Abstract
Several arenaviruses, chiefly Lassa virus (LASV) and Junin virus in West Africa and Argentina, respectively, cause hemorrhagic fever (HF) disease in humans that is associated with high morbidity and significant mortality. The investigation of antiviral strategies to combat HF arenaviruses is hampered by the requirement of biosafety level 4 (BSL-4) facilities to work with these viruses. These biosafety hurdles could be overcome by the use of recombinant single-cycle infectious arenaviruses. To explore this concept, we have developed a recombinant lymphocytic choriomeningitis virus (LCMV) (rLCMVΔGP/GFP) where we replaced the viral glycoprotein (GP) with the green fluorescent protein (GFP). We generated high titers of GP-pseudotyped rLCMVΔGP/GFP via genetic trans complementation using stable cell lines that constitutively express LCMV or LASV GPs. Replication of these GP-pseudotyped rLCMVΔGP/GFP viruses was restricted to GP-expressing cell lines. This system allowed us to rapidly and reliably characterize and quantify the neutralization activities of serum antibodies against LCMV and LASV within a BSL-2 facility. The sensitivity of the GFP-based microneutralization assay we developed was similar to that obtained with a conventionally used focus reduction neutralization (FRNT) assay. Using GP-pseudotyped rLCMVΔGP/GFP, we have also obtained evidence supporting the feasibility of this approach to identify and evaluate candidate antiviral drugs against HF arenaviruses without the need of BSL-4 laboratories.
Arenaviruses include several causative agents of hemorrhagic fever (HF) disease in humans, which is associated with high morbidity and significant mortality. Members of this virus family are classified into Old World and New World arenaviruses (4). Two Old World arenaviruses, Lassa virus (LASV) and Lujo virus (LUJV), and five New World arenaviruses, Junin virus (JUNV), Machupo virus (MACV), Sabia virus (SABV), Guanarito virus (GTOV), and Whitewater Arroyo virus (WWAV), are known to cause HF disease in humans, with LASV posing the highest public health concern among HF arenaviruses (5, 10). On the other hand, evidence indicates that the globally distributed prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is likely a neglected human pathogen of clinical significance in congenital infections (1). Moreover, LCMV infection of immunocompromised individuals can result in severe disease and death (8, 19). Public health concerns posed by human-pathogenic arenaviruses are aggravated by the lack of FDA-licensed vaccines and by current antiarenaviral therapy being limited to an off-label use of the nucleoside analog ribavirin that is only partially effective. Moreover, to be effective, ribavirin therapy requires early and intravenous administration and is often associated with significant side effects (29).
Arenaviruses are enveloped viruses with a bisegmented negative-strand (NS) RNA genome. Each genomic RNA segment, L (ca. 7.3 kb) and S (ca. 3.5 kb), uses an ambisense coding strategy to direct the synthesis of two polypeptides in opposite orientations. The S RNA encodes the viral glycoprotein precursor (GPC) and the nucleoprotein (NP). GPC is posttranslationally cleaved by the cellular site 1 protease (S1P) to yield the two mature virion glycoproteins GP1 and GP2, which form the spikes that decorate the virus surface and mediate receptor recognition and cell entry. The L RNA encodes the viral RNA (vRNA)-dependent RNA polymerase (RdRp, or L polymerase) and the small RING finger protein Z, which is the arenavirus counterpart of the M protein found in many other NS RNA viruses.
Research on HF arenaviruses has been hampered by a requirement of biosafety level 4 (BSL-4) to handle live forms of these agents. Therefore, the study of HF arenaviruses would be facilitated by the development of valid surrogate systems that could be used under less-strict biosafety conditions to circumvent the cost and intrinsic complications associated with the use of BSL-4 facilities. Progress in this area has already been made by using recombinant retroviruses pseudotyped with the GP of HF arenaviruses (6, 15, 32). This approach, however, is limited to the investigation of arenavirus cell entry, without addressing the contribution of the arenavirus L, NP, and Z gene products to virus fitness and virulence, as well as virus-host interactions underlying mechanisms of disease.
Generation of single-cycle infectious viruses, where a reporter gene replaces one of the essential viral genes and production of infectious progeny is achieved via genetic trans complementation, has been documented for several BSL-3/4 viruses, including highly pathogenic strains of influenza virus (17) and Ebola virus (11). These single-cycle reporter-expressing viruses have been proved to be safe candidates for identifying virus-specific neutralizing antibodies and for studying several aspects of the virus replication cycle. In this study, we have investigated the application of this concept to HF arenaviruses. For this, we generated a recombinant single-cycle infectious LCMV (rLCMV) by substituting green fluorescent protein (GFP) for the virus GP gene (rLCMVΔGP/GFP) and used trans-complementation by a cell line constitutively expressing LASV GP to generate high titers of GFP-expressing LCMV pseudotyped with LASV GP (rLCMVΔGP/GFP-LASVGP), whose replication was restricted to cells expressing LASV GP. We present evidence supporting the use of rLCMVΔGP/GFP pseudotyped with GPs from HF arenaviruses for the development of safe, rapid, and sensitive assays for detection and quantification of neutralizing antibodies (NAbs) against HF arenaviruses, as well as identification of potential inhibitors of HF arenavirus GP-mediated attachment and cell entry. We also provide evidence that single-cycle infectious recombinant arenaviruses expressing an appropriate reporter gene could provide a robust BSL-2 platform to identify and characterize antiviral drugs against HF arenaviruses.
MATERIALS AND METHODS
Cell lines and viruses.
Vero (African green monkey kidney epithelial; ATCC CCL-81) and BHK-21 (baby hamster kidney fibroblast; ATCC CCL-10) cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), supplemented with l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml). For infections, cells were maintained in a mixture (1:1) of Opti-MEM and their normal medium in the absence of hygromycin B (referred to as infection medium). Cells were grown at 37°C in a 5% CO2 atmosphere. Wild-type (WT) LCMV (Armstrong strain; ARM5-3b) was produced by infecting BHK-21 cells (multiplicity of infection [MOI] = 0.01 to 0.1) and harvesting the supernatant on days 3 and 4 postinfection. Virus was titrated by immunofocus assay, as described previously (2).
Generation of GP-expressing stable cell lines.
Vero and BHK-21 cell lines constitutively expressing LCMV GP or LASV GP were generated by cotransfection of the respective GP-expressing pCAGGs plasmids together with the hygromycin resistance expression plasmid pCB7 (17) (7:1 ratio) by using Lipofectamine 2000. Forty-eight hours posttransfection, cells were plated into 10-cm culture dishes at low density and cell clones were selected in the presence of 500 μg/ml of hygromycin B. Cell clones were screened for GP expression by immunofluorescence, flow cytometry, and Western blotting (WB) using both GP type-specific and cross-reactive monoclonal (MAb) and polyclonal (PAb) antibodies and for their ability to rescue replication of rLCMVΔGP/GFP. GP-expressing Vero and BHK-21 cell lines were maintained in DMEM supplemented with 10% FBS, penicillin/streptomycin (P/S), and 200 to 500 μg/ml of hygromycin B.
Generation of rLCMVΔGP/GFP-expressing viruses.
Reverse genetics methods for rescue of recombinant LCM viruses (rLCMV) have been previously described (28) (7). Plasmid pPOLI-S ΔGP/GFP was generated by replacing the GP gene of LCMV with that for green florescent protein (GFP). This plasmid contained the viral RNA S segment flanked at its 5′ and 3′ termini by the murine POLI promoter and terminator sequences, respectively (7). To rescue rLCMVΔGP/GFP, BHK-21 cells were cotransfected with pPOLI plasmids expressing the viral genomic RNA segment L (pPOLI-L) and pPOLI-S ΔGP/GFP, together with plasmids expressing the minimal trans-acting factors NP (pCAGGs-NP) and L (pCAGGs-L) and pCAGGs expressing LCMV GP (pCAGGs-GP), required to generate infectious GP-pseudotyped rLCMVΔGP/GFP virion particles. At 72 h posttransfection, tissue culture supernatants were passaged into LCMV GP-expressing BHK-21 cells to amplify rLCMVΔGP/GFP pseudotyped with LCMV GP (pLCMV). LASV GP-pseudotyped rLCMVΔGP/GFP (pLASV) was generated in the same manner but using LASV GP-expressing BHK-21 cells.
Viral growth kinetics and viral titrations.
Growth kinetic analyses of WT LCMV and GP-pseudotyped rLCMVΔGP/GFP viruses were performed in parental and GP-stably expressing BHK-21 cell lines. Subconfluent cell monolayers were infected at a low MOI (0.1). After 90 min of adsorption at 37°C, the virus inoculum was removed, cells were washed, and infection medium was added. Fluorescence images were obtained for pLCMV at the indicated time postinfection (p.i.), and the supernatants from infected cells were collected, clarified by centrifugation, and titrated. WT LCMV supernatants were titrated on Vero cells. GP-pseudotyped rLCMVΔGP/GFP supernatants were titrated on LCMV GP-expressing Vero cells. Virus titers (focus-forming units [FFU]/ml) in tissue culture supernatants were determined by immunofocus assay. Briefly, Vero or GP-expressing Vero cells were infected with 10-fold serial dilutions of WT or GP-pseudotyped rLCMVΔGP/GFP, respectively, and fixed at 24 h p.i. with 4% formaldehyde in 1× PBS, permeabilized with 0.2% Triton X-100 in 1× PBS, and blocked with 2.5% bovine serum albumin (BSA) in 1× PBS. LCMV WT-infected cells were identified by staining with an anti-NP MAb (1.1.3) and a fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG secondary antibody. GP-pseudotyped rLCMVΔGP/GFP-infected cells were identified by GFP expression.
Virus purification.
BHK-21 and GP-expressing BHK-21 cells were infected with WT LCMV or GP-pseudotyped rLCMVΔGP/GFP at a low MOI (0.01). At 3 days p.i., tissue culture supernatants were harvested and clarified at 2,000 rpm for 10 min at 4°C. Infected cells were pelleted, washed with 1× PBS, and stored at −20°C. A further clarification step of virus-containing cell supernatants was performed at 10,000 rpm for 30 min at 4°C in a Beckman Coulter Optima L-90K ultracentrifuge using an SW-32Ti rotor. Finally, two-step virus purification on a 20% sucrose cushion in NTE buffer (100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA) was performed at 35,000 rpm for 2.5 h at 4°C in a Beckman Coulter Optima L-90K ultracentrifuge using an SW-41 rotor. The pelleted virus was resuspended in 80 to 100 μl of 1× PBS and stored at 4°C.
Indirect immunofluorescence and fluorescence-activated cell sorting (FACS) analysis. (i) Indirect immunofluorescence.
Screening and characterization of Vero and BHK-21 stable cell lines expressing GPs were performed using the following anti-GP specific MAbs: 83.6, an arenavirus cross-reactive anti-GP2 (31); 36.1, anti-GP1 LCMV (16); and L52-74-7A, anti-GP1 LASV (kindly provided by Randal J. Schoepp [12]). In addition, we also used guinea pig and rhesus monkey sera to LCMV and LASV, respectively. Monoclonal antibody 1.1.3 against LCMV NP was used as hybridoma supernatants and has been previously described (7). Subconfluent monolayers of normal and GP-expressing Vero and BHK-21 cells growing on cover glass were fixed with 4% formaldehyde. For intracellular staining, cells were permeabilized with 0.2% Triton X-100 at room temperature, followed by a blocking step with 2.5% BSA in 1× PBS for 16 h at 4°C or 1 to 2 h at room temperature. After blocking, cells were incubated with the indicated antibodies for 1 h at 37°C. Cells were then washed with 1× PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) and goat anti-mouse IgG-Alexa Fluor 488 for MAbs, protein A-FITC for anti-LCMV PAb, and goat anti-rhesus IgG(H+L)-FITC for anti-LASV PAb for 30 min at 37°C. Microscope cover glasses with stained cells were mounted with Mowiol on glass slides and analyzed by florescence microscopy using a 63× oil immersion objective. Images were colored using the Adobe Photoshop CS4 (v11.0) software program.
(ii) FACS analysis.
Single-cell preparations of Vero and BHK-21 cells were blocked with 2.5% BSA in 1× PBS containing 1% normal heat-inactivated goat serum for 30 min on ice and then incubated with MAb 36.1 or a murine IgG1 (mIgG1) isotype control for 1 h on ice. Cells were washed twice with 1× PBS and incubated with goat F(ab′)2 fragment of anti-mouse IgG-FITC in blocking buffer for 30 min on ice. Cells were washed twice with 1× PBS, fixed with 0.5% paraformaldehyde in 1× PBS, and suspended in 1× PBS before analysis using an Accuri C6 flow cytometer and the Flowjo software program (v8.8.7).
Protein gel electrophoresis and Western blot analysis.
Parental and GP-expressing Vero and BHK-21 cell lines were lysed with 1% NP-40 cell lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% [vol/vol] Nonidet P-40, and complete protease inhibitor cocktail) for 30 to 60 min at 4°C. Cell lysates were clarified by centrifugation at 13,000 rpm in a benchtop centrifuge for 10 min at 4°C. Total protein in the cleared lysates was determined using the Micro BCA protein assay kit (Pierce). Total protein (100 μg) from whole-cell lysates was separated using denaturing SDS-PAGE and transferred to nitrocellulose membranes. After blocking with 10% nonfat dry milk in 1× PBS, membranes were reacted with MAbs 83.6 and L52-74-7A for detection of GPs or with an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PAb (Vero) or an anti-GAPDH MAb (BHK-21) for normalization of protein expression levels. After overnight incubation at 4°C, the membranes were probed with sheep anti-mouse IgG-horseradish peroxidase (HRP) or donkey anti-rabbit IgG-HRP. Protein bands were visualized using the ECL protein detection system (Denville Scientific, Inc.). For Western blot analysis of virus-infected cells, cells were lysed with 1% NP-40 cell lysis buffer and clarified, and the total protein content was estimated as previously described. Total protein (100 μg) from whole-cell lysates was separated by denaturing SDS-PAGE and probed with anti-LCMV guinea pig serum for LCMV NP, anti-GFP MAb (JL-8; Clontech) and GAPDH MAb. After overnight incubation at 4°C, the membranes were probed with rabbit anti-guinea pig IgG(H+L)-HRP for PAb and sheep anti-mouse IgG-HRP for MAbs. Protein bands were visualized using the ECL protein detection system.
Transmission electron microscopy.
Purified virus was fixed with 2.5% glutaraldehyde for 5 min at room temperature, diluted in filtered water, and adsorbed onto Formvar-carbon-coated 200-mesh copper grids for 2 min at room temperature. After 20 s of drying, the grids were negatively stained with a filtered aqueous solution of 2% uranyl acetate (pH 4.2 to 4.5) for 1 min at room temperature. After excess stain was blotted, dried grids were imaged using a Hitachi (Tokyo, Japan) 7650 transmission electron microscope (TEM) with an attached Gatan 11-megapixel Erlangshen (Pleasanton, CA) digital camera at 80-kv acceleration voltage, 3.5-s exposure, and magnification ×200,000 (×200K) to ×300K. Representative micrographs were obtained, and virion diameter (n = 27) was analyzed using Digital micrograph software. For immunoelectron microscopy (IEM), fixed virus was adsorbed onto Formvar-carbon-coated 200-mesh nickel grids for 30 min and placed on droplets of 1% BSA for 30 min. Primary antibodies (36.1 MAb and anti-LASV rhesus serum) diluted in 1% BSA were reacted for 2 h at room temperature. The grids were rinsed with 1× PBS and transferred onto droplets of either goat anti-mouse IgG (12-nm-gold-tagged secondary; 1:40) (Jackson ImmunoResearch, West Grove, PA) for MAb labeling or protein A (15-nm-gold-tagged; 1:25 diluted) for PAb labeling (Electron Microscopy Sciences, Hatfield, PA) for 1 h at room temperature. The grids were rinsed with 1× PBS and postfixed in 2% phosphate-buffered glutaraldehyde for 15 min, rinsed in distilled water, and negatively stained with an aqueous solution of 2% uranyl acetate for 1 min at room temperature. The grids were examined and photographed as previously described.
FRNT assay.
Parental or GP-expressing Vero cell lines were infected with WT or GP-pseudotyped rLCMV (25 FFU/well; 24-well plate format) for 90 min at 37°C and overlaid with 0.5% agarose (Seakem ME) containing Eagle minimal essential medium (EMEM) with l-glutamine and penicillin-streptomycin without phenol red. At days 2, 3, and 7 postinfection, agarose plugs were removed and the infected cells were fixed using 4% formaldehyde in 1× PBS for 1 to 2 h at room temperature. Plates were imaged for green florescent foci with the Kodak image station (4000MM Pro molecular imaging system; Carestream Health, Inc., NY) and Kodak molecular imaging software (v5.0.1.30). Fixed cells were permeabilized using 0.01% Triton X-100 for 15 to 20 min at room temperature and washed twice with 1× PBS. Plaques/foci were immunostained with anti-LCMV NP MAb (1.1.3) in 1% normal horse serum using a Vectastain ABC kit (Vector Laboratories, CA). For focus reduction neutralization assay (FRNT) assays, approximately 100 FFU of pLCMV or pLASV was mixed with serial dilutions of MAb or PAb, incubated for 60 min at 37°C for immune complex formation, and adsorbed onto subconfluent Vero LCMV GP-expressing cell monolayers for 90 min at 37°C and then overlaid, infected for 3 days, and focus immunostained as described above.
GFP-based microneutralization assay and antiviral activity evaluation.
The GFP-based microneutralization assay was done as described for hemagglutinin (HA)-pseudotyped GFP-expressing influenza viruses (17). Antibody samples were incubated at 56°C for 45 min to inactivate complement. Purified or hybridoma supernatant MAbs or antisera (PAbs) were serially (2-fold or as indicated) diluted in the infection medium at 50 μl (final volume)/well in 96-well microtiter plates and incubated for 60 min at 37°C with a predetermined amount of GP-pseudotyped GFP-expressing rLCMVs to obtain ∼70 to 100% GFP-positive cells in the Vero LCMV GP cell line (MOI = ∼1 to 2). These preformed immune complexes were adsorbed onto cell monolayers (in triplicate) for 90 min at 37°C. The immune complexes were removed, and the cells were maintained in the infection medium. Forty-eight to ninety-six hours postinfection, depending on the amount of virus used, the medium was aspirated, and the cell monolayers were washed and replenished with 1× PBS to remove any background green fluorescence before imaging in fluorescence microscopy. Green fluorescence intensity was measured with a GFP microplate reader. Ribavirin antiviral activity was assessed by treating Vero LCMV GP-expressing cell monolayers for 60 min at 37°C with serial dilutions of ribavirin, followed by infection with GP-pseudotyped rLCMVΔGP/GFP (MOI = ∼1 to 2) in the presence of the indicated drug concentrations. Levels of infection were determined based on GFP expression. Cell viability was evaluated by bright-field microscopy and comparison of ribavirin- and mock-treated cells.
Statistical analysis.
GFP-based microneutralization assays were performed in triplicate. The mean and standard deviation (SD) were calculated using the Microsoft Excel software program. Percent neutralization or inhibition was fitted to a sigmoidal dose-response curve using the GraphPad Prism software program.
RESULTS
Generation and characterization of GP-expressing stable cell lines.
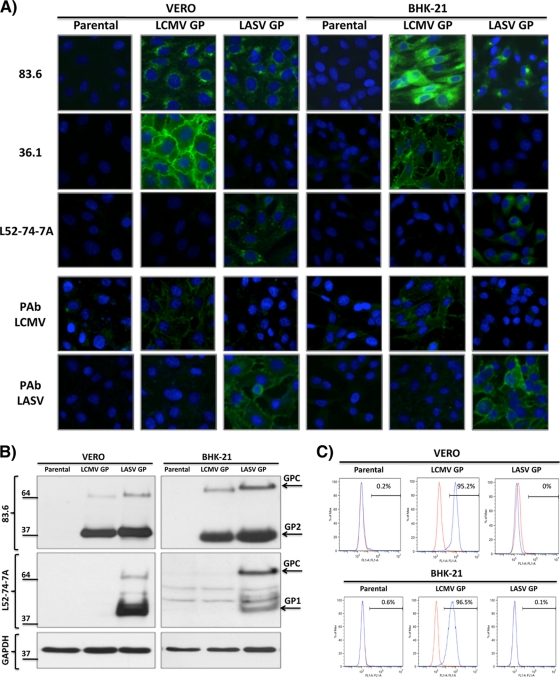
We used procedures similar to those we employed to produce influenza HA-expressing cell lines (17) to generate Vero and BHK-21 cells expressing LCMV and LASV GP to complement the growth of rLCMVΔGP/GFP. Briefly, Vero or BHK-21 cells were cotransfected with the GP-expressing pCAGGs plasmids together with the hygromycin resistance plasmid pCB7 (17), and stable cell clones were selected by addition of hygromycin B. Parental and GP-expressing Vero and BHK-21 cell lines were characterized by immunofluorescence (Fig. 1 A), Western blotting (WB) (Fig. 1B), and fluorescence-activated cell sorting (FACS) analysis (Fig. 1C) by using type-specific or cross-reactive MAb and PAb for LCMV and LASV GPs. The Vero and BHK-21 parental cell lines did not react with any of the antibodies. As expected, the cross-reactive MAb 83.6 (anti-GP2) recognized the LCMV and LASV GP-expressing Vero and BHK-21 cell lines. In contrast, LCMV (36.1) and LASV (L52-74-7A) GP1-specific MAbs stained only the LCMV and LASV GP-expressing cell lines, respectively. We obtained similar results with sera against LCMV (PAb LCMV) and LASV (PAb LASV). In Western blotting, MAb 83.6 recognized both LCMV and LASV GP, whereas MAb L52-74-7A recognized only LASV GP (Fig. 1B). Consistent with previous findings (33), the conformation-sensitive and specific LCMV-GP MAb 36.1 did not recognize viral GPs in WB but stained the surface only of LCMV-GP-expressing cells (Fig. 1A and C).
FIG. 1.
Characterization of the GP-expressing Vero and BHK-21 cells. (A) Immunofluorescence assay (IFA). Parental and GP-expressing Vero and BHK-21 cells were tested for GP expression by IFA with the cross-reactive MAb 83.6 to GP2, LCMV-GP1-specific MAb 36.1, and LASV-GP1-specific MAb L52-74-7A. Nuclei were stained with DAPI. (B) Western blot assay. Cell lysates from parental and GP-expressing Vero and BHK-21 cells were prepared and analyzed by Western blotting with the indicated MAbs. Expression levels of GAPDH were used for normalization. Positions of LCMV and LASV GPC, GP1, and GP2 bands are indicated by arrows. (C) FACS analysis. Parental and GP-expressing Vero and BHK-21 cells were subjected to FACS analysis with the 36.1 MAb. The red line indicates staining with the mIgG1κ isotype control. The blue line indicates staining with MAb 36.1.
Rescue of GP-deficient, GFP-expressing rLCMV (rLCMVΔGP/GFP).
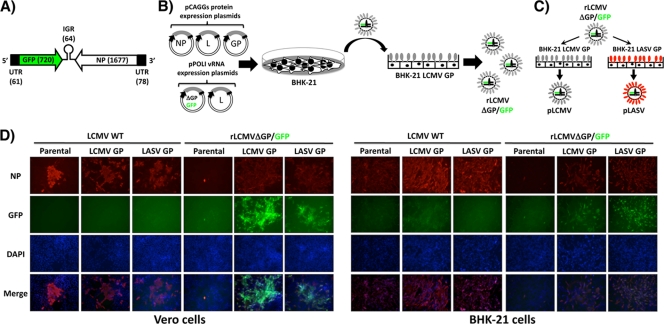
To generate rLCMVΔGP/GFP, we cotransfected BHK-21 cells with the vRNA expression plasmids pPOLI-S ΔGP/GFP (Fig. 2 A) and pPOLI-L, together with the pCAGGs-based expression plasmids for NP, L, and GP (Fig. 2B). At 72 h posttransfection, tissue culture supernatants were used to infect fresh monolayers of BHK-21 cells expressing LCMV GP to rescue LCMV GP-pseudotyped rLCMVΔGP/GFP (pLCMV). To generate LASV GP-pseudotyped rLCMV (pLASV), we infected LASV-GP-expressing cells with pLCMV (Fig. 2C). GFP expression and the use of specific antibodies to detect the viral NP confirmed the virus rescue (Fig. 2D). To further confirm the dependence of pLCMV growth on GP-expressing cells, we infected parental and GP-expressing Vero and BHK-21 cell lines (MOI ranging from 0.01 to 1) with pLCMV, and at 72 h p.i., we prepared single-cell suspensions and analyzed them by flow cytometry for GFP expression. We observed over 90% GFP-positive cells for GP-expressing infected cells and <1% GFP-positive cells for parental cells.
FIG. 2.
Generation of rLCMVΔGP/GFP. (A) Schematic representation of the GFP/NP vRNA S segment. The GFP was substituted for GP within the LCMV S segment. Sizes (in nucleotides) of GFP and NP open reading frames (ORFs) are indicated in parentheses. (B) Schematic representation of the virus rescue. BHK-21 cells were transfected with plasmids expressing viral NP, L, and GP together with the pPOLI vRNA plasmids for L and S (ΔGP/GFP). Seventy-two hours posttransfection, tissue culture supernatants were used to infect BHK-21 cells constitutively expressing LCMV-GP, and virus rescue was confirmed by the appearance of GFP foci. (C) Production of GP-pseudotyped rLCMVΔGP/GFP. pLCMV was used to infect BHK-21 cells constitutively expressing either LCMV-GP or LASV-GP to produce pLCMV and pLASV, respectively. (D) Multiplication of rLCMVΔGP/GFP in LCMV and LASV GP-expressing cell lines. Vero and BHK-21 parental and GP-expressing cells were infected (MOI = 0.1) with WT and pLCMV. Infected cells were detected using an LCMV anti-NP MAb, 1.1.3 (NP), GFP expression (GFP), and nuclear staining (DAPI) by fluorescence microscopy. Merged images from NP (red), GFP (green), and DAPI (blue) stainings are indicated at the bottom of each panel.
Growth properties of rLCMVΔGP/GFP.
We next assessed the growth properties of GP-pseudotyped rLCMVΔGP/GFP in parental and GP-expressing Vero and BHK-21 cells and compared them to that of WT LCMV. For this, we infected (MOI = 0.1) parental and GP-expressing BHK-21 cells with either LCMV WT or pLCMV, and at the indicated time p.i., we captured representative images (Fig. 3 A) and determined levels of infectious virus in tissue culture supernatants (Fig. 3B). LCMV WT grew to similar titers in parental and GP-expressing BHK-21 cells. In contrast, rLCMVΔGP/GFP was detected at high titers in tissue culture supernatants from GP-expressing BHK-21 cells but not in tissue culture supernatants from parental BHK-21 cells. Accordingly, results from WB analysis showed that at 72 h after infection with pLCMV, detectable levels of viral NP and GFP were observed only in GP-expressing BHK-21 cells, while viral NP expression (but not GFP) was detected in parental and GP-expressing cell lines infected with LCMV WT (Fig. 3C). Parental and GP-expressing cells infected at a MOI of 3 with pLCMV or pLASV showed similar levels of GFP, indicating that parental and GP-expressing cells were equally susceptible to infection with GP-pseudotyped rLCMVDGP/GFP. As predicted, LCMV WT produced plaques to similar extents in all cell lines (Fig. 3D). Consistent with our previous findings, pLCMV generated plaques or foci in GP-expressing but not in parental Vero cells (Fig. 3E). As expected, plaques derived from pLCMV were fluorescent (GFP positive) (Fig. 3E).
FIG. 3.
Growth properties of rLCMVΔGP/GFP. (A) Kinetics of virus propagation. Parental and GP-expressing BHK-21 cells were infected (MOI = 0.1) with pLCMV in triplicate. At indicated times postinfection, representative images were taken using fluorescence microscopy. (B) Kinetics of infectious progeny production. Tissue culture supernatants from panel A were titrated by indirect immunofluorescence (WT LCMV) with the NP MAb 1.1.3 or by GFP expression (rLCMVΔGP/GFP) on parental or LCMV GP-expressing Vero cells, respectively. (C) Viral NP and GFP expression levels. Parental and GP-expressing BHK-21 cells were infected (MOI = 0.1) with LCMV WT and pLCMV. At 72 h p.i., cell lysates were prepared and subjected to Western blot analysis using antibodies to NP, GFP, and as loading control to GAPDH. (D and E) Plaque assay. Subconfluent monolayers of parental and GP-expressing Vero cells were infected with WT LCMV (D) or pLCMV (E) and overlaid with semisolid agar medium. At days 2, 3, and 7 p.i., cells were immunostained with the LCMV NP MAb. rLCMVΔGP/GFP plaques were also visualized with a Kodak image station. Representative images of at least three independent experiments are shown.
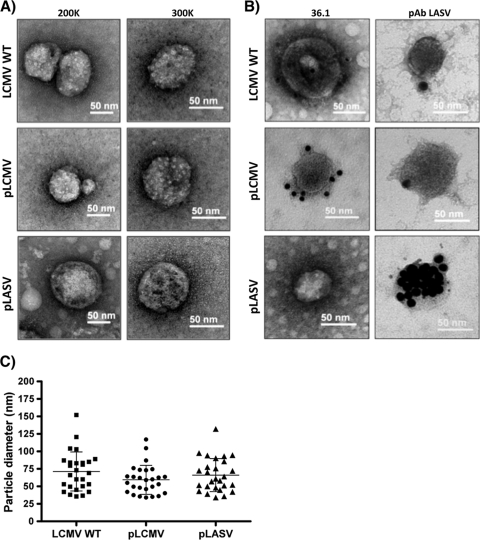
Electron microscopic characterization of LCMV and LASV GP-pseudotyped rLCMVΔGP/GFP (pLCMV and pLASV).
We next evaluated whether virion particle morphology or size, or both, differed between LCMV WT and GP-pseudotyped rLCMVΔGP/GFP. For this, we concentrated virion particles present in tissue culture supernatants from GP-expressing BHK-21 cells infected with pLCMV or parental BHK-21 cells infected with LCMV WT and examined them by TEM using negative staining (Fig. 4 A). GP-pseudotyped rLCMVΔGP/GFP showed an overall roughly spherical morphology and particle size similar to those of LCMV WT. Likewise, immunogold surface staining using the LCMV-GP1-specific MAb 36.1 or rhesus serum for LASV (anti-LASV PAb) revealed a similar pattern of GP surface staining between LCMV WT and GP-pseudotyped rLCMVΔGP/GFP (Fig. 4B). The LCMV GP1-specific MAb 36.1 detected only the GP present at the surface of pLCMV, whereas the anti-LASV PAb strongly stained the surface of pLASV, showing only weak cross-reactivity for GP present at the surface of pLCMV or LCMV WT. Quantitative analysis of 27 virion particles located in different fields revealed comparable sizes for LCMV WT and GP-pseudotyped rLCMVΔGP/GFP (Fig. 4C). Average diameters (means ± SD) were 71.3 ± 27.9 nm, 59.3 ± 20.7, nm and 66.1 ± 23.7 nm for LCMV WT, pLCMV, and pLASV, respectively, which are in good agreement with published data for LCMV (3) (18).
FIG. 4.
Morphology and size of GP-pseudotyped rLCMVΔGP/GFP virion particles. (A) Morphological comparison of GP-pseudotyped rLCMVΔGP/GFP and WT LCMV virion particles. WT LCMV and GP-pseudotyped rLCMVΔGP/GFP virion particle morphology was assessed by EM negative staining. Representative images of viral preparations at magnifications ×200K and ×300K are shown. (B) Immunoelectron microscopy of the WT LCMV and GP-pseudotyped rLCMVΔGP/GFP virion particles. Same viral preparations from panel A were surface stained with the LCMV-GP1-specific 36.1 MAb hybridoma supernatant (1/8 dilution) and the anti-LASV rhesus monkey serum (1/100 dilution) and counterstained with 12-nm-gold-labeled goat anti-mouse IgG antibody and 15-nm-gold-labeled protein A, respectively. Wild-type and LCMV or LASV GP-pseudotyped viruses are indicated. (C) Virus particle size. Diameters of WT and GP-pseudotyped rLCMVΔGP/GFP virion particles (n = 27) were measured, and average diameters were compared. The y axis indicates the diameters (nm) of the viral particles.
GFP-based microneutralization assay for characterization of neutralizing antibodies against GP-pseudotyped rLCMVΔGP/GFP.
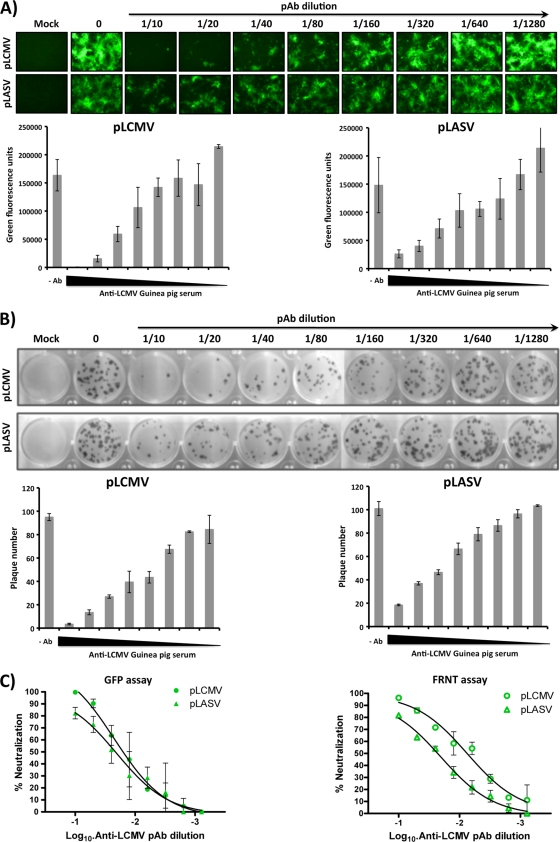
Conventional assays to detect neutralizing antibodies against HF arenaviruses involve handling live viruses, which requires BSL-4 facilities. Assays based on the use of GP-pseudotyped retroviruses (6, 15, 32) do not recapitulate all the steps of the arenavirus life cycle and may not accurately assess the true neutralizing capability of antibodies in the presence of other arenaviral proteins. We therefore examined whether our GP-pseudotyped rLCMVΔGP/GFP system could be developed as a novel, reliable, and safe approach to characterizing neutralizing antibodies against arenaviruses. The use of GFP expression as a surrogate of neutralization activity allowed us to use an automated plate reader with a fluorescence microplate format, which makes this assay simple and rapid. In this assay, neutralizing antibodies were identified based on a reduction in GFP expression after infection of GP-expressing cell lines with respective GP-pseudotyped rLCMVΔGP/GFP. To validate our approach, we studied the neutralizing abilities of a guinea pig antiserum for LCMV (PAb-LCMV) (Fig. 5). The complement was inactivated to evaluate IgG-mediated virus neutralization in all cases. To demonstrate the specificity of the assay, we included both pLCMV and pLASV. Results based on GFP expression indicated that PAb-LCMV neutralized pLCMV more efficiently than pLASV (Fig. 5A). These results correlated well with those obtained using the traditional focus reduction neutralization test (FRNT) (Fig. 5B). Comparison of the two assays indicated that our GFP-based microneutralization assays recapitulated a neutralization profile and neutralization order similar to those obtained with the classical FRNT assay (Fig. 5C).
FIG. 5.
A novel GFP-based microneutralization assay for the detection of arenavirus-neutralizing antibodies. (A) GFP-based microneutralization assay. LCMV or LASV GP-pseudotyped, GFP-expressing rLCMV (MOI ∼1 to 2) was preincubated with the indicated dilutions of the anti-LCMV guinea pig serum for 1 h at 37°C. The virus-antibody immune complex mixture was used to infect LCMV-GP-expressing Vero cells (96-well format, triplicate), and the presence of neutralizing antibodies was identified with fluorescence microscopy and quantified using a GFP microplate reader. (B) Focus reduction neutralization (FRNT) assay. The same GP-pseudotyped rLCMVs were incubated with the indicated dilutions of the LCMV PAb, and the presence of neutralizing antibodies was determined using the classical FRNT assay in LCMV GP-expressing Vero cells (24-well format, triplicate). Representative scans and quantification of plaque numbers are indicated. (C) Comparison of the GFP-based microneutralization assay and the conventional FRNT assay.
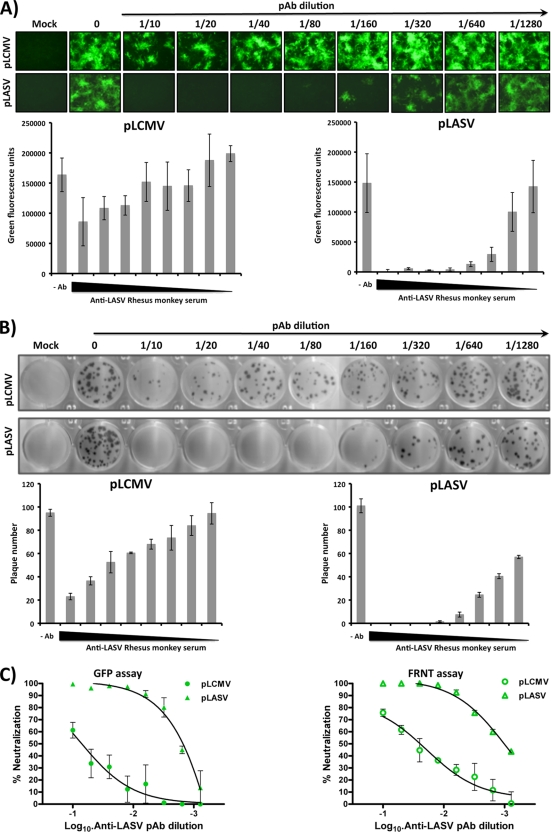
We conducted a similar experiment with an anti-LASV rhesus serum (PAb-LASV) (Fig. 6). Compared to PAb-LCMV, PAb-LASV exhibited stronger specific neutralization of the pLASV at a wide range of PAb dilutions (1/10 to 1/160) tested (Fig. 6A). Interestingly, PAb-LASV exhibited only very weak neutralizing activity against pLCMV in both GFP-based (Fig. 6A) and FRNT-based (Fig. 6B) assays. It is possible that binding of PAb-LCMV to other components besides GP on the virus particles contributed to its ability to neutralize both pLCMV and pLASV. Our GFP-based microneutralization and FRNT assays performed comparably (Fig. 6C). We also tested the GFP-based neutralization assay using previously described LCMV-GP1 (36.1)- and LASV-GP1 (L52-74-7A)-specific MAbs. LCMV-GP1 MAb 36.1 neutralized pLCMV but not pLASV (Fig. 7). LASV-GP1-specific MAb L52-74-7A did not neutralize pLCMV. Unexpectedly, under our experimental conditions, MAb L52-74-7A failed to neutralize pLASV even at the highest concentration tested, although it has been previously reported to have neutralizing activity (27).
FIG. 6.
Identification and quantification of LASV-neutralizing antibodies using the GFP-based microneutralization assay. Experiments were done as for Fig. 5 but using an anti-LASV rhesus monkey serum. (A) Representative images of the GFP-based microneutralization assay obtained by fluorescence microscopy and quantification with the GFP microplate reader. (B) FRNT scans and quantification of the plaque numbers. (C) Comparison of the neutralization properties of the LASV polyclonal sera in the GFP-based microneutralization assay and the classical FRNT assay with GP-pseudotyped LCMV (pLCMV) and LASV (pLASV) rLCMVΔGP/GFP.
FIG. 7.
GFP-based microneutralization assays with the MAb 36.1. Experiments were performed as described in the legend for Fig. 5 but using the LCMV (36.1) GP-specific MAbs. pLCMV and pLASV were incubated with the indicated concentrations of the LCMV-GP1 MAb 36.1 prior to infection of LCMV-GP-expressing Vero cells. The percentage of neutralization of pLCMV or pLASV in the GFP-based microneutralization assay is indicated.
Use of GP-deficient, GFP-expressing rLCMV to identify antiviral drugs against arenavirus.
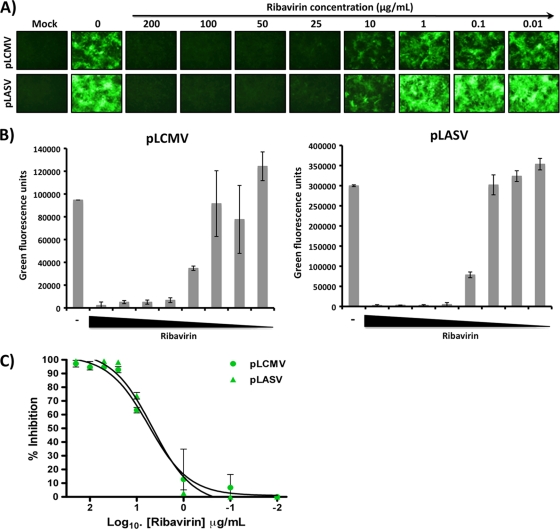
Our GP-pseudotyped rLCMVΔGP/GFP viruses are single-cycle infectious viruses, able to execute all the steps of the virus life cycle and undergo multiple rounds of infection only in GP-expressing cells. We therefore reasoned that they would be suitable for assessing the activity of antiviral drugs affecting different stages of the virus life cycle. To provide a proof of concept for this idea, we examined the effect of the nucleoside analog ribavirin on replication of GP-pseudotyped rLCMVΔGP/GFP in GP-expressing cells (Fig. 8). The exact mechanisms whereby ribavirin inhibits arenavirus replication are not fully understood, but previous studies have demonstrated that concentrations as low as 25 μg/ml reduced the virus yield and specific antigen formation to undetectable levels (24). Consistent with previous reports (26), we observed approximately 50% inhibition of viral replication (48 h p.i.) when ribavirin was present at ∼10 μg/ml and undetectable GFP levels with 25 μg/ml (Fig. 8A and B). Ribavirin inhibited to similar efficiencies the replication of pLCMV and pLASV (Fig. 8C).
FIG. 8.
GP-pseudotyped rLCMVΔGP/GFP as a tool to detect antiviral drugs against arenaviruses. LCMV GP-expressing Vero cells (96-well format, triplicate) were infected (MOI of ∼1 to 2) with the GP-pseudotyped LCMV or LASV rLCMVΔGP/GFP. One hour before infection and for 48 h postinfection, cells were treated with the indicated concentrations of ribavirin. (A) Representative fluorescence images. (B) Quantification of GFP expression using a GFP microplate reader. (C) Percentage of ribavirin-induced virus inhibition.
DISCUSSION
In this work we have documented the development of single-cycle infectious GFP-expressing rLCMVΔGP that could be efficiently pseudotyped with LCMV or LASV GP and that grew to high titers in cells constitutively expressing either LCMV or LASV GP. This newly developed system should facilitate the safe, rapid, and sensitive detection of virus-specific neutralizing antibodies and antiviral drugs against pathogenic arenaviruses without the requirement of high-biosafety-level facilities.
Consistent production of large quantities of high titers of GP-pseudotyped rLCMVΔGP/GFP via transient transfection with GP of rLCMVΔGP/GFP-infected cells posed technical difficulties likely due to variability in transfection efficiency and GP expression. We therefore examined whether the use of cell lines constitutively expressing the GP of interest could overcome this problem. Constitutive high levels of GP expression might trigger the cellular endoplasmic reticulum (ER) stress response and subsequent cell toxicity, which would negatively impact the feasibility of this approach. However, we succeeded in obtaining both Vero and BHK-21 cell lines that expressed constitutively high levels of GP that consistently permitted production of high titers of GP-pseudotyped rLCMVΔGP/GFP.
Both WT LCMV and GP-pseudotyped rLCMVΔGP/GFP exhibited the same particle morphology and size, and immunoelectron microscopy (immuno-EM) studies with specific antibodies to LCMV or LASV clearly supported the specificity of the GP-pseudotyped virus. These findings indicated that our GP-pseudotyping approach did not lead to noticeable structural changes in the virions. In addition, GP-pseudotyped rLCMVΔGP/GFP exhibited only slight differences in growth kinetics compared to WT LCMV, which likely reflected limited GP expression levels in the GP-expressing cell lines compared to the GP levels produced in WT LCMV-infected cells. Notably, our rLCMVΔGP/GFP showed both genetic and phenotypic stability in GP-expressing cell lines. During several passages at low or high MOI, no mutations affecting GFP expression or virus viability were detected. As predicted, both reporter gene expression and viral production were restricted to GP-expressing cell lines.
These results indicated that GP-pseudotyped rLCMVΔGP/GