Abstract
The capsids of herpesviruses, which comprise major and minor capsid proteins, have a common icosahedral structure with 162 capsomers. An electron microscopic study shows that Epstein-Barr virus (EBV) capsids in the nucleus are immunolabeled by anti-BDLF1 and anti-BORF1 antibodies, indicating that BDLF1 and BORF1 are the minor capsid proteins of EBV. Cross-linking and electrophoresis studies of purified BDLF1 and BORF1 revealed that these two proteins form a triplex that is similar to that formed by the minor capsid proteins, VP19C and VP23, of herpes simplex virus type 1 (HSV-1). Although the interaction between VP23, a homolog of BDLF1, and the major capsid protein VP5 could not be verified biochemically in earlier studies, the interaction between BDLF1 and the EBV major capsid protein, viral capsid antigen (VCA), can be confirmed by glutathione S-transferase (GST) pulldown assay and coimmunoprecipitation. Additionally, in HSV-1, VP5 interacts with only the middle region of VP19C; in EBV, VCA interacts with both the N-terminal and middle regions of BORF1, a homolog of VP19C, revealing that the proteins in the EBV triplex interact with the major capsid protein differently from those in HSV-1. A GST pulldown study also identifies the oligomerization domains in VCA and the dimerization domain in BDLF1. The results presented herein reveal how the EBV capsid proteins interact and thereby improve our understanding of the capsid structure of the virus.
The capsids of herpesviruses have an icosahedral structure, with 12 pentons at its vertices and 150 hexons on the surface of the icosahedron with a triangulation number of 16 (5, 32, 41). The capsid structure of herpes simplex virus type 1 (HSV-1) is the best studied of those of all herpesviruses and is usually used as a model for interpreting the capsid structure of other herpesviruses. In HSV-1, the capsomers, which comprise the major capsid protein, VP5, are connected by 320 triplexes that are formed by two minor capsid proteins, VP19C and VP23 (23, 32, 48). Earlier studies have established that the triplex contains a monomer VP19C and a dimer VP23 (34, 39, 40, 45). Moreover, VP26 sits on top of the six VP5 molecules of the hexons, forming a ring on the capsomers (12, 42, 44, 46). So far, the function of VP26 is unclear, as this protein is dispensable for capsid assembly and the lack of this protein does not seem to affect the viability of the virus (13). On the other hand, the homolog of VP26 in Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) is required for capsid assembly (21, 36).
An earlier cryo-electron microscopic (cryo-EM) work revealed the structures of HSV-1 capsid proteins and concluded that VP5 contains 24 α-helices, of which two long helices are located on the top of the protein that interacts with VP26 (45); seven lay horizontally in the central region and participate in oligomerization and the formation of pentameric and hexameric capsomers (6, 45). Furthermore, the structure of the two floor domains in VP5 (3) that form the capsid shell and the structure of capsid proteins in several bacteriophages, including HK97, P22, and T4, are strikingly similar, indicating that the structures are evolutionarily conserved (4). The cryo-EM work also demonstrated that the region from amino acids 27 to 71 in VP5, which is located in one of the floor domains (3), interacts with the scaffolding protein (15, 47). An earlier study found that the HSV-1 minor capsid proteins form a triplex, which contains a dimer VP23 and a monomer VP19C (39). The cryo-EM study also confirmed the presence of such a triplex on the capsid floor (1, 31, 45). Earlier investigations have established that 320 of these triplexes connect the capsomers on the capsid to form the protein shell (32, 48). The results of EM studies also suggested that VP23 is a molten protein whose conformation changes, since the two VP23 molecules in the triplex have different conformations (23, 37). Furthermore, although an interaction between VP23 and VP5 is verifiable by cryo-EM (37), it cannot be demonstrated biochemically (33) or in a yeast two-hybrid study (14), further indicating that the conformation of VP23 changes during capsid assembly.
In EBV, the major capsid protein is the viral capsid antigen (VCA). The minor capsid proteins are predicted to be BDLF1 and BORF1 based on their sequence homology to VP23 and VP19C (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf), respectively (9, 22). An earlier study demonstrated that a mutant virus that lacks one of these proteins cannot produce viral particles (9). Additionally, Henson et al. (21) showed that the expression of BDLF1 and BORF1 along with the other components of the capsid is required for the assembly of the EBV capsid in insect cells. The present investigation finds that VCA interacts with both minor capsid proteins in a triplex and elucidates the interaction domains in these proteins.
MATERIALS AND METHODS
Cell lines and activation of the EBV lytic cycle.
Akata, EBV-negative Akata, and P3HR1 cells were cultured in RPMI 1640 medium in 75-cm2 flasks (Corning). 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) in 10-cm2 culture dishes (Corning). Both media were supplemented with 10% fetal calf serum. P3HR1 cells were treated with 3 ng/ml of 12-O-tetradecanoylphorbol-13-acetate (TPA) and 3 mM sodium butyrate to induce the EBV lytic cycle (8, 10).
Plasmids.
The VCA gene, BcLF1, was amplified by PCR, using maxi-EBV DNA (11) as a template, with primers VCA-F (5′-CCGGGTACCATGGCCTCAAATGAGGGT) and VCA-R (5′-CCGGATATCAAAAACCACCTTATTTCC) (F for forward and R for reverse), which contain a KpnI site and an EcoRV site, respectively, at the 5′ terminus. Amplified DNA fragment was digested by KpnI and EcoRV and inserted into the KpnI-EcoRV sites in pcDNA-HA2 to construct pcDNA-VCA. Plasmids pcDNA-BORF1, pTag-BORF1, and pGEX-BORF1, which expressed HA-BORF1, Flag-BORF1, and GST-BORF1, were constructed by inserting a BORF1 DNA fragment that was amplified using primers BORF1-F (5′-CGCGGATCCGACGCCATGAAGGTCCAG-3′) and BORF1-R (5′-GGCCTCGAGCCTCCTCCTCCTGTTTCC-3′), which contain a BamHI and a XhoI site, respectively, at the 5′ terminus, into the BamHI and XhoI sites in pcDNA-HA2, pTag-2B, and pGEX-4T1. A BDLF1 fragment was amplified by PCR using primers BDLF1-F (5′-CCGGAATTCATGGATTTGAAAGTGGTA-3′) and BDLF1-R (5′-CCGCTCGAGTTATCTTAACCAGCAAGT-3′), digested with EcoRI and XhoI, and inserted into the EcoRI and XhoI sites in pcDNA-HA2, pTag-2B, and pGEX-4T1 to generate pcDNA-BDLF1, pTag-BDLF1, and pGEX-BDLF1, respectively. DNA fragments that encoded the regions in VCA from amino acids 1 to 199 [V(1-199)], 197 to 474 [V(197-474)], 476 to 658 [V(476-658)], 659 to 857 [V(659-857)], 854 to 1032 [V(854-1032)], 1028 to 1213 [V(1028-1213)], and 1208 to 1382 [V(1208-1382)] were amplified by PCR. These DNA fragments were inserted into the BamHI-XhoI sites in pGEX-4T1 and pcDNA-HA2 to express proteins that were fused with glutathione S-transferase (GST) and tagged with a hemagglutinin (HA) sequence at the N terminus, respectively. A plasmid that encoded VCA that lacked the region from amino acids 659 to 857 was generated by inverse PCR, using pcDNA-VCA as a template and primers mVCA-F (5′-GTTCACAAAGGCAAGCCTG) and mVCA-R (5′-GACAATTTGGAGAATGGGAC) (m for mutant). Plasmids expressing a GST fusion protein that contained a segment of BORF1 from amino acids 1 to 121 [O(1-121)], 122 to 249 [O(122-249)], and 244 to 365 [O(244-365)] were constructed by inserting a PCR-amplified DNA fragment into the BamHI-XhoI sites of pGEX-4T1. Plasmids that expressed fragments of BDLF1 that contained the region in BDLF1 from amino acids 1 to 112 [D(1-112)], 105-207 [D(105-207)], and 201 to 302 [D(201-302)] were constructed using the same method.
