Abstract
Many human papillomavirus (HPV)-positive high-grade lesions and cancers of the uterine cervix harbor integrated HPV genomes expressing the E6 and E7 oncogenes from chimeric virus-cell mRNAs, but less is known about HPV integration in head and neck cancer (HNC). Here we compared viral DNA status and E6-E7 mRNA sequences in HPV-16-positive HNC tumors to those in independent human keratinocyte cell clones derived from primary tonsillar or foreskin epithelia immortalized with HPV-16 genomes. Three of nine HNC tumors and epithelial clones containing unintegrated HPV-16 genomes expressed mRNAs spliced from HPV-16 SD880 to SA3358 and terminating at the viral early gene p(A) signal. In contrast, most integrated HPV genomes in six HNCs and a set of 31 keratinocyte clones expressed HPV-16 major early promoter (MEP)-initiated mRNAs spliced from viral SD880 directly to diverse cellular sequences, with a minority spliced to SA3358 followed by a cellular DNA junction. Sequence analysis of chimeric virus-cell mRNAs from HNC tumors and keratinocyte clones identified viral integration sites in a variety of chromosomes, with some located in or near growth control genes, including the c-myc protooncogene and the gene encoding FAP-1 phosphatase. Taken together, these findings support the hypothesis that HPV integration in cancers is a stochastic process resulting in clonal selection of aggressively expanding cells with altered gene expression of integrated HPV genomes and potential perturbations of cellular genes at or near viral integration sites. Furthermore, our results demonstrate that this selection also takes place and can be studied in primary human keratinocytes in culture.
Mucosal high-risk (HR) human papillomavirus (HPV) types are found in nearly 100% of carcinomas of the uterine cervix, many anogenital cancers, and ∼25% of head and neck cancers (HNC) (9, 13, 42). The most common HR HPV is type 16, found in over 50% of cervical cancers and 90 to 95% of HPV-positive HNCs. In persistent infection, HPV genomes are maintained as circular, unintegrated plasmids that persist in the nuclei of infected cells (see Fig. 1A). In contrast, HR HPV DNA found in cervical cancer specimens is frequently disrupted and integrated in the cellular genome in a way that potentially alters the expression program of the viral early gene region and/or the host genome (2, 5, 8, 21, 23, 24, 31, 43). The integrated HR HPV fragments in high-grade precancerous cervical lesions, cervical carcinomas, and derived cell lines express the viral transforming genes E6 and E7 from chimeric virus-cell mRNAs, while the downstream early genes that are required for viral replication and regulated viral gene expression are disrupted or silenced (33, 37, 47, 55). Further, chimeric virus-cell mRNAs may be more stable than the viral early mRNAs, which harbor a 3′ destabilization motif (20, 53). In addition, some integration sites in cervical cancer and precursor lesions have been found near potential growth control genes (11, 40, 55).
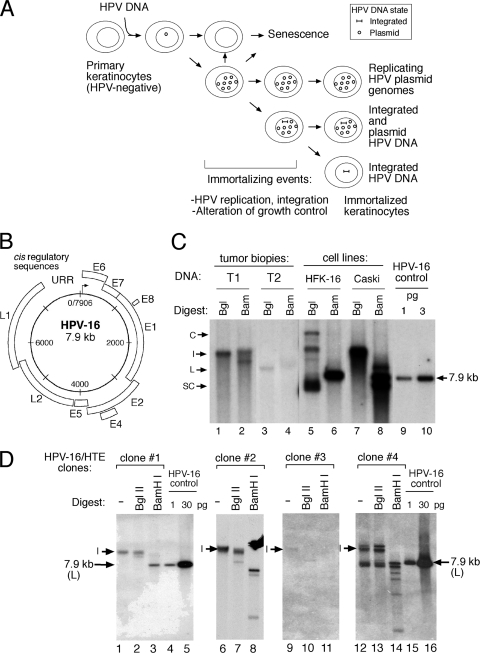
FIG. 1.
(A) Model of the physical status of the HPV genome within the context of viral persistence, alteration of growth phenotypes, and host cell immortalization. (B and C) Organization of the HPV-16 genome (B) or detection of HPV-16 genomes in HNC tumor biopsy specimens by Southern blotting of total DNA (C). (D) Physical status of viral genomes in clonal HPV-immortalized tonsillar epithelial cells (HPV-16/HTE) as determined by Southern blotting of total DNA digested with a “no-cutter” BglII or “single-cutter” BamHI enzyme. HPV DNA forms are as follows: I, integrated; C, concatameric; S, supercoiled; L, linearized. Faster-migrating HPV fragments are derived from digestion of integrated HPV DNAs.
Advances in keratinocyte culture techniques have permitted the study of the life cycle of HR HPV types that replicate efficiently and persist in primary human keratinocytes, such as HPV-16, HPV-31 (32, 34, 35), and to a more limited extent HPV-18 (28, 36). However, relatively little is known about the progression of the infected cell toward a malignant phenotype in vivo or in culture. While some cervical cancers have been shown to harbor extrachromosomal HPV DNA, it is apparent that integration of viral DNA accompanied by disruption of the integrity of the HR HPV genomes represents a common, albeit not absolutely obligatory, step associated with carcinogenic progression associated with the genital tract (42, 57). The majority of cervical carcinomas caused by the two most common HR HPV types, HPV-16 and -18, contain integrated viral sequences (52) that express E6-E7 from chimeric, spliced virus-cell transcripts. However, the genetic structure and expression of viral gene products from integrated versus extrachromosomal HPV genomes in HNC have not been thoroughly defined (4, 22, 25). Furthermore, the integration process has not been systematically studied in primary human keratinocytes—the natural host for HPV infection. Conclusions about the consequences of integration in the host cell have been largely based on studies of established cell lines derived from cervical cancers, such as SiHa, HeLa, and CaSki, or lines derived from other sources (12), cell lines derived from cervical intraepithelial neoplasia (CIN) lesions (W12E [19] or CIN612 [14]), or spontaneously immortalized epithelial cells (such as NIKS 3]) that exhibit inherent chromosomal instability and altered growth phenotypes compared to primary mucosal keratinocytes.
In this study, we compared the physical state of HPV DNAs, mRNA sequences, and spliced structures in nine HPV-16-containing HNC tumors to those found in sets of isogenic clones of primary human keratinocytes derived from tonsillar or foreskin epithelia immortalized by the introduction of plasmid HPV-16 genomes (28, 49). These culture reagents and assays provide the necessary tools to answer questions regarding viral integration and oncogenic progression that otherwise might not be effectively addressed with clinical material alone and offer an effective model system for examining HR HPV integration and its role in HPV-associated carcinogenesis.
