Abstract
UL69 of human cytomegalovirus (HCMV) encodes a pleiotropic transactivator protein and has a counterpart in every member of the Herpesviridae family thus far sequenced. However, little is known about the conservation of the functions of the nuclear phosphoprotein pUL69 in the homologous proteins of other betaherpesviruses. Therefore, eukaryotic expression vectors were constructed for pC69 of chimpanzee cytomegalovirus, pRh69 of rhesus cytomegalovirus, pM69 of murine cytomegalovirus, pU42 of human herpesvirus 6, and pU42 of elephant endotheliotropic herpesvirus. Indirect immunofluorescence experiments showed that all pUL69 homologs expressed by these vectors were localized to the cell nucleus. Coimmunoprecipitation experiments identified homodimerization as a conserved feature of all homologs, whereas heterodimerization with pUL69 was restricted to its closer relatives. Further analyses demonstrated that pC69 and pRh69 were the only two homologs that functioned, like pUL69, as viral-mRNA export factors. As we had reported recently that nucleocytoplasmic shuttling and interaction with the cellular DExD/H-box helicases UAP56 and URH49 were prerequisites for the nuclear-mRNA export activity of pUL69, the homologs were characterized with regard to these properties. Heterokaryon assays demonstrated nucleocytoplasmic shuttling for all homologs, and coimmunoprecipitation and mRNA export assays revealed that the interaction of UAP56 and/or URH49 with pC69 or pRh69 was required for mRNA export activity. Moreover, characterization of HCMV recombinants harboring mutations within the N-terminal sequence of pUL69 revealed a strong replication defect of viruses expressing pUL69 variants that were deficient in UAP56 binding. In summary, homodimerization and nucleocytoplasmic shuttling activity were identified as conserved features of betaherpesviral pUL69 homologs. UAP56 binding was shown to represent a unique characteristic of members of the genus Cytomegalovirus that is required for efficient replication of HCMV.
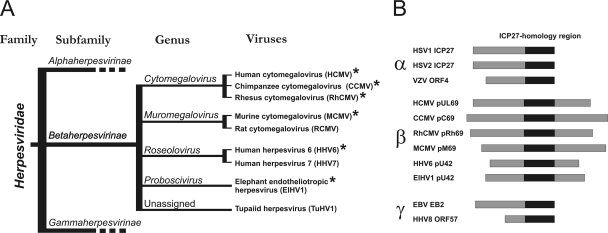
On the basis of biological properties, genome structure, and comparisons of primary amino acid sequences, the family Herpesviridae has been subdivided into three subfamilies, the Alpha-, Beta- and Gammaherpesvirinae (Fig. 1 A) (40). Separation of these subfamilies is estimated to have proceeded approximately 180 to 200 million years ago (12), with development of genera within each subfamily occurring by more recent events (32). The subfamily Betaherpesvirinae contains four genera, Cytomegalovirus, Muromegalovirus, Roseolovirus, and Proboscivirus (Fig. 1A). The most intensively studied betaherpesviruses belong to the genus Cytomegalovirus and include human cytomegalovirus (HCMV; species, Human herpesvirus 5), rhesus cytomegalovirus (RhCMV; species, Macacine herpesvirus 3), and chimpanzee cytomegalovirus (CCMV; species, Panine herpesvirus 2). The genus Muromegalovirus comprises murine cytomegalovirus (MCMV; species, Murid herpesvirus 1) and rat cytomegalovirus (RCMV; species, Murid herpesvirus 2). Human herpesvirus 6 (HHV6; species, Human herpesvirus 6) and human herpesvirus 7 (HHV7; species, Human herpesvirus 7) belong to the genus Roseolovirus and, like HCMV, infect humans. Elephant endotheliotropic herpesvirus (ElHV; species, Elephantid herpesvirus 1) is the founder member of the genus Proboscivirus (14). There are additional viruses within the Betaherpesvirinae subfamily that have not yet been classified into genera (Fig. 1A).
FIG. 1.
Taxonomy of herpesviruses encoding members of the ICP27 family. (A) Schematic phylogeny of the Herpesviridae family. Representative homologs from the viruses included in the functional studies are indicated by asterisks (12). (B) Schematic alignment of alpha-, beta-, and gammaherpesviral proteins in the ICP27 family. The most highly conserved region (ICP27 homology region) is highlighted by a black bar.
Many conserved proteins have been identified among members of the Herpesviridae family, and they are assumed to fulfill similar functions during the course of infection. Among these is a family of homologous proteins (the ICP27 family) whose members function as posttranscriptional activators that facilitate the nuclear export of intronless mRNAs (45, 46, 56). Members of the ICP27 family are present in every mammalian or avian herpesvirus sequenced to date, thus underlining their functional importance. Proteins belonging to this family are depicted in Fig. 1B and include, among others, the alphaherpesviral proteins ICP27 of herpes simplex virus type 1 (HSV-1; species, Human herpesvirus 1) and ORF4 of varicella-zoster virus (VZV; species, Human herpesvirus 2) (25, 42, 47), the betaherpesviral protein pUL69 of HCMV (60, 61), and the gammaherpesviral protein EB2 of Epstein-Barr virus (EBV; species, Human herpesvirus 4) (6, 20). Interestingly, although the overall function of characterized ICP27 family members as posttranscriptional regulators and viral-mRNA export factors appears to be well conserved, specific motifs (e.g., the RNA-binding motif and nuclear localization and export signals) have diverged considerably (46, 49, 56). Indeed, overall amino acid sequence identity among members is low (∼20%). The region containing the C-terminal 200-amino-acid (aa) residues of HSV-1 ICP27 is the best conserved (∼35%) and corresponds to a central domain within pUL69 and other betaherpesviral homologs. This domain is called the ICP27 homology region (Fig. 1B) (59). Thus, pUL69 and its betaherpesviral homologs are characterized by a unique C-terminal extension that is absent from the alphaherpesviral and gammaherpesviral proteins (Fig. 1B) (10, 59, 59).
HCMV pUL69 is a nuclear phosphoprotein that consists of 744-amino-acid residues and has a molecular mass of ∼105 to 116 kDa. It is expressed during the early late phase of viral replication (60). By analogy to its counterparts in HSV-1 (ICP27) and EBV (EB2), pUL69 functions as a transactivator of gene expression (6, 39, 60). One well-conserved characteristic of pUL69 is its ability, like that of HSV-1 ICP27 and human herpesvirus 8 (HHV8; species, Human herpesvirus 8) ORF57, to self-associate and form high-molecular-mass complexes (28, 31, 62). Self-interaction is mediated by the conserved ICP27 homology region present in all members of the family and represents the only common function so far assigned to this domain. Moreover, pUL69, like all well-characterized members of this family, acts as a posttranscriptional transactivator that facilitates the nuclear export of mRNAs via its capability to shuttle between the nucleus and the cytoplasm and recruit components of the cellular-mRNA export machinery (8, 9, 11, 29, 58). Importantly, in the case of HCMV pUL69, loss of mRNA export activity occurs either when the protein-protein interaction with UAP56 (a cellular DExD/H-box RNA helicase involved in mRNA export) is inhibited or when its export by the CRM1-independent pathway is blocked (27, 29).
The aim of the current study was to carry out a functional characterization of betaherpesviral pUL69 homologs in order to identify conserved and unique features of this protein family and thus to gain further insights into how these viral proteins are able to access and manipulate the cellular machinery for viral-mRNA export. In order to include homologs from each of the four betaherpesviral genera, we decided to analyze pUL69 (HCMV), pC69 (CCMV), and pRh69 (RhCMV) as representatives of the genus Cytomegalovirus, pM69 (MCMV) as a representative of the genus Muromegalovirus, HHV6A pU42 as a representative of the genus Roseolovirus, and ElHV1 pU42 as a representative of the genus Proboscivirus. We show that homodimerization and nucleocytoplasmic shuttling activity are conserved features of the betaherpesviral pUL69 protein family, whereas binding to the mRNA export factor UAP56 is confined to members of the genus Cytomegalovirus and correlates with the activity of the respective proteins in facilitating the nuclear export of unspliced RNA. Our observation that UAP56-binding-deficient viruses exhibit a strong replication defect ultimately leads to the conclusion that UAP56 interaction is crucial for efficient replication of HCMV.
MATERIALS AND METHODS
Oligonucleotides and plasmid constructs.