Antibodies.
Rat monoclonal anti-HA and mouse monoclonal anti-Flag antibodies were purchased from Roche and Sigma-Aldrich, respectively. Rabbit anti-GST antibody was purchased from Santa Cruz Biotechnology. Rabbit anti-IgG antibody was purchase from DakoCytomation. Horseradish peroxidase (HRP)-conjugated anti-mouse IgG and anti-rat IgG antibodies were purchased from KPL. Donkey anti-rabbit IgG antibody that was conjugated with 6-nm gold particles was purchased from EMS. Anti-BDLF1 and anti-BORF1 antibodies were generated by this laboratory in rabbits using GST-BDLF1 and GST-BORF1 that were overexpressed and purified from Escherichia coli. The antibodies were purified using His-BORF1 and His-BDLF1 based on a method described elsewhere (43). Monoclonal anti-FenB antibody was reported elsewhere (43).
Antibody labeling of EBV capsid.
Akata cells were treated with TPA and sodium butyrate for 96 h. The cells were then washed with phosphate-buffered saline (PBS) and fixed with 2.5% glutaraldehyde and 2% paraformaldehyde for 30 min. The cells were further fixed in 2% osmium tetroxide in PBS for 15 min, stained with 1% uranyl acetate for 1 h, and embedded in Spurr's resin. After thin sectioning, the cells were immunolabeled using affinity-purified rabbit anti-BDLF1 and anti-BORF1 antibodies, which were detected using donkey anti-rabbit IgG secondary antibody that was conjugated to 6-nm gold particles. EBV capsid was observed with a JEOL JEM-1200 electron microscope that was operated at an acceleration voltage of 100 kV. Moreover, randomness of the distribution of gold particles in EM images was analyzed by the method of Mayhew et al. (28, 29).
GST pulldown assay.
GST fusion proteins were expressed in E. coli BL21(DE3) and purified using glutathione-Sepharose 4B beads (7). Proteins that were pulled down by the beads were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting (25).
Immunoprecipitation.
A lysate was prepared from 293T cells that had been cotransfected with pcDNA-BORF1 and pTag-BDLF1, pcDNA-VCA and pTag-BORF1, or pcDNA-VCA and pTag-BDLF1 using Lipofectamine 2000 (Invitrogen). Proteins in the lysate were immunoprecipitated using rat anti-HA and mouse anti-Flag M2 antibodies (27). After protein A/G agarose beads (Oncogene) were mixed with the lysate for 1 h at 4°C, the beads were collected by centrifugation and washed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.8], 0.1 M NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.5% NP-40, 0.1% sodium deoxycholate, and 1 mM phenylmethylsulfonyl fluoride). Proteins that were bound to the beads were extracted using electrophoresis sample buffer and analyzed by immunoblotting with anti-HA, anti-Flag, anti-BORF1, or anti-BDLF1 antibodies.
Purification of Flag-tagged proteins and native gel electrophoresis.
293T cells (5 × 105) that had been transfected with pTag2B-BDLF1 or pTag2B-BORF1 were lysed with 1 ml lysis buffer that contained 1 mM EDTA, 5 mM dithiothreitol, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride in phosphate-buffered saline. Flag-tagged proteins in the lysates were purified using M2 beads (Sigma) by the method of Park and Jung (35). Purified proteins in electrophoresis loading buffer, which contained 150 mM Tris-HCl (pH 6.8), 20% glycerol, and 10% β-mercaptoethanol, were separated by gel electrophoresis under native conditions using Tris-Tricine buffer (24).
Cross-linking.
Glutaraldehyde (Ted Pella) (2.3%; 5 μl) was added to 100 μl of solution that contained 5 μg Flag-BDLF1 in 20 mM HEPES (pH 7.5). After incubation for 3 min at 37°C, the reaction was stopped by adding 10 μl of 1 M Tris-HCl (pH 8.0). Proteins were subsequently analyzed by immunoblotting after SDS-polyacrylamide gel electrophoresis (7).
Sucrose gradient sedimentation.
A 25 to 65% continuous sucrose gradient was prepared in Beckman SW41 centrifuge tubes using a gradient station (BioComp). The lysate from 293T cells that had been transfected with pcDNA-VCA or pcDNA-mVCA was prepared using RIPA buffer. His-FenB that had been purified from E. coli BL21(pFB200) (43) was added to the 293T lysates as a control. The lysate mixtures were loaded onto the gradient and centrifuged at 30,000 rpm for 24 h at 4°C using a Beckman SW41 rotor. The gradient was fractionated into 10 fractions using the gradient station (Biocomp); proteins in the fractions were separated by SDS-PAGE and analyzed by immunoblotting using anti-HA and anti-FenB antibodies.
RESULTS
Presence of BDLF1 and BORF1 on EBV capsid.