MATERIALS AND METHODS
Cell culture, clinical material, and plasmid constructions.
Primary human foreskin (HFK) and tonsillar epithelial (HTE) keratinocyte cultures were prepared from neonatal foreskins or tonsillectomy-derived tissues as described previously (28, 32, 44, 49). All keratinocyte cultures were grown on irradiated J2 fibroblast feeder cells in E medium, containing 0.5 μg/ml hydrocortisone, 0.1 nM cholera toxin, 5 μg/ml transferrin, 5 μg/ml insulin, 2 nM 3,3′-5-triodo-l-thyronine, and 5 ng/ml epidermal growth factor (18). Total DNA was extracted from frozen HNC tumor biopsy specimens derived from various oropharyngeal sites as described previously (26, 45), and the physical status of the HPV DNA was determined by Southern blotting. The CIN-derived, HPV-16 W12E subclone (harboring extrachromosomal HPV-16) and replication-competent HPV-16 W12 plasmid genome were gifts from Paul Lambert (19).
Transcription, replication, and immortalization assays.
Prior to transfection, the wild-type (wt) HPV genome was first released from the bacterial pUC vectors by restriction digestion of HPV-16 W12 plasmid DNA with BamHI and then religated and purified over plasmid purification columns (MaxiKit; Qiagen, Valencia, CA) as described previously (30). For RNase protection assays, 3 μg of recircularized DNA was transfected into primary keratinocyte cell cultures. Total RNA was harvested 24 h later using RNAqueous kits (Ambion, Austin, TX) and analyzed in RNase protection assays using PCR-generated probes as described previously (31) or by 3′ random amplification of cDNA ends (3′RACE) (amplification of papillomavirus oncogene transcripts [APOT]) (58). Total DNA samples were extracted (QIAamp DNA blood kit; Qiagen, Valencia, CA) and digested, and HPV-16 was linearized with BamHI or BglII endonuclease before Southern blotting was done (30). For colony formation assays, 2 × 106 low-passage (8 to 10 population doublings [PDs] postexplant) primary human keratinocytes per 100-mm by 20-mm dish were transfected with 2 μg of recircularized HPV DNA and 1 μg of pRSV-neo. Control cultures transfected with pCMV-ßgal showed reproducible transfection efficiencies of 15 to 25%. Cells were transferred at dilutions onto an irradiated J2 fibroblast feeder 1 day later, selected in 100 to 200 μg G418/ml E medium for 5 days, and allowed to grow for another 15 to 20 days without selection. Individual colonies were subcultured using cloning cylinders from dishes with <40 colonies. Total cellular DNA and total RNA were harvested from clonal cultures to assess HPV persistence and transcription as described above.
Mapping chimeric virus/cellular mRNA and DNA structures.
Amplified 3′RACE products derived from virus-cell chimeric mRNAs were electrophoretically resolved on a 1% agarose gel, excised, purified (QIAkwik PCR purification kit; Qiagen, Valencia, CA), and sequenced (University of Iowa DNA Core Laboratories). Chimeric HPV/cell sequences were then compared to available human genome data (GenBank) by using BLASTN v.2.2 (NIH). HPV-cellular DNA junctions were amplified from total DNA isolated from selected clonal HPV-immortalized keratinocytes by Splinkerette PCR mapping (17), and sequence analysis of purified amplicons was done as described above.
Southern, Northern, and Western blotting.
For Southern blotting, 2 μg of whole-cell DNAs of HPV-induced clonal cultures was resolved on 1.0% agarose gels, depurinated in 0.25 M HCl, and blotted directly onto positively charged nylon membranes (Hybond-XL; Amersham Biosciences Corp., Piscataway, NJ) by alkaline transfer with 0.4 N NaOH. Blots were then hybridized at 65°C with probes (1.5 × 106 cpm/ml hybridization buffer) containing an equimolar cocktail of PCR-amplified segments of HPV-16 (nucleotides [nt] 6226 to 3873/4471 to 6000) [α-32P]dATP/dCTP labeled by random priming (HotPrime kit; GenHunter Corp., Nashville, TN). A titration of the respective linearized HPV DNA was included to determine viral copy numbers per cell. For Northern blotting, a total of 8 μg of total RNA was electrophoretically resolved on a 1% agarose gel. Probes for c-myc and involucrin were prepared from the respective PCR-amplified fragments labeled by random priming (HotPrime kit; GenHunter Corp., Nashville, TN). For Western blotting, a total of 30 μg of cell lysates derived from primary human keratinocytes, clonal HPV-immortalized keratinocyte clones, HeLa cells, and murine fibroblasts were resolved via SDS-PAGE and blotted with monoclonal antisera specific to human c-myc (Santa Cruz Biotechnology, Santa Cruz, CA) using standard protocols. Blots were then stripped and rehybridized with monoclonal beta-actin antiserum (Sigma, St. Louis, MO) as a control.
Clonal cell line characterization.
Growth phenotypes were determined by plating 7 × 105 to 1 × 106 cells onto 100-mm by 20-mm dishes in the presence of irradiated fibroblasts, refeeding the cultures daily with E medium within a 3- and 7-day period, and determining the final count of keratinocytes after differential removal of feeder fibroblasts in a hemocytometer to determine PD times. The PD times (in days) were calculated as follows: PD = [log (0.5)/log(nt − n0)/T], where nt is the number of cells on day t, n0 is the initial number of cells, and T is the total number of days in culture. Karyotypes were determined by arresting cells grown without feeders for 24 to 36 h with colcemide prior to G banding of chromosomes and digital analysis of structural and numerical abnormalities of representative mitoses by the Cytogenetics Laboratory, Department of Pediatrics, University of Iowa Hospitals and Clinics.
RESULTS
Detection of extrachromosomal and integrated HPV-16 genome structure and expression in HNC tumors.
Total DNA and RNA were extracted from a set of nine HNC tumor biopsy specimens. We initially examined the physical status of the viral DNA in these tumor samples. Consistent with our previous studies, which reported genomic mapping of HPV-16 and HPV-18 DNA in cervical carcinoma biopsy specimens (31), the majority of tumor biopsy specimens harbored integrated HPV-16 DNA (Fig. 1 C, lanes 1 to 4, and Table 1), similar to HPV integration patterns observed in some cell lines, such as Caski, which harbors multiple tandem copies of the HPV-16 genome (Fig. 1C, lanes 7 and 8). We also examined the physical status of HPV-16 genomes in immortalized HTE cells, which similarly displayed a range of viral integration patterns via Southern blotting (Fig. 1D). Chimeric HPV E6-E7 transcripts were subsequently detected in total RNA extracted from HNC tumors by 3′RACE (APOT), followed by sequence analysis of purified amplification products (Fig. 2 A to C) corresponding to viral/cellular transcripts in tumor samples harboring integrated or extrachromosomal HPV DNA (Table 1).
