Abstract
Members of the human APOBEC3 family of editing enzymes can inhibit various mobile genetic elements. APOBEC3A (A3A) can block the retrotransposon LINE-1 and the parvovirus adeno-associated virus type 2 (AAV-2) but does not inhibit retroviruses. In contrast, APOBEC3G (A3G) can block retroviruses but has only limited effects on AAV-2 or LINE-1. What dictates this differential target specificity remains largely undefined. Here, we modeled the structure of A3A based on its homology with the C-terminal domain of A3G and further compared the sequence of human A3A to those of 11 nonhuman primate orthologues. We then used these data to perform a mutational analysis of A3A, examining its ability to restrict LINE-1, AAV-2, and foreign plasmid DNA and to edit a single-stranded DNA substrate. The results revealed an essential functional role for the predicted single-stranded DNA-docking groove located around the A3A catalytic site. Within this region, amino acid differences between A3A and A3G are predicted to affect the shape of the polynucleotide-binding groove. Correspondingly, transferring some of these A3A residues to A3G endows the latter protein with the ability to block LINE-1 and AAV-2. These results suggest that the target specificity of APOBEC3 family members is partly defined by structural features influencing their interaction with polynucleotide substrates.
The APOBEC3 family of polynucleotide cytidine deaminases is collectively endowed with the ability to restrict a large panel of genetic invaders, from endogenous retroelements to several DNA and RNA viruses. The human genome encodes seven APOBEC3 proteins, each of them able to inhibit a particular set of mobile elements. APOBEC3A (A3A) is a nucleocytoplasmic editing enzyme expressed mainly in primary monocytes and keratinocytes (30, 37, 44). It can block endogenous retroelements such as the long terminal repeat (LTR)-retrotransposon intracisternal A particle (IAP) and the non-LTR retroelements LINE-1 and Alu (7, 12), and it also is active against the parvoviruses adeno-associated virus type 2 and minute virus of mouse (AAV-2 and MVM, respectively) (12, 42). In contrast, it is inactive against HIV in cell lines (6, 18), although the knockdown of A3A in monocytes increases their susceptibility to this virus (44). Finally, sequence marks compatible with A3A-mediated editing can be detected on the genome of papilloma virus, which infects cutaneous and mucosal keratinocytes (54).
Although many of the molecular details of A3A-mediated restriction remain unknown, the cytidine deaminase primarily perturbs the genome of its targets, and editing seems to be at the heart of many of its effects. In the presence of A3A, replicating viral genomes are decreased in AAV producer cells (12), which mirrors the reduced levels of LINE-1 reverse transcripts observed in A3A-expressing cells (43). Catalytically defective A3A mutants are largely inactive against LINE-1 and AAV (12), although mutants devoid of detectable in vitro deaminase activity have been identified that still can restrict the parvovirus (42). While A3A normally is unable to target HIV, it can induce high levels of editing on the viral genome and block its replication once forced into the retroviral capsid (1, 18). Similarly, the overexpression of A3A results in high-frequency C→U mutations in papillomavirus genomes (54) and transfected plasmid DNA (52). Since A3A acts only on single-stranded DNA (ssDNA), as demonstrated in in vitro deaminase assays (12), the editing of papillomavirus and plasmid DNA likely takes place during transient single-stranded phases.
The present work aimed at investigating the molecular determinants of A3A involved in recognizing the genome of its targets. For this, we combined structural modeling with phylogenetic analyses and functional studies. This led to the identification of residues essential not only for editing but also for restricting AAV, LINE-1, and foreign DNA, with critical positions forming a potential single-stranded DNA-docking groove connected to the catalytic center. Moreover, our data suggest that amino acid differences in this region between APOBEC3 family members influence their respective spectra of restriction.
MATERIALS AND METHODS
Orthologous gene amplification in nonhuman primates and molecular analysis.
APOBEC3A orthologue coding sequences from primates were generated by the amplification and sequencing of genomic DNA from bonobo (Pan paniscus), gorilla (Gorilla gorilla), Bornean orangutan (Pongo pygmaeus), Lar gibbon (Hylobates lar), nomascus (Hylobates leucogenys), siamang (Hylobates syndactylus), and cotton-top tamarin (Saguinus Oedipus). African green monkey [Cercopithecus (chlorocebus) aethiops] and common marmoset (Callithrix jacchus jacchus) sequences were obtained by the amplification and sequencing of cDNA from kidney and owl monkey (Aotus trivirgatus) from liver cDNA. All GenBank accession numbers are listed in Table S2 in the supplemental material. To obtain the chimpanzee (Pan troglodytes) A3A sequence, we downloaded the genome assembly (panTro2) from the University of California-San Cruz genome browser. BLAT suite.34 (27) was used with the human coding sequences (CDSs) as the template to retrieve homology sequences with the parameters “−t dnax −q dnax” (i.e., translated DNA). BLAT of the querying sequence was performed chromosome by chromosome using the positive-strand chain to identify the homologous sequence with the maximum match value. Exons were amplified by primers designed for the flanking intron regions (for genomic DNA) and for 3′- and 5′-untranslated regions (UTRs) (for cDNA). HotStarTaq master mix (Qiagen) was used for PCR amplification. All primer sequences used for amplification are presented in Table S1 in the supplemental material. Sequences were aligned using MUSCLE (15). Coding regions were aligned according to their corresponding amino acid sequences using the European Molecular Biology Open Software Suite package (47).
Constructs.
The pCMV expression plasmid expressing the C-terminal hemagglutinin (HA)-tagged form of A3A (4) was a kind gift from M. Malim (King's College, London, United Kingdom). The plasmid pcDNA3.1(+)-A3A-HA and the plasmid expressing the C-terminal moiety of A3G (pCMV-A3G197-384-HA) have been described previously (9, 12). Point mutations were introduced using QuikChange site-directed mutagenesis (Stratagene), and each mutant was verified by sequencing reactions. The plasmid required for the LINE-1 retrotransposition assay was a kind gift from T. Heidmann (p220.CMV-L1.2BneoTNF) (16). Plasmids expressing AAV Rep/Cap proteins (pXX2), the adenovirus helper proteins (pXX6), and the recombinant AAV (rAAV) vector plasmid (pACLALuc) have been described previously (42).
Cell culture and transfection.
HeLa HA cells were passaged regularly in minimum essential medium supplemented with nonessential amino acids (Gibco) in addition to 10% fetal calf serum, antibiotics, and 2 mM glutamine, and they were transfected with Fugene 6 (Roche) at low density in six-well plates using the manufacturer's instructions. 293T and human osteosarcoma U2OS cell lines were purchased from the American Tissue Culture Collection. 293A (HEK-293A) cells were purchased from Q Bio-gene, Carlsbad, CA. Cells were passaged regularly in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum and antibiotics.
LINE-1 replication assays.
For the LINE-1 retrotransposition assays, 1 × 105 to 2 × 105 HeLa HA cells were seeded in six-well plates and transfected 16 h later using p220.CMV-L1.2BneoTNF (16) and pCMV5 or plasmid encoding APOBEC3 in duplicates, using the indicated doses. If one plasmid was titrated down, pCMV5 empty backbone was used to keep constant the total amount of transfected DNA. After overnight incubation, cells were washed and plated into 10-cm dishes or six-well plates. Three days posttransfection, the cells in 10-cm dishes were selected with neomycin (G418; Gibco) at 2 mg/ml for 2 days and then at 0.5 mg/ml for 5 to 7 more days. Colonies then were fixed with ethanol, stained with crystal violet, counted, and averaged from duplicates. The six-well plates were collected at the time of selection for Western blot or reverse transcription-PCR (RT-PCR) analysis. For immunoblotting, lysates were prepared by standard methods and the protein concentration was quantified by bicinchoninic acid assay (Bio-Rad). Three μg of protein extract was loaded per well onto polyacrylamide gels and analyzed as previously described (9). To test whether A3A interferes with resistance to neomycin or affects cell growth/survival, we performed a similar assay, except that the LINE-1 reporter construct was replaced by the peGFP.C1 plasmid (Clontech) coding for resistance to neomycin along with green fluorescent protein (GFP). Cells were plated 16 h posttransfection in 10-cm dishes and selected 2 days later with neomycin for 10 more days. In parallel, cells also were plated in 24-well plates, serially diluted, and maintained without selection for the same amount of time. Colonies then were fixed, stained, and counted. For the hygromycin resistance assay, we used the same conditions as those in a standard LINE-1 retrotransposition assay, except that cells were selected with hygromycin (200 μg/ml; Roche) rather than neomycin.
Production of recombinant AAV.
Recombinant AAV production assays were performed as previously described (42). Briefly, 80% confluent 293T cells were cotransfected in six-well plates with pXX6 (1.5 μg), pXX2 (0.5 μg), pACLALuc (0.5 μg), and APOBEC3 expression vector (1 μg, unless otherwise stated) or pcDNA3.1(+) control vector. When less than 1 μg of APOBEC3 plasmid was tested, the total amount of effector DNA was maintained at 1 μg by the addition of pcDNA3.1(+). Transfections were performed in duplicate or triplicate using polyethyleneimine (41). Cells were harvested 72 h posttransfection, and one-fourth of each sample was removed for immunoblotting. The other three-fourths of the sample were used to generate rAAVLuc virus lysates by freeze-thaw cycles followed by centrifugation. Virus lysates were used to transduce 293A cells, and luciferase activity was quantified after 48 h in a TopCount NXT scintillation and luminescence counter (PerkinElmer) using Steady-Glo luciferase substrate reagent (Promega). Background luminescence from mock-transfected cells was subtracted automatically.
Semiquantitative PCR of LINE-1 RNA and cDNA.
Briefly, 3 days posttransfection (concurrently with the start of neomycin selection), we extracted total cellular DNA or RNA from transiently transfected cells using an DNAeasy or RNAeasy mini kit, respectively (Qiagen). DNA was used as the template for semiquantitative PCR using primers specific for the spliced formed of the neomycin resistance gene (NeoBF, 5′ AAGAACTCGTCAAGAAGGCG 3′; NeoMR2, 5′ GAAGAGCATCAGGGCCTCGC 3′) (see Fig. 3B). p220.CMV-L1.2BneoTNF and peGFP.N1 plasmids were used as negative and positive controls of PCR, respectively. RNA was reverse transcribed using Superscript III reverse transcriptase (Invitrogen) and poly(dT) as the primer. The use here of PCR primers spanning the exon junction allows us to discriminate the sense from the antisense transcript (see Fig. 3B). Quantification was done by the serial dilution of the template cDNA. Equal loading was verified by running control PCR on actin DNA in each condition (Actin_F, 5′ TCACCCACACTGTGCCCATCTACGA 3′; Actin_R, 5′ CAGCGGAACCGCTCATTGCCAATGG 3′). Water served as a negative control of PCR.
In vitro deaminase assay.
For in vitro UDG-dependent deaminase assays, A3A and its mutants were synthesized in vitro using the TNT coupled wheat germ extract system (Promega) with T7 polymerase. Translation reactions were performed in 50-μl reaction mixtures that included 1 μg of pcDNA3.1(+) plasmid encoding HA-tagged A3A by following the manufacturer's protocol. Reaction mixtures were incubated for 90 min at 30°C. After incubation, translation reaction mixtures were centrifuged at 10,000 × g for 1 min, and 10 μl of the supernatant was removed for immunoblot analysis. To determine deaminase activity, 20 μl of supernatant was incubated with a 5′-end fluorescein isothiocyanate (FITC)-labeled single-stranded deoxyoligonucleotide (0.4 μM) in a final reaction volume of 30 μl containing 40 mM Tris, pH 8.0, 10% glycerol, 40 mM KCl, 50 mM NaCl, 5 mM EDTA, and 1 mM dithiothreitol (DTT). The reactions were incubated at 37°C for 20 h, stopped by being heated to 90°C for 5 min, cooled on ice, and then centrifuged at 10,000 × g for 1 min. Twenty μl of the supernatant then was incubated with uracil DNA glycosylase (New England Biolabs) in buffer containing 20 mM Tris, pH 8.0, 1 mM DTT for 1 h at 37°C and treated with 150 mM NaOH for 30 min h at 37°C. Samples then were incubated at 95°C for 5 min and 4°C for 2 min and separated by 15% Tris-borate-EDTA (TBE)-urea-PAGE. Gels were directly analyzed using an FLA-5100 scanner (Fuji). The PAGE-purified ssDNA oligonucleotide (Invitrogen) used for the deaminase assays contains a single cytosine in the A3A-specific target trinucleotide 5′-TCA (FITC-5′-TATTATTATTATTATTATTCATTTATTTATTTATTTATTT-3′).
Structural model and sequence alignment.
The APOBEC3A human sequence (residues 13 to 195; UniProtKB [2] code P31941) was defined as the target sequence, while the recently resolved human APOBEC3G C-terminal domain (23) (PDB [3] no. 3IR2 [50]) served as a template. The two proteins show good sequence identity (65%). Sequence alignment was done with the align2d function of the MODELLER program (48), and homology models were built with the same program. One thousand models were calculated and scored with the MODELLER objective function and ANOLEA energy (38) using the default 5-residue window averaging. A zinc ion was added to the active site of the model before energy minimizing it with the CHARMM program (8) and the CHARMM22 all-atom force field (36), consisting of 100 steps of the steepest descent. A harmonic restraint of 5 kcal/mol/Å2 toward their initial position was imposed on all heavy atoms, except for the zinc site residues H70, E72, C101, and C106, which were restrained to display the same distances to the zinc ion as those in the X-ray structure. The FoldX program (22) was used to perform an in silico alanine scan by mutating each residue to an alanine and estimating the change of folding energy. Molecular visualization and structural alignments were done using Chimera (45).
RESULTS
Structural modeling and phylogenetic analysis of A3A.
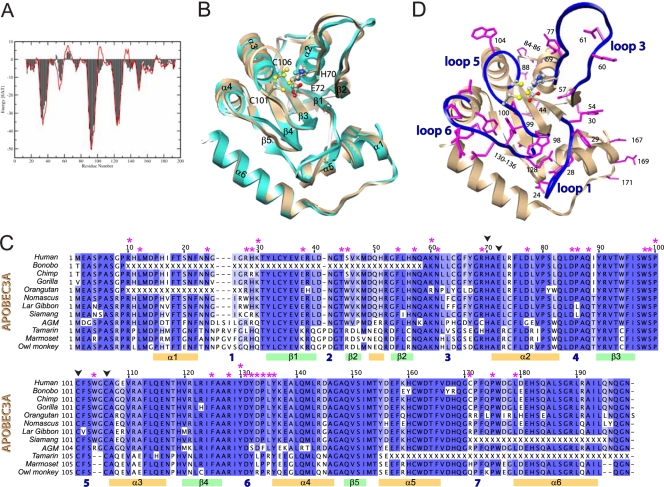
A3A is a single-domain cytidine deaminase, the three-dimensional structure of which has not been determined experimentally, but it is phylogenetically close enough to the A3G C-terminal domain (32) (65% percent sequence identity) for the latter to serve as the basis for structural modeling. We thus generated a model of A3A (residues 13 to 195) (available online as supplementary material) based on the recent crystal structure of the A3G C terminus (50) (residues 191 to 384). This template was preferred because it shows better crystallographic refinement statistics and lower α carbon B factors than earlier structures (24). Sequence alignment was done with the MODELLER program (48) (Fig. 1). One thousand models were generated and scored with ANOLEA (38), estimating the folding free energy of each amino acid in its respective environment. The models were clustered based on their structure, and the one with the best ANOLEA score belonging to the largest cluster was chosen for further refinement. A zinc ion was added to the active site before energy minimization. A harmonic restraint toward their initial position was imposed on all heavy atoms, except for the residues of the catalytic site, H70, E72, C101, and C106, which were restrained to display the same distances to the zinc ion as those in the X-ray structure. The resulting A3A predicted structure had a good ANOLEA score profile (Fig. 2 A) that was comparable to that of the A3G template. Similarly to the A3G C terminus, the core of A3A is predicted to be comprised of five anti-parallel β-sheets supporting three α-helices on each side (Fig. 2B). The A3A zinc-coordinating residues reside on loops 3 and 5, which connect β-sheets 2 and 3 to α-helices 2 and 3, respectively. The biggest structural differences between the A3G template and the A3A model are seen in loops 1 and 3, as well as in residues W104 and G105, which are part of helix 3 in the A3G C terminus and of loop 5 in A3A.
FIG. 1.
Alignment of human A3A and A3G C terminus. Color bars below the alignment reflect predicted secondary-structure elements (α-helices in orange and β-strands in green), while loop domains are simply numbered (in blue) from 1 to 7. Red stars indicate residues selected for site-directed mutagenesis on human A3A; black arrowheads indicate zinc-coordinating residues.
FIG. 2.
Homology modeling of human A3A. (A) Comparison of the ANOLEA scores of the A3A model (red) and the A3G X-ray structure 3IR2 (black). (B) Superposition of the A3A model (brown) and the A3G C-terminal X-ray structure (light blue). The zinc atom (yellow) and residues required for zinc coordination are illustrated. (C) Alignment of human A3A together with 11 novel primate orthologues, with mutated residues analyzed for restriction activity marked with an asterisk. Color bars below the alignment reflect predicted secondary-structure elements. Missing codons due to amplification failure are indicated with an X. Dark blue, >80% identity; blue, >60%; light blue, >40%. Alignment was performed using MUSCLE. Magenta stars indicate residues selected for site-directed mutagenesis on human A3A; black arrowheads indicate zinc-coordinating residues. AGM, African green monkey. (D) Residues selected for site-directed mutagenesis (magenta) mapped on the A3A homology model. The four loops flanking the catalytic site are shown in blue.
As an additional basis for subsequent mutagenesis/function studies, we examined the phylogenesis of A3A genes from human and 11 nonhuman primates, reflecting 40 million years of evolution. For this, we sequenced PCR products amplified from genomic DNA and/or reverse transcribed from total cellular RNA and combined them with the BLAT program on complete genomes when available. The alignment of the 12 collected sequences revealed overall high levels of conservation among primate A3A orthologues (72.0% pairwise identity), with a relatively lower degree of conservation (58.7% pairwise identity) for the region comprising amino acids 25 to 67 (Fig. 2C). While residues 25 to 31, 41 to 44, and 57 to 67 within this region of A3A are predicted to partake in loops 1, 2, and 3, respectively, in APOBEC2 and APOBEC3C (A3C) residues 45 to 56 correspond to a β-sheet that mediates homodimer formation (46, 51).
Functional study of A3A-mediated LINE, AAV, and foreign DNA restriction.
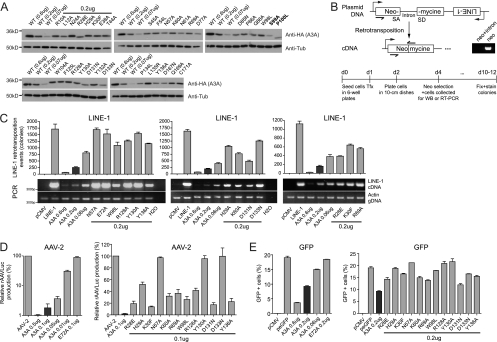
Based on our model of A3A and the phylogenetic analysis of A3A orthologues, we performed a mutagenesis-based functional study of A3A, selecting target residues according to the following criteria: (i) sequence conservation, (ii) predicted location in loop domains rather than more structured regions, and (iii) low probability to participate in structural stability, as estimated by in silico alanine scanning (data not shown). We focused on sequence conservation rather than variability, because we aimed to identify conserved features of restriction implicated in ancient genetic conflicts, such as that with LINE-1 retrotransposons. Due to the endogenous nature of these elements, it is unlikely that they evolved as fast as exogenous retroviruses, which further justifies our focus on conservation rather than divergence. We also capitalized on functional data previously obtained with other APOBEC3 family members that locate residues of functional importance on loop domains. Residues on loops 1, 5, and 6 of the A3G N terminus contribute to RNA-dependent packaging and antiviral activity against HIV (9, 25), while residues on homologous loop 5 of the A3F N terminus or on A3C are critical for retroviral inhibition (51, 56). Thirty-four residues were mutated, most of which resided within predicted loop 1, 3, 5, or 6 of A3A (Fig. 2C and D). The steady-state level of expression of the resulting mutants was evaluated by the Western blotting of cellular extracts from transiently transfected HeLa cells (Fig. 3A). Two of the mutations (S99A and P100L) led to poorly expressed proteins (Fig. 3A, boldface characters), while the rest of the mutants exhibited wild-type or near-wild-type steady-state levels of expression (Fig. 3 A, plain characters). All well-expressed A3A derivatives further exhibited wild-type patterns of cytoplasmic and nuclear localization, as assessed by indirect immunofluorescence (Fig. 4). Mutants for which expression levels and localization were similar to those of wild-type A3A were further analyzed for functionality.
FIG. 3.
LINE-1, AAV, and foreign DNA restriction by wild-type and mutant A3A. (A) Levels of expression of transiently expressed A3A at three different doses for the wild type, while mutants were tested at the intermediate dose of plasmid DNA (0.2 μg), as analyzed by Western blotting using HA- and tubulin-specific antibodies. Two mutants with a steady-state level of expression that was reduced by 3-fold or more compared to that of the wild type are highlighted in boldface (top blot, far right). (B) Top, exon junction-spanning primers (black half arrows) selectively amplify the spliced neomycin resistance cassette obtained after LINE-1-mediated reverse transcription. Neo, neo-carrying plasmid; neo+intron, LINE-1 reporter plasmid; SD, splice donor; SA, splice acceptor. Bottom, experimental setup of a typical LINE-1 retrotransposition assay. Tfx, transfection. (C) LINE-1 retrotransposition efficiency as measured by semiquantitative PCR analysis of reverse-transcribed cDNA (bottom) or by the selection of neomycin-resistant colonies (top) in the absence or presence of the wild type or stably expressed yet functionally defective A3A mutants. Means and standard errors are representatives of independent duplicates. The production of rAAVLuc (D) and expression of GFP (E) in the presence or absence of the A3A derivatives also are shown. The amount of A3A-expressing plasmids used is indicated. In panel D, results from three independent experiments are presented as the averages of the relative values compared to those of mock transfections with pcDNA3.1(+).
FIG. 4.
Subcellular localization of wild-type A3A and functionally defective mutants. APOBEC3 protein localization was determined by the indirect immunofluorescence of HA-tagged APOBEC3 proteins (HA) expressed in U2OS cells. Cells were costained with DAPI to visualize nuclei (Merge). U2OS cells were grown on glass coverslips and transfected with 0.8 μg APOBEC3 expression vector using Lipofectamine 2000 (Invitrogen). After 24 h, cells were fixed with 3% paraformaldehyde and extracted with 0.5% Triton X-100 in PBS. Cells were incubated with anti-HA MAb 16b12 (1:2,000) (Covance) and a 1:2,000 dilution of goat anti-mouse-conjugated Alexa Fluor 568 (Invitrogen) and DAPI (Sigma Aldrich). The coverslips were mounted in Fluoromount G (Southern Biotech), and images were acquired using a Leica TCS SP2 confocal microscope.
We functionally characterized the library of A3A mutants through assays testing three of its known restriction activities, namely, the blockade of LINE-1 retrotransposition (12), the inhibition of AAV replication (12), and the decrease in expression from transfected plasmids due to the degradation of foreign DNA (52). A commonly used LINE-1 retrotransposition assay relies on the transfection of a LINE-1-expressing plasmid into HeLa cells, leading to the single-round retrotransposition-dependent generation of neomycin-resistant colonies (16, 39) (Fig. 3B). Out of 32 A3A mutants tested, 12 were significantly defective for LINE-1 inhibition (Fig. 3C), while the others retained wild-type or near-wild-type activity (data not shown). Mutations targeting conserved residues in the catalytic site also inactivated the protein (Fig. 3C and data not shown). To characterize the functional defect of these mutants, we tested their ability to interfere with LINE-1 cDNA synthesis. As for the wild type, there was a strict correlation between levels of LINE-1 cDNA and a decrease in the number of induced G418-resistant colonies (Fig. 3C). This corroborates a previous study indicating that A3A induces a decrease in LINE-1 replicative intermediates (43).
We then tested our panel of A3A mutants for their ability to block the parvovirus AAV-2. Briefly, recombinant AAV vector particles were generated by the cotransfection of a luciferase-encoding AAV-derived vector plasmid, together with a packaging plasmid and a plasmid expressing the necessary adenoviral helper proteins (12, 57). The production of infectious rAAV particles was determined by measuring luciferase activity in target cells infected with the resulting vector preparation. As previously reported, luciferase activity was decreased in a dose-dependent manner when A3A was coexpressed in AAV-producing cells (Fig. 3D), and the mutation of the conserved catalytic-site residues (H70, E72, C101, and C106) resulted in proteins that had lost the restriction activity (Fig. 3D and data not shown). Furthermore, out of 28 mutants tested, we found that the 12 defective for LINE-1 inhibition were the only ones significantly impaired for AAV-2 restriction (Fig. 3D and data not shown).
A3A was recently reported to mediate the UNG2-dependent degradation of foreign DNA (52). We additionally tested the ability of our functionally defective mutants to interfere with plasmid DNA under conditions similar to the ones used by Stenglein et al. (52). Briefly, a small amount of GFP-expressing plasmid (0.05 μg) was cotransfected with the dose of A3A-expressing plasmid used in the LINE-1 retrotransposition assays, and cells were analyzed 5 days later for GFP expression. Under these conditions, the percentage of GFP-positive cells was decreased by A3A in a dose-dependent manner (Fig. 3E, left). A3A mutants that were defective for LINE-1 and AAV inhibition also failed to interfere with GFP expression (Fig. 3E, right), whereas mutants with wild-type phenotypes in the L1 and AAV assays (Y132A and L135R) were as effective as wild-type activity in the GFP assay (data not shown).
The effect of a given dose of A3A DNA on the recovery of GFP-positive cells after the transfection of a GFP-expressing plasmid was significantly milder than the effect measured on LINE-1 or AAV-2 (compare Fig. 3C, D, and E). Still, we asked whether, under our experimental conditions, bona fide A3A-mediated restriction of LINE-1 retrotransposition occurred, rather than indirect interference with expression from the LINE-1 plasmid. For this, we compared the impact of A3A expression on the synthesis of LINE-1 transcription and reverse transcription products using primers that distinguish intron-deleted reverse-transcribed DNA from its intron-containing plasmid parent (Fig. 3B). Since the measured RNA could be expressed from recently integrated LINE-1 elements rather than from the parental plasmid DNA, we performed the LINE-1 assay in the presence and absence of reverse transcriptase inhibitors, and we collected the cells 3 days after transfection. While A3A reduced the amount of LINE-1 reverse transcripts in a dose-dependent manner and in a linear relationship with the reduction in the number of G418-resistant colonies (Fig. 5 A), the cytidine deaminase had no significant effect on the levels of LINE-1 RNA transcript, regardless of the presence of reverse transcriptase inhibitors (Fig. 5A). This suggests that A3A activity on the LINE-1 reverse transcription product is specific and does not originate from initial differences in LINE-1 RNA levels. Furthermore, and in agreement with previous reports (6, 35, 40), we found that, compared to the W98L mutant, wild-type A3A selectively decreased the number of LINE-1-induced neomycin-resistant colonies, with only a limited effect on the recovery of G418-resistant colonies after the transfection of a plasmid simply expressing the neo cDNA or on cell growth/survival in the absence of antibiotic selection (Fig. 5B). However, when we tested the effect of A3A expression on antibiotic resistance (hygromycin) originating directly from the LINE-1-expressing plasmid, we found that the number of hygromycin-resistant colonies was significantly reduced in the presence of wild-type A3A at three different doses (Fig. 5B). Again, at a given dose, the effect of A3A on LINE-1 retrotransposon-induced G418-resistant colonies (Fig. 5B, left, 30- and 8-fold at doses of 0.6 and 0.2 μg, respectively, of A3A plasmid DNA) was markedly more pronounced than the corresponding decrease in LINE-1 plasmid-induced hygromycin-induced resistant colonies (Fig. 5B, right, 10- and 2.7-fold). Taken together, these results indicate that the experimental approach used here measures the bona fide blockade of LINE-1 reverse transcription and AAV replication by A3A. Our results also appear to confirm that, under some circumstances, A3A can lead to decrease expression from transfected plasmids, presumably due to the reported cytidine deaminase-mediated destabilization of transfected DNA (52). It appears that the same residues of A3A are critical for all three of these restrictive effects.
FIG. 5.
Inhibition of LINE-1 retrotransposition by A3A partially correlates with interference with plasmid DNA. (A) LINE-1 transcription and reverse transcription products on day 2 posttransfection, in the presence or absence of A3A and nucleoside reverse transcriptase inhibitors (NRTI). AZT, 40 μM; 3TC, 20 μM. Actin DNA served as a loading control and water as a negative control (H2O). Samples run in the absence of reverse transcriptase failed to yield any signal (not illustrated). (B) Determination of the number of neomycin- or hygromycin-resistant colonies in the presence of the LINE-1 reporter or a neo-carrying plasmid, as well as cell viability in the absence of G418. The transfection conditions among the four assays were essentially the same, except that the LINE-1-expressing plasmid in the first assay was replaced by a neo-carrying plasmid in the latter two assays. Black columns correspond to the A3A plasmid dose used in most functional assays to test A3A mutants.
Functionally important A3A residues map to a common DNA-capturing groove.
On our structural model, the positions determined as important for LINE-1, AAV, and foreign DNA restriction map to regions flanking the catalytic site. The side chains of most of these residues are predicted to point outwards to form a groove-shaped continuous surface connected to the active-site pocket, with the noticeable exception of R69, whose side chain points in a direction opposite that of the predicted groove (Fig. 6 A). Furthermore, the groove is formed by residues that have a charged side chain (R28/128, K30/60, and D131/133), contain an aromatic group (W98, H29, and Y130/136), or are neutral yet polar (N57). This is similar to what was previously reported for the antiviral activity of other cytidine deaminases (9, 51) and suggests that this groove serves to accommodate single-stranded DNA molecules. To probe this issue, we applied the molecular docking algorithm EADock (20-21) to model binding of the A3A target sequence 5′-TTCA (12) onto the A3A structure. We included the following spatial constraints derived from experimental evidence: (i) the C-3 position must be compatible with deamination, using spatial constraints similar to the ones obtained from the crystal structure of the bacterial deaminase tadA in complex with RNA (34), (ii) the T1 and T2 bases contact D131 and D133; this assumption is based on studies of A3G and A3F, where homologous aspartate residues influence the target sequence at positions −1 and −2 relative to the edited cytidine (24, 31), and (iii) if possible, A4 should interact with residues H29, K30, and K60, which are delimiting the other side of the active-site pocket relative to D131/133. The docking yielded several DNA configurations fulfilling the imposed constraints, including the one with the most favorable energy, represented in Fig. 6B.
FIG. 6.
Molecular docking of a single-stranded DNA template and cytidine deaminase assay. (A) Residues important (orange) or dispensable (magenta) for A3A restriction activity mapped on the structural model. Yellow, zinc-coordinating residues. (B) Constrained molecular docking of a 5′-TTCA oligonucleotide (cyan) onto the A3A model using EADocks. (C) Deaminase activity of A3A and mutant proteins generated by in vitro-coupled transcription-translation (IVT) was analyzed in UDG-dependent deaminase assays. Arrows indicate substrate deoxyoligonucleotide and cleaved deaminated product. Bottom panel, Western blot of one-fifth of the IVT reaction mixtures using HA-specific antibody.
This model suggested that, with the exception of R69, the residues identified as functionally important for restriction partake in the binding and/or editing of polynucleotidic substrates. To probe this hypothesis, we tested the editing capacity of the corresponding mutants in an in vitro cytidine deaminase assay using a single-stranded DNA oligonucleotide containing 5′-TTCA as the target sequence (12) (Fig. 6C). Wild-type A3A resulted in a high ratio of edited/cleaved substrate to nonedited substrate, while the reverse was observed with the catalytic-site E72Q mutant, as previously reported (12). The majority of other tested mutants were completely (N57A, R128A, Y130A, and D133N) or partially (H29A, K30F, K60A, R69A, W98L, and Y136A) defective for deaminase activity. Only two of the mutants that were defective for restriction retained close-to wild-type levels of cytidine deaminase activity in vitro (R28E and D131N). Globally, a good parallel was noted between editing and restriction activities for the A3A derivatives tested here.
Structure/sequence comparison between A3A and the A3G C terminus.
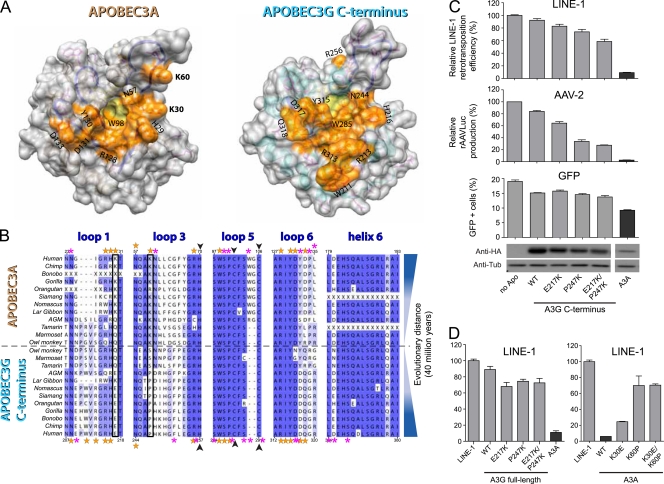
A3A and the A3G C-terminal domain share several characteristics, including a high degree of amino acid identity (65%), a catalytically active site (12, 13), and similar subcellular localizations in both the cytoplasm and the nucleus (7). However, while A3A can block the replication of multiple mobile genetic elements, including LINE-1 and AAV, the C-terminal domain of A3G on its own is not endowed with any known restriction function (7, 12). To investigate the structural features that might account for these functional differences, and since A3A activities tightly correlate with the structure of a predicted DNA docking/editing groove, we examined the corresponding domain of A3G. For this, we projected on the A3G C terminus crystal structure (50) 80 mutants previously screened by Chen et al. for cytidine deamination (13, 14). Several residues critical for editing located within loops that flanked the active site, forming a continuous surface with the shape of a groove reminiscent of that delineated for A3A (Fig. 7 A). Functionally important amino acids residing outside these loops pointed toward the protein core, suggesting that they stabilize the structure (not illustrated). In contrast, residues identified as having little influence on editing mapped to other parts of the protein. In the A3G C terminus, most of the residues important for editing identified were charged (R213/215/256 and D317), aromatic (W211/285, H216, and Y315), or neutral yet polar (N208/218 and Q318). However, two major differences emerged between the A3A and A3G deaminases: (i) in the A3G C terminus, one side of the groove, flanked by W211, R213, and by Q318, was more restrained than in A3A, and (ii) in A3A, residues K30 and K60 delineated a clamp-like structure at the outlet of the groove. K30 and K60 residues were absent from the A3G C terminus, and no other amino acid could reconstitute such a clamp-like structure in this domain.
FIG. 7.
Structure and primate sequence comparisons reveal a distinct DNA-binding groove on human A3A and the A3G C terminus. (A) Space-filling visualization showing the surface formed by the residues important for editing A3A or that are important for editing and are present on loops 1, 3, 5, and 6 for the A3G C terminus (orange), placed on the homology model or the X-ray structure. The catalytic-site pocket is highlighted in yellow. (B) Alignment of the 12 primate A3A orthologues with their A3G C-terminal counterparts, focusing on the four loops represented in Fig. 1D and on helix 6, the C-terminal-most helix of A3A. Orange and magenta stars indicate residues with or without functional importance, respectively; black frames indicate K30 and K60; black arrowheads indicate zinc-coordinating residues. Missing codons due to amplification failure are indicated with an X. Note that human A3G editing-defective mutants on helix 6 (at positions 364, 375, 378, and 379 [13]) are not depicted. (C) LINE-1 and AAV-2 replication efficiency and GFP expression in the presence of derivatives of the A3G C-terminal domain (residues 197 to 384). Means and standard errors are representative of the results for independent duplicates from three (LINE), two (AAV), or one experiment (GFP). The bottom shows expression levels of the A3G C-terminal derivatives and A3A, as evaluated by Western blot analysis. (D) LINE-1 retrotransposition efficiency, depicted as described for panel C, except that full-length A3G (residues 1 to 384; left) or A3A (right) was used.
We further compared novel primate A3A sequences to those of the 12 cognate A3G C-terminal homologues recovered from various databases. As expected, generally less conservation was observed in loops than in secondary-structure elements, which was particularly evident for loops 1, 3, and 6 (Fig. 7B; also see Fig. S1 in the supplemental material). The majority of positions identified as important for A3A activity corresponded to ones determined to be of equal functional importance in human A3G and were largely conserved throughout the A3A and A3G primate orthologues (human A3A residues H29, N57, R69, W98, R128, Y130, and D131 [orange stars in Fig. 7B] and Y136 [not depicted]). R28 presented the lowest level of conservation among APOBEC3 homologues, in agreement with its limited impact on A3A-editing and restriction functions. On the other hand, the two lysines at positions 30 and 60 that contributed to the exclusive shape of the putative DNA-binding groove of A3A were specific and generally conserved (K30 or Q30 and a single primate with R60) among primate A3A orthologues, yet this was not found in primate A3G C-terminal domains. D133 was another position at which marked differences were noted between primate A3A and A3G C-terminal homologues, but its conservation among primate A3A orthologues was lower than that of K30 and K60.
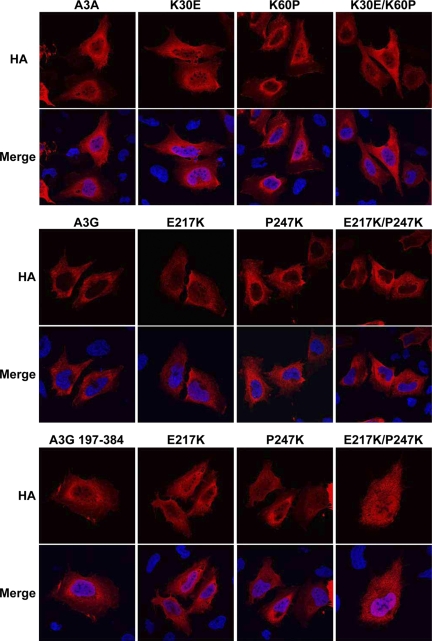
To understand the influence of the A3A-specific residues K30 and K60 in restriction, we transferred those amino acids to the homologous region of the A3G C-terminal domain (corresponding to E217 and P247), either one by one or in combination, and tested the activity of the resulting A3G derivatives in the three assays. As seen in Fig. 7C (top), while swapping one residue had only a little influence on the LINE-1 restriction activity of the A3G C terminus, the simultaneous replacement of E217 and P247 by lysines conferred to this protein a strong ability to block the retroelement, albeit still less efficiently than A3A. In addition, the A3G C-terminal chimera could block AAV-2 replication, although in this case most of the activity resulted from the P247K replacement alone (Fig. 7C, middle). On the other hand, all A3G C-terminal constructs had a minor impact on the recovery of GFP-positive cells after plasmid transfection (Fig. 7C, bottom). While these swapping mutants also increased full-length A3G restriction activity against LINE-1 (Fig. 7D, left), reverse mutations on A3A interfered with its ability to block the LINE-1-mediated induction of neo-resistant colonies (Fig. 7D, right). It is noteworthy that we could not reveal a parallel between subcellular localization and restriction activity with any tested A3A or full-length A3G chimera (Fig. 8).
FIG. 8.