Oligonucleotide primers used for this study were purchased from Biomers GmbH (Ulm, Germany). The sequences of all primers are listed in Table 1. Expression constructs for betaherpesviral pUL69 homologs CCMV pC69 (GenBank accession number AF480884; gi 19881028) (13), RhCMV pRh69 (GenBank accession number AY186194; gi 31377878) (21), MCMV pM69 (GenBank accession number U68299; gi 1688100) (37), HHV6A pU42 (GenBank accession number X92436; gi 1044869) (18), and ElHV1 pU42 (GenBank accession number AF322977; gi 109639413) (16) were generated by either a two-step nested PCR followed by a specific PCR (CCMV pC69 and ElHV1 pU42) or by direct amplification of the respective open reading frame (ORF) from genomic DNA (RhCMV pRh69, MCMV pM69, and HHV6A pU42). The respective amplicons were digested with adequate restriction enzymes followed by ligation into a pcDNA3.1-derived vector that facilitates expression in fusion with either a FLAG (pHM971) or a Myc (pHM1580) epitope (23). Site-directed mutagenesis of the UAP56 interaction motif within FLAG-C69 and FLAG-Rh69 was performed using a QuikChange site-directed mutagenesis kit as instructed by the manufacturer (Stratagene). The resulting constructs, as well as FLAG-UL69mutUAP56 (29), were subcloned into the Myc expression vector pHM1580, yielding Myc-UL69mutUAP56, Myc-C69mutUAP56, and Myc-Rh69mutUAP56. The integrity of all newly generated plasmids was confirmed by automated DNA sequence analysis. FLAG-UL69, Myc-UL69, and FLAG-URH49 were constructed as described earlier (28, 29). Use of the chloramphenicol acetyltransferase (CAT) reporter plasmid pDM128/CMV/RRE and the human immunodeficiency virus type 1 (HIV-1) Rev (pcRev) expression plasmid (kindly provided by J. Hauber, Hamburg, Germany) was described elsewhere (54). FLAG-UAP56 was a generous gift of K. Nagata (35) and was subcloned into pHM1580 as described earlier (29). Plasmids pCFN-βGal and pCFNrev-βGal, used as controls in interspecies heterokaryon assays, were obtained from M. Dobbelstein (Göttingen, Germany) (15, 41, 41) and were used as described previously (27). UL69 expression plasmids mUAP, ΔR1ΔRS, and ΔR2ΔRS, encoding N-terminal mutations of UL69 that either abrogated the RNA-binding activity and/or the interaction of UL69 with UAP56, were described in previous studies (29, 55).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| 5CCMVC69nestedO | GCCGCGCTTTCCGCGTGCCC |
| 5CCMVC69nestedI | ATATATTAGCAGGCCGCGTG |
| 5CCMV69EcoRV | GCTAGATATCCATGGAGCTGCACGGGCGTG |
| 5RhCMV69BamH1 | CAGTGGATCCTCATTCGAACGCGAAGAACGTGC |
| 5MCMV69EcoRV | ATCGGATATCCATGCTGCGGACCGGTGTCAAGAGACG |
| 5HHV6U42bamH1 | CAGTGGATCCTATCCTCGTGGAGTAAAACGCTC |
| 5ELHVU42nestedO | CAGCTAGATGATCTGTCCCC |
| 5ELHVU42nestedI | GGGAACAGCAACGCCAGCAG |
| 5ELHVU42Bam H1 | GCATGGATCCATGGAATCTGGAAGGCGG |
| 3CCMVC69nestedO | GAACATCCGAATCGGGGTGG |
| 3CCMVC69nestedI | GGGAGAGAGACAGGCAAGTC |
| 3CCMV69XhoI | CAGTCTCGAGTCAGTACTCGTCCATGTCGTCGCTG |
| 3RhCMV69XhoI | CAGTCTCGAGCTACAAATATCCCTCTTCATCGTC |
| 3MCMV69XbaI | ATCGTCTAGACTAGTCAGTCAGAGTCCATCTCGCTGTAGG |
| 3HHV6U42XhoI | CAGTCTCGAGTTATTCTGAGTCAGAAGAACATG |
| 3ELHVU42nestedO | CATAGTATAATGATCCGAGG |
| 3ELHVU42nestedI | CGCTACACATTATAAATTGC |
| 3ELHVU42EcoRI | GCATGAATTCCTACAGCGTCATATCGCTC |
| C69mutUAPc | CCTCAGCGAGCGAGAAGCCGCCGCGGCCGCGGCACGGCGCTTCTG |
| C69mutUAPnc | CAGAAGCGCCGTGCCGCGGCCGCGGCGGCTTCTCGCTCGCTGAGG |
| Rh69mutUAPc | GCGACCGAGAACGCCGTGCTCGGCGGGCACGGCGCTTCTGC |
| Rh69mutUAPnc | GCAGAAGCGCCGTGCCCGCCGAGCACGGCGTTCTCGGTCGC |
| 5GalKUL69as2forw | GAGCTGCACTCACGCGGCCGTCACGACGCTCCGTCGTTATCTT |
| CCCTGAGCCTGTTGACAATTAATCATCGGCA | |
| 3GalKUL69as360rev | GTTGACGGGTAGCCCGTGCAGGTGGTGGTACTTCATGTAGCT |
| GAGCGTTTCTCAGCACTGTCCTGCTCCTT | |
| 5UL69aa1 | ATGGAGCTGCACTCACGCGGCCGTC |
| 3UL69aa367 | CTCCAGATAGGGATCGTGCGGGTTG |
| n-termofUL69 | CAACGCCAAAAACGTCCAC |
| c-termofUL69 | CACCAAGCTCAGGCACGC |
| 3UL69aa440Eco | GCATGAATTCGGCCATGGCGCGGTGCATGG |
Cell culture, viruses, and plasmid transfections.
HEK293T cells were cultivated in Dulbecco's minimal essential medium (DMEM) containing 10% fetal calf serum. HeLa cells and primary human foreskin fibroblasts (HFFs) were cultivated in Eagle's minimal essential medium with 5% fetal calf serum. Transfections were performed at a cell confluence of approximately 80% via the calcium phosphate coprecipitation procedure described earlier (23, 60). The cytomegalovirus strain employed in this study as a control for growth kinetics analyses was obtained by reconstitution of infectious viruses using the bacterial artificial chromosome (BAC) pHB5, which contains the genomic sequence of the HCMV laboratory strain AD169 (5). Stocks of wild-type and recombinant viruses were prepared and titrated by IE1p72 fluorescence exactly as described previously (4). Multistep growth curve analyses and quantifications of viral DNA in supernatants from infected cells were performed as described in a previous study (30).
Indirect immunofluorescence analysis.
HeLa cells were grown on coverslips in 6-well dishes (3.0 × 105 cells/well) and transfected by standard calcium phosphate coprecipitation 1 day after seeding. Two days later, cells were fixed with 4% paraformaldehyde (10 min at room temperature) and permeabilized using phosphate-buffered saline (PBS)-0.2% Triton X-100 (20 min at 4°C). After being incubated with the appropriate primary antibody (MAb-FLAG M2; Sigma-Aldrich, Deisenhofen, Germany; MAb-Myc 1-9E10.2; ATCC) for 45 min at 37°C, cells were excessively washed and subsequently incubated for 30 min at 37°C with fluorescein isothiocyanate (FITC)- and/or Cy3-conjugated secondary antibodies (Dianova, Hamburg, Germany). Counterstaining of the cell nuclei was achieved by DAPI (4′,6-diamidino-2-phenylindole) containing Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Immunofluorescence data were analyzed by an Axiovert 135 (Zeiss, Jena, Germany) or a Leica TCS SP5 (Leica, Wetzlar, Germany) microscope at magnifications of ×400 and ×630 (23).
Western blotting and CoIP assay.
For Western blot analysis, transfected HEK293T cells were lysed, diluted in sodium dodecyl sulfate (SDS)-Laemmli buffer, and boiled at 95°C for 5 min (23). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 12.5% polyacrylamide gels and transferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany), followed by chemiluminescence detection according to the manufacturer's protocol (ECL Western blotting detection kit; Amersham Pharmacia Europe, Freiburg, Germany). Coimmunoprecipitation (CoIP) analysis was performed as described previously (2). Briefly, HEK293T cells were transfected in 6-well plates (5.0 × 105 cells/well) via calcium phosphate coprecipitation. Two days posttransfection, cells were lysed for 20 min at 4°C in 800 μl of CoIP buffer (50 mM Tris-HCl [pH 8.0], 150 to 300 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg/ml of aprotinin, 2 μg/ml of leupeptin, and 2 μg/ml of pepstatin). After centrifugation, the supernatant was incubated with the appropriate antibody (MAb-FLAG M2; Sigma-Aldrich, Deisenhofen, Germany; MAb-Myc 1-9E10.2; ATCC) coupled to protein A Sepharose beads for 1.5 h at 4°C. The Sepharose beads were collected and washed five times in CoIP lysis buffer. Antigen-antibody complexes were recovered by boiling in SDS sample buffer and analyzed via Western blotting.
Nucleocytoplasmic shuttling assay.
To examine the nucleocytoplasmic shuttling activity of betaherpesviral pUL69 homologs, interspecies heterokaryon analyses were performed (36). After transfected HeLa cells were fused with nontransfected murine NIH 3T3 cells by incubation with PEG3500, proteins were allowed to shuttle for 4 h exactly as described previously (27). Thereafter, cells were fixed by 4% paraformaldehyde and heterokaryons were subjected to standard indirect immunofluorescence analyses as described above, using mouse anti-β-galactosidase (β-Gal) antibody (MAb-β-gal; Roche, Mannheim, Germany) and a FLAG-tagged specific antibody (RAb-FLAG; Sigma-Aldrich, Deisenhofen, Germany) as primary antibodies.
Nuclear mRNA export assay for betaherpesviral pUL69 homologs.
A nuclear-mRNA export assay based on the export activity of transiently expressed pUL69 or betaherpesviral pUL69 homologs was performed by transfection of HEK293T cells as described previously (29). Chloramphenicol acetyltransferase (CAT) reporter assays were performed by the use of the plasmid pDM128/CMV/RRE, which codes for chloramphenicol acetyltransferase within an artificial intron-containing mRNA (17). To quantify CAT protein expression, a CAT enzyme-linked immunosorbent assay (Roche, Mannheim, Germany) was used. Determinations were performed in triplicate.
BAC mutagenesis.
For the construction of UL69 recombinants, a two-step recombination strategy using the galactokinase gene as a selection marker was employed (57). For the first recombination, the galK gene with 5′ and 3′ flanking regions homologous to UL69 sequences was amplified from plasmid pgalK (57), using the primers 5GalKUL69as2forw and 3GalKUL69as360rev. In order to accomplish the homologous recombination, the PCR fragment was then transformed into Escherichia coli (strain SW102), which already harbored the BAC pHB5. Cells were then plated on minimal medium (M63) agar plates containing 0.2% galactose and chloramphenicol and incubated at 32°C for 5 days. The colonies that appeared were streaked twice on MacConkey agar plates containing 0.2% galactose and chloramphenicol. Bacterial colonies appearing in bright red on MacConkey plates, and hence positive for the use of galactose, were used for the second recombination. For this recombination, the UL69 wild-type sequence, as well as UL69 mutant sequences, were amplified by PCR using primers 5UL69aa1 and 3UL69aa367 together with plasmids pHM160, mUAP, ΔR1ΔRS, and ΔR2ΔRS as templates (29, 55, 60). In order to select for bacteria with loss of the galK gene, the transformed bacteria were plated on M63 agar plates containing 0.2% 2-deoxygalactose and chloramphenicol. Bacterial colonies appearing on these plates were subjected to further characterization (see below). Reconstitution of recombinant cytomegaloviruses using purified BAC DNA was performed as described previously (30).
Viral nucleic acid isolation and analysis.
BAC DNA was isolated either by standard alkaline lysis minipreparation from 5-ml liquid cultures or by using a NucleoBond BAC 100 kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. Subsequently, the integrity of BAC DNA was analyzed by digestions with restriction enzymes and separation by 0.7% agarose gel electrophoresis followed by Southern blot analysis as described in previous studies (4, 30). For a further characterization of recombinant BACs by PCR, reactions were performed with primer pairs n-termofUL69 and c-termofUL69 or n-termofUL69 and 3UL69aa440Eco, resulting in either amplification of the entire UL69 open reading frame, including flanking sequences, or the N-terminal part of UL69, respectively. Subsequently, the nucleotide sequence of the resulting PCR fragments was determined by automated sequence analysis (ABI, Weiterstadt, Germany) in order to confirm the correct insertion of intended mutations as well as to exclude the presence of accidental mutations within UL69.
RESULTS
Expression analyses of pUL69 and homologous proteins of the Betaherpesvirinae.
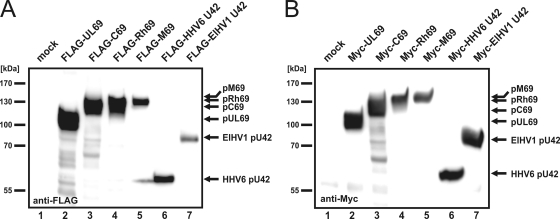
Due to the lack of antibodies directed against any betaherpesviral pUL69 homolog, the starting point of our analyses was the engineering of N-terminally epitope-tagged eukaryotic expression constructs for a representative selection of betaherpesviral pUL69 homologs. While FLAG- UL69 and Myc-UL69 were already available in our laboratory, FLAG- and Myc-tagged full-length constructs of CCMV C69, RhCMV Rh69, MCMV M69, and U42 of HHV6 and ElHV1 were successfully synthesized for the first time. In order to verify the functionality and to determine the expression patterns of the respective constructs, they were transfected into HEK293T cells, and 2 days later protein expression was analyzed by Western blotting using anti-FLAG or anti-Myc antibodies (Fig. 2 A and B). Since FLAG and Myc epitopes are short polypeptides, the molecular mass of each protein was not significantly altered; therefore, FLAG- pUL69 and Myc-pUL69, which served as transfection and expression controls, could be detected with their characteristic isoforms of 105 to 116 kDa (Fig. 2A and B, lanes 2) (61). Intriguingly, open reading frame C69, which encodes 920 amino acids, was translated into a protein of approximately 125 kDa, while Rh69, which encodes 777 amino acids, was translated into a protein of 130 kDa (Fig. 2A and B, lanes 3 and 4). U42 of HHV6, which encodes 515 amino acids, was detected in its FLAG- and Myc-tagged versions at about 60 kDa (Fig. 2A and B, lanes 6). Most surprisingly, pM69, with an intermediate length of 843 amino acids, was determined to have the highest molecular mass of any betaherpesviral pUL69 homolog, with an apparent size of approximately 135 to 140 kDa (Fig. 2A and B, lanes 5). Finally, ORF U42 of ElHV1, which encodes 660 amino acids, was translated into a protein of about 80 kDa (Fig. 2A and B, lanes 7). Thus, the protein expression of all pUL69 homologs could be confirmed by these experiments.
FIG. 2.
Expression analysis of pUL69 and its betaherpesviral homologs. (A and B) HEK293T cells were transfected with plasmids encoding N-terminally FLAG-tagged (A) or Myc-tagged (B) versions of pUL69 (lane 2), pC69 (lane 3), pRh69 (lane 4), pM69 (lane 5), and pU42 of HHV6 (lane 6) and ElHV1 (lane 7), as indicated. Transfection of an empty vector served as a specificity control (mock, lanes 1). Cells were lysed under denaturing conditions at about 36 h posttransfection. Lysates were separated by SDS-PAGE, and protein expression was analyzed by Western blotting using anti-FLAG- or anti-Myc-specific antibodies. Molecular size markers are indicated (kDa).
Nuclear localization of pUL69 and homologous betaherpesviral proteins.
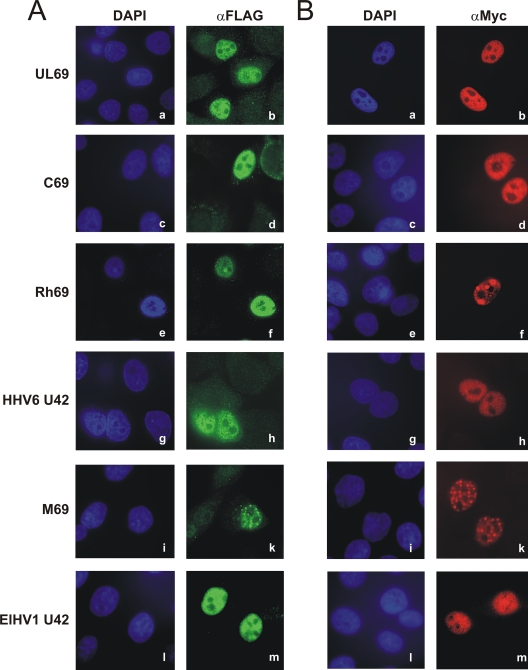
As HCMV pUL69 was already known to accumulate within the nuclei of infected and transfected cells (60), indirect immunofluorescence analyses of the betaherpesviral pUL69 homologs pC69, pRh69, pM69, HHV6 pU42, and ElHV1 pU42 were carried out in order to determine their subcellular distributions in HeLa cells. As illustrated in Fig. 3, all investigated FLAG- and Myc-tagged homologs exhibited intranuclear accumulation (Fig. 3A and B, c to m), as was also observed for pUL69, which served as an internal control (Fig. 3A and B, a and b). The same diffuse distribution of pUL69, pC69, pRh69, pM69, and pU42 of HHV6 and ElHV1 throughout the nucleus except for the nucleoli could also be detected in HFF cells (data not shown), thereby further underlining the specificity of the results presented here. Interestingly, in some cells pRh69 and pM69 (Fig. 3A and B, f and k) were concentrated in distinct foci within the nucleus, an observation that is consistent with previous findings of pUL69 aggregates in infected cells (60). Nuclear localization was conclusively identified to be conserved within the subfamily Betaherpesvirinae within the ICP27 family and most likely throughout the Herpesviridae.
FIG. 3.
Nuclear localization of pUL69 and homologous betaherpesviral proteins. (A and B) HeLa cells were transfected with plasmids for FLAG-tagged (A) or Myc-tagged (B) versions of pUL69 (a and b), pC69 (c and d), pRh69 (e and f), HHV6 pU42 (g and h), pM69 (i and k) or ElHV1 pU42 (l and m), as indicated on the left. Thirty-six hours posttransfection, cells were fixed and analyzed for intracellular distribution of the respective proteins by indirect immunofluorescence, using anti-FLAG-specific (A) or anti-Myc-specific (B) antibodies. As a secondary antibody, anti-mouse-FITC (A) or anti-mouse-Cy3 (B) was used, and cell nuclei were counterstained with DAPI.
Homo- and heterodimerization of betaherpesviral pUL69 homologs.
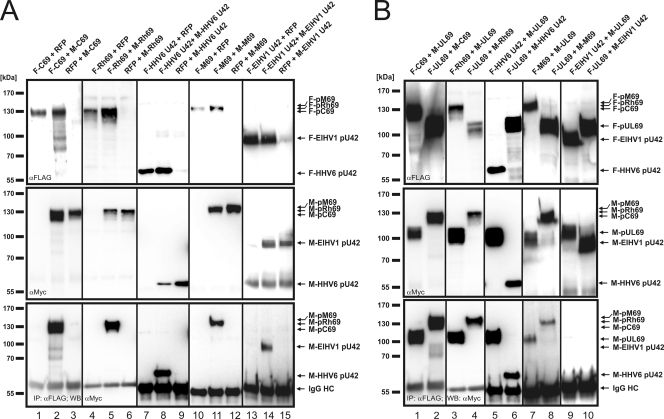
Because homodimerization was reported as a well-conserved characteristic of all studied members of the ICP27 family (28, 31, 62), we analyzed whether this was also true for the betaherpesviral counterparts. For this analysis, HEK293T cells were cotransfected with a combination of two plasmids, a FLAG- and a Myc-tagged version of C69, Rh69, M69, and U42 of HHV6 or ElHV1, either in combination or together with a construct encoding the red fluorescent protein (RFP), as indicated in Fig. 4 A. Two days later, cells were harvested and protein expression was verified by Western blotting using anti-FLAG- or anti-Myc-specific antibodies (Fig. 4A, upper and middle panels). Next, FLAG-tagged proteins were immunoprecipitated by an anti-FLAG antibody before nonbound protein complexes were removed by extensive washing steps. After electrophoretic separation of the protein complexes, coprecipitated Myc-tagged versions of pC69, pRh69, pM69, and pU42 of HHV6 or ElHV1 could be detected by Western blotting (Fig. 4A, lower panel, lanes 2, 5, 8, 11, and 14). The results were considered specific since no coprecipitation was observed when the FLAG-tagged protein was replaced with RFP (Fig. 4A, lanes 3, 6, 9, 12, and 15). These results show that homodimerization is a conserved feature of pUL69 homologs within the subfamily Betaherpesvirinae.
FIG. 4.
Homo- and heterodimerization of betaherpesviral pUL69 homologs. (A and B) Coimmunoprecipitation analyses. (A) For homodimerization analyses, HEK293T cells were cotransfected with a combination of plasmids for RFP and FLAG- or Myc-tagged pC69 (lanes 1 and 3), pRh69 (4 and 6), HHV6 pU42 (7 and 9), pM69 (10 and 12), and ElHV1 pU42 (13 and15) or a combination of plasmids for FLAG- and Myc-tagged pUL69 homologs (lanes 2, 5, 8, 11, and 14). (B) For heterodimerization analyses of pC69 (lanes 1 and 2), pRh69 (3 and 4), HHV6 pU42 (5 and 6), pM69 (7 and 8), and ElHV1 pU42 (9 and 10), HEK293T cells were cotransfected with the respective FLAG-tagged pUL69 homolog and Myc-pUL69 (lanes 1, 3, 5, 7, and 9) or vice versa (lanes 2, 4, 6, 8 and 10). (A and B) Two days posttransfection, cells were harvested and lysed and immunoprecipitation was performed, using anti-FLAG antibodies. After electrophoresis, coprecipitated proteins were visualized by Western blotting using anti-Myc antibodies (A and B, lower panels). Immunoglobulin heavy chain (IgG HC) served as an internal control for the presence of the precipitating antibody.
Since betaherpesviral proteins exhibit considerable amino acid conservation of the ICP27 homology region in the centers of the proteins, we reasoned that an analysis of the capacity to heterodimerize with pUL69 might identify critical amino acids required for this interaction. To investigate this, HEK293T cells were cotransfected with vectors encoding FLAG-tagged pC69, pRh69, pM69, or pU42 of HHV6 or ElHV1 together with Myc-UL69, or vice versa, as indicated in Fig. 4B. Two days later, cells were harvested and protein expression was monitored by Western blotting (Fig. 4B, upper and middle panels). After immunoprecipitation of FLAG-tagged proteins, coprecipitated proteins were identified by Western blotting using an anti-Myc antibody. As illustrated in the lower panel of Fig. 4B, a protein-protein interaction with HCMV pUL69 could be detected for pC69, pRh69, HHV6 pU42, and pM69 (Fig. 4B, lanes 1 to 8). Interestingly, our study revealed that ElHV1 pU42 was unable to heterodimerize with pUL69 (Fig. 4B, lanes 9 and 10). Moreover, nonspecific interactions of FLAG- or Myc-tagged pUL69 homologs could be excluded, since no bands were visible on the coimmunoprecipitation blot when RFP was cotransfected with any of the FLAG- or Myc-tagged proteins (Fig. 4A and data not shown).
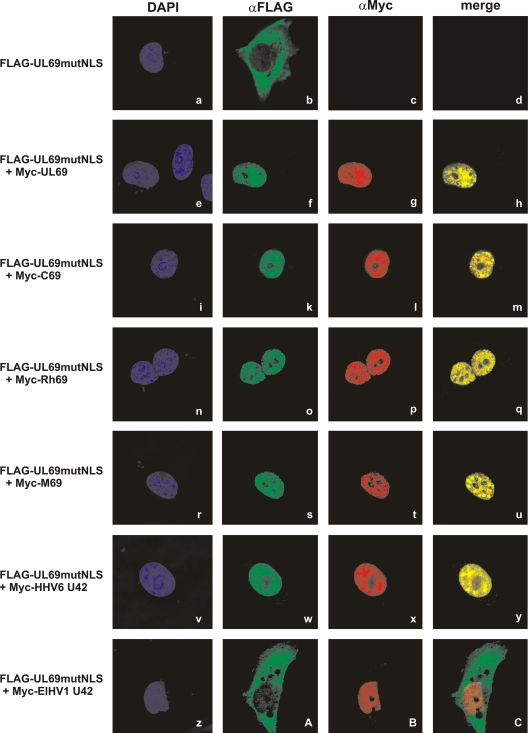
To further validate these results in a different cell type and by using an alternative experimental approach, we took advantage of the construct FLAG-UL69mutNLS, which carries four alanine substitutions within its bipartite nuclear localization signal (NLS) and therefore displays a predominantly cytoplasmic distribution. Importantly, we demonstrated previously that the cytoplasmic pUL69mutNLS protein is recruited to the nucleus via homodimerization with cotransfected wild-type pUL69 (28). Here, we wanted to analyze whether pUL69mutNLS is analogously relocalized when coexpressed with any of the betaherpesviral homologs. For this analysis, HeLa cells were transfected with FLAG-UL69mutNLS alone (Fig. 5 a to d) or in combination with Myc-tagged UL69 (Fig. 5e to h), C69 (Fig. 5i to m), Rh69 (Fig. 5n to q), M69 (Fig. 5r to u), HHV6 U42 (Fig. 5v to y), or ElHV1 U42 (Fig. 5z to C). Two days later, cells were fixed and the subcellular localization of the proteins was examined by indirect immunofluorescence microscopy. As shown before, all Myc-tagged proteins containing a functional NLS localized to the cell nuclei, whereas FLAG-UL69mutNLS displayed a cytoplasmic distribution (Fig. 3B and 5b). Interestingly, coexpression of pUL69, pC69, pRh69, pM69, or HHV6 pU42 with pUL69mutNLS resulted in nuclear localization of both proteins (Fig. 5h, m, q, u, and y). In contrast, pU42 of ElHV1 was not able to relocalize pUL69mutNLS from the cytoplasms into the nuclei of transfected cells (Fig. 5A). This confirms our results obtained by coimmunoprecipitation analyses. Thus, we conclude that heterodimerization with HCMV pUL69 is a conserved feature of homologs belonging to the more closely related genera Cytomegalovirus, Muromegalovirus, and Roseolovirus but not of that of the more distantly related genus Proboscivirus.
FIG. 5.
Nuclear recruitment of a pUL69-NLS mutant by heterodimerization with pUL69 homologs. HeLa cells were either transfected solely with a plasmid for FLAG-tagged pUL69 harboring point mutations within its nuclear localization signal (Flag-UL69mutNLS; a to d) or in combination with Myc-tagged pUL69 (e to h) or one of the pUL69 homologs (i to C). Two days later, indirect immunofluorescence analyses were performed using primary antibodies RAb-FLAG and MAb-Myc and anti-rabbit FITC or anti-mouse Cy3 as a secondary antibody. For visualization of cell nuclei, costaining of cells with DAPI was carried out.
Stimulation of nuclear export of unspliced RNAs is restricted to the cytomegaloviral proteins pUL69, pC69, and pRh69.
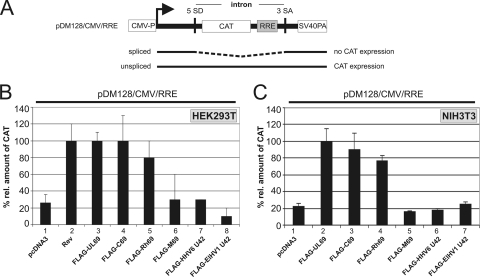
One major characteristic of the members of the ICP27 family characterized so far is their ability to function as viral-mRNA export factors. In order to investigate if representative betaherpesviral pUL69 homologs can likewise facilitate the nuclear export of unspliced mRNAs, we utilized a functional reporter assay that was developed by Hope and colleagues to monitor RNA export in vivo (24). This assay, which is based on the pDM128/CMV/RRE reporter plasmid harboring the CAT coding sequence inserted into an intron, has previously been used to demonstrate the nuclear export of unspliced RNA by either pUL69 or the homologous protein of Epstein-Barr virus, EB2 (17, 22, 29). Here, HEK293T cells were cotransfected with the pDM128/CMV/RRE reporter (Fig. 6 A) and vectors encoding the well-characterized mRNA export factors HIV-1 Rev (54) and HCMV pUL69 (29) or one of the FLAG-tagged betaherpesviral pUL69 homologs, pC69, pRh69, pM69, or pU42 of HHV6 or ElHV1, as indicated in Fig. 6B. Two days later, cells were harvested and the amount of CAT protein was quantified using a CAT-enzyme-linked immunosorbent assay (ELISA) (Fig. 6B). In accordance with our previous results, cotransfection of the empty pcDNA3 vector had only minor effects on the level of CAT protein expression, while expression was significantly increased upon coexpression of HIV-1 Rev or HCMV pUL69 (Fig. 6B, compare lane 1 with lanes 2 and 3). Coexpression of pDM128/CMV/RRE with pC69 or pRh69 resulted in an increased CAT protein level similar to that observed when pUL69 was present (Fig. 6B, lanes 4 and 5), implying that these two betaherpesviral proteins can, in analogy to their HCMV counterpart, function as viral-mRNA export factors. However, neither pM69, HHV6 pU42, nor ElHV1 pU42 was able to promote the accumulation of unspliced CAT mRNAs in the cytoplasm, as can be deduced from CAT protein levels that were comparable to those of the negative control (Fig. 6B, lanes 6 to 8).
FIG. 6.