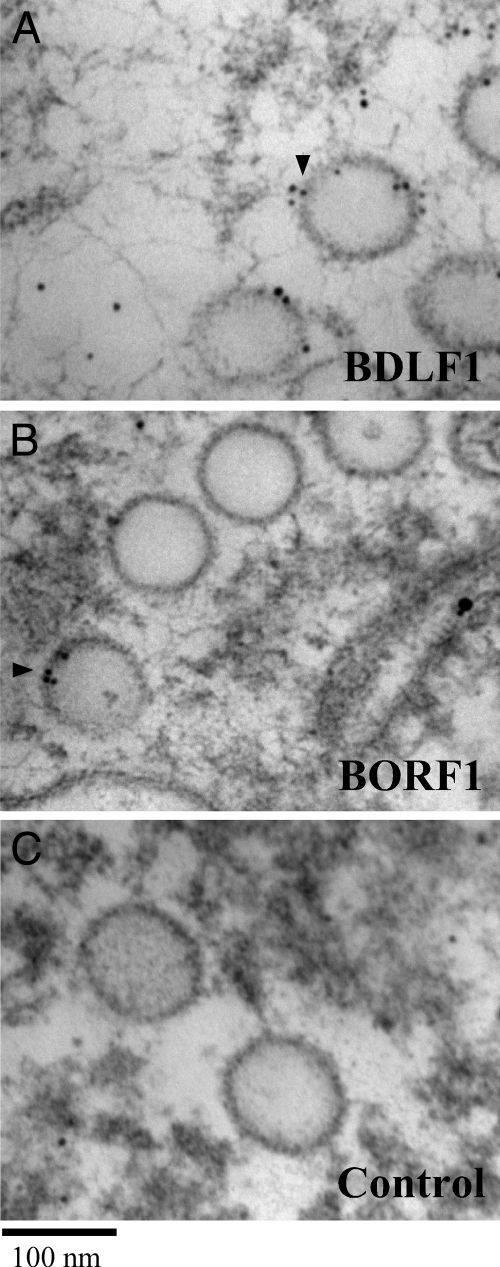
BDLF1 and BORF1 exhibit a low degree of sequence identify with VP23 and VP19C, respectively (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf), and the presence of these two proteins on the EBV capsid was not demonstrated experimentally. In this study, the EBV capsids in Akata cells after lytic induction for 4 days were observed microscopically. EBV capsids were found in the nucleus and had diameters of 80 to 100 nm (Fig. 1). These capsids were labeled using 6-nm gold particles after the cells were sliced into thin sections and treated with anti-BDLF1 (Fig. 1A), anti-BORF1 (Fig. 1B), and anti-IgG (Fig. 1C) antibodies, revealing the presence of BDLF1 and BORF1 on the EBV capsid. The distributions of gold particles in the EM images were further analyzed by an established method (28, 29). Distribution analysis of 200 gold particles in the image obtained from samples treated with anti-IgG antibody revealed that 57 gold particles were on the capsid and 143 were in the remaining area of the nucleus (Table 1). However, cells treated with anti-BDLF1 antibody showed a different distribution type. Gold particles preferentially appeared on the capsid; of the 200 gold particles analyzed, 164 particles were found on the capsid and 36 particles in the remaining area of the nucleus. The relative labeling index (RLI) of the gold particles found on the capsid was 2.88, i.e., significantly higher than that present in the remaining area of the nucleus, 0.25 (Table 1). Preferential labeling of the capsid by gold particles was also observed in the sample treated with anti-BORF1 antibody; 192 of the 200 gold particles counted were on the capsid. An RLI value for labeling of the capsids by anti-BORF1 antibody was 4.27 (Table 1). According to the χ2 test, the distribution of gold particles on the capsids significantly differs from that of random distribution with P values lower than 0.001 (Table 1).
FIG. 1.
Electron micrographs of EBV capsid. EBV capsids were stained with anti-BDLF1 (A), anti-BORF1 (B), and anti-IgG (C), followed by an donkey anti-rabbit IgG antibody conjugated to 6-nm gold beads (black arrowheads). Bar, 100 nm.
TABLE 1.
Localizations of gold-labeled anti-BDLF1 and anti-BORF1 antibodies in the nucleus in Akata cells
| Characteristic | Value for characteristic |
|||||
|---|---|---|---|---|---|---|
| Anti-BDLF1 |
Anti-BORF1 |
|||||
| Nucleusd | Capsid | Total | Nucleus | Capsid | Total | |
| No. of gold particles | 36 | 164 | 200 | 8 | 192 | 200 |
| Expected gold counta | 143 | 57 | 200 | 155 | 45 | 200 |
| Relative labeling indexb | 0.25 | 2.88 | 1 | 0.05 | 4.27 | 1 |
| χ2 valuec | 80.06 | 200.86 | 280.92 | 139.41 | 480.20 | 619.61 |
Particle numbers labeled by anti-IgG antibody.
The relative labeling index is the number of gold particles divided by the number of expected gold particles. A relative labeling index of >1 indicates preferential labeling.
The χ2 test was performed on the (number of gold particles − number of expected gold particles)2/(number of expected particles). According to this test, the distribution of gold particles on capsids was significantly different from that of a random distribution (P < 0.001).
Zone in the nucleus excluding the areas where capsids are.
Interactions among VCA, BORF1, and BDLF1 in vitro.
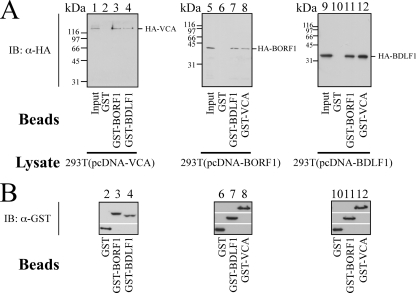
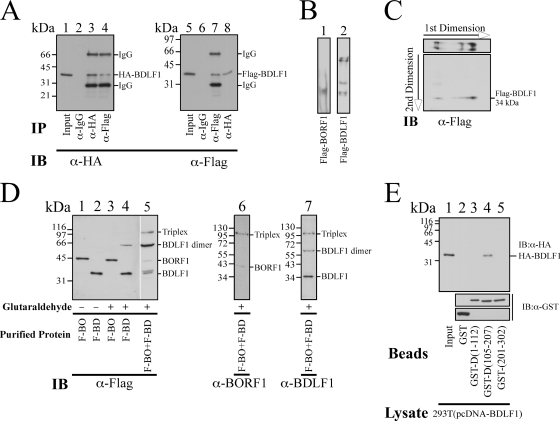
After the presence of BDLF1 and BORF1 in the EBV capsid was demonstrated, a GST pulldown assay using bacterially expressed GST, GST-BDLF1, GST-BORF1, and GST-VCA was conducted to investigate whether BDLF1 and BORF1 interact with each other and with VCA in vitro. Assay results revealed that HA-VCA in the 293T(pcDNA-VCA) lysate (Fig. 2A, lane 1) was pulled down by GST-BORF1 and GST-BDLF1 that were bound to glutathione-Sepharose beads (Fig. 2A, lanes 3 and 4) but not by GST-glutathione-Sepharose beads (Fig. 2A, lane 2), indicating that HA-VCA interacts with both BDLF1 and BORF1. A similar study revealed that GST-BDLF1- and GST-VCA-glutathione-Sepharose beads, but not GST-glutathione-Sepharose beads, pulled down HA-BORF1 from the 293T(pcDNA-BORF1) lysate (Fig. 2A, lanes 6, 7, and 8), showing that HA-BORF1 interacts with BDLF1 and VCA. Additionally, GST-BORF1- and GST-VCA-glutathione-Sepharose beads (Fig. 2A, lanes 11 and 12), but not GST-glutathione-Sepharose beads (Fig. 2A, lane 10) pulled down HA-BDLF1 from the 293T(pcDNA-BDLF1) lysate (Fig. 2A, lanes 10, 11, and 12), revealing that BDLF1 interacted with BORF1 and VCA. Our results further demonstrated that the amounts of GST and GST fusion proteins binding to glutathione-Sepharose beads that were used in the pulldown study were approximately equal (Fig. 2B).
FIG. 2.