TABLE 1.
HPV integration status in HNC tumor tissues
| Tumor | HPV DNAa | mRNA splice structureb | GenBank accession no.b | Chromosome no. | Gene locusb |
|---|---|---|---|---|---|
| T1 | I | 880^human chromosome 13 | AL354696.11 | 13 | MTRF-1 |
| T2 | I | 880^human chromosome 6 | AL353138.10 | 6 | HNRPA3 |
| T3 | I | 880^human chromosome 13 | L11910.1 | 13 | Rb gene |
| T4 | I | 880^human chromosome 11 | AC009635.5 | 11 | Unknown |
| T5 | I | 880^human chromosome 7 | AC002467 | 7 | Unknown |
| T6 | I | 880^human chromosome 6 | AL135839 | 6 | SGK1 |
| T7 | E | 880^3358 HPV | |||
| T8 | E | 880^3358 HPV | |||
| T9 | E | 880^3358 HPV |
Physical status (I, integrated; E, extrachromosomal) of HPV DNA, determined by Southern blotting as illustrated in Fig. 1C.
Chimeric mRNA-derived amplification products were sequenced to determine mRNA structure (type A) and compared to the NIH GenBank database by BLASTN search to identify chromosomal integration loci: HNRPA3, heterogenous ribonucleoprotein A3; SGK1, serum glucocorticoid regulated kinase 1; MTRF1, mitochondrial translation release factor.
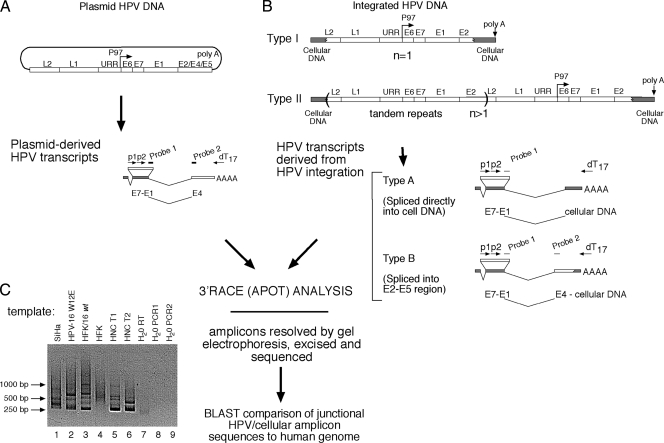
FIG. 2.
Analysis of HPV-16 integration patterns. (A) Schematic of the distinctive structure of viral transcripts derived from intact HPV-16 plasmid genomes. (B) Structures of viral transcripts (type A or B) derived from either type I or type II integrated HPV DNA. (C) Example of 3′RACE (APOT) analysis of resolved amplicons derived from total RNA extracted from SiHa, HPV-16 W12E, HPV-16/HFK, primary (HPV-negative) HFK cells, and HNC tumor biopsy specimens T1 and T2.
Tumors that harbored persistent levels of extrachromosomal HPV plasmid genomes were found to transcribe early genes from viral early gene mRNAs that initiated at the HPV-16 early promoter (MEP), P97, used the viral splice donor (SD) site at nt 880 and splice acceptor (SA) at nt 3358, and terminated at the major early viral poly(A) site at nt 4215 (Fig. 2A). In contrast, integration of the viral DNA can involve incorporation of a single copy of the viral genome (type I) or integration of multiple genomic copies in a tandem array (type II). Both type I and type II DNA integration patterns can generate distinct chimeric virus/cell transcripts (type A and type B mRNAs) that initiate at the P97 promoter under the control of the ubiquitously preserved viral regulatory region and terminate at polyadenylation sites within cellular genes (Fig. 2B) (20, 24).
P97-initiated mRNAs generated from integrated viral DNA sequences commonly utilize the HPV SD 880 in conjunction with SA sites located within adjacent cellular DNA, terminating in cellular poly(A) addition sites (type A mRNAs) (Fig. 2B). Alternatively, some integrated HPV structures express mRNA transcripts that splice from the SD 880 to the viral splice acceptor at 3358 (SA 3358) and then continue directly into cellular sequences through the virus/cell DNA junction (type B mRNA). Both type A and type B mRNAs can be produced from the same integrated HPV genomes, provided the HPV SA sites (e.g., SA 3358) are intact, as determined by viral and/or cellular splice site affinities.
We analyzed the structure of chimeric HPV/cellular mRNA isolated from HNC tumor biopsy specimens and clonal HPV-immortalized keratinocytes by 3′RACE analysis which specifically amplified HPV transcripts in tumor samples and HPV-16 control cell lines (Fig. 2C, lanes 2, 5, and 6) but not in HPV-negative controls (Fig. 2C, lanes 4 and 7 to 9). All chimeric HPV/cell mRNA sequences were compared to human sequences in the GenBank database (NIH) using BLASTN v.2.2 (Tables 1 to 3 ).
TABLE 2.