Nuclear mRNA export by betaherpesviral pUL69 homologs. (A) Schematic representation of the pDM128/CMV/RRE reporter plasmid. The CAT reporter gene (CAT) and the HIV-1 Rev responsive element (RRE) are localized within an intron flanked by inefficiently used HIV-1 splicing sites (5′SD and 3′SA). Also depicted are the CMV promoter (CMV-P) and the SV40 late poly(A) signal (SV40PA). (B and C) CAT mRNA export assay of pUL69 and homologous proteins. The diagram shows the relative amounts of CAT protein measured 36 h posttransfection of HEK293T (B) or NIH 3T3 (C) cells with a mixture of the reporter pDM128/CMV/RRE and plasmids expressing either FLAG-pUL69 (panel A, lane 3; panel B, lane 2) or FLAG-tagged betaherpesviral pUL69 homologs as indicated at the bottoms of the diagrams (panel A, lanes 4 to 8; panel B, lanes 3 to 7). HIV-1 Rev served as a positive control (panel A, lane 2) and pcDNA3.1 as a negative control (panels A and B, lanes 1). The relative amounts of CAT protein are given as percentages relative to that of HIV-1 Rev (panel B, lower panel) or HCMV pUL69 (panel C, lower panel), and standard deviations of at least three experiments are shown as bars.
However, these CAT mRNA export assays were performed in human HEK293T cells and might therefore not be suitable for all pUL69 homologs of the Betaherpesvirinae subfamily with different host species, for example, pM69 of MCMV or ElHV1 pU42, due to the lack of species-specific host cofactors. In order to exclude this possibility, analogous CAT reporter assays were performed in murine NIH 3T3 cells (Fig. 6C). Interestingly, coexpression of pDM128/CMV/RRE and HCMV pUL69, as a positive control, or CCMV pC69 and RhCMV pRh69 again yielded a significant increase in the CAT protein level in NIH 3T3 cells and therefore underlined their ability to function as viral-mRNA export factors, even in cells from a different host species. In contrast, coexpression of MCMV pM69, HHV6 pU42, or ElHV1 pU42 with the reporter plasmid in murine host cells did not increase the CAT protein level, thus confirming the results obtained with HEK293T cells. This finding argues against a dependency on species-specific host cofactors. In summary, our results show that the cytomegaloviral proteins pUL69, pC69, and pRh69 exert mRNA export activity irrespective of the cell system utilized, while, in contrast, the betaherpesviral counterparts pM69 and HHV6 or ElHV1 pU42 were not able to promote the nuclear export of unspliced mRNA in this particular reporter assay.
Nucleocytoplasmic shuttling of HCMV pUL69 and its betaherpesviral homologs.
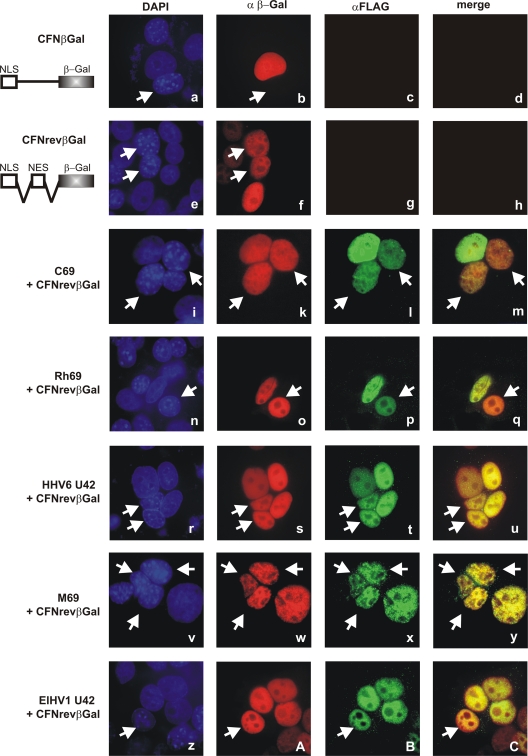
It has become apparent that some pUL69 homologs within the subfamilies Alphaherpesvirinae and Gammaherpesvirinae, including HSV-1 ICP27, EBV EB2, HHV8 ORF57, and herpesvirus saimiri (HVS) ORF57, continuously shuttle between the nucleus and the cytoplasm independently of virus-encoded cofactors (3, 19, 33, 34, 44, 48, 50). Such nucleocytoplasmic shuttling activity was also confirmed for HCMV pUL69 and represents a prerequisite for its mRNA export activity (27, 29). To analyze if representative betaherpesviral pUL69 homologs exert nucleocytoplasmic shuttling activity, which might account for their different behavior in the mRNA export assay presented before, interspecies heterokaryon assays were performed as originally described by Piñol-Roma and colleagues (36). HeLa cells were cotransfected with vectors encoding FLAG-pUL69 or its betaherpesviral counterpart FLAG-pC69, FLAG-pRh69, HHV6-pU42, MCMV-pM69, or ElHV1-pU42 and the reporter pCFNrevβGal, which served as a shuttling positive control. Two days later, HeLa cells were fused to nontransfected murine NIH 3T3 cells, resulting in heterokaryons comprising the cell nuclei of both species. Four hours after fusion, cells were fixed and the subcellular localization of transfected proteins was analyzed by indirect immunofluorescence analyses using polyclonal anti-FLAG and monoclonal anti-β-Gal antibodies (Fig. 7). In order to distinguish between murine and human nuclei, cells were counterstained with DAPI, thereby identifying the murine nuclei by their characteristic punctate pattern (Fig. 7, white arrows). For a control, HeLa cells were transfected with one of two control plasmids termed CFNrev-βGal and CFN-βGal. CFNrev-βGal encodes β-galactosidase fused to the NLS of simian virus 40 large-T antigen (SV40 T-Ag) as well as to the nuclear export signal (NES) of HIV-1 Rev (Fig. 7, second panel, NLS-NES-βGal). Consequently, this construct served as a shuttling positive control and β-galactosidase was detected in both types of nuclei within a heterokaryon (Fig. 7e and f), as it was for HCMV pUL69 (data not shown). The second plasmid, CFN-βGal, expresses β-galactosidase fused to the NLS of SV40 T-Ag; however, it lacks an NES (Fig. 7, first panel, NLS-βGal), thus serving as a nonshuttling control and hence located exclusively in human nuclei but not in murine nuclei of a heterokaryon (Fig. 7a and b). In analogy, CFNrev-βGal, serving as an internal positive control for each approach, could always be detected in murine and human cell nuclei (Fig. 7k, o, s, w, and A). Accordingly, our heterokaryon analyses revealed that the cytomegaloviral proteins pC69 and pRh69 were detected in both types of cell nuclei in interspecies heterokaryons and therefore displayed nucleocytoplasmic shuttling activity analogous to that of HCMV pUL69 (Fig. 7i to m for pC69 and n to q for pRh69). To our surprise, the more distantly related pM69, as well as HHV6 and ElHV1 pU42, which were negative in the mRNA export assays, were also able to accumulate within murine cell nuclei and were therefore evaluated as nucleocytoplasmic shuttling proteins (Fig. 7r to u for pM69, v to y for HHV6 pU42, and z to C for ElHV1 pU42). In summary, our heterokaryon analyses identified pC69, pRh69, pM69, and HHV6 and ElHV1 pU42 as nucleocytoplasmic shuttling proteins that share this functional activity with their HCMV counterpart. This result not only identified nucleocytoplasmic shuttling as a well-conserved feature throughout the Herpesviridae family but further suggests that this activity may not be sufficient for mRNA export.
FIG. 7.
Nucleocytoplasmic shuttling of betaherpesviral pUL69 homologs as analyzed by interspecies heterokaryon assays. HeLa cells were cotransfected with CFN-βGal (a to d), CFNrev-βGal (e to h), or a combination of CFNrev-βGal and the indicated pUL69 homolog (i to C). CFNrev-βGal served as a shuttling positive control, as it contains β-galactosidase fused to the NLS of the SV40 large T antigen and the NES of HIV-1 Rev. CFN-βGal also codes for β-galactosidase fused to the SV40 NLS but lacks a functional NES and therefore displays the shuttling negative control (41). Interspecies heterokaryons were formed by fusion of transfected HeLa cells with murine NIH 3T3 cells. Four hours later, cells were fixed and the localization of transfected proteins was assessed by indirect immunofluorescence using an antibody specific for β-galactosidase and RAb-FLAG for detection of pUL69 homologs. Upon counterstaining by DAPI, murine nuclei (indicated by arrows) displayed a characteristic punctate pattern and could thereby easily be discriminated from the evenly stained human nuclei.
Interaction with UAP56/URH49 is a prerequisite for stimulation of mRNA export by pUL69, pC69, and pRh69.
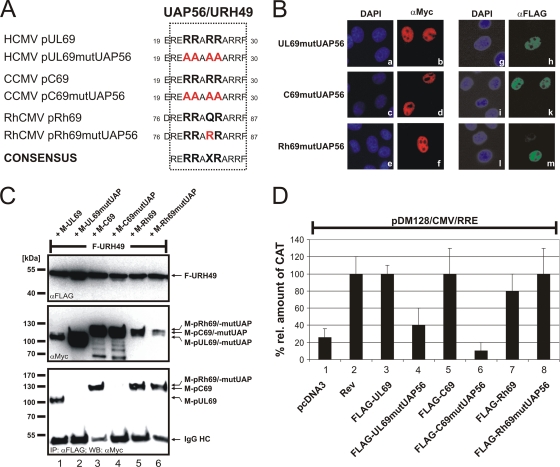
Our group reported previously that interaction of pUL69 with the cellular DExD/H-box RNA helicase UAP56 or URH49 is a prerequisite for its mRNA export activity (29). Since UAP56 is well conserved throughout the vertebrates it was tempting to speculate that an UAP56/URH49 interaction might correlate with the mRNA export activity of the cytomegaloviral pUL69 homologs. Initial bioinformatic approaches revealed that the UAP56/URH49 interaction motif of pUL69 (29) is 100% or 91% conserved within the cytomegaloviral proteins pC69 and pRh69, respectively (Fig. 8 A), but is absent in pM69, HHV6 pU42, and ElHV1 pU42, thereby supporting the assumption that a UAP56 interaction accounts for the mRNA export activities of cytomegaloviral proteins pUL69, pC69, and pRh69. To test this hypothesis, amino acid exchange mutants of pC69 and pRh69 were constructed with alterations of the properties of each protein to a loss of function or a gain of function with regard to the UAP56/URH49 interaction. In the case of pUL69 and pC69, the UAP56 interaction motif was altered by site-directed mutagenesis of four codons for essential arginines, thereby aiming to disrupt the protein-protein interaction (Fig. 8A). The UAP56/URH49 interaction motif within pRh69 varies from the defined interaction motif by a single amino acid, which was altered by site-directed mutagenesis to yield the UAP56/URH49 interaction motif as contained within pUL69 (Fig. 8A). Since the UAP56/URH49 interaction motif of pUL69 overlaps in part with its NLS and since the NLSs of pC69 and pRh69 have not yet been mapped in detail, the subcellular localization of each mutant was determined first. For this experiment, HeLa cells were transfected with expression constructs encoding FLAG-tagged or Myc-tagged mutants of pUL69, pC69, or pRh69. Two days later, indirect immunofluorescence analyses revealed an intranuclear localization of each FLAG-tagged (Fig. 8B, g to m) or Myc-tagged (Fig. 8B, a to f) mutant.
FIG. 8.
UAP56/URH49 interaction is crucial for cytomegaloviral mRNA export. (A) Amino acid alignment of the UAP56/URH49 interaction motif of HCMV pUL69 and the corresponding sequences of CCMV pC69 and RhCMV pRh69. To generate gain-of-function or loss-of-function mutants, site-directed mutagenesis was performed as indicated and the resulting constructs were subsequently analyzed in the experiments described below. (B) Nuclear localization of amino acid exchange constructs indicated in panel A. HeLa cells were transfected with FLAG- or Myc-tagged mutants of pUL69, pC69, or pRh69, as indicated. Two days later, cells were fixed and the subcellular localization of proteins was determined by indirect immunofluorescence analyses using anti-FLAG (g to m) or anti-Myc (a to f) antibodies as described before. DAPI was used for counterstaining of the cell nuclei. (C) URH49-binding capacity of constructs indicated in panel A. Coimmunoprecipitation analyses of cell lysates from HEK293T cells that were cotransfected with FLAG-URH49 (lanes 1 to 6) and Myc-tagged wild-type pUL69, pC69, or pRh69 (lanes 1, 3, and 5) or point-mutated versions of these proteins (lanes 2, 4, and 6). Two days posttransfection, cells were lysed and immunoprecipitation was performed, using anti-FLAG antibodies. After electrophoresis, coprecipitated proteins were visualized by Western blotting with anti-Myc antibody (lower panel). Immunoglobulin heavy chain (IgG HC) served as an internal control for the presence of the precipitating antibody. (D) CAT mRNA export assay of wild-type pUL69, pC69, and pRh69 and the respective mutants carrying mutations in the UAP56/URH49 interaction motif. The diagram shows the relative amounts of CAT protein measured 36 h after transfection of HEK293T cells with a mixture of the reporter pDM128/CMV/RRE and plasmids expressing either wild-type pUL69 (lane 3), pC69 (lane 5), or pRh69 (lane 7) or mutants pUL69mutUAP (lane 4), pC69mutUAP (lane 6), and pRh69mutUAP (lane 8). HIV-1 Rev served as a positive control (lane 2) and pcDNA3.1 as a negative control (lane 1). The relative amounts of CAT protein are given as percentages relative to that of HIV-1 Rev. Standard deviations of at least three experiments are shown as bars.
Next, the UAP56/URH49-binding capacity of mutant and wild-type proteins was investigated by CoIP. HEK293T cells were cotransfected with FLAG-URH49 and Myc-tagged genuine and mutated versions of pUL69, pC69, or pRh69 (Fig. 8C). Two days later, cells were lysed and immunoprecipitation was performed using anti-FLAG antibodies. As determined by staining of the CoIP Western blot with anti-Myc antibody, URH49 was able to coprecipitate wild-type pUL69 and pC69 (Fig. 8C, lower panel, lanes 1 and 3) but not the respective loss-of-function mutants (Fig. 8C, lower panel, lanes 2 and 4). Interestingly, URH49 coprecipitated not only the gain-of-function mutant pRh69mutUAP but also the wild-type pRh69 carrying one amino acid substitution within the UAP56/URH49 interaction motif of pUL69 (Fig. 8C, lower panel, lanes 5 and 6). Thus, our results identified a UAP56/URH49 consensus motif (RERRAXRARRF) within the cytomegaloviral proteins pUL69, pC69, and pRh69 that was essential for the URH49 interaction.
Next, we analyzed by CAT mRNA export assays whether the UAP56/URH49 interaction was required for stimulation of nuclear-mRNA export by the cytomegaloviral proteins. In accordance with our previous results, cotransfection of the CAT reporter pDM128/CMV/RRE with HCMV pUL69, CCMV pC69, or RhCMV pRh69 resulted in an increased level of CAT protein, similar to that observed after coexpression of the positive control HIV-1 Rev (Fig. 8D, lanes 2, 3, 5, and 7). However, neither pUL69mutUAP56 nor pC69mutUAP56, both of which harbor point mutations abrogating the UAP56 interaction, was able to promote the accumulation of unspliced CAT mRNA in the cytoplasm (Fig. 8D, lanes 4 and 6). In the case of pRh69mutUAP56, the predicted gain-of-function mutation of the UAP56 interaction motif slightly increased its ability to export unspliced CAT mRNA compared to that of wild-type pRh69 (Fig. 8D, lanes 7 and 8). In summary, this set of experiments indicates that interaction with the cellular DExD/H-box RNA helicases UAP56 and URH49 is a prerequisite for the mRNA export activity of the cytomegaloviral proteins pUL69, pC69, and pRh69.
Interaction with UAP56/URH49 but not RNA binding of pUL69 is essential for efficient replication of human cytomegalovirus.
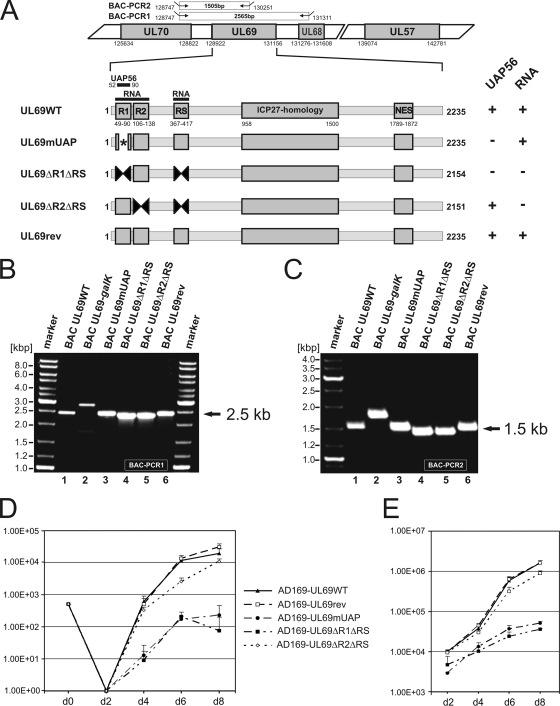
Due to our observation that the UAP56-binding site of pUL69 is functionally conserved in pC69 and pRh69, we hypothesized that this protein interaction might play a critical role during HCMV replication. In order to investigate this possibility, we decided to generate several recombinant viruses harboring mutations within the N-terminal coding sequence of pUL69. Since we had observed in previous studies that the UAP56-binding site overlaps with a tripartite arginine-rich RNA-binding motif of pUL69 (55), three different UL69 mutations were introduced into the HCMV genome via BAC recombination. UL69mUAP corresponds to a previously described mutant of UL69 that carries alanine substitutions within the UAP56-binding domain, thus abrogating the interaction with UAP56 (29). UL69ΔR1ΔRS and UL69ΔR2ΔRS are internal deletion mutants of arginine-rich regions, both of which have been shown to lack RNA-binding activity (55). In addition, however, ΔR1ΔRS is also negative for UAP56 binding, since the arginine-rich region R1 overlaps with the UAP56-binding site (Fig. 9A). The structural integrity of mutant and revertant viruses was confirmed by restriction enzyme and Southern blot analyses (data not shown) as well as by PCR amplification of UL69 genomic sequences (Fig. 9B and C) followed by nucleotide sequence determination of the entire UL69 ORF. After reconstitution of infectious viruses, multistep growth curve analyses of wild-type, mutant, and revertant AD169 viruses were performed in parallel. For these analyses, HFFs were infected in triplicate (multiplicity of infection [MOI], 0.1) with viral inocula normalized for equal immediate-early (IE) units. After harvesting of the supernatants at 2, 4, 6, and 8 days postinfection (p.i.), they were subjected to IE1p72 immunofluorescence titration. Figure 9D shows that we could detect a clear decrease in the release of progeny virions of the AD169-UL69mUAP and AD169-UL69ΔR1ΔRS viruses, both of which lack a functional UAP56-binding motif. The growth properties of the revertant virus AD169-UL69rev were not altered compared to those of the wild-type AD169 virus (Fig. 9D), while the AD169-UL69ΔR2ΔRS virus expressing an RNA-binding-deficient pUL69 exhibited a slight but significant delay in the release of progeny virions. Since pUL69 has been shown to be a constituent of viral particles (61), the mutations introduced into pUL69 might affect the infectivity of HCMV virions. To exclude this possibility, DNA was extracted from the viral supernatants obtained from the growth curve experiment of Fig. 9D, and viral genomic equivalents were determined by real-time PCR. As shown in Fig. 9E, the amounts of DNA in the viral supernatants clearly paralleled the release of progeny virus as observed by growth curve analyses (compare Fig. 9D and E). Taken together, these results strongly suggest that the interaction of pUL69 with UAP56/URH49 is required for the efficient replication of human cytomegalovirus.
FIG. 9.
Growth kinetics of recombinant AD169-derived viruses carrying UL69 mutations that either affect the UAP56-binding capacity and/or the RNA-binding capacity of pUL69. (A to C) Construction and verification of recombinant HCMVs expressing either mutant or wild-type pUL69. (A) Schematic diagram illustrating the structure of recombinant viruses that were generated by homologous recombination in Escherichia coli. In a first step, the selection marker galK was inserted into UL69 of BAC pHB5. Subsequently, the selection marker was replaced by either a wild-type or a mutated UL69 sequence. The upper half of panel A shows the genomic region of HCMV strain AD169 containing UL69 (numbers refer to nucleotide positions of AD169; BAC-PCR1 and BAC-PCR2 refer to PCR amplifications that were performed to confirm the integrity of recombinant viruses). The lower half of panel A illustrates functional domains of UL69 and indicates the localization of mutations/deletions that were introduced into recombinant viruses (numbers refer to nucleotide positions of the UL69 open reading frame). UAP56, UAP56-binding motif of UL69; RNA, RNA-binding domain of UL69, comprising the arginine-rich motifs R1, R2, and RS; ICP27 homology, ICP27 homology region of UL69; NES, nuclear export signal. On the right of panel A, the UAP56- and/or RNA-binding capacities of pUL69 variants are indicated. (B and C) For verification of the correct recombination sites within the HCMV genome, PCR analyses of bacterial clones harboring the indicated BACs were performed using oligonucleotides specific for UL70/UL68 (B) or UL70/UL69 (C), respectively. The localizations of primers used for amplification are shown in the upper half of panel A (BAC-PCR1 and BAC-PCR2). (D) Growth kinetics of recombinant-AD169-derived viruses. In order to determine the replication capacities of the recombinant viruses AD169-UL69WT (wild-type), AD169-UL69rev (revertant), AD169-UL69mUAP (alanine substitution mutant of UAP56-binding motif), AD169-UL69ΔR1ΔRS (deletion mutant of arginine-rich motifs R1 and RS), and AD169-UL69ΔR2ΔRS (deletion mutant of arginine-rich motifs R2 and RS), multistep growth curve analyses were performed. HFFs were infected with equal IE units (MOI, 0.1) of wild-type, revertant, or mutant viruses. The viral supernatants were harvested at the indicated time points (days [d]), followed by the determination of viral titers. Each infection was performed in triplicate, and the standard deviations are depicted by error bars. (E) Quantification of viral genomes in the supernatants of infected HFF cells by real-time PCR. Aliquots of the supernatants obtained for the multistep growth curves shown in panel D were treated with proteinase K, incubated at 56°C for 1 h, and subsequently denatured at 95°C. Five microliters of each lysate was subjected to real-time PCR to quantify the genomic equivalents in the supernatants of the recombinant viruses. Evaluations were performed in triplicate for each of the three infections per virus. Standard deviations are indicated.
DISCUSSION
The human cytomegalovirus protein encoded by ORF UL69 belongs to a family of regulatory factors that is conserved among all herpesviruses and includes the proteins ICP27 of herpes simplex virus type 1, EB2 of Epstein-Barr virus, and ORF57 of Kaposi's sarcoma-associated herpesvirus. For all characterized members of this protein family, a function as posttranscriptional activators that facilitate the nuclear export of mRNA has been documented. This function is mediated via the capability of these proteins to shuttle between the nucleus and the cytoplasm and to interact with components of the cellular mRNA export machinery (for reviews, see references 46, 49, and 56). Interestingly, most of the protein domains important for mRNA export stimulation (e.g., the interaction site for cellular mRNA export factors and the nuclear export signal) have been mapped to regions of the herpesviral-mRNA export factors that show either no sequence conservation or a low level of sequence conservation. For this reason, we hypothesized that characterization of the closely related proteins within the Betaherpesvirinae subfamily homologous to pUL69 might reveal further insights into the evolution of regulatory protein domains.
Due to the lack of reactive antibodies against any betaherpesviral pUL69 homolog, the starting point of our analyses was the construction of epitope-tagged eukaryotic expression constructs for pC69 of chimpanzee cytomegalovirus, pRh69 of rhesus cytomegalovirus, pM69 of murine cytomegalovirus, and pU42 of human herpesvirus type 6 or elephant endotheliotropic herpesvirus. Interestingly, Western blot analyses revealed that the apparent molecular masses of the respective proteins (C69, ∼125 kDa; Rh69, ∼130 kDa; M69, ∼135 to 140 kDa; HHV6 U42, ∼60 kDa; and ElHV1 U42, ∼90 kDa) differed from the predicted molecular masses calculated by the ExPASy proteomics server (C69, 100.7 kDa; Rh69, 87.5 kDa; M69, 93.0 kDa; HHV6 U42, 59.8 kDa; ElHV1 U42, 71.4 kDa). This result was not without precedent, since the predicted and apparent molecular masses of pUL69 also differ considerably and we reported previously that phosphorylation by cellular and viral kinases contributes to aberrant migration in SDS-PAGE (38, 53, 61). However, the presence of additional posttranslational modifications is likely, since a recent study predicted SUMOylation of pUL69, although the respective attachment sites for SUMO were not characterized in detail (43). Furthermore, as recently demonstrated for HSV-1 ICP27 (51, 52), pUL69 protein function might also be regulated via methylation. It is therefore feasible to speculate that at least some of the betaherpesviral homologs are also posttranslationally modified in the context of human cells, thus contributing to the discrepancies between predicted and observed molecular masses.
Even though in silico analyses failed to identify a classical NLS within pUL69 or any of its homologs and the alignment of the nonconventional bipartite NLS of pUL69 (29) with protein sequences of the homologs revealed 94.6% and 47.1% sequence identity with pC69 and pRh69, respectively, whereas HHV6 pU42 (20.6%) and pM69 (20.6%) and ElHV1 pU42 (29.4%) displayed only insignificant sequence homology, we determined a nuclear distribution for every protein. Remarkably, pRh69 and pM69 concentrated in distinct foci within the nucleus, which resembled nuclear pUL69 aggregates observed in transfected or infected cells treated with protein kinase inhibitors (38, 53, 60). In previous studies, we addressed the question of whether this speckled staining pattern of pUL69 could be explained by colocalization with nuclear splice domain components like SC35; however, this was not detected for pUL69 (38, 60) nor for pRh69 and pM69 (data not shown). Currently, we are investigating whether specific cellular proteins contribute to the formation of these aggregates. Thus, given the low level of NLS conservation within the betaherpesviral pUL69 protein family, in order to be translocated to the nucleus these proteins must either be capable of interacting with human importins or have at least one common cellular interaction partner.
Like HCMV pUL69, all of its betaherpesviral homologs share a region with a high degree of conservation throughout the three herpesviral subgroups, the so-called ICP27 homology region. The only common function assigned to the ICP27 homology region so far is self-interaction, which has been demonstrated for ICP27 and pUL69 (28, 62). By alignment of the pUL69 self-interaction domain (aa 269 to 574) with protein sequences of betaherpesviral pUL69 homologs, pC69 of CCMV was determined to be the homolog displaying the highest score for sequence identity (77.1%). In contrast, pRh69, pM69, HHV6 pU42, and ElHV1 pU42 displayed significantly less amino acid sequence identity (<36%). Hence, our findings that homodimerization is a common feature of the betaherpesviral UL69 proteins and, more strikingly, that pC69, pRh69, pM69, and HHV6 pU42 heterodimerize with pUL69 should ultimately help to narrow down the homodimerization and heterodimerization domains within each pUL69 homolog and to identify highly conserved amino acid residues that are required for these interactions.
One important feature of the ICP27 protein family is their capacity to function as viral-mRNA export factors, as described in the introduction. We performed an established CAT reporter assay originally developed by Hope and colleagues to monitor mRNA export in vivo (24) to show that only the cytomegaloviral proteins pUL69, pC69, and pRh69, analogous to HIV-1 Rev, were able to export unspliced CAT mRNA to the cytoplasm, while pM69, HHV6 pU42, and ElHV1 pU42 were negative in this assay. It is noteworthy that these assays were performed in human HEK293T and HeLa cells (data not shown). Thus, one might assume that pC69 and pRh69, which diverged more recently from HCMV pUL69 during evolution, might still interact with human cellular cofactors necessary for this export activity. In contrast, MCMV pM69 and ElHV1 pU42, which diverged earlier from HCMV pUL69, might not be able to recognize and interact with human cofactors, even though they could possess mRNA export activity within their natural hosts. Note that pM69 also failed to export this specific reporter RNA in the context of murine NIH 3T3 cells, while pUL69, pC69, and pRh69 remained active, thereby suggesting that pM69, at least in the context of this RNA export assay, does not function as a viral-mRNA export factor, in contrast to its cytomegaloviral counterparts. As HHV6 pU42 was also negative in the context of human cells, one might speculate that HHV6, as a lymphotropic human pathogen, requires cellular cofactors that are not expressed in tissue culture cells like HeLa or HEK293T. Further experiments using virus-specific host cell types need to be performed before a final conclusion can be reached.
In a previous study, we reported that nucleocytoplasmic shuttling activity was a prerequisite for pUL69-mediated mRNA export (29). Therefore, we were interested in analyzing whether the betaherpesviral homologs also exhibit a nucleocytoplasmic shuttling activity. Importantly, an in silico search for a classical leucine-rich NES did not detect a matching sequence in any of the betaherpesviral pUL69 homologs. This finding was not without precedent, as we reported previously that the C terminus of pUL69 contains a nonclassical bipartite CRM1-independent NES that is located between amino acids 597 and 624 (27). However, alignment of the pUL69 NES with sequences of the other betaherpesvirus family members failed to identify a motif with strong homology. The highest NES sequence identity of 39.3% was detected for pM69 of MCMV, whereas even the more closely related cytomegaloviral pUL69 homologs pC69 and pRh69 displayed a sequence homology of only approximately 30%. Interestingly, however, results from our heterokaryon analyses clearly showed that in addition to the cytomegaloviral proteins pUL69, pC69, and pRh69, the more distantly related factors pM69, HHV6 pU42, and ElHV1 pU42 exhibited nucleocytoplasmic shuttling activity. Future studies will aim at a precise definition of the amino acid sequence motifs as well as the cellular factors that facilitate the nuclear export of each member of the betaherpesviral pUL69 protein family. Nevertheless, our findings suggest that nucleocytoplasmic shuttling activity is not sufficient for the observed mRNA export activity of the cytomegaloviral proteins pUL69, pC69, and pRh69.
Because we detected in our previous studies that HCMV facilitates the cytoplasmic accumulation of unspliced mRNA via recruitment of the cellular DExD/H-box RNA helicases UAP56 and URH49 (29), it was tempting to investigate whether this was also true for pC69 and pRh69. Site-directed mutagenesis was performed, and subsequent coimmunoprecipitation analyses in concert with CAT mRNA reporter assays clearly demonstrated that the UAP56/URH49 interaction of pUL69, pC69, or pRh69 was absolutely essential for their mRNA export activity. Since the evolutionary conservation of the UAP56/URH49-binding site suggests an important function of this protein interaction motif for viral replication, recombinant human cytomegaloviruses carrying mutations within pUL69 that have previously been shown to either abrogate the interaction with UAP56 and/or to interfere with the RNA-binding capacity of pUL69 (29, 55) were constructed. Growth curve analyses clearly revealed a severe replication defect of two viruses harboring UAP56-binding site mutations, while a revertant virus and a virus expressing an RNA-binding-deficient pUL69 mutant replicated comparably to wild-type virus. This result strongly suggests that the interaction of pUL69 with UAP56/URH49 is essential for efficient viral replication. While the manuscript was under review, Kronemann and colleagues reported on the generation and characterization of a recombinant cytomegalovirus expressing the UAP56-binding-deficient alanine substitution mutant mUAP of pUL69 (26). Surprisingly, the growth curve analyses performed in this study revealed that the UAP56-binding-deficient virus replicated to levels even greater than those of the wild-type virus (26). The reason for this obviously contradictory result is presently not clear and requires further investigation but may be related to the fact that Kronemann and colleagues introduced the UL69 mutations into an already altered AD169-derived virus that carries an additional expression cassette for green fluorescent protein and a puromycin resistance gene, thereby deleting the open reading frame UL21.5 (7, 26). Furthermore, we consider it highly unlikely that the introduction of an adventitious mutation accounts for the impaired replication associated with the lack of UAP56-binding activity as observed in the present study, since the respective phenotype was observed with two different viruses (UL69mUAP and UL69ΔR1ΔRS) that were generated in independent recombination reactions.
In summary, in spite of the low level of amino acid sequence conservation within the betaherpesviral pUL69 protein family, our study characterized homodimerization and nucleocytoplasmic shuttling activity as conserved features of the betaherpesviral proteins homologous to pUL69, and we were able to identify UAP56/URH49-binding capacity as a unique feature of the cytomegaloviral proteins pUL69, pC69, and pRh69 that is essential for their mRNA export activity and for efficient replication of HCMV. Interestingly, a recent study demonstrated that in addition to its effect on mRNA export, pUL69 facilitates the translation of mRNAs by excluding 4EBP1 from the cap-binding complex (1). Thus, it is tempting to speculate that nucleocytoplasmic shuttling, which would be required for such a cytoplasmic function to modulate protein translation, is highly conserved within the betaherpesviruses while interaction with UAP56 to access the cellular mRNA export pathway has evolved only in the cytomegaloviral members of the pUL69 protein family. Further experiments will be required to address this hypothesis.
Acknowledgments
We thank G. Hayward (Baltimore, MD) for CCMV DNA and M. Mach and F. Neipel (Erlangen, Germany) for genomic RhCMV DNA and HHV6A cosmids. We also thank B. Ehlers (Berlin, Germany) for kindly providing ElHV1 DNA and Peter Lischka (AiCuris GmbH & Co. KG, Wuppertal, Germany) for stimulating discussions.
This study was supported by the Deutsche Forschungsgemeinschaft (SFB 796) and the Elitenetzwerk Bayern (BIGSS scholarship to Barbara Zielke).
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Aoyagi, M., M. Gaspar, and T. E. Shenk. 2010. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc. Natl. Acad. Sci. U. S. A. 107:2640-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 3.Bello, L. J., et al. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 4.Berndt, A., H. Hofmann-Winkler, N. Tavalai, G. Hahn, and T. Stamminger. 2009. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 83:12881-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buisson, M., et al. 1989. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J. Virol. 63:5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, I. D., F. Shanahan, and P. J. Farrell. 1994. Epstein-Barr virus SM protein. Virology 205:217-227. [DOI] [PubMed] [Google Scholar]
- 11.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 13.Davison, A. J., et al. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 14.Davison, A. J., et al. 2009. The order Herpesvirales. Arch. Virol. 154:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehlers, B., et al. 2001. Genetic and ultrastructural characterization of a European isolate of the fatal endotheliotropic elephant herpesvirus. J. Gen. Virol. 82:475-482. [DOI] [PubMed] [Google Scholar]
- 17.Farjot, G., et al. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gompels, U. A., et al. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin, D. J., K. T. Hall, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 1999. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 73:10519-10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruffat, H., et al. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen, S. G., L. I. Strelow, D. C. Franchi, D. G. Anders, and S. W. Wong. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77:6620-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiriart, E., et al. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. U. S. A. 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inchauspe, G., and J. M. Ostrove. 1989. Differential regulation by varicella-zoster virus (VZV) and herpes simplex virus type-1 trans-activating genes. Virology 173:710-714. [DOI] [PubMed] [Google Scholar]
- 26.Kronemann, D., S. R. Hagemeier, D. Cygnar, S. Phillips, and W. A. Bresnahan. 2010. Binding of the human cytomegalovirus (HCMV) tegument protein UL69 to UAP56/URH49 is not required for efficient replication of HCMV. J. Virol. 84:9649-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lischka, P., M. Thomas, Z. Toth, R. Mueller, and T. Stamminger. 2007. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J. Gen. Virol. 88:405-410. [DOI] [PubMed] [Google Scholar]
- 29.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorz, K., et al. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik, P., and J. B. Clements. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32:5553-5569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 35.Momose, F., et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piñol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 37.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rechter, S., et al. 2009. Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 and modulate its nuclear localization and activity. J. Biol. Chem. 284:8605-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, S. A., and D. M. Knipe. 1988. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 62:3814-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizmann, B., et al. 1992. The family Herpesviridae: an update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 41.Roth, J., and M. Dobbelstein. 1997. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J. Virol. 71:8933-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salsman, J., N. Zimmerman, T. Chen, M. Domagala, and L. Frappier. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS. Pathog. 4:e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri-Goldin, R. M. 2001. Nuclear export of herpes virus RNA. Curr. Top. Microbiol. Immunol. 259:2-23. [PubMed] [Google Scholar]
- 46.Sandri-Goldin, R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241-5256. [DOI] [PubMed] [Google Scholar]
- 47.Sandri-Goldin, R. M., and G. E. Mendoza. 1992. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 6:848-863. [DOI] [PubMed] [Google Scholar]
- 48.Semmes, O. J., et al. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sergeant, A., H. Gruffat, and E. Manet. 2008. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 13:3798-3813. [DOI] [PubMed] [Google Scholar]
- 50.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Souki, S. K., P. D. Gershon, and R. M. Sandri-Goldin. 2009. Arginine methylation of the ICP27 RGG box regulates ICP27 export and is required for efficient herpes simplex virus 1 replication. J. Virol. 83:5309-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souki, S. K., and R. M. Sandri-Goldin. 2009. Arginine methylation of the ICP27 RGG box regulates the functional interactions of ICP27 with SRPK1 and Aly/REF during herpes simplex virus 1 infection. J. Virol. 83:8970-8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, M., et al. 2009. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 90:567-578. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, S. L., et al. 1998. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J. Virol. 72:2935-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toth, Z., P. Lischka, and T. Stamminger. 2006. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 34:1237-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toth, Z., and T. Stamminger. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939-2949. [DOI] [PubMed] [Google Scholar]
- 57.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, B. J., et al. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA via the cellular mRNA export pathway. Biochem. J. 387:295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkler, M., T. aus dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhi, Y., K. S. Sciabica, and R. M. Sandri-Goldin. 1999. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 257:341-351. [DOI] [PubMed] [Google Scholar]