants expressing DN-MyD88 and DN-MyD88-DN-TRIF, IL-6 expression was not affected by those DN factors. The efficacies of both mutations (DN-MyD88 and DN-MyD88-DN-TRIF) were quite similar. We validated with control experiments that pathogen-induced NF-κB induction was significantly reduced in both transfectants, down to 56% and 63% for DN-MyD88-TRIF and DN-MyD88, respectively.
FIG. 4.
Impact of functional knockdown of TLR signaling on induced cytokine gene expression. Shown are data for the alteration of TNF-α, IL-1A, and IL-6 mRNA concentrations in WT MAC-T cells or those stably transfected with vectors expressing DN-MyD88 and DN-MyD88-DN-TRIF mutants after challenge with heat-inactivated E. coli particles. Mean fold induction values (ordinate, ±SEM) relative to the mRNA concentrations measured in unstimulated MAC-T cells (set as 1) were derived from one experiment representative of three, which was assayed in triplicate. Asterisks indicate that the mRNA concentrations measured for the DN mutant transfectants are statistically different (*, P ≤ 0.05 [by t test]) from those for WT MAC-T cells at the respective time point after E. coli challenge.
We did not conduct the same experiment using S. aureus as a stimulus since these cells show generally a much weaker response after stimulation with E. coli than that recorded for pbMEC (cf. Fig. 2 and 4). S. aureus generally fails to significantly stimulate the expression of any of the three master cytokines (e.g., IL-1A, TNF-α, and IL-6) in those cells.
DISCUSSION
A molecular understanding of the principles causing the different etiologies of clinical and subclinical mastitis is particularly important to eventually develop innovative strategies to prevent and treat subclinical mastitis. Animal models are less practical for those analyses due to the long-known impact of interindividual variations (8, 11) and the impossibility to analyze gene expression in tissue samples in adequate numbers of animals. Comparative cytokine profiling elicited by different pathogen species during trials of mastitis was done only with milk or from milk cells (1, 2, 4, 5, 40). At the outset of the study we measured specific inflammation markers in E. coli-infected udders from several cows and confirmed the known (8) strong influence of the individual animal upon the extent and kinetics of the activation of immune response. As an alternative, we used pbMEC cultures as a model for udder tissue in order to compare the extents and kinetics of the immune responses elicited by E. coli or S. aureus pathogens. We validated that the heat inactivation of both pathogen species renders preparations with the same bioactivity as that obtained after inactivating them with UV light (see Fig. S2 in the supplemental material). We applied 107 particles/ml in any challenge and, thus, approximately 30 particles per host cell. This represented a robust stimulus considering that 104 to 106 CFU per ml of milk has frequently been retrieved as peak values in cows after experimental infection with live pathogens of either species (4, 5; see reference 8 and references therein).
We report the key observations that heat-inactivated S. aureus particles induce a retarded response and regulate the expression of less than ∼20% of E. coli-regulated genes but that the stimuli are applied at the same time in the same concentration. However, S. aureus particles are as swiftly recognized as those from E. coli, as is clear from the equal kinetics of the strong induction of CYP1A1 expression by both pathogens (cf. Tables S4A and S4B in the supplemental material). The pathogen-specific response is differentiated already during the first hour. At this early time point, E. coli regulates almost 10-fold-more genes than does S. aureus. Thus, the heat-inactivated S. aureus particles, without any secreted virulence factors, cause a striking retardation in the immune response of MEC. They either fail to activate key inducers that set in motion the defense program of the MEC or trigger a delaying reaction. S. aureus but not E. coli particles almost fail to activate IL-1A and TNF-α expression while still substantially inducing IL-6 expression. Also, the induction of this gene is retarded compared to that with the E. coli stimulation. Conceivably, increasing the challenge dose might speed up the induction of IL-6 expression, following our own experiences regarding the dosage-dependent kinetics of cytokine expression in this cell system (17). However, increasing the challenge intensity would conceivably also accelerate the induction kinetics subsequent to the E. coli stimulus. Thus, the data altogether show that S. aureus elicits a much weaker and slower immune response than E. coli but that it nevertheless robustly induces chemokines serving for leukocyte recruitment.
Differential induction of IL-1 and TNF-α is crucial for the pathogen-specific response of pbMEC.
Many of the genes that are strongly and specifically activated by the E. coli challenge are part of a regulatory network controlled by IL-1 and TNF-α. Both factors are known as proinflammatory master cytokines and ubiquitous mediators of inflammation (10). Their receptors are found on almost any cell type. Moreover, the signaling cascade downstream of the IL-1 receptor 1 shares many factors in common with the signaling cascade downstream of the pathogen-sensing TLR receptors. By projecting known pathways of IL-1- and TNF-α-signaling genes onto the list of E. coli-regulated genes (Fig. 5A), we find that, conceivably, this axis stimulates the expression of the antimicrobial and chemotactic proteins S100A8 and S100A9; the β-defensin LAP; the antiapoptotic factors BIRC2, BIRC3, and BCL2A1; the complement factors C2 and CFB; as well as enzymes degrading the extracellular matrix (MMP9) or the key regulator of prostaglandin synthesis (PTGS2). All these genes are known to be induced by IL-1 in keratinocytes (60), a subtype of epithelial cells. In addition, TNF-α is one of the prime signals inducing apoptosis in eukaryotic cells. The counterregulation of this death signal in the infected cell is the induction of antiapoptotic factors like BIRC2, BIRC3, and BCL2A1 (45). We found these factors to be induced in E. coli-stimulated pbMEC.
FIG. 5.
Proposed signaling pathways being activated in MEC through challenges with heat-inactivated particles of E. coli or S. aureus pathogens. (A) E. coli immediately induces the early expression of the proinflammatory master cytokines IL-1α and IL-1β as well as TNF-α. The activation of these cytokines is MyD88 dependent. IL-1β and TNF-α are synthesized and secreted and may bind to their cognate receptors (IL1R and TNFR). This subsequently leads to the activation of NF-κB and AP1, which in turn induces the expression of the secondary response genes (e.g., BCL2A1, BIRC1/2, LAP, S100A8/9, CFB, COX2, CX3CL1, and MMP9). In addition, TNF-α is also one major mediator of apoptosis. Intracellular IL-1α may also function as an endogenous transcription regulator inside the cell of its synthesis (22). (B) S. aureus and E. coli stimulations both lead to an early activation of NFKBIZ. This transcriptional activator is known to be essential for the TLR-mediated induction of IL-6 expression. We assume that the induction of IL-6 expression in MEC is MyD88 independent. Secreted IL-6 binds to the IL-6 receptor (IL6R), resulting in the activation of both JAK and MAPK signaling cascades. The activation of the JAK cascade leads to the formation of an active ISGF3 complex, which may induce the expression of genes harboring ISRE sites in their promoters. MAPKs activated through IL-6 signaling induce the activation of NF-IL-6, also known as the transcription factor C/EBPβ. The transcription induced by ISGF3 and C/EBPβ dominates the secondary response to S. aureus.
In contrast, the S. aureus challenge fails to induce IL-1A and only marginally (1% of the E. coli level) induces the expression of TNF-α. Our hybridization data are fully supported by the qRT-PCR measurements. We validated for IL-1A that these pathogen-specific, grossly differing mRNA levels are also reflected at the protein level (Fig. 2). It was reported previously that S. aureus induces a significant but very weak (<5% of the E. coli level) induction of TNF-α in these model cells (58). Thus, the S. aureus-specific response does not feature the autocrine and/or paracrine potentiation of the inflammatory response that apparently is so characteristic of the E. coli-elicited response.
A bovine pbMEC culture system was recently used to compare the pathogen-specific induction kinetics of some cytokine-encoding genes (15). Those authors also used particles of heat-inactivated E. coli and S. aureus pathogens. They also generally found a stronger induction of the key factors TNF-α, IL-1β, and IL-6 by E. coli in the period from 1 h to 24 h p.i. However, those authors observed a stronger induction by S. aureus than by E. coli during the first hour. This is at variance with our data. We note that the induction kinetics for these factors are grossly different between both studies. We observed a strong downregulation of the mRNA concentration for all these cytokines, commencing 1 to 3 h p.i., as has often been found (17, 26, 49, 58) but which was not recorded in the that study (15). We cannot conclusively reconcile these differences. There are differences in the establishment and handling of the pbMEC cultures between these studies. Significantly, we used only once-passaged cells, while those authors used cells after three passages. We have previously shown that the immune reactivity of pbMEC is grossly altered after two and three passages (16). Differences in the pathogen strains might also be relevant. Indeed, live pathogens of one strain of S. aureus, but not two others, induced TNF-α and IL-1β more strongly than did an E. coli strain 3 h after coculture with pbMEC (26).
Induction of IL-6 dominates the response of pbMEC toward S. aureus.
The only master cytokine to be significantly induced by the S. aureus challenge in pbMEC was IL-6. The binding of this factor to its receptor causes the activation of the Janus tyrosine kinases (JAK), eventually activating STAT1 and STAT3 (13, 19). The phosphorylation of STAT3 results in its homo- or heterodimerization and nuclear translocation. The association of STAT3 with different transcription factors and coactivators mediates the transcription of many different target genes (29). STAT1 binds as a homodimer to the gamma interferon-activating sequence or trimerizes with STAT2 and IRF9 to form ISGF3 (interferon-stimulated transcription factor 3). This factor complex is capable of binding to interferon-stimulated response element (ISRE) DNA sequence motifs (13). We found that most of the genes being induced late after challenge with either S. aureus or E. coli feature ISRE binding sites in their promoters. Most of these genes, such as MX1, MX2, and OAS1, are known to be induced by IFN and JAK/STAT signaling (18).
The activation of the IL-6 receptor also activates mitogen-activated protein kinases (MAPKs). Signaling involving these kinases leads to the activation of the transcription factor NF-IL-6, also known as C/EBPβ (19). We observed that CCL5, SAA3, and HP are most strongly and lately induced after both the E. coli challenge and the S. aureus challenge. All three genes are known to be regulated by C/EBPβ (24, 46, 53). This highlights the pivotal role of this transcription factor in the inflammatory response of pbMEC toward both pathogen species.
Our data together suggest that IL-6 signaling dominates the response of pbMEC to S. aureus but also orchestrates the late response after an E. coli challenge. We observed, as an S. aureus challenge-specific feature, increased concentrations of IFN-β1- and IFN-β2-encoding mRNAs. IFN-β is known to inhibit the release of the proinflammatory cytokine IL-12 through increasing IL-10 synthesis in peripheral blood mononuclear cells in humans (55). In addition, IFN-β is also thought to downregulate the IFN-γ-induced expression of MHC class II on antigen-presenting cells (23) and lower the expression of adhesion molecules and MMPs (30). We found, in good agreement, that the S. aureus challenge, unlike the E. coli challenge, did not induce the expression of MHC factors or of MMPs and the intercellular adhesion molecules (ICAMs) in pbMEC.
Impairment of TLR signaling still allows for induced IL-6 expression.
Our experiments to functionally knock down TLR signaling in MAC-T cells were based on the expression of the trans-dominant negative factor DN-MyD88 and the dual factor DN-MyD88-DN-TRIF. The efficacy of these constructs to completely block TLR2- or TLR4-mediated signaling in HEK293 cells was previously validated (58). We exploited these constructs as tools to specifically examine if the synthesis of the key factor IL-6 of the MEC defense program against S. aureus would still be possible in the absence of MyD88 signaling. We found that the expression of either construct in MAC-T cells does not affect the induction of IL-6 synthesis in response to an E. coli challenge. A MyD88-independent activation of IFN-β and IL-6 expression was previously observed for macrophages (28) and intestinal epithelial cells (12). Our present results for the MyD88-independent induction of IL-6 expression with the previously reported S. aureus-mediated prevention of NF-κB activation in pbMEC (58) collectively suggest that S. aureus may indeed subvert the MyD88-dependent route of TLR signaling in MEC. Our data show that heat-inactivated particles of this pathogen species induce an early inhibitor of NF-κB activation (TNFAIP3) as well as the atypical NF-κB factor NFKBIZ. This factor is crucial for induced IL-6 expression (57) but is also known to block TNF-α synthesis (31).
A blockage or failure to activate the MyD88-dependent branch of TLR signaling by S. aureus in MEC is conceivably the reason why strong effector genes of innate immunity are not activated early after infection of the udder in this very abundantly present cell type. Such genes include bactericidal β-defensins (59) and inducible nitric oxide synthase (iNOS) but also genes encoding membrane-protecting acute-phase proteins, such as SAA3 or HP. Particularly, the failure to activate those genes encoding bactericidal factors (β-defensins and iNOS) in MEC, representing the dominant cell type of the healthy milk parenchyma and the host's front line for fighting off invading pathogens, presents a reasonable explanation for the impaired pathogen clearance encountered in cases of subclinical or persistent mastitis caused by those Gram-positive bacteria (4, 35, 51). These mild infections eventually result in the persistent presence of the pathogens within the udder. In contrast, the immune response elicited by E. coli includes a massive induction of those genes, highlighting the principal importance of the MEC to eliminate invading microbes.