Chimeric RNA from clonal HPV-16-positive keratinocytes
| Clone no. | Splice structurea | GenBank accession no.b | Chromosome | Gene locusb |
|---|---|---|---|---|
| HTE-01 | 880^3358-3548-human chromosome 8 | NT_008046.16 | 8 | c-myc |
| HTE-02 | 880^human chromosome 5 | NT_006713.15 | 5 | Integrin α-2 |
| HTE-03 | 880^human chromosome 2 | NW_001838868 | 2 | Unknown |
| HTE-04 | 880^3358-3469-human chromosome 2 | NW_001838863.1 | 2 | Unknown |
| HTE-05 | 880^human chromosome 8 | NT_008046.16 | 8 | c-myc |
| HTE-06 | 880^human chromosome 3 | NT_005612.16 | 3 | Plexin D1 |
| HTE-07 | 880^human chromosome 1 | NT_004487.19 | 1 | Monooxygenase 3, isoform 1 |
| HTE-08 | 880^human chromosome 8 | NT_008046.16 | 8 | c-myc |
| HFK-01 | 880^human chromosome 4 | NT_016354.9 | 4 | FAP-1/PTPN13 |
| HFK-01 | 880^3358-3633-human chromosome 4 | NT_016354.9 | 4 | FAP-1/PTPN13 |
| HFK-02 | 880^human chromosome 12 | NW_001838060.2 | 12 | Methionine sulfoxide reductase B3 |
| HFK-03 | 880^human chromosome 4 | NT_016354.9 | 4 | FAP-1/PTPN13 |
| HFK-04 | 880^human chromosome 6 | NW_001838981 | 6 | |
| HFK-05 | 880^human chromosome 6 | NT_007299.13 | 6 | |
| HFK-06 | 880^human chromosome 7 | AC005246 | 7 | Atrophin-1-interacting gene |
| HFK-07 | 880^3358-3776^human chromosome 6 | NT_025741.15 | 6 | Laminin alpha 2 antisense |
| HFK-08 | 880^human chromosome 12 | NT_029419.12 | 12 | HMG AT-hook, isoform b |
| HFK-09 | 880^human chromosome 6 | NT_025741.15 | 6 | Laminin alpha 2, isoform b |
| HFK-10 | 880^human chromosome 1 | NT_032977.9 | 1 | |
| HFK-11 | 880^human chromosome 12 | NW_001838060.2 | 12 | Methionine sulfoxide reductase B3 |
| HFK-12 | 880^human chromosome 12 | NW_001838060.2 | 12 | Methionine sulfoxide reductase B3 |
| HFK-13 | 880^human chromosome 8 | NW_001839136.1 | 8 | Protein serine kinase H2 |
| HFK-14 | 880^human chromosome 10 | NT_030059.13 | 10 | |
| HFK-15 | 880^human chromosome 8 | NT_008046.16 | 8 | c-myc |
| HFK-16 | 880^3358-3631-human chromosome 9 | NT_008470.19 | 9 | |
| HFK-17 | 880^human chromosome 4 | NT_016354.19 | 4 | BMP-2 inducible kinase, isoform a |
| HFK-18 | 880^human chromosome 12 | NT_029419.12 | 12 | HMG AT-hook, isoform b |
| HFK-19 | 880^human chromosome 8 | NT_008046.16 | 8 | c-myc |
| HFK-20 | 880^3358-3489^human chromosome 5 | NT_034772.6 | 5 | Fer tyrosine kinase |
| HFK-21 | 880^human chromosome X | NT_079573.4 | X | |
| HFK-22 | 880^human chromosome 12 | NW_001838060.2 | 12 | HIV-1 Rev binding protein 2 |
| HFK-23 | 880^human chromosome 13 | NT_024524.14 | 13 |
Chimeric mRNA-derived amplification products from HPV-immortalized human tonsillar (HTE) or foreskin (HFK) keratinocytes were sequenced to determine structure.
Chimeric mRNA-derived amplification products were compared to the GenBank database by BLASTN search; for example, FAP-1 is FAS-activated phosphatase 1 (PTPN13). Clonal HPV+/HTE cultures were derived from 3 independent experiments, while HPV+/HFK clones were derived from 12 independent experiments.
TABLE 3.
HPV integration can occur during extended culturing
| Clonea | 32 PDs (t = 0 days) |
∼120 PDs (t = 180 days) |
Splice structureb | Gene locus | ||||
|---|---|---|---|---|---|---|---|---|
| HPV DNA status | PD time (days) | Karyotype | HPV DNA status | PD time (days) | Karyotype | |||
| A1 | I | 1.4 | 2N {46,XY} | I | 1.4 | 2N {46,XY, add(3)(p26),i(8)(q10), +9} | 880^h-chromosome 6 | HMG AT Hook |
| A2 | P | 1.3 | 2N {46,XY} | I | 1.4 | 2N {46,XY, i(8)(q10)} | 880^3358 | |
| A3 | I | 1.7 | 2N {46,XY} | I | 1.9 | Near 4N {84∼89, XXY,-Y,-4,-7,i(8)(q10),del(11)(p11.2), −13,−14,−15,−16,+20} | 880^h-chromosome 13 | Unknown |
| A4 | P | 1.9 | 2N {46,XY} | I | 1.9 | 2N {46,XY, i(8),(q10),+21} | 880^unknown | |
| A5 | I | 2.7 | 2N {46,XY} | I | 1.3 | >2N {52,XY, +5,+6,+9,+14,+20,+21} | 880^h-chromosome 6 | Laminin α2 |
| A6 | P | 3.0 | 2N {46,XY} | P | 1.5 | 2N {46,XY, add(5)(p13), add(6)(p25),add[15](q22)} | NDc | |
A set of six clonal HPV-16-immortalized human foreskin keratinocytes was cultured for 180 days, or an average of 120 population doublings (PDs), while monitoring the population doubling rate (PD) in days, viral DNA status (P, plasmid; I, integrated) by Southern blotting, and the host karyotype.
Chimeric mRNAs were analyzed by 3′RACE and BLASTN v.2.2 comparison to human cellular sequences (GenBank). h-chromosome, human chromosome.
ND, not determined.
In HPV-16 HNC tumors, sequence analysis of the amplified products derived from the chimeric virus-cell mRNAs revealed viral integration at multiple loci within the human genome, with the majority conforming to a type A mRNA structure—splicing from the HPV SD 880 to various cellular SA sites. One integration event occurred within the retinoblastoma (Rb) gene (Table 1). Analysis of the chimeric HPV/Rb mRNA in this biopsy specimen demonstrated that it is spliced from the HPV-16 SD 880 site to a cellular SA site upstream of intron 17 in an antisense orientation within the Rb gene. Any effects of expression of this potential antisense Rb message on keratinocyte growth, however, remain to be determined.
Detection of extrachromosomal and integrated HPV-16 expression in clonal HPV-16-immortalized keratinocytes.
We recently reported the characterization of HPV-immortalized keratinocyte clones derived from transfection of intact, recircularized HPV genomes into primary keratinocytes of human foreskin (HFK), cervix (HCK), and tonsillar epithelial (HTE) origins (28). These cultures harbor persistent levels of extrachromosomal HR-HPV plasmid genomes (31) that permitted the characterization of phenotypic changes in the host cells, such as growth patterns and genomic stability, that might otherwise be masked in heterogeneic mass cultures or tissue biopsy specimens. Clonal keratinocyte cultures harboring integrated HPV genomes were used to characterize HPV integration events in HPV+/HTE clones in this study. Similar to the chimeric virus/cell mRNA analysis of HNC tumor biopsy specimens in Table 1, 3′RACE analysis of eight HPV-16 HTE clonal cultures revealed distinct HPV-cell mRNAs (spliced chimeric type A or type B mRNA structures) associated with a variety of cellular genes, including the c-myc protooncogene locus in three clones (Table 2, HTE-01, -05, and -08) and the plexin D1 and integrin α2 genes (Table 2, HTE-06 and HTE-02, respectively).