Subcellular localization of wild-type A3A, full-length A3G, and the C terminus of A3G and the indicated mutants.
DISCUSSION
We performed here a structure/function study of the determinants dictating A3A functions, and we identified 12 residues at the core of A3A-editing and restriction activities. Mutations at these residues affect the blockade of LINE-1 retrotransposition, the inhibition of AAV replication, and the decrease in gene expression from foreign plasmid DNA without affecting the expression levels or subcellular localization of the protein. Structural modeling and molecular docking guided by experimental observations predicted that these residues form a groove that can accommodate a single-stranded DNA molecule. The restriction and editing defects of the mutants coincided with their inability to prevent the accumulation of LINE-1 reverse transcription products, using experimental conditions where no effect of A3A on LINE-1 RNA transcript levels or G418 resistance induced by a neomycin phosphotransferase-expressing plasmid could be detected (Fig. 5). However, some effect on transfected LINE-1 DNA could be noted, which translated in a moderate, albeit significant, decrease in the recovery of hygromycin-resistant colonies. Why this was observed in the absence of an impact on the amounts of plasmid-generated LINE1 RNA is unclear. However, it is worth noting that the bulk of reverse transcription-competent LINE-1 RNA likely is produced shortly after transfection from unintegrated circular plasmid DNA. In contrast, the induction of hygromycin-resistant colonies requires that the plasmid DNA not only migrates to the nucleus but also becomes linearized and integrated, and hence it follows a path where it may be more exposed to A3A-induced degradation. It is clear that investigating thoroughly the molecular mechanism of A3A-induced LINE-1 inhibition would greatly benefit from a system allowing for the production of retrotransposition-competent LINE-1 from an integrated locus rather than from a transfected plasmid. Remarkably, mutants found here to alter the ability of A3A to block LINE-1 were equally defective for AAV-2 inhibition and the perturbation of gene expression from plasmid DNA. It was demonstrated that A3A acts on AAV by decreasing levels of viral replicative intermediates (12). Our data support a model whereby all three effects of A3A are linked to interference with the synthesis or stability of some DNA intermediate. The absence of the editing activity of A3A on double-stranded DNA in vitro (12) and the restricted size of the putative DNA-binding groove revealed by our structural model suggest that this DNA intermediate is a single-stranded molecule.
We recently reported that A3A mutants with no detectable levels of deaminase activity in vitro still can block AAV-2 replication (42). These data suggest that A3A acts through distinct or simply additive effects. Supporting evidence for multiple yet overlapping functions in APOBEC3 proteins is the segregation of the DNA editing from the RNA-binding activities of the single-domain cytidine deaminase A3C (51). Similarly, it could be that the editing-independent blockade of HIV replication observed with high levels of A3G (5, 26, 33) stems from the sequestration of single-stranded viral DNA products by the cytidine deaminase (26). Accordingly, A3A could interfere with LINE-1/AAV genome replication by interfering with DNA replication before inducing its editing and degradation. In the absence of a high-resolution structure of an APOBEC3 protein bound to its substrate, the modalities of the interaction of these molecules with nucleic acids remain speculative. Distinct DNA-binding models have been proposed for the A3G C terminus based on mutagenesis and/or chemical shift perturbations. One of these models (24) is in agreement with the putative DNA-binding groove predicted by our study (Fig. 6B and 7A) and with results obtained from the constrained molecular docking of the 5′-TTCA target sequence on A3A, while another model suggests that residues on the A3G C-terminal loop 4 participate in DNA binding (17). Since several highly conserved residues on A3A loop 4 do not contribute to A3A restriction activities on LINE/AAV (data not shown), this loop probably is not relevant to the restriction function of this protein. Moreover, the two helices linking the active site to loop 4 (helix 2 and 3) do not harbor residues with protruding positive charges or aromatic groups that could interact with single-stranded polynucleotides. Although the aromatic residue F75 found to be essential for A3A deaminase activity but not AAV-2 restriction (42) resides on helix 2, it points inside the protein and likely participates in the stability of secondary-structure elements, as predicted by in silico alanine scanning (data not shown). Since catalytic-site residues also map on helix 2, mutation at position F75 may affect the local structure of the enzyme and reduce the editing activity while leaving intact other putative DNA-binding functions. In agreement, we found here that the mutation of R69, which is adjacent to the zinc-coordinating residue H70, had a more significant impact on editing than on LINE-1 or AAV restriction. Alternatively, F75L may alter the ability of A3A to access its target by preventing its interaction with an unknown partner.
Despite the absence of the intrinsic restriction activity of the C-terminal domain of human A3G, the comparison of A3A and A3G C terminus sequences from different primates reveals that residues that may contribute to the restriction activities of A3A are highly conserved between primate A3A and the A3G C-terminal homologous sequences. Two remarkable exceptions are the residues K30 and K60 of human A3A, which generally are conserved among A3A primate sequences yet are absent from A3G C-terminal domain orthologues. Remarkably, introducing simultaneously the two residues into the C-terminal half of human A3G conferred on this molecule the ability to block LINE-1 replication (Fig. 7C). Only one of the two mutations (P247K) seemed to be sufficient for the gain of activity against AAV-2, a result in line with a previous experiment that showed that the transfer of a patch of residues on A3A loop 3 (including K60) to the A3G C terminus was sufficient to confer antiviral activity against AAV-2 (42). Since we failed to recapitulate the complete restriction activity of wild-type A3A, additional residues are likely to be of functional importance. It is expected that less conserved residues or residues present in secondary structure elements play an additional role in the specific activity of A3A. In agreement, a recent study found that a series of residues under positive selection influences murine APOBEC3 restriction activity against Friend virus (49). In addition, extra residues, such as the three-amino-acid patch found specifically on human A3G C terminus loop 1 but not on A3A or more ancient A3G homologues, could limit the restriction activity of the enzyme regardless of the presence of other beneficial amino acids. Interestingly, residues homologous to human A3A K30 and K60 are absent from human A3C (51), which is far less active than A3A against LINE-1 (7, 12, 28, 43) or AAV-2 (12).
One potential consequence of these structural variations is a difference in affinity for DNA binding. Although this affinity still needs to be determined for A3A, the surprisingly high Kd (dissociation constant) (>10 μM) of the A3G C terminus for single-stranded DNA (11) suggests that this domain needs to be present at a high concentration close to the target DNA for efficient editing and antiviral activity, as is the case with full-length A3G in HIV particles (58). The purification of wild-type and mutant A3A and the evaluation of their respective Kds for DNA binding will be required to validate this hypothesis. Residues on loop 6 influence A3G C terminus sequence preference at positions −1 and −2 relative to the edited cytidine (10, 29). It has been shown that swapping the loop between different APOBEC homologues can modify their editing sequence preference accordingly (29). Surprisingly, swapping functionally important residues of loop 6 (D133 and Y136) from A3A into the A3G C-terminal domain failed to confer any gain-of-function activity against LINE-1 (not illustrated). Another study found that replacing the variable region of A3A loop 6 with the one from the A3G N terminus is sufficient to allow the targeting of A3A to HIV virions (19). On A3G, these residues are critical for binding to specific small cellular RNAs and for virion incorporation (9, 25, 55). Although it is tempting to speculate that the identity of the target sequence plays less of a role for the editing-dependent restriction of LINE-1 elements than for specific binding to certain small cellular RNAs, this hypothesis will need to be tested by a thorough analysis of the influence of loop 6 residues on cytidine deamination target site preference. Since human A3A and A3G C-terminal domains are predicted to have a common phylogenetic origin (32), these data suggest that the specific restriction activities of A3A evolved after the divergence from the common ancestor gene but before the emergence of New World monkeys and was maintained throughout primate evolution. In agreement, preliminary data suggest that at least one New World monkey orthologue (marmoset A3A) is endowed with restriction activity against LINE-1 (data not shown). This hypothesis needs to be challenged by more in-depth vertical studies of APOBEC3 molecules.
The ability of A3G and A3C to bind to small cellular RNAs is one major determinant for packaging into retroviral particles and subsequent antiviral activity (9, 51, 53). Future studies will indicate whether the loss of editing and restriction activities of the A3A mutants identified here stem from decreased binding affinity for single-stranded DNA. This finding would be consistent with a model in which critical positions in APOBEC3 proteins influence the modalities of their interactions with polynucleotidic sequences, thereby determining their spectrum of action against mobile genetic elements.
Supplementary Material
Acknowledgments
This work was supported by grants from the Strauss Foundation and the Infectigen Association to D.T. and the Swiss National Science Foundation to D.T. and O.M. This work also was funded by fellowships from the Instituto de Salud Carlos III/Consejo Superior de Investigaciones Científicas/Salk Institute and the Lynn Streim Postdoctoral Endowment Fellowship to I.N. Work in the Weitzman laboratory was partially supported by a Pioneer Developmental Chair and by a grant from the U.S. National Institutes of Health (AI74967).
We thank John V. Moran for the HeLa HA cells. For computational resources we thank the Vital-IT team at the Swiss Institute of Bioinformatics. The modeling part of this work was performed within the Protein Modeling Facility (PMF) of the University of Lausanne. Molecular graphic images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. Special thanks to Nadine Zangger for sequence alignments and Charlène Raclot and Sandra Offner for technical assistance.
Footnotes
Published ahead of print on 1 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aguiar, R. S., N. Lovsin, A. Tanuri, and B. M. Peterlin. 2008. Vpr.A3A chimera inhibits HIV replication. J. Biol. Chem. 283:2518-2525. [DOI] [PubMed] [Google Scholar]
- 2.Apweiler, R., et al. 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38:D142-D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, H., K. Henrick, H. Nakamura, and J. L. Markley. 2007. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35:D301-D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd, H. P., et al. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks, B. R., et al. 1983. Charmm-a program for macromolecular energy, minimization, and dynamics calculations. J. Computat. Chem. 4:187-217. [Google Scholar]
- 9.Bulliard, Y., et al. 2009. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 83:12611-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter, M. A., E. Rajagurubandara, P. Wijesinghe, and A. S. Bhagwat. 2010. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amsterdam) 9:579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelico, L., C. Prochnow, D. A. Erie, X. S. Chen, and M. F. Goodman. 2010. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 285:16195-16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., et al. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 13.Chen, K. M., et al. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452:116-119. [DOI] [PubMed] [Google Scholar]
- 14.Chen, K. M., et al. 2007. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 581:4761-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnault, C., J. F. Casella, and T. Heidmann. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 30:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa, A., et al. 2009. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 28:440-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goila-Gaur, R., M. A. Khan, E. Miyagi, S. Kao, and K. Strebel. 2007. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology 4:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gooch, B. D., and B. R. Cullen. 2008. Functional domain organization of human APOBEC3G. Virology 379:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosdidier, A., V. Zoete, and O. Michielin. 2009. Blind docking of 260 protein-ligand complexes with EADock 2.0. J. Comput. Chem. 30:2021-2030. [DOI] [PubMed] [Google Scholar]
- 21.Grosdidier, A., V. Zoete, and O. Michielin. 2007. EADock: docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins 67:1010-1025. [DOI] [PubMed] [Google Scholar]
- 22.Guerois, R., J. E. Nielsen, and L. Serrano. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320:369-387. [DOI] [PubMed] [Google Scholar]
- 23.Harjes, E., et al. 2009. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holden, L. G., et al. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456:121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huthoff, H., F. Autore, S. Gallois-Montbrun, F. Fraternali, and M. H. Malim. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwatani, Y., et al. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, W. J. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinomoto, M., et al. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35:2955-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohli, R. M., et al. 2009. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284:22898-22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koning, F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaRue, R. S., et al. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 34.Losey, H. C., A. J. Ruthenburg, and G. L. Verdine. 2006. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13:153-159. [DOI] [PubMed] [Google Scholar]
- 35.MacDuff, D. A., Z. L. Demorest, and R. S. Harris. 2009. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 37:1854-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell, A. D., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586-3616. [DOI] [PubMed] [Google Scholar]
- 37.Madsen, P., et al. 1999. Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein apobec-1. J. Investig. Dermatol. 113:162-169. [DOI] [PubMed] [Google Scholar]
- 38.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 39.Moran, J. V., et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 40.Muckenfuss, H., et al. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 41.Narvaiza, I., et al. 2006. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J. Virol. 80:12236-12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narvaiza, I., et al. 2009. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niewiadomska, A. M., et al. 2007. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81:9577-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng, G., et al. 2007. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood 110:393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen, E. F., et al. 2004. UCSF chimera-A visualization system for exploratory research and analysis. J. Computat. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 46.Prochnow, C., R. Bransteitter, M. G. Klein, M. F. Goodman, and X. S. Chen. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445:447-451. [DOI] [PubMed] [Google Scholar]
- 47.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 48.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 49.Sanville, B., et al. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 6:e1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shandilya, S. M., et al. 2010. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stauch, B., et al. 2009. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc. Natl. Acad. Sci. U. S. A. 106:12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenglein, M. D., M. B. Burns, M. Li, J. Lengyel, and R. S. Harris. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svarovskaia, E. S., et al. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 54.Vartanian, J. P., D. Guetard, M. Henry, and S. Wain-Hobson. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230-233. [DOI] [PubMed] [Google Scholar]
- 55.Wang, T., et al. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 81:13112-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, T., C. Tian, W. Zhang, P. T. Sarkis, and X. F. Yu. 2008. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 375:1098-1112. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, H., et al. 2007. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 360:247-256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.