The sequential order of chemokine expression in pathogen-challenged pbMEC correlates well with general principles of leukocyte infiltration.
E. coli challenge elicits a massive induction of many cytokine-encoding genes. This was reported previously after long time (24 h) of stimulation of those cells (16). We now report that this outstanding group of immune factors is strongly induced in pbMEC by S. aureus as well. It is general knowledge that these factors recruit leukocytes to the site of inflammation (9, 42) and that neutrophils dominate during the initial influx. Later during inflammation, leukocytes of the monocyte/macrophage lineage replace the neutrophil as the predominant leukocyte. In good agreement with this, we found in our pbMEC model that the neutrophil-recruiting factors CXCL1, CXCL2, CXCL3, and IL-8 are very early and strongly induced by the E. coli challenge but also by the S. aureus stimulus. Only at later times did both stimuli induce those genes encoding the chemoattractants CCL2 and CCL5, known to recruit monocytes, T cells, eosinophils, and basophils. Thus, the pbMEC cell model faithfully recapitulates the complex time pattern for the consecutive induction of various chemokines recruiting the different immune cells to the infected udder.
Our observed weak but significant induction of chemokine expression in S. aureus-stimulated pbMEC is consistent with the previously reported observation of a significant recruitment of leukocytes into the milk of S. aureus-infected animals (35).
The regulation of the chemokine CCL20 is peculiar. We found its expression to be extremely strong and induced early after E. coli stimulation but only very weakly after S. aureus stimulation. CCL20 is known to interact with the CCR6 receptor expressed on dendritic cells (DC) and some T cells. However, it also shares structural and functional characteristics with β-defensins (21). Both CCL20 and β-defensins display antimicrobial activity and also bind to the CCR6 receptor. Their strong induction in response to E. coli challenge conceivably represents a protective mechanism against invading bacteria. This mechanism is not induced by a challenge with S. aureus.
Regulation of the pathogen-specific inflammatory response in pbMEC models the in vivo response.
E. coli stimulation elicited a very strong induction of immune genes, including cytokines and antimicrobial and acute-phase response genes, in our pbMEC model cells. This was also found by global transcriptome profiling studies of mammary glands after infection with live E. coli pathogens in both the udder of cows (16, 32) and the mammary glands of mice (61). However, our pbMEC cultures did not display a downregulation of genes contributing to fatty acid metabolism, which is so overt from those studies analyzing mammary glands after E. coli infections. Such limitations of the pbMEC model were described previously (16).
Our observation of a much weaker induction of inflammatory mediators by S. aureus-infected udders has frequently been observed for cows suffering from S. aureus mastitis (4, 39, 40). Moreover, S. aureus infection was previously found to cause only modest changes (<4-fold-changed mRNA concentrations) in the global transcriptome of milk cells from goats (36). Those authors recorded 177 regulated genes, comprising many leukocyte-specific genes. Interestingly, among the chemokine-encoding genes, only CXCL6 was found to be regulated in those milk cells. Levels of IL-1A mRNA concentrations were slightly enhanced (2-fold) at 12 h p.i. Overall, however, those results compare well with our observation of 138 genes found to be regulated in our pbMEC subsequent to the S. aureus challenge. A cell type-specific difference appears to be the weak induction of chemokine synthesis in milk cells (comprising neutrophils, lymphocytes, and macrophages), which is so prominent in MEC.
As mentioned above, a previously described animal infection model compared the time courses of udder infection with E. coli strain 1303 and S. aureus strain 1027 (35). These are the same pathogen strains used in our study. Experimental infections of udders with those pathogens demonstrated a homogeneous response of E. coli-inoculated animals with strong clinical symptoms (fever and leukopenia) characteristic of an acute inflammatory response (35). The response to S. aureus inoculations was milder and more heterogeneous, with a lack of clinical symptoms and no induction of TNF-α expression (58). This is consistent with our observations of pbMEC stimulated with heat-inactivated particles of the same pathogen strains. The failure to significantly induce IL-1 and the only marginal induction of TNF-α observed for S. aureus-stimulated pbMEC comply with the lack of systemic inflammation in S. aureus-infected animals. Reduced TNF-α and IL-1 secretion during mastitis caused by S. aureus and other Gram-positive bacteria has frequently been observed (39, 40; see reference 1 for a survey).
Conclusion.
We used global transcriptome profiling to analyze the pathogen-specific inflammatory response of primary mammary epithelial cells against heat-inactivated particles of two different species of mastitis pathogens, which, in vivo, in the udders of cows, reliably cause different etiopathologies. Considering the extent and time course of the host reaction, we find as key observations a characteristic early expression of TNF-α and IL-1 in E. coli-challenged cells, the likely causes of the severe inflammatory symptoms often observed for udder infections with E. coli. These strong inductions are not elicited by S. aureus particles, which slightly induce the expression of IL-6. This cytokine dominates the weak secondary response to S. aureus. Our knowledge gained in vitro bears on immune processes going on in vivo, in the udder, since key aspects of our results comply with previously reported clinical findings for animals suffering from acute E. coli or subclinical S. aureus mastitis.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical support from Angelika Deike, Bärbel Pletz, and Anne Bernd.
This work was supported by the Deutsche Forschungsgemeinschaft through program FOR585, grant Se 326/14-3. We have neither other financial relations to declare nor any other relationships constituting conflict of interests.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 29 November 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bannerman, D. D. 2009. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J. Anim. Sci. 87:10-25. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, D. D., et al. 2004. Innate immune response to intramammary infection with Serratia marcescens and Streptococcus uberis. Vet. Res. 35:681-700. [DOI] [PubMed] [Google Scholar]
- 3.Bannerman, D. D., M. J. Paape, W. R. Hare, and J. C. Hope. 2004. Characterization of the bovine innate immune response to intramammary infection with Klebsiella pneumoniae. J. Dairy Sci. 87:2420-2432. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman, D. D., et al. 2004. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin. Diagn. Lab. Immunol. 11:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannerman, D. D., M. J. Paape, and A. Chockalingam. 2006. Staphylococcus aureus intramammary infection elicits increased production of transforming growth factor-α, β1, and β2. Vet. Immunol. Immunopathol. 112:309-315. [DOI] [PubMed] [Google Scholar]
- 6.Barbosa-Cesnik, C., K. Schwartz, and B. Foxman. 2003. Lactation mastitis. JAMA 289:1609-1612. [DOI] [PubMed] [Google Scholar]
- 7.Brightbill, H. D., et al. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 8.Burvenich, C., V. Van Merris, J. Mehrzad, A. Diez-Fraile, and L. Duchateau. 2003. Severity of E. coli mastitis is mainly determined by cow factors. Vet. Res. 34:521-564. [DOI] [PubMed] [Google Scholar]
- 9.Coussens, L. M., and Z. Werb. 2002. Inflammation and cancer. Nature 420:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinarello, C. A. 2000. Proinflammatory cytokines. Chest 118:503-508. [DOI] [PubMed] [Google Scholar]
- 11.Dosogne, H., et al. 1997. Increased surface expression of CD11b receptors on polymorphonuclear leukocytes is not sufficient to sustain phagocytosis during Escherichia coli mastitis in early postpartum dairy cows. Vet. Immunol. Immunopathol. 60:47-59. [DOI] [PubMed] [Google Scholar]
- 12.Friis, L. M., M. Keelan, and D. E. Taylor. 2009. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect. Immun. 77:1553-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhartz, C., et al. 1996. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J. Biol. Chem. 271:12991-12998. [DOI] [PubMed] [Google Scholar]
- 14.Goldammer, T., et al. 2004. Mastitis increases mammary mRNA abundance of β-defensin 5, Toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin. Diagn. Lab. Immunol. 11:174-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griesbeck-Zilch, B., H. H. D. Meyer, C. H. Kuhn, M. Schwerin, and O. Wellnitz. 2008. Staphylococcus aureus and Escherichia coli cause deviating expression profiles of cytokines and lactoferrin messenger ribonucleic acid in mammary epithelial cells. J. Dairy Sci. 91:2215-2224. [DOI] [PubMed] [Google Scholar]
- 16.Günther, J., et al. 2009. Assessment of the immune capacity of mammary epithelial cells: comparison with mammary tissue after challenge with Escherichia coli. Vet. Res. 40:31. doi: 10.1051/vetres/2009014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Günther, J., S. Liu, K. Esch, H. J. Schuberth, and H. M. Seyfert. 2010. Stimulated expression of TNF-α and IL-8, but not of lingual antimicrobial peptide reflects the concentration of pathogens contacting bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 135:152-157. [DOI] [PubMed] [Google Scholar]
- 18.Haller, O., G. Kochs, and F. Weber. 2007. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 18:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich, P. C., et al. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, J. R., J. A. Corbett, G. Kwon, C. A. Marshall, and M. L. McDaniel. 1996. Nitric oxide regulates interleukin 1 bioactivity released from murine macrophages. J. Biol. Chem. 271:22672-22678. [DOI] [PubMed] [Google Scholar]
- 20a.Hintermair, V. 2007. Comparative analysis of Bos taurus and Homo sapiens DNA microarrays. M.S. thesis. Ludwig Maximilian University, Munich, Germany.
- 21.Hoover, D. M., et al. 2002. The structure of human MIP-3α/CCL20: linking antimicrobial and CCR6 receptor binding activities with human β-defensins. J. Biol. Chem. 277:37647-37654. [DOI] [PubMed] [Google Scholar]
- 22.Hu, B., et al. 2003. A nuclear target for interleukin-1α: interaction with the growth suppressor necdin modulates proliferation and collagen expression. Proc. Natl. Acad. Sci. U. S. A. 100:10008-10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh, H. K., J. Oger, and K. Dorovinizis. 1995. Interferon-β downregulates interferon-γ-induced class II MHC molecule expression and morphological changes in primary cultures of human brain microvessel endothelial cells. J. Neuroimmunol. 60:63-73. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M. O., H. S. Suh, C. F. Brosnan, and S. C. Lee. 2004. Regulation of RANTES/CCL5 expression in human astrocytes by interleukin-1 and interferon-β. J. Neurochem. 90:297-308. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, H., T. Kawai, and S. Akira. 2009. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388:621-625. [DOI] [PubMed] [Google Scholar]
- 26.Lahouassa, H., E. Moussay, P. Rainard, and C. Riollet. 2007. Differential cytokine and chemokine responses of bovine mammary epithelial cells to Staphylococcus aureus and Escherichia coli. Cytokine 38:12-21. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, T., and D. W. Gilroy. 2007. Chronic inflammation: a failure of resolution? Int. J. Exp. Pathol. 88:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leber, J. H., et al. 2008. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, D. E., and J. E. Darnell. 2002. STATs: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 30.Liuzzi, G. M., T. Latronico, A. Fasano, G. Carlone, and P. Riccio. 2004. Interferon-β inhibits the expression of metalloproteinases in rat glial cell cultures: implications for multiple sclerosis pathogenesis and treatment. Mult. Scler. 10:290-297. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo, S., S. Yamazaki, K. Takeshige, and T. Muta. 2007. Crucial roles of binding sites for NF-κB and C/EBPs in IκB-ζ-mediated transcriptional activation. Biochem. J. 405:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitterhuemer, S., et al. 2010. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genomics 11:138. doi: 10.1186/1471-2164-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mogensen, T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muzio, M., J. Ni, P. Feng, and V. M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278:1612-1615. [DOI] [PubMed] [Google Scholar]
- 35.Petzl, W., et al. 2008. Escherichia coli, but not Staphylococcus aureus triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet. Res. 39:18. [DOI] [PubMed] [Google Scholar]
- 36.Pisoni, G., et al. 2010. Differentially expressed genes associated with Staphylococcus aureus mastitis in dairy goats. Vet. Immunol. Immunopathol. 135:208-217. [DOI] [PubMed] [Google Scholar]
- 37.Poltorak, A., et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 38.Rainard, P., and C. Riollet. 2006. Innate immunity of the bovine mammary gland. Vet. Res. 37:369-400. [DOI] [PubMed] [Google Scholar]
- 39.Rainard, P., A. Fromageau, P. Cunha, and F. Gilbert. 2008. Staphylococcus aureus lipoteichoic acid triggers inflammation in the lactating bovine mammary gland. Vet. Res. 39:52. doi: 10.1051/vetres:2008034. [DOI] [PubMed] [Google Scholar]
- 40.Riollet, C., P. Rainard, and B. Poutrel. 2000. Differential induction of complement fragment C5a and inflammatory cytokines during intramammary infections with Escherichia coli and Staphylococcus aureus. Clin. Diagn. Lab. Immunol. 7:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riollet, C., P. Rainard, and B. Poutrel. 2001. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J. Dairy Sci. 84:1077-1084. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, G., and G. Majno. 1977. Acute inflammation. A review. Am. J. Pathol. 86:183-276. [PMC free article] [PubMed] [Google Scholar]
- 43.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 44.Seegers, H., C. Fourichon, and F. Beaudeau. 2003. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 34:475-491. [DOI] [PubMed] [Google Scholar]
- 45.Siwkowski, A. M., et al. 2004. Effects of antisense oligonucleotide-mediated depletion of tumor necrosis factor (TNF) receptor 1-associated death domain protein on TNF-induced gene expression. Mol. Pharmacol. 66:572-579. [DOI] [PubMed] [Google Scholar]
- 46.Son, D. S., P. F. Terranova, and K. F. Roby. 2010. Interaction of adenosine 3′,5′-cyclic monophosphate and tumor necrosis factor-α on serum amyloid A3 expression in mouse granulosa cells: dependence on CCAAT-enhancing binding protein-β isoform. Endocrinology 151:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the Sv-40 early region promoter. J. Mol. Appl. Genet. 1:327-342. [PubMed] [Google Scholar]
- 48.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strandberg, Y., et al. 2005. Lipopolysaccharide and lipoteichoic acid induce different innate immune responses in bovine mammary epithelial cells. Cytokine 31:72-86. [DOI] [PubMed] [Google Scholar]
- 50.Swanson, K. M., et al. 2009. Transcriptome profiling of Streptococcus uberis-induced mastitis reveals fundamental differences between immune gene expression in the mammary gland and in a primary cell culture model. J. Dairy Sci. 92:117-129. [DOI] [PubMed] [Google Scholar]
- 51.Taponen, S., and S. Pyörälä. 2009. Coagulase-negative staphylococci as cause of bovine mastitis—not so different from Staphylococcus aureus? Vet. Microbiol. 134:29-36. [DOI] [PubMed] [Google Scholar]
- 52.Ulitsky, I., et al. 2010. Expander: from expression microarrays to networks and functions. Nat. Protoc. 5:303-322. [DOI] [PubMed] [Google Scholar]
- 53.Uskokovic, A., et al. 2009. Differences between molecular mechanisms involved in the regulation of haptoglobin gene expression during the acute phase response and dietary restriction. Folia Biol. 55:107-115. [DOI] [PubMed] [Google Scholar]
- 54.Vangroenweghe, F., I. Lamote, and C. Burvenich. 2005. Physiology of the periparturient period and its relation to severity of clinical mastitis. Domest. Anim. Endocrinol. 29:283-293. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., M. Chen, K. P. Wandinger, G. Williams, and S. Dhib-Jalbut. 2000. IFN-β-1b inhibits IL-12 production in peripheral blood mononuclear cells in an IL-10-dependent mechanism: relevance to IFN-β-1b therapeutic effects in multiple sclerosis. J. Immunol. 165:548-557. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Z. J., R. A. Irizarry, R. Gentleman, F. Martinez-Murillo, and F. Spencer. 2004. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909-917. [Google Scholar]
- 57.Yamamoto, M., et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430:218-222. [DOI] [PubMed] [Google Scholar]
- 58.Yang, W., et al. 2008. Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-κB in mammary epithelial cells and to quickly induce TNFα and interleukin-8 (CXCL8) expression in the udder. Mol. Immunol. 45:1385-1397. [DOI] [PubMed] [Google Scholar]
- 59.Yang, W., A. Molenaar, B. Kurts-Ebert, and H. M. Seyfert. 2006. NF-κB factors are essential, but not the switch, for pathogen-related induction of the bovine β-defensin 5-encoding gene in mammary epithelial cells. Mol. Immunol. 43:210-225. [DOI] [PubMed] [Google Scholar]
- 60.Yano, S., T. Banno, R. Walsh, and M. Blumenberg. 2008. Transcriptional responses of human epidermal keratinocytes to cytokine interleukin-1. J. Cell. Physiol. 214:1-13. [DOI] [PubMed] [Google Scholar]
- 61.Zheng, J., A. D. Watson, and D. E. Kerr. 2006. Genome-wide expression analysis of lipopolysaccharide-induced mastitis in a mouse model. Infect. Immun. 74:1907-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.