We detected a comparable diversity in the integration structures in 23 individual HPV-16/HFK clones harboring integrated HPV-16 genomes (Table 2). We noted a wide range of cellular loci linked to integration of the viral genome, including two additional clones harboring c-myc integrations, the majority of which generated type A chimeric virus/cell mRNA transcripts, splicing from HPV SD 880 into various cellular SA sites. Interestingly, we also detected two clones (Table 2, HFK-01 and -03) with integration events linked to the 4q21 locus (similar to a previously reported HPV-16 integration in a genital carcinoma [54]) and the Fas-activated phosphatase 1 (FAP-1) (PTPN13, PTPL1) gene, a critical cell growth regulator which inhibits Fas-induced apoptosis (1). The chimeric HPV-16/FAP-1 mRNA (type A) from HFK-03 spliced from the HPV-16 SD 880 to an SA site upstream of exons 43 to 48 at the 3′ end of the FAP-1 gene, thus preserving the 317 amino acid catalytic domain in frame. A second FAP-1-associated integration in keratinocyte clone HFK-01 produced both type A and type B chimeric mRNAs, both of which incorporated the same SA site located upstream of the FAP-1 open reading frame (ORF) (Table 2). This integrant could potentially generate an mRNA capable of expressing an HPV/FAP-1 fusion protein or alternatively influence the expression of FAP-1 or JNK3 (located upstream) by altering the activity of their shared bidirectional promoter. Additional HPV integration loci detected among these HPV+/keratinocyte clones included the methionine sulfoxide reductase, laminin α2, atrophin-1 interacting protein, and serine/tyrosine kinase genes.
Further, we mapped the DNA structures of HPV/cellular integration junctions in a subset of the clonal cell lines to confirm the results obtained from the chimeric mRNA (3′RACE) sequence analyses. Using “splinkerette” amplification and comparative BLAST sequence analysis of the DNA junctions, we detected chimeric DNA consistent with templates capable of expressing the chimeric virus/cellular mRNA, such as in clone HFK-14, which harbors an HPV-16-human chromosome 10 integration pattern, i.e., HPV-16 (nt 1127 to 1691)-chromosome 10, which corresponds to a template capable of encoding a spliced HPV-16 880^human chromosome 10 transcript, as detected in the same clonal keratinocytes (Table 2). Similarly, clone HTE-06 contained a DNA integration junction corresponding to HPV-16 (nt 1988 to 2612)-chromosome 3, capable of encoding the same chimeric mRNA detected by 3′RACE analysis.
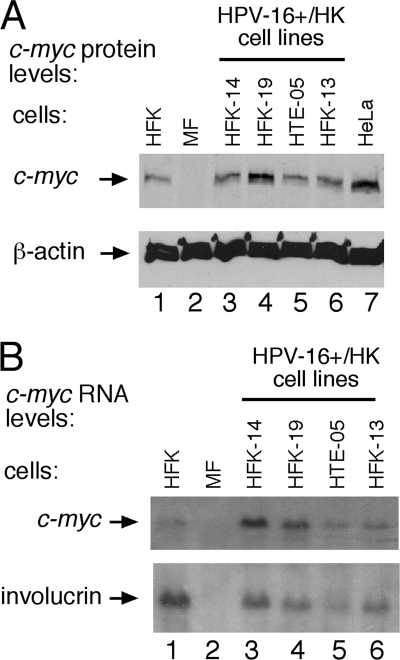
Although some HPV integration sites appear to be distally upstream of the c-myc gene and its promoter, previous studies have shown that chromosomal alteration of sequences distal to the c-myc gene can dysregulate c-myc expression, as in Burkitt's lymphoma (56). Furthermore, HPV integration near the c-myc gene in some invasive genital carcinomas can be associated with overexpression of c-myc (11, 39) (Fig. 3 A, compare lanes 1 and 7). In this study, some of the cultures harboring c-myc integrations (Table 2) displayed increased c-myc protein levels (Fig. 3A, lanes 4 and 7) compared to those of HPV-negative primary keratinocytes (Fig. 1A, lane 1) by Western blotting, though elevated c-myc protein levels were not strictly linked to HPV integration within the 8q24 locus (Fig. 3A, compare lanes 3 to 6). Not surprisingly, c-myc RNA levels were elevated in some clones, as shown by Northern blotting (Fig. 3B, compare lane 1 to lanes 3 and 4), although this was not strictly linked to elevated c-myc protein levels or 8q24 disruption (Fig. 3A). These results are consistent with those in previous reports analyzing variable myc expression in genital and head and neck carcinomas and in established cell lines derived from various sources (15, 39).
FIG. 3.
c-myc expression is altered in some HPV-16-immortalized integrated clonal human keratinocytes (HK). (A) c-myc protein levels were quantified by Western blot analysis of 30 μg of total protein extracted from clonal HPV+/HK cell lines (defined in Table 2) with integration at chromosome 10 (lane 3, clone HFK-14) and in chromosome 8 (c-myc) (lanes 4 to 6) compared to c-myc levels in primary HFK cells. Murine fibroblast (MF) cell extracts were used to demonstrate human c-myc antiserum specificity. (B) c-myc RNA levels quantified by Northern blot analysis of total RNA isolated from clonal HPV+/HFK cell lines and control cultures. The blot was stripped and rehybridized with a probe specific for cellular involucrin RNA. HeLa cells were used as a control for elevated c-myc levels compared to those of HFK (primary foreskin keratinocytes). Murine fibroblasts were used as a negative c-myc control.
HPV-mediated immortalization, integration, and host genomic instability.
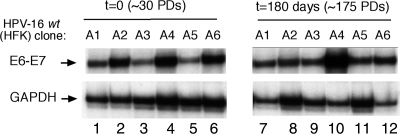
To determine if progressive integration events and genomic instability could be detected in our clonal HPV-immortalized keratinocytes in the course of extended culturing, we passaged six independently derived HPV-16/HFK clones for an additional 6 months (an average of an additional 100 population doublings [PDs], ranging from 60 to 175 PDs) (Table 3). Three of these cultures initially harbored replicating HPV-16 plasmids (Table 3, clones A2, A4, and A6), while the other three contained integrated HPV-16 genomes (Table 3, clones A1, A3, and A5). The growth rate, E6/E7 transcription, host karyotype, and physical status of the viral DNA were determined for each culture prior to and following extended culturing. Initially (at t = 0), the majority of the six clonal cultures were normal diploid, with the exception of clone A5, which was derived from a donor isolate harboring preexisting trisomy 21. Two of the three clonal cultures (Table 3, clones A2 and A4) that contained replicating plasmids subsequently integrated during extended culturing (t = 180 days), confirming that viral integration was possible but not a predetermined outcome under our culture conditions. The majority of the clonal cultures remained diploid with the exception of two HPV-16/HFK clones (Table 3, clones A3 and A5), which were found to be nearly tetraploid and greater than diploid, respectively, and each contained several chromosomal alterations not necessarily linked to integration of the viral genome as shown by 3′RACE analysis (Table 3). These results demonstrated that while these clonal cultures harbored the capacity to acquire alterations to the host genome over time, such genetic instability was not a prerequisite for HPV immortalization or a predetermined outcome in early stages of HPV persistence.
