Abstract
The host adaptation of influenza virus is partly dependent on the sialic acid (SA) isoform bound by the viral hemagglutinin (HA). Avian influenza viruses preferentially bind the α-2,3 SA and human influenza viruses the α-2,6 isoform. Each isoform is predominantly associated with different surface epithelial cell types of the human upper airway. Using recombinant HAs and human tracheal airway epithelial cells in vitro and ex vivo, we show that many avian HA subtypes do not adhere to this canonical view of SA specificity. The propensity of avian viruses to adapt to human receptors may thus be more widespread than previously supposed.
Influenza virus causes significant respiratory disease in the human population, but the natural reservoir is aquatic birds (41). Human influenza virus pandemics arise when viruses bearing a novel hemagglutinin (HA) protein are introduced into the population as transmissible viruses either through direct zoonoses from birds or swine or by reassortment with a circulating human virus (15). Little or no immunity in the global population to a novel HA protein results in widespread infection and the associated potential for significant morbidity and mortality. Human pandemics in the 20th century were caused by viruses with HA subtypes H1, H2, and H3 (15) and in the 21st century by a novel form of H1 originating in swine (25). However, there are 16 HA subtypes circulating in the avian population, inviting the question of what the potential may be for lesser-studied HA subtypes to cause human disease. HA is an envelope glycoprotein of influenza virus and facilitates viral attachment and subsequent entry of the virion into host cells via interaction with sialic acid (SA) moieties on the cell surface (24). Terminal sialic acid linkage to the penultimate sugar of the glycan chain occurs via carbon 3 or carbon 6, giving the two isomers α-2,3 and α-2,6 SA, respectively (32). The human upper respiratory tract, the primary site of influenza virus replication in the human host, has abundant SA of the α-2,6 isomer, while in the avian gut, the site of replication in avian hosts, the α-2,3 conformer predominates, as demonstrated by isoform-specific lectin staining (22, 23, 38). Among a variety of factors that influence tropism, a major host adaptation facilitating avian influenza virus infection and transmission among humans is mutation in the receptor binding pocket of the HA protein to allow efficient α-2,6 SA rather than α-2,3 SA binding (19, 21, 33). This has not occurred in the highly pathogenic H5N1 subtype, despite nearly 500 people having been infected. Some H7 avian viruses and certain H9 mutant strains transmit in mammalian model systems and demonstrate an expansion of receptor binding capacity for α-2,6 SA (26, 39). Eurasian and North American H6 subtypes can infect mice and ferrets without prior adaptation (9), and swine lineage H4 viruses that possess the α-2,6 SA binding motif L226/S228 efficiently infect human airway primary epithelial cells (2). The extent to which other HA subtypes interact with different SA isoforms and consequently have the potential for human infection is unclear, and receptor screening for this could be a beneficial component of pandemic planning. Although screening could use viruses, the biohazard posed by an emerging virus may be unknown. In addition, cell-based propagation of influenza virus isolates immediately imposes selection on the virus population for the predominant receptor present (7, 11). At the level of a suitable substrate, the exact form of α-2,6 sialyated glycan(s) bound by influenza virus in the human respiratory tract is undetermined and may depend on topology (4, 28). Red blood cells and synthetic sialic acid mimics do not accurately reflect this microenvironment or the type and distribution of glycans present (8, 14, 16, 42, 43, 44). We and others have previously reported that the ciliated cells in well-differentiated human airway epithelium (HAE) or ex vivo human tracheal sections showed a lectin staining pattern indicating predominant expression of α-2,3 SA, whereas lectin staining of nonciliated cells indicated α-2,6 SA (31, 37). Moreover, the binding of soluble HA to the different cell types in these epithelia reflected the SA binding preference of the HA derived from glycan binding profiles (29, 30). Viruses that transmit between humans, including pandemic H1N1 virus and 1918 virus, have been shown to attach more readily to nonciliated cells of the upper respiratory tract than avian viruses (21, 28, 33, 34). Here, we assess the binding of a panel of lesser-studied avian HA subtypes to cells derived from the human upper respiratory tract.