Interactions among VCA, BORF1, and BDLF1 in vitro. (A and B) GST-glutathione-Sepharose (lanes 2, 6, and 10), GST-VCA-glutathione-Sepharose (lanes 8 and 12), GST-BORF1-glutathione-Sepharose (lanes 3 and 11), and GST-BDLF1-glutathione-Sepharose (lanes 4 and 7) beads were added to the 293T(pcDNA-VCA) (lanes 1 to 4), 293T(pcDNA-BORF1) (lanes 5 to 8), and 293T(pcDNA-BDLF1) (lanes 9 to 12) lysates. Lanes 1, 5, and 9 were loaded with 5%, 1%, and 1% of cell lysate, respectively. Proteins that bound to the beads were extracted and analyzed by immunoblotting (IB) with anti-HA (α-HA) and anti-GST (α-GST) antibodies.
Interaction among BDLF1, BORF1, and VCA in vivo.
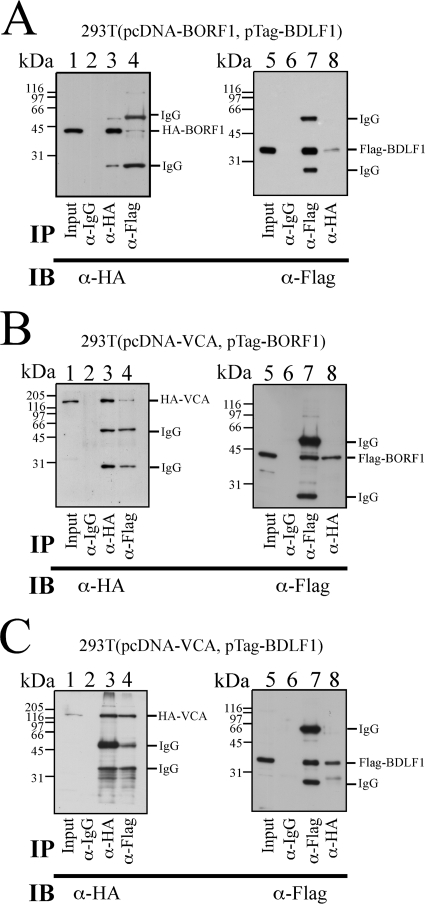
After demonstrating that BDLF1, BORF1, and VCA interact in vitro, this study further investigated whether the three proteins interact with each other in vivo. To achieve this, 293T cells were cotransfected with pcDNA-BORF1 and pTag-BDLF1 to express HA-BORF1 and Flag-BDLF1, respectively. Immunoblotting using anti-HA antibody revealed that HA-BORF1 in the lysate was immunoprecipitated by anti-HA antibody (Fig. 3A, lane 3) and coimmunoprecipitated with Flag-BDLF1 by anti-Flag antibody (Fig. 3A, lane 4). However, anti-IgG antibody did not immunoprecipitate HA-BORF1 (Fig. 3A, lane 2). An experiment using the same lysate revealed that Flag-BDLF1 was immunoprecipitated using anti-Flag antibody (Fig. 3A, lane 7) and coimmunoprecipitated with HA-BORF1 by anti-HA antibody (Fig. 3A, lane 8). However, Flag-BDLF1 was not immunoprecipitated by anti-IgG antibody (Fig. 3A, lane 6), showing that BDLF1 interacts with BORF1. A separate set of experiments using the 293T(pcDNA-VCA, pTag-BORF1) lysate revealed that HA-VCA was immunoprecipitated by anti-HA antibody (Fig. 3B, lane 3) and coimmunoprecipitated with Flag-BORF1 by anti-Flag antibody (Fig. 3B, lane 4); Flag-BORF1 was immunoprecipitated by anti-Flag antibody and coimmunoprecipitated with HA-VCA by anti-HA antibody (Fig. 3B, lanes 7 and 8). Neither HA-VCA nor Flag-BORF1 was immunoprecipitated by anti-IgG antibody (Fig. 3B, lanes 2 and 6), indicating that HA-VCA interacted with Flag-BORF1. HA-VCA was also immunoprecipitated by anti-HA antibody (Fig. 3C, lane 3) and coimmunoprecipitated with Flag-BDLF1 by anti-Flag antibody from the 293T(pcDNA-VCA, pTag-BDLF1) lysate (Fig. 3C, lane 4). Flag-BDLF1 was immunoprecipitated by anti-Flag antibody (Fig. 3C, lane 7) and coimmunoprecipitated with HA-VCA by anti-HA antibody (Fig. 3C, lane 8) from the same lysate. A parallel experiment indicated that anti-IgG antibody immunoprecipitated neither HA-VCA nor Flag-BDLF1 (Fig. 3C, lanes 2 and 6), indicating that HA-VCA interacted with Flag-BDLF1. Furthermore, the HRP-conjugated anti-rat IgG antibody used in this study recognized nonspecifically heavy and light chains of mouse IgG, explaining why two IgG bands appeared if proteins were immunoprecipitated using mouse anti-Flag antibody and immunoblotted using rat anti-HA and HRP-conjugated anti-rat IgG antibodies. However, the IgG bands were undetected if immunoprecipitation was performed using rat anti-HA and immunoblotting were performed using mouse anti-Flag and HRP-conjugated anti-mouse IgG antibodies.
FIG. 3.
Interactions among VCA, BORF1, and BDLF1 in vivo. (A to C) Lysates were prepared from 293T cells that were cotransfected with pcDNA-BORF1 and pTag-BDLF1 (A), pcDNA-VCA and pTag-BORF1 (B), pcDNA-VCA and pTag-BDLF1 (C). Proteins in the lysate were immunoprecipitated (IP) and subsequently analyzed by immunoblotting (IB) with anti-HA (α-HA) and anti-Flag antibodies. Rabbit anti-IgG antibody was used in IP as a negative control. Input lanes were loaded with 1% of cell lysate. Because the HRP-conjugated anti-rat IgG antibody lacked specificity, two mouse IgG bands were detected if a lane was loaded with proteins immunoprecipitated and immunoblotted with anti-Flag and anti-HA antibodies, respectively.
Mapping the interaction domains in VCA, BORF1, and BDLF1.
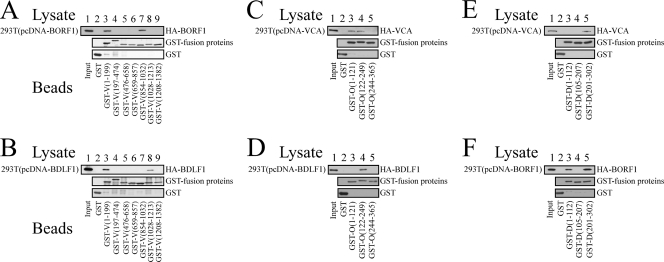
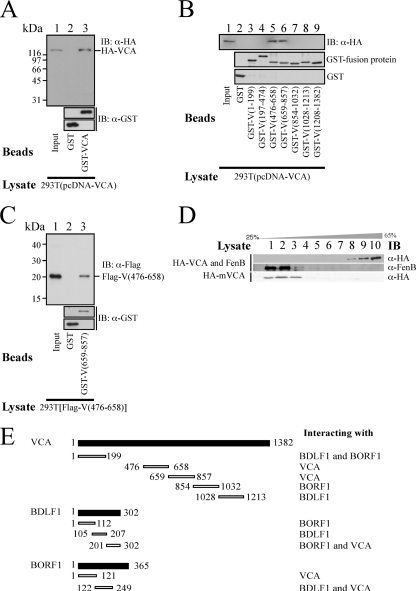
After demonstrating the interaction among BDLF1, BORF1, and VCA in vitro and in vivo, this study further conducted GST pulldown assay to elucidate the interaction domains in these three proteins. Immunoblot analysis revealed that bacterially expressed GST fusion proteins that contained the VCA regions from amino acids 1 to 199 [GST-V(1-199)] and 854 to 1032 [GST-V(854-1032)], when the proteins were bound to glutathione-Sepharose beads, pulled down HA-BORF1 from the 293T(pcDNA-BORF1) lysate (Fig. 4A, lanes 3 and 7). However, neither GST nor GST fusion proteins that contained other regions in VCA pulled down HA-BORF1, indicating that HA-BORF1 interacted with the regions in VCA from amino acids 1 to 199 and 854 to 1032. The results of a similar experiment revealed that GST-V(1-199)- and GST-V(1028-1213)-glutathione-Sepharose beads pulled down HA-BDLF1 from the 293T(pcDNA-BDLF1) lysate (Fig. 4B, lanes 3 and 8), indicating that the regions from amino acids 1 to 199 and 1028 to 1213 in VCA interacted with HA-BDLF1. Additionally, GST fusion proteins that contain a segment of BORF1 from amino acids 1 to 121 [GST-O(1-121)], 122 to 249 [GST-O(122-249)], and 244 to 365 [GST-O(244-365)] were used to analyze the regions in BORF1 that interacted with VCA (Fig. 4C) and BDLF1 (Fig. 4D). Immunoblot analysis revealed that GST-O(1-121)- and GST-O(122-249)-glutathione-Sepharose beads pulled down HA-VCA from the 293T(pcDNA-VCA) lysate (Fig. 4C, lanes 3 and 4) and GST-O(122-249) pulled down HA-BDLF1 from the 293T(pcDNA-BDLF1) lysate (Fig. 4D, lane 4). GST fusion proteins that contained segments of BDLF1 from amino acids 1 to 112 [GST-D(1-112)], 105 to 207 [GST-D(105-207)], and 201 to 302 [GST-D(201-302)] were used to delineate the regions in BDLF1 that interacted with VCA and BORF1. Immunoblot analysis revealed that GST-D(201-302)-glutathione-Sepharose beads pulled down HA-VCA (Fig. 4E, lane 5) from the 293T(pcDNA-VCA) lysate and that GST-D(1-122)- and GST-D(201-302)-glutathione-Sepharose beads pulled down HA-BORF1 from the 293T(pcDNA-BORF1) lysate (Fig. 4F, lanes 3 and 5). Moreover, this study demonstrated that the quantities of the GST fusion proteins binding to the glutathione-Sepharose beads were approximately equal (Fig. 4).
FIG. 4.
Mapping interaction domains in BDLF1, BORF1, and VCA. VCA, BDLF1, and BORF1 were deleted and fused with GST. Fusion proteins were expressed and purified from E. coli. GST pulldown assay was conducted by adding GST fusion proteins that were bound to glutathione-Sepharose beads to the lysate that had been prepared from 293T cells that had been transfected with pcDNA-BORF1 (A and F), pcDNA-BDLF1 (B and D), and pcDNA-VCA (C and E). Proteins pulled down by the beads were detected by immunoblotting with anti-HA and anti-GST antibodies after SDS-polyacrylamide gel electrophoresis. Proteins on the glutathione-Sepharose beads were also extracted, and the quantities of proteins on the beads were examined by immunoblotting using anti-GST antibody. D, O, and V in the protein names, represent BDLF1, BORF1, and VCA, respectively. The numbers in the protein names indicate the positions of amino acids.
Analysis of the BDLF1-BORF1 complex.
Because the HSV-1 minor capsid proteins form a triplex, this study more closely examined whether BDLF1 and BORF1 form a similar structure. Accordingly, pcDNA-BDLF1 and pTag-BDLF1 were cotransfected into 293T cells to express HA-BDLF1 and Flag-BDLF1, respectively (Fig. 5A, lanes 1 and 5). Immunoprecipitation analysis revealed that anti-HA antibody not only immunoprecipitated HA-BDLF1 (Fig. 5A, lane 3) but also coimmunoprecipitated Flag-BDLF1 (Fig. 5A, lane 8). Additionally, anti-Flag antibody immunoprecipitated Flag-BDLF1 (Fig. 5A, lane 7) and coimmunoprecipitated HA-BDLF1 (Fig. 5A, lane 4). Meanwhile, anti-IgG antibody did not immunoprecipitated either HA-BDLF1 or Flag-BDLF1 (Fig. 5A, lanes 2 and 6), indicating that Flag-BDLF1 interacted with HA-BDLF1.
FIG. 5.
Dimerization of BDLF1 and interaction between BDLF1 and BORF1. (A) 293T cells were cotransfected with pcDNA-BDLF1 and pTag-BDLF1. Lysates were prepared from transfected cells, and proteins in the lysates were immunoprecipitated (IP) and detected by immunoblotting (IB) with anti-HA (α-HA) and anti-Flag antibody. Rabbit anti-IgG antibody was used in IP as a negative control. Lanes 1 and 5 were loaded with 1% of cell lysate. (B) Flag-BORF1 (lane 1) and Flag-BDLF1 (lane 2), purified using M2 antibody agarose beads from 293T cells, were separated on a polyacrylamide gel under nondenaturing conditions. (C) Flag-BDLF1 was first separated in a native gel (1st dimension) and then separated by SDS-PAGE (2nd dimension). (D) Purified Flag-BDLF1 (F-BD) and Flag-BORF1 (F-BO) (lanes 1 and 2), each at a concentration of 50 μg/ml, were treated with glutaraldehyde (+) and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting. Lanes 1 and 2 were loaded with Flag-BORF1 and Flag-BDLF1 not treated (−) with glutaraldehyde, respectively. A mixture of Flag-BDLF1 and Flag-BORF1 was treated with glutaraldehyde and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-Flag (lane 5), anti-BORF1 (lane 6), and anti-BDLF1 (lane 7) antibodies. (E) GST-glutathione-Sepharose (lane 2), GST-D(1-112)-glutathione-Sepharose (lane 3), GST-D(105-207)-glutathione-Sepharose (lane 4), and GST-D(201-302)-glutathione-Sepharose beads were added to the 293T(pcDNA-BDLF1) lysate. Proteins bound to the beads were analyzed by IB using anti-HA and anti-GST antibodies. The input lane (lane 1) was loaded with 1% of the lysate.