We previously demonstrated that integration of the HPV genome did not necessarily result in an increase in early viral gene expression (28, 31). We therefore examined early viral gene expression after extended culturing, as determined in RNase protection assays. Consistent with our previous reports, transcription from the E6/E7 P97 promoter did not increase in all cultures during this assay, nor did integration of the viral genome necessarily result in an increase in steady-state levels of major early, E6/E7 transcripts (Fig. 4). Similar to our results with syngeneic clonal cell lines previously generated in cervical keratinocytes (49), telomerase activity was already elevated in HPV-immortalized HFKs at low passaging at the beginning of the assay (at t = 0) and remained so during the course of this assay with no direct correlation to the advent of genomic instability of the host cells (data not shown).
FIG. 4.
Clonal HPV-immortalized keratinocytes do not necessarily acquire altered viral transcription phenotypes after extended culturing. Total RNA extracted from a set of six clonal HPV-positive keratinocytes prior to and following extended culturing was analyzed by RNase protection assays. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control.
DISCUSSION
In this study, we compared the structure and expression of HPV-16 genomes in HPV-associated HNC tumors to those in HPV-immortalized keratinocyte clones by restriction enzyme analysis and mRNA sequencing. Our results indicate that while some HNC tumors harbor unintegrated HPV-16 plasmid genomes, HPV sequences are integrated in the majority of these cancers in a manner similar to findings for cervical cancers and precursor lesions. We also found that integrated HPV-16 genomes in 31 independent, primary human tonsillar or foreskin keratinocyte clones immortalized by the introduction of complete, circular HPV-16 plasmid genomes gave rise to chimeric virus-cell mRNA transcripts with the same features as those found in cervical and head and neck cancers. Furthermore, HPV integrations in HNC and in culture often took place within or near growth control genes, some of which have the potential to influence the cell growth phenotype.
Taken together, these findings support the hypothesis that HPV integration in cancers of the cervix and of the head and neck is a stochastic process that results in the clonal selection of aggressively expanding cells. Based on our results from HNCs and keratinocyte clones together with those from cervical lesions, this selective advantage may not be due just to altered viral gene expression from the integrated HPV genomes but frequently may be augmented by perturbations of cellular genes at or near viral integration sites. Our results imply that this clonal selection of expanding cells also takes place in and can be studied under experimental conditions in primary human keratinocytes in culture.
HPV-16-associated head and neck cancers harbor and express either extrachromosomal or integrated viral genomes.
Although six of nine HNC tumors contained integrated HPV-16 sequences expressing the viral E6 and E7 oncogenes as chimeric virus-cell mRNAs, the remaining three HNC tissues were found to harbor extrachromosomal HPV-16 plasmid genomes by both Southern blot and viral mRNA sequence analyses.
Previous conclusions about the physical status of HPV genomes in HNC have been based on indirect methods. A comparison of the levels of mRNAs of the preserved 5′ E6 oncogene region and the common 3′ region of viral transcripts encompassing the HPV E2 ORF (48) discerns HPV transcripts stemming from unintegrated, extrachromosomal HPV plasmid genomes (which transcribe both the 5′ “E6” and 3′ “E2” mRNA segments) from chimeric virus-cell mRNAs transcribed from integrated HPV genomes (in which 3′ E2 is absent). However, Fig. 2A further illustrates why testing ratios of the HPV E6 and E2 segments in HPV-harboring lesions at the level of DNA amplification as reported (38) is unsuitable for the detection of unintegrated HPV genomes. This E6/E2 DNA ratio approach will identify many but not all of the less common type I integration events that involve the disruption of a single circular HPV plasmid genome as long as there is an interruption of the 3′ early (or “E2”) gene segment chosen as the test amplicon. Not all HPV type I integrations match this assumption, since some of those found in cervical cancers harbor smaller deletions in the E2 or E1 ORF (31). The HPV E6/E2 DNA ratio cannot discern complete, unintegrated circular HPV DNA plasmids from the integrated tandem head-to-tail arrays of complete but linearized HPV DNA genomes in type II integrations (20, 31).
As in other studies, the number of suitable fresh-frozen HNC samples we were able to examine was limited. Nevertheless, our analysis of HNC tumor DNA and HPV-16 mRNA sequencing conclusively show that, similar to preneoplastic cervical lesions and cancers (52, 55), HPV-16 integration in HNC is not a prerequisite for carcinogenesis, since HPV-16 genomes persist and are transcribed as unintegrated plasmids in some tumors. Determining whether the prevalence of unintegrated and integrated HPV-16 genomes in HNC differs from that in cervical lesions will require larger epidemiologic studies.
HPV-16 mRNAs in immortalized primary human tonsillar or foreskin keratinocytes show features indistinguishable from those in head and neck and cervical cancers.
The structure of chimeric virus-cell mRNAs expressed from the integrated HPV-16 genomes in HNC corresponded to those previously seen in cervical cancers, precursor lesions, and derived cell lines in integrants derived from HPV-16-positive CIN W12 cells (20) (Fig. 2A). All of the HNC chimeric mRNAs were spliced from the viral splice donor (SD) site at nt 880 directly into cellular splice acceptor (SA) sites as type A transcripts. Chimeric virus-cell mRNAs found in 31 independent HPV-16-immortalized keratinocyte clones of tonsillar or foreskin epithelial origin followed the same pattern: 6 of 8 tonsillar and 16 of 23 foreskin keratinocyte clones (71% in all) expressed type A chimeric E6-E7 mRNAs spliced from SD880 to and terminating in cellular sequences. The remaining two tonsillar and four foreskin clones harbored type B E6-E7 mRNA spliced from SD880 to HPV-16 SA3358, followed by a direct virus-cell DNA junction with the HPV-16 breakpoint between nt 3469 and 3776 and terminating in cellular sequences. Interestingly, integration in one clone, HFK-01, gave rise to both type A and type B E6-E7 mRNAs, a possibility not observed previously in cervical lesions (27).