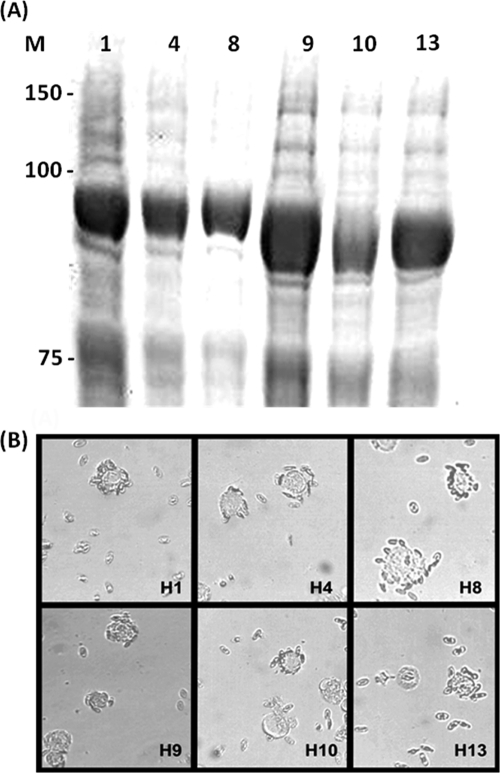
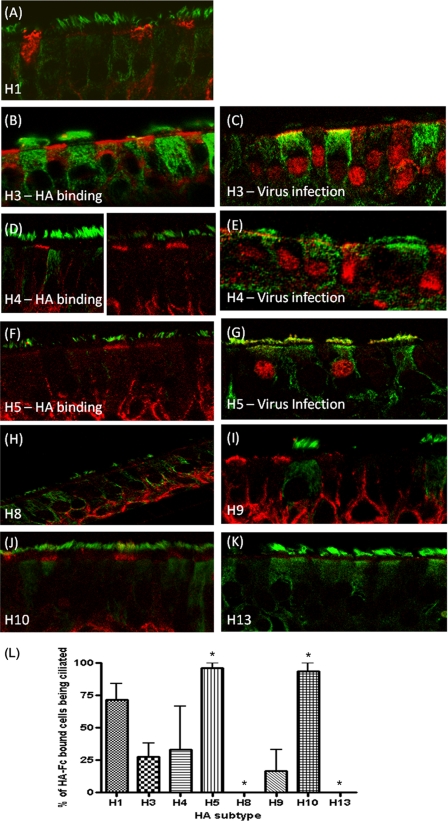
Avian influenza virus HA proteins comprising the subtypes H4 (A/Pintail/Alberta/210/99; GenBank accession no. AY633356.1), H5 (A/Vietnam/1194/04; GenBank accession no. AY651333), H8 (A/Turkey/Ontario/6118/1968; GenBank accession no. J02089.1), H9 (A/Hong Kong/1073/99; GenBank accession no. AB080226.1), H10 (A/Shorebird/DE/10/2004; GenBank accession no. CY005930.1), and H13 (A/Shorebird/DE/2004; GenBank accession no. CY006001.1) and two human influenza virus HA proteins, H1 (A/New York/221/03; GenBank accession no. CY002984.1) and H3 (A/Panama/2007/99; GenBank accession no. DQ865956.1), were generated as described previously (1, 5, 18). Expression levels were similar and all HAs were competent for SA binding by hemadsorption with either turkey or guinea pig red blood cells (Fig. 1). Formalin-fixed sections of HAE, containing a mixture of ciliated, nonciliated, and mucin-containing “goblet” cells (31), were incubated with HA-Fc and an anti-human Fc conjugate (Fig. 2) (1). Human influenza virus H3 HA bound mainly nonciliated cells (Fig. 2B), while avian H5 HA bound exclusively ciliated cells (Fig. 2F) as before (1). H10 displayed a binding profile similar to that of the H5 HA, while H8 and H13 failed to bind to any cells in HAE. Avian H4 and H9 HAs, however, showed considerable binding to nonciliated cell types (Fig. 2D and I). Quantification of the relative binding for each HA was obtained by blinded counting of the number of each cell type bound from five different fields of view and subjected to analysis of variance (ANOVA) (Fig. 2L). As the cellular tropism indicated by the binding of some HAs, notably H4, was unexpected, live HAE cultures were also infected with a selection of viruses whose HA genes were identical to those used for expressed protein binding assays. Cultures were inoculated by A/Panama/2007/99 (H3N2) and two recombinant viruses bearing the H4 and H5 HAs with the internal genes of A/PR/8/34 and fixed 24 h later, and histological sections were prepared and immunostained with an antinucleoprotein serum. The pattern of infection exactly mirrored the binding phenotype shown by the recombinant HA-Fc proteins. For example, the H4 virus clearly infected some nonciliated cells, whereas those cells infected by the H5 virus always possessed cilia (Fig. 2E and G). In order to confirm that HAE cultures, a tractable model of human upper airway epithelia (17), were representative of tracheal epithelial surfaces, HA-Fc proteins were also used to probe ex vivo sections of human tracheal airway tissue. The binding phenotypes observed were subtly different from those observed for HAE sections. H3 HA bound nonciliated cells to ciliated cells with a 2:1 ratio (Fig. 3 