Flag-BORF1 and Flag-BDLF1 were purified using M2 agarose beads from 293T cells that had been individually transfected with pTag-BORF1 and pTag-BDLF1. The proteins were then separated by polyacrylamide gel electrophoresis under nondenaturing conditions (Fig. 5B). Immunoblotting with anti-Flag antibody revealed a single Flag-BORF1 band (Fig. 5B, lane 1). Unlike Flag-BORF1, Flag-BDLF1 migrated to multiple positions in the gel (Fig. 5B, lane 2). These protein bands were subsequently analyzed in a second dimension under denaturing conditions using an 8% SDS-polyacrylamide gel. Immunoblotting with anti-Flag antibody revealed that these bands migrated to the 34-kDa position (Fig. 5C), revealing that BDLF1 aggregated and formed complexes.
Glutaraldehyde was also added to the solution containing purified Flag-BDLF1 and Flag-BORF1. SDS-polyacrylamide gel electrophoresis and immunoblotting revealed that after glutaraldehyde was added to solution that contained 50 μg/ml Flag-BORF1, glutaraldehyde did not cross-link BORF1 and the migration pattern of Flag-BORF1 in an SDS-polyacrylamide gel was unchanged following treatment (Fig. 5D, lanes 1 and 3), suggesting that the protein did not form a complex. In contrast with Flag-BORF1, Flag-BDLF1, at the same concentration, shifted from the 34-kDa position to the 68-kDa position in the gel following glutaraldehyde treatment (Fig. 5D, lane 4), suggesting that Flag-BDLF1 formed a dimer. When glutaraldehyde was added to a mixture that contained both Flag-BDLF1 and Flag-BORF1, a protein band of 108 kDa was observed (Fig. 5D, lane 5), which is consistent with the size of a complex that contains a dimer Flag-BDLF1 and a monomer Flag-BORF1. Proteins in the blot were further detected using anti-BDLF1 and anti-BORF1 antibodies. The results indicated that anti-BORF1 antibody detected a faint 40-kDa BORF1 band and the 108-kDa triplex band, but not the BDLF1 monomer and dimer bands (Fig. 5D, lane 6). Meanwhile, anti-BDLF1 antibody detected monomer and dimer BDLF1, as well as the triplex, but not the BORF1 band at the 40-kDa position (Fig. 5D, lane 7).
GST pulldown assay was also performed in this study to identify the dimerization domain in BDLF1. GST-D(105-207)-glutathione-Sepharose beads (Fig. 5E, lane 4), but not GST-, GST-D(1-112)- or GST-D(201-302)-glutathione-Sepharose beads, pulled down HA-BDLF1 (Fig. 5E, lanes 2, 3, and 5), revealing that the dimerization domain in BDLF1 was between amino acids 105 and 207 (Fig. 6E). The deletion derivatives of GST-BDLF1 bound to glutathione-Sepharose beads were also extracted. The intensities of the protein bands were nearly the same (Fig. 5E), indicating that the lack of a pulldown of HA-BDLF1 by GST-D(1-112) and GST-D(201-302) was not attributed to the lack of sufficient amounts of GST fusion proteins on glutathione-Sepharose beads.
FIG. 6.
Oligomerization of VCA. (A) Bacterially expressed GST and GST-VCA proteins were bound to glutathione-Sepharose beads and mixed with the 293T(pcDNA-VCA) lysate. Proteins on the beads were pulled down and analyzed by immunoblotting using anti-HA and anti-GST antibodies. (B) GST fusion proteins that contained a segment of VCA were purified from E. coli, bound to glutathione-Sepharose beads, and added to the 293T(pcDNA-VCA) lysate. Proteins that were pulled down by the beads were analyzed by immunoblotting using anti-HA and anti-GST antibodies. Lane 1 was loaded with 1% of cell lysate. (C) A similar pulldown assay was conducted using GST-glutathione-Sepharose (lane 2), GST-V(659-857)-glutathione-Sepharose beads, and the 293T[pTag-V(476-658)] lysate. The input lane (lane 1) was loaded with 1% of cell lysate. (D) FenB was added to the 293T(pcDNA-VCA) lysate, which was then centrifuged through a 25 to 65% sucrose gradient. Following centrifugation, fractionated proteins were analyzed by immunoblotting with anti-VCA and anti-FenB antibodies. A similar experiment was conducted using a lysate that contained mutant VCA, HA-mVCA, which lacked the region between amino acids 659 and 857. After fractionation, proteins in the sucrose solution were separated by SDS-PAGE and analyzed by immunoblotting with anti-HA antibody. (E) The interaction domains, including the dimerization domain in BDLF1 and the oligomerization domain in VCA, are summarized and illustrated. The numbers are the positions of amino acids.
Oligomerization of VCA.
Because the major capsid protein of herpesviruses forms pentameric or hexameric capsomers, GST pulldown assay was performed using GST- and GST-VCA-glutathione-Sepharose beads to determine how VCA oligomerizes. Immunoblot analysis using anti-HA antibody revealed that although HA-VCA was not pulled down by GST-glutathione-Sepharose beads (Fig. 6A, lane 2), the protein was pulled down by GST-VCA-glutathione-Sepharose beads (Fig. 6A, lane 3), indicating that VCA aggregates together and forms a complex. The results of the assay also revealed that GST-V(476-658)- and GST-V(659-857)-glutathione-Sepharose beads pulled down HA-VCA (Fig. 6B, lanes 5 and 6). However, GST fusion proteins that contained the N-terminal and C-terminal regions in VCA, including GST-V(1-199), GST-V(197-474), GST-V(854-1032), GST-V(1028-1213), and GST-V(1208-1382), did not pull down HA-VCA (Fig. 6B). A parallel experiment showed that GST-glutathione-Sepharose beads did not pull down HA-VCA (Fig. 6B, lane 2), showing that the regions from amino acids 476 to 658 and 659 to 857 participated in VCA oligomerization. A similar GST pulldown study revealed that GST-V(659-857) pulled down Flag-V(476-658) from the 293T[pTag-V(476-658)] lysate (Fig. 6C, lane 3), suggesting that VCA oligomerized through an interaction between the two regions. Since the regions from amino acids 476 to 658 and 659 to 857 interact with each other, sucrose gradient sedimentation analysis was performed to elucidate the effect of a deletion in the region between amino acids 659 and 857 on VCA oligomerization. In this study, FenB (43), which has a molecular mass of 154 kDa, similar to that of VCA, was used as a reference and added to the 293T(pcDNA-VCA) lysate before it was loaded on a 25 to 65% sucrose gradient. Following ultracentrifugation, the gradient was fractionated and analyzed by immunoblotting. The results revealed that FenB was distributed on the top of the gradient between fractions 1 and 3 (Fig. 6D). A mutant VCA, HA-mVCA, which lacked the region between amino acids 659 and 857, also sedimented between fractions 1 and 3 (Fig. 6D). However, VCA sedimented toward the bottom of the tube between fractions 8 and 10 (Fig. 6D), revealing the dependence of the two regions from amino acids 476 to 658 and 659 to 857 for VCA oligomerization. Figure 6E summarizes the interaction domains in BDLF1, BORF1, and VCA.
DISCUSSION
The capsids of herpesviruses have a common icosahedral structure that contains 12 pentameric and 150 hexameric capsomers. In HSV-1, these capsomers contain the major capsid protein, VP5, and are connected to each other by triplexes that contain two minor capsid proteins, VP19C and VP23 (34, 40, 45). The structure of the HSV-1 capsid, which was elucidated by cryo-EM and X-ray crystallographic analyses (3, 4, 6, 45), is often used as a model of the structure of other herpesviruses. Although the EM results yield useful information on the structure of the capsid, the method has its limitation and leaves the exact nature of the interaction of the capsid proteins with each other mostly unclear. The only information available on the interactions of HSV-1 capsid proteins was obtained from yeast two-hybrid studies (1, 14, 18).
VCA has been established immunologically as the major capsid protein in EBV (16, 30) and has been used as a diagnostic marker for EBV-related cancers, such as nasopharyngeal carcinoma (17, 20, 26). The minor capsid proteins of EBV have been predicted to be BDLF1 and BORF1 (9, 22), based on their sequence identities of 19.2% and 16.6% with VP23 and VP19C, respectively (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf). However, the identities of these two proteins as the minor capsid proteins are not completely certain, because their sequence identities are low; in particular, the sequence identity between BORF1 and VP19C is only 16.6%, making the regions of alignment with the identical amino acid sparse (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf) and making the status of BORF1 as one of the capsid proteins uncertain. Therefore, this study investigates whether BDLF1 and BORF1 are indeed components of the EBV capsid.
An electron microscope was used to study the EBV capsids in the nuclei of Akata cells 4 days after lytic induction. Capsids were found in the nuclei (Fig. 1). They had a diameter of 80 to 100 nm, which is consistent with the size of the EBV capsids observed in another study (21). In this study, anti-BDLF1 and anti-BORF1 antibodies were used to immunolabel the capsids, which were detected using a donkey anti-rabbit secondary antibody that was conjugated to 6-nm gold particles (Fig. 1). Statistical analysis also reveals that distribution of gold particles is not random and is preferentially located on EBV capsids (Table 1), indicating that BDLF1 and BORF1 are components of the EBV capsid.
Earlier studies have shown that the two minor capsid proteins of HSV-1 formed a triplex, in which VP19C is a monomer and VP23 is a dimer (34, 39, 45). This study finds that BDLF1 and BORF1 also form a similar triplex structure. The evidence that BDLF1 forms a dimer is that HA-BDLF1 and Flag-BDLF1 are coimmunoprecipitated (Fig. 5A), multiple BDLF1 bands are detected under native conditions after gel electrophoresis (Fig. 5B and C), the size of purified Flag-BDLF1 is doubled from 34 kDa to 68 kDa upon cross-linking with glutaraldehyde (Fig. 5D, lane 4), and the dimerization domain is mapped to the region between amino acids 105 and 207 (Fig. 5E, lane 4). Additionally, after purified Flag-BORF1 and Flag-BDLF1 were mixed and cross-linked by glutaraldehyde, a protein band of 108 kDa, which is consistent with the size of a complex that contains a Flag-BDLF1 dimer and a Flag-BORF1 monomer, is detected, suggesting that BORF1 and BDLF1 form a triplex. In this triplex, both N- and C-terminal regions in BDLF1 interact with the region between amino acids 122 and 249 in BORF1 (Fig. 6E). In fact, in the HSV-1 triplex, the N- and C-terminal regions in VP23, from amino acids 22 to 67 and 103 to 310, also interact with the middle region from amino acid 193 to 310 in VP19C (1, 34), indicating a structural similarity between the two triplexes.
Although the minor capsid proteins from EBV and HSV-1 form similar triplexes, these triplexes may interact differently with their respective major capsid proteins. The evidence was obtained from an earlier yeast two-hybrid study, which demonstrated that the middle region in VP19C from amino acids 242 to 330 interacts with VP5 (1, 14). However, this study reveals that both the N-terminal and middle regions in BORF1 from amino acids 1 to 121 and 122 to 249, respectively, interact with VCA (Fig. 6E). Additionally, although a cryo-EM study revealed that the HSV-1 VP23 in the triplex and VP5 interact and both lie on the capsid floor (31, 45), a physical interaction between VP23 and VP5 could not be verified biochemically in vitro or in vivo (33). Hence, VP23 has been suggested to be a molten protein that undergoes a conformational change before capsid assembly (23). In the case of EBV, this investigation establishes that VCA interacts with the C-terminal region from amino acids 201 to 302 in BDLF1 (Fig. 6E). This study also suggests that the BORF1 region between amino acids 122 and 249 may contain two subregions that interact with two regions in BDLF1 (Fig. 6E).
As is generally known, the major capsid protein oligomerizes to form capsomers (6, 31, 45, 48). A sucrose gradient study reveals that VCA, which was expressed in 293T cells, sediments toward the bottom of a 25 to 65% sucrose gradient (Fig. 6D). The distribution pattern of VCA in the gradient differs from that of FenB, which has a molecular mass similar to that of VCA (43), and a mutant VCA, HA-mVCA, that lacks one of the oligomerization domains from amino acids 659 to 857 (Fig. 6D). Furthermore, this study finds that VCA cannot enter a 6% polyacrylamide gel under native conditions (data not shown), suggesting that VCA aggregates into large complexes. The distribution of VCA in the gradient and the electrophoresis results reveal that VCA oligomerizes into large complexes. The fact that purified HSV-1 capsid proteins form a capsid in vitro (22, 45) also supports the claim that VCA may self-oligomerize into capsomers. Additionally, the results of the sucrose gradient sedimentation study reveal little VCA on the top of the gradient, implying that most of the VCA in the solution is oligomerized (Fig. 6D). Because loading a sufficient amount of VCA onto the gradient is required for detecting the protein in the gradient by immunoblotting, the VCA concentration loaded onto the gradient is high, possibly promoting oligomerization of the protein. Additionally, the VP5 protein of HSV-1 has two floor domains (3), which have a structure similar to that of the capsid protein of several bacteriophages, including HK97, P22, and T4 (4). On the basis of the sequence identity between VP5 and VCA, these floor domains in EBV are predicted to be located in the N-terminal and C-terminal regions from amino acid 1 to 313 and 1020 to 1297, respectively (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf). Structure modeling using Swiss-Model Workspace (2, 19, 38) reveals that the floor domains in VP5 and VCA are similar, implying that the structure of the domains in EBV is conserved in evolution (4).
The results of a GST pulldown assay reveal that the region from amino acids 476 to 658 interacts with the region from amino acids 659 to 857 in VCA (Fig. 6C and E), suggesting that VCA molecules interlock to form a capsomer via an interaction between these two regions. In fact, earlier studies showed that similar regions in VP5 also interact (45), suggesting the similarity between VP5 and VCA in oligomerization. Additionally, cryo-EM and X-ray crystallographic studies revealed that two VP5 molecules in an HSV-1 capsomer come into contact via a group of seven short parallel α-helices in the middle region (6, 45). These α-helices closely interact with each other to constrict the channels in the centers of the pentons and hexons (6, 45). Sequence analysis reveals that the oligomerization domain in VCA is the region in the protein with the highest sequence identity with VP5 (see Fig. S1 posted at http://gibms.cgu.edu.tw/ezfiles/41/1041/attach/38/pta_2420_1729080_05410.pdf). The same analysis also reveals several α-helices between amino acids 578 and 821, which contain the oligomerization domains in VCA, suggesting that VCA may oligomerize in a manner similar to VP5. Furthermore, VCA may interact differently to form pentons and hexons because in VP5, Glu-684 and Arg-691 in one VP5 molecule interact with Arg-660 and Glu-620 in an adjacent VP5, respectively, to form hexons; Glu-684 and Arg-691 in one VP5 interact with His-944 and Asp-946 in another VP5, respectively, to form pentons (6).
This investigation demonstrates that BDLF1 and BORF1 form a triplex similar to that of HSV-1. The interaction of the EBV triplex with VCA suggests that the function of the triplex is similar to that of HSV-1 in connecting capsomers on a capsid. This study also shows the interaction domains among the EBV capsid proteins, which are valuable in elucidating the process of EBV capsid assembly.
Acknowledgments
This research is supported by the National Science Council of the Republic of China, Taiwan (NSC99-3112-B-182-005), and Chang-Gung Memorial Hospital (CMRPD160113).
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Adamson, W. E., D. McNab, V. G. Preston, and F. J. Rixon. 2006. Mutational analysis of the herpes simplex virus triplex protein VP19C. J. Virol. 80:1537-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 3.Baker, M. L., et al. 2003. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J. Mol. Biol. 331:447-456. [DOI] [PubMed] [Google Scholar]
- 4.Baker, M. L., W. Jiang, F. J. Rixon, and W. Chiu. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, T. S., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1990. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J. Virol. 64:563-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, B. R., M. L. Baker, F. J. Rixon, W. Chiu, and F. A. Quiocho. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L. K., et al. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803-38812. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, Y. F., et al. 2007. A comprehensive library of mutations of Epstein Barr virus. J. Gen. Virol. 88:2463-2472. [DOI] [PubMed] [Google Scholar]
- 10.Davies, A. H., R. J. Grand, F. J. Evans, and A. B. Rickinson. 1991. Induction of Epstein-Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J. Virol. 65:6838-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., J. C. Akpa, and S. Person. 2003. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J. Virol. 77:391-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 14.Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. [DOI] [PubMed] [Google Scholar]
- 15.Desai, P., and S. Person. 1999. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261:357-366. [DOI] [PubMed] [Google Scholar]
- 16.Dolken, G., M. H. Moar, and G. Klein. 1979. Detection of the Epstein-Barr virus-associated antigens EA (early antigen) and VCA (viral capsid antigen) by direct or indirect binding of iodinated antibodies to antigen immobilized in polyacrylamide gel. Eur. J. Cancer 15:821-824. [DOI] [PubMed] [Google Scholar]
- 17.Fachiroh, J., S. J. Stevens, S. M. Haryana, and J. M. Middeldorp. 2010. Combination of Epstein-Barr virus scaffold (BdRF1/VCA-p40) and small capsid protein (BFRF3/VCA-p18) into a single molecule for improved serodiagnosis of acute and malignant EBV-driven disease. J. Virol. Methods 169:79-86. [DOI] [PubMed] [Google Scholar]
- 18.Fossum, E., et al. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Gunven, P., G. Klein, G. Henle, W. Henle, and P. Clifford. 1970. Epstein-Barr virus in Burkitt's lymphoma and nasopharyngeal carcinoma. Antibodies to EBV associated membrane and viral capsid antigens in Burkitt lymphoma patients. Nature 228:1053-1056. [DOI] [PubMed] [Google Scholar]
- 21.Henson, B. W., E. M. Perkins, J. E. Cothran, and P. Desai. 2009. Self-assembly of Epstein-Barr virus capsids. J. Virol. 83:3877-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannsen, E., et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkitadze, M. D., et al. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirschner, A. N., J. Omerovic, B. Popov, R. Longnecker, and T. S. Jardetzky. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J. Virol. 80:9444-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, Y. H., Y. F. Chiu, W. H. Wang, L. K. Chang, and S. T. Liu. 2008. Activation of the ERK signal transduction pathway by Epstein-Barr virus immediate-early protein Rta. J. Gen. Virol. 89:2437-2446. [DOI] [PubMed] [Google Scholar]
- 26.Levine, P. H., et al. 1981. The reliability of IgA antibody to Epstein-Barr virus (EBV) capsid antigen as a test for the diagnosis of nasopharyngeal carcinoma (NPC). Cancer Detect. Prev. 4:307-312. [PubMed] [Google Scholar]
- 27.Liu, S. T., et al. 2006. Sumoylation of Rta of Epstein-Barr virus is preferentially enhanced by PIASxbeta. Virus Res. 119:163-170. [DOI] [PubMed] [Google Scholar]
- 28.Mayhew, T. M. 2009. Quantifying immunogold localization patterns on electron microscopic thin sections of placenta: recent developments. Placenta 30:565-570. [DOI] [PubMed] [Google Scholar]
- 29.Mayhew, T. M., C. Muhlfeld, D. Vanhecke, and M. Ochs. 2009. A review of recent methods for efficiently quantifying immunogold and other nanoparticles using TEM sections through cells, tissues and organs. Ann. Anat. 191:153-170. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, J. L., C. M. Doyle, M. V. Land, and P. L. Devine. 1998. Comparison of commercial ELISA for detection of antibodies to the viral capsid antigen (VCA) of Epstein-Barr virus (EBV). Dis. Markers 13:245-249. [DOI] [PubMed] [Google Scholar]
- 31.Newcomb, W. W., et al. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 32.Newcomb, W. W., et al. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson, P., et al. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 75:1091-1099. [DOI] [PubMed] [Google Scholar]
- 34.Okoye, M. E., G. L. Sexton, E. Huang, J. M. McCaffery, and P. Desai. 2006. Functional analysis of the triplex proteins (VP19C and VP23) of herpes simplex virus type 1. J. Virol. 80:929-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, S. G., and G. Jung. 2001. Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J. Virol. 75:6962-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perkins, E. M., et al. 2008. Small capsid protein pORF65 is essential for assembly of Kaposi's sarcoma-associated herpesvirus capsids. J. Virol. 82:7201-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 73:6821-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 72:3944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trus, B. L., et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 41.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. U. S. A. 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingfield, P. T., et al. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J. Virol. 71:8955-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, C. Y., et al. 2007. Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases. J. Biol. Chem. 282:5608-5616. [DOI] [PubMed] [Google Scholar]
- 44.Yu, X., et al. 2005. Three-dimensional localization of the smallest capsid protein in the human cytomegalovirus capsid. J. Virol. 79:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Z. H., et al. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, Z. H., et al. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, Z. H., et al. 1998. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc. Natl. Acad. Sci. U. S. A. 95:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, Z. H., B. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]