It is somewhat surprising that the virus-cell E6-E7 mRNAs expressed in immortalized keratinocyte clones with integrated HPV-16 are indistinguishable from those found in HNC and cervical cancer. The integration events observed in the keratinocyte clones are not due to transfection-induced rearrangements. The circular HPV-16 genomes introduced into the cells required initial genome amplification (Fig. 1A) since replication-defective viral genomes with mutations in HPV-16 E1, E2, or cis sequences required for replication failed to immortalize primary keratinocytes in parallel (30). We conclude that immortalization of keratinocytes in culture by HPV involves the same selective pressures as those observed in the development of HPV-associated clinical lesions.
The selective growth advantage potentially conferred on the cell by integrated HR HPV E6-E7 transcripts has traditionally been ascribed to the absence of the full-length product of the viral E2 ORF, the E2-TA transactivator protein (7, 51), or the absence of the E8^E2 replication/transcription repressor (29, 50). All the type A mRNA integrants eliminate the downstream gene region from the E6-E7 mRNAs. Furthermore, type B E6-E7 mRNA-expressing integrants harbor 3′-truncated E2 ORFs with no intact C-terminal DNA binding domain and therefore cannot express a functional repressor of the MEP, P97 (Fig. 2A). However, there is limited information defining the structure of mRNAs encoding E2-TA and no direct data quantifying the level of its expression in HPV persistence or cancer. While limiting levels of E2-TA and/or E8^E2 proteins appear to tightly control viral gene expression and genome replication during HPV persistence, steady-state levels of mRNA encoding E2-TA, E8^E2 (29), and P14-initiated E1 (30) in HPV-16 persistence may be significantly lower than those of E6-E7 mRNA, thus escaping detection in cells with unintegrated, persistent circular or integrated HPV genomes. An additional consequence of HPV integration may be the elimination of the viral 3′ AU-rich destabilization sequence from the chimeric HPV E6-E7-cell mRNA transcripts (20, 53). Nevertheless, we did not discern a clear difference in steady-state E6-E7 mRNA transcript levels between syngeneic keratinocyte clones harboring unintegrated or integrated HPV-16 genomes (Fig. 4) (31).
HPV integration within or near cellular genes may provide additional selective advantages to the cell in cancer progression and in cell culture.
Consistent with previous observations of HPV integration patterns in cervical cancers and precursor lesions, HPV integrations in HPV-associated HNCs and clonal HPV-immortalized keratinocytes occurred in a wide variety of loci on different human chromosomes. Some of the cellular genes at the integration loci are involved in the control of the cell growth phenotype: the retinoblastoma susceptibility gene in HNC T3, 2 integrations in the FAP-1/PTPN13 gene, and recurrent HPV-16 integration at the c-myc locus in 5 out of 31 (16%) HPV-positive keratinocyte clones, also repeatedly detected in cervical cancers (6, 40). While HPV integration associated with these and other cellular genes and associated chimeric mRNA structures is intriguing given their potential involvement in cell immortalization or tumor progression (16, 46), the functional role of these viral integration events in the course of oncogenic progression remains to be defined.
HPV and host genomic instability.
Previous studies have suggested that HPV integration plays a role in genomic instability of the HPV-infected keratinocyte (10, 40, 41). However, the karyotypes of the HPV-immortalized keratinocyte clones harboring extrachromosomal or integrated viral genomes exhibited a normal diploid cell complement with no gross structural chromosomal abnormalities (28). In contrast, HPV-positive cell cultures derived from cervical lesions displayed significant chromosomal aberrations (28). This could be the outcome of accumulation of such alterations as a function of time during the proliferation of the lesion in vivo and additional growth in culture. Consistent with this model, integration of the viral genome could conceivably introduce additional genomic instability. However, the stochastic accumulation of chromosomal alterations as a result of an extended host cell life span is sufficient to account for the observed genomic instability in these long-term cultures (Table 3) and potentially in HPV-associated lesions. We conclude that the individual, syngeneic keratinocyte clones immortalized by unintegrated or integrated HPV genomes serve as a sensitive model for studying virus-cell interactions in the progression of HPV-associated lesions to HNC.
Acknowledgments
We thank Linda Rubenstein for her help with collecting tissue samples and epidemiologic information from HNC patients.
This work was supported by merit awards from the Department of Veterans Affairs to L.P.T. and T.H.H. and by USPHS/National Institutes of Health/NIDCR grants R01 DE015944 and R21 DE017212 to E.M.S.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Abaan, O. D., and J. A. Toretsky. 2008. PTPL1: a large phosphatase with a split personality. Cancer Metast. Rev. 27:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alazawi, W., M. Pett, B. Arch., L. Scott, T. Freeman, M. A. Stanley, and N. Coleman. 2002. Changes in cervical keratinocyte gene expression associated with integration of human papillomavirus 16. Cancer Res. 62:6959-6965. [PubMed] [Google Scholar]
- 3.Allen-Hoffmann, B. L., et al. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Investig. Dermatol. 114:444-455. [DOI] [PubMed] [Google Scholar]
- 4.Braakhuis, B. J., et al. 2004. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J. Natl. Cancer Inst. 96:998-1006. [DOI] [PubMed] [Google Scholar]
- 5.Choo, K. B., C. M. Chen, C. P. Han, W. T. Cheng, and L. C. Au. 1996. Molecular analysis of cellular loci disrupted by papillomavirus 16 integration in cervical cancer: frequent viral integration in topologically destabilized and transcriptionally active chromosomal regions. J. Med. Virol. 49:15-22. [DOI] [PubMed] [Google Scholar]
- 6.Couturier, J., X. Sastre Garau, S. Schneider Maunoury, A. Labib, and G. Orth. 1991. Integration of papillomavirus DNA near myc genes in genital carcinomas and its consequences for proto-oncogene expression. J. Virol. 65:4534-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cripe, T. P., et al. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 6:3745-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, A. P., R. Reid, M. Campion, and A. T. Lörincz. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 65:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Souza, G., et al. 2007. Case-control study of human papillomavirus and oropharyngeal cancer. N. Engl. J. Med. 356:1944-1956. [DOI] [PubMed] [Google Scholar]
- 10.Duensing, S., and K. Munger. 2003. Centrosomes, genomic instability, and cervical carcinogenesis. Crit. Rev. Eukaryot. Gene Expr. 13:9-23. [DOI] [PubMed] [Google Scholar]