B and I). In contrast, 81% of all cells bound by avian H5 were ciliated (Fig. 3D and I). Human H1 HA bound more nonciliated than ciliated cells, similar to H3 (Fig. 3A and I). None of the avian virus HAs bound to as many cells in the tracheal sections as did the human virus HAs. However, as before, H4 and H9 displayed a binding phenotype similar to that of the human HAs; that is, they bound mainly to nonciliated cells but still with a significant proportion (7 and 34%) of binding occurring on ciliated cells (Fig. 3C, F, and I). The H8 HA, which had failed to bind to sections of HAE, bound to tracheal sections, predominantly to ciliated cells (57%) (Fig. 3E), akin to the H10 HA (54.5%) (Fig. 3G). As with the HAE cultures, the H13 HA failed to bind to any cell type in the tracheal sections (Fig. 3H) despite being competent for hemadsorption (Fig. 1B). The differences observed with the trachea section model were not significant by the statistical test used, but the trend observed was the same as for the HAE section binding. Tracheal sections from different donors are more variable in the types and distribution of sialic acid on their surface, as the age of the subject, genetic makeup, and health status at donation may all have an effect. In contrast, HAE cultures are stricter in their cell-tropic sialic acid linkage due to the uniform stimulus applied during the in vitro differentiation procedure. In addition, a glycan with terminal sialic acid present on human tracheal epithelium may not be faithfully represented in HAE cultures.
FIG. 1.
Expression and function of HAs as HA-Fc fusion proteins. (A) HA-Fc fusion proteins detected by Coomassie blue staining of a 10% SDS-PAGE gel of insect cell supernatant following recombinant baculovirus infection. (B) Red blood cell binding as a measure of biological competence. The assays used turkey red blood cells with the exception of H1, which used guinea pig. HAs from A/Panama/2007/99 (H3) and A/Vietnam/1194/04 (H5) have been described previously (1). M, molecular size markers.
FIG. 2.
Recombinant HA binding and virus infection of human airway cultures. (A, B, D, F, and H to K) HAE culture sections were probed with HA-Fc proteins from human and avian influenza virus strains. Ciliated cells were identified using anti-acetylated α-tubulin (green), and the HA-Fc proteins were visualized with anti-human Fc (red). (C, E, and G) HAE cultures were infected with a multiplicity of infection (MOI) of 1 for 24 h with H3N2 A/Panama/2007/99, H4/PR8, or H5/PR8 and then probed with anti-NP serum (red). Images are representative of multiple probed sections. (L) The percentage of recombinant HA protein-bound epithelial cells that were ciliated (±standard error of the mean [SEM]). ANOVA was applied relative to H3. *, P < 0.05.
FIG. 3.
Recombinant HA binding to ex vivo human tracheal sections. (A to H) Ex vivo human trachea sections were probed with HA-Fc proteins. Ciliated cells were identified using anti-acetylated α-tubulin (green), and the HA-Fc proteins were visualized with anti-human Fc (red). Images are representative of multiple probed sections. (I) The percentage of recombinant HA protein-bound epithelial cells that were ciliated (±SEM). ANOVA was applied relative to H3. *, P < 0.05.
Of the panel of avian HAs investigated here, the H4 and H9 HAs exhibited human influenza virus-like binding profiles. This may be explained for the H9 HA by the fact that the sequence chosen contains a 226L residue previously described as facilitating α-2,6 SA binding and transmission in ferrets (37, 39). No such change was present in the H4 sequence used, yet it preferentially bound nonciliated cell types known to display α-2,6 SA, a phenotype of potential human transmissibility (28, 33, 34). Both H8 and H10 HA proteins also bound nonciliated cells in the human tracheal epithelium sections, although the predominant target cells were ciliated. Binding patterns were not an artifact of the experimental system, as they correlated with actual virus infection where tested. While entry of H4 virus to HAE cells does not guarantee a full cycle of replication and spread, these observations suggest that the potential for a human-tropic virus emerging from an avian reservoir may be greater than previously supposed. To properly assess the pandemic potential, not only SA binding phenotype but also the relative abundance of each subtype in the wild should be considered. Surveys of avian influenza virus in the wild bird populations of Europe and North America estimate an incidence range of from 2 to 30% (3, 6, 10, 12, 13, 20, 27, 36, 40), and in Europe, subtypes H12, H6, and H4 are the most prevalent (Fig. 4). The binding and infection pattern exhibited by H4 in our assays taken together with studies of H4 viruses already established in pigs (2) suggest that this subtype in particular may be poised for a species jump. The use of tagged HAs sourced directly without virus passage and the available HAE culture system may represent a tractable and informative tool for assessing pandemic potential among influenza viruses.
FIG. 4.
Avian influenza virus diversity in the wild bird population. Graphical representations of recent avian influenza virus surveillance in the wild bird populations of predominantly Europe and America, categorized by subtype (as reported in references 3, 6, 10, 12, 13, 20, 27, 36, and 40).
Acknowledgments
This work was funded by a grant from the United Kingdom Medical Research Council (G0500597).
All confocal work was undertaken in the Henry Wellcome Imaging Centre at St. Mary's Imperial College London.
Footnotes
Published ahead of print on 24 November 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ayora-Talavera, G., et al. 2009. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One 4:e7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A. C., M. G. Busch, A. I. Karasin, N. Bovin, and C. W. Olsen. 2008. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for alpha2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J. Virol. 82:8204-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busquets, N., et al. 2010. Influenza A virus subtypes in wild birds in North-Eastern Spain (Catalonia). Virus Res. 149:10-18. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekaran, A., et al. 2008. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 26:107-113. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., X. Xu, and I. M. Jones. 2007. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferro, P. J., et al. Multiyear surveillance for avian influenza virus in waterfowl from wintering grounds, Texas coast, U.S.A. Emerg. Infect. Dis. 16:1224-1230. [DOI] [PMC free article] [PubMed]
- 7.Gambaryan, A. S., et al. 1997. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology 232:345-350. [DOI] [PubMed] [Google Scholar]
- 8.Gamblin, S. J., et al. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838-1842. [DOI] [PubMed] [Google Scholar]
- 9.Gillim-Ross, L., et al. 2008. Avian influenza H6 viruses productively infect and cause illness in mice and ferrets. J. Virol. 82:10854-10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Globig, A., et al. 2009. Ducks as sentinels for avian influenza in wild birds. Emerg. Infect. Dis. 15:1633-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govorkova, E. A., S. Kodihalli, I. V. Alymova, B. Fanget, and R. G. Webster. 1999. Growth and immunogenicity of influenza viruses cultivated in Vero or MDCK cells and in embryonated chicken eggs. Dev. Biol. Stand. 98:39-51, 73-74. [PubMed] [Google Scholar]
- 12.Gronesova, P., P. Kabat, A. Trnka, and T. Betakova. 2008. Using nested RT-PCR analyses to determine the prevalence of avian influenza viruses in passerines in western Slovakia, during summer 2007. Scand. J. Infect. Dis. 40:954-957. [DOI] [PubMed] [Google Scholar]
- 13.Hanson, B. A., et al. 2005. Avian influenza viruses and paramyxoviruses in wintering and resident ducks in Texas. J. Wildl. Dis. 41:624-628. [DOI] [PubMed] [Google Scholar]
- 14.Harvey, R., A. C. Martin, M. Zambon, and W. S. Barclay. 2004. Restrictions to the adaptation of influenza A virus H5 hemagglutinin to the human host. J. Virol. 78:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591-600. [DOI] [PubMed] [Google Scholar]
- 16.Ilyushina, N. A., E. A. Govorkova, T. E. Gray, N. V. Bovin, and R. G. Webster. 2008. Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog. 4:e1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kesimer, M., et al. 2009. Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am. J. Physiol. Lung Cell. Mol. Physiol. 296:L92-L100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathewson, A. C., et al. 2008. Interaction of severe acute respiratory syndrome-coronavirus and NL63 coronavirus spike proteins with angiotensin converting enzyme-2. J. Gen. Virol. 89:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munster, V. J., et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pappas, C., et al. 2010. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS One 5:e11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai, S. P., and C. W. Lee. 2010. Species and age related differences in the type and distribution of influenza virus receptors in different tissues of chickens, ducks and turkeys. Virol. J. 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinya, K., et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435-436. [DOI] [PubMed] [Google Scholar]
- 24.Skehel, J. J., and D. C. Wiley. 2002. Influenza haemagglutinin. Vaccine 20(Suppl. 2):S51-S54. [DOI] [PubMed] [Google Scholar]
- 25.Smith, G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 26.Song, H., H. Wan, Y. Araya, and D. R. Perez. 2009. Partial direct contact transmission in ferrets of a mallard H7N3 influenza virus with typical avian-like receptor specificity. Virol. J. 6:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spackman, E., et al. 2009. Characterization of low pathogenicity avian influenza viruses isolated from wild birds in Mongolia 2005 through 2007. Virol. J. 6:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srinivasan, A., et al. 2008. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 105:2800-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, J., O. Blixt, J. C. Paulson, and I. A. Wilson. 2006. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 4:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, J., et al. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404-410. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, C. I., W. S. Barclay, M. C. Zambon, and R. J. Pickles. 2006. Infection of human airway epithelium by human and avian strains of influenza a virus. J. Virol. 80:8060-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traving, C., and R. Schauer. 1998. Structure, function and metabolism of sialic acids. Cell Mol. Life Sci. 54:1330-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumpey, T. M., et al. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655-659. [DOI] [PubMed] [Google Scholar]
- 34.van Riel, D., et al. 2010. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 176:1614-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Riel, D., et al. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallensten, A., et al. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13:404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan, H., and D. R. Perez. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. 81:5181-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan, H., and D. R. Perez. 2006. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan, H., et al. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3:e2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, R., et al. 2008. Examining the hemagglutinin subtype diversity among wild duck-origin influenza A viruses using ethanol-fixed cloacal swabs and a novel RT-PCR method. Virology 375:182-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada, S., et al. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378-382. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Z. Y., et al. 2007. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen, H. L., et al. 2009. Changes in H5N1 influenza virus hemagglutinin receptor binding domain affect systemic spread. Proc. Natl. Acad. Sci. U. S. A. 106:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]