- 11.Dürst, M., C. M. Croce, L. Gissmann, E. Schwarz, and K. Huebner. 1987. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. U. S. A. 84:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego, M. I., and P. A. Lazo. 1995. Deletion in human chromosome region 12q13-15 by integration of human papillomavirus DNA in a cervical carcinoma cell line. J. Biol. Chem. 270:24321-24326. [DOI] [PubMed] [Google Scholar]
- 13.Gillison, M. L., and D. R. Lowy. 2004. A causal role for human papillomavirus in head and neck cancer. Lancet 363:1488-1489. [DOI] [PubMed] [Google Scholar]
- 14.Gray, E., et al. 2010. In vitro progression of human papillomavirus 16 episome-associated cervical neoplasia displays fundamental similarities to integrant-associated carcinogenesis. Cancer Res. 70:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha, P. K., et al. 2003. A transcriptional progression model for head and neck cancer. Clin. Cancer Res. 9:3058-3064. [PubMed] [Google Scholar]
- 16.Hoover, A. C., et al. 2009. Impaired PTPN13 phosphatase activity in spontaneous or HPV-induced squamous cell carcinomas potentiates oncogene signaling through the MAP kinase pathway. Oncogene 28:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn, C., et al. 2007. Splinkerette PCR for more efficient characterization of gene trap events. Nat. Genet. 39:933-934. [DOI] [PubMed] [Google Scholar]
- 18.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J. Virol. 73:1835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalantari, M., E. Blennow, B. Hagmar, and B. Johansson. 2001. Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn. Mol. Pathol. 10:46-54. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. H., et al. 2007. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int. J. Cancer 120:1418-1425. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa, K., et al. 1996. Genomic organization of human papillomavirus type 18 in cervical cancer specimens. Jpn. J. Cancer Res. 87:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaes, R., et al. 1999. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 59:6132-6136. [PubMed] [Google Scholar]
- 25.Klussmann, J. P., et al. 2009. Genetic signatures of HPV-related and unrelated oropharyngeal carcinoma and their prognostic implications. Clin. Cancer Res. 15:1779-1786. [DOI] [PubMed] [Google Scholar]
- 26.Klussmann, J. P., et al. 2001. Prevalence, distribution, and viral load of human papillomavirus 16 DNA in tonsillar carcinomas. Cancer 92:2875-2884. [DOI] [PubMed] [Google Scholar]
- 27.Kraus, I., et al. 2008. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 68:2514-2522. [DOI] [PubMed] [Google Scholar]
- 28.Lace, M. J., et al. 2009. Human papillomavirus (HPV) type 18 induces extended growth in primary human cervical, tonsillar, or foreskin keratinocytes more effectively than other high-risk mucosal HPVs. J. Virol. 83:11784-11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lace, M. J., J. R. Anson, G. S. Thomas, L. P. Turek, and T. H. Haugen. 2008. The E8^E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J. Virol. 82:10841-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lace, M. J., J. R. Anson, L. P. Turek, and T. H. Haugen. 2008. Functional mapping of the human papillomavirus type 16 E1 cistron. J. Virol. 82:10724-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lace, M. J., et al. 2009. Upstream regulatory region alterations found in human papillomavirus type 16 (HPV-16) isolates from cervical carcinomas increase transcription, ori function, and HPV immortalization capacity in culture. J. Virol. 83:7457-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J. H., et al. 2004. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc. Natl. Acad. Sci. U. S. A. 101:2094-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matovina, M., I. Sabol, G. Grubisic, N. M. Gasperov, and M. Grce. 2009. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol. Oncol. 113:120-127. [DOI] [PubMed] [Google Scholar]
- 34.McCance, D. J., R. Kopan, E. Fuchs, and L. A. Laimins. 1988. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. U. S. A. 85:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 36.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nambaru, L., B. Meenakumari, R. Swaminathan, and T. Rajkumar. 2009. Prognostic significance of HPV physical status and integration sites in cervical cancer. Asian Pac. J. Cancer Prev. 10:355-360. [PubMed] [Google Scholar]
- 38.Peitsaro, P., B. Johansson, and S. Syrjanen. 2002. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 40:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peter, M., et al. 2006. MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene 25:5985-5993. [DOI] [PubMed] [Google Scholar]
- 40.Peter, M., et al. 2010. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J. Pathol. 221:320-330. [DOI] [PubMed] [Google Scholar]
- 41.Pett, M. R., et al. 2004. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res. 64:1359-1368. [DOI] [PubMed] [Google Scholar]
- 42.Psyrri, A., and D. DiMaio. 2008. Human papillomavirus in cervical and head-and-neck cancer. Nat. Clin. Pract. Oncol. 5:24-31. [DOI] [PubMed] [Google Scholar]
- 43.Pyeon, D., et al. 2007. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 67:4605-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rheinwald, J. G., and H. Green. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6:331-343. [DOI] [PubMed] [Google Scholar]
- 45.Ritchie, J. M., et al. 2003. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int. J. Cancer 104:336-344. [DOI] [PubMed] [Google Scholar]
- 46.Roodink, I., et al. 2005. Plexin D1 expression is induced on tumor vasculature and tumor cells: a novel target for diagnosis and therapy? Cancer Res. 65:8317-8323. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz, E., et al. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 48.Smeets, S. J., et al. 2007. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int. J. Cancer 121:2465-2472. [DOI] [PubMed] [Google Scholar]
- 49.Sprague, D. L., et al. 2002. Telomerase activation in cervical keratinocytes containing stably replicating human papillomavirus type 16 episomes. Virology 301:247-254. [DOI] [PubMed] [Google Scholar]
- 50.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinokurova, S., et al. 2008. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 68:307-313. [DOI] [PubMed] [Google Scholar]
- 53.Vinther, J., M. W. Rosenstierne, K. Kristiansen, and B. Norrild. 2005. The 3′ region of human papillomavirus type 16 early mRNAs decrease expression. BMC Infect. Dis. 5:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wentzensen, N., et al. 2002. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene 21:419-426. [DOI] [PubMed] [Google Scholar]
- 55.Wentzensen, N., S. Vinokurova, and M. von Knebel Doeberitz. 2004. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 64:3878-3884. [DOI] [PubMed] [Google Scholar]
- 56.Zeidler, R., et al. 1994. Breakpoints of Burkitt's lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer 9:282-287. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, Z. M., and C. C. Baker. 2006. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 11:2286-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziegert, C., et al. 2003. A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene 22:3977-3984. [DOI] [PubMed] [Google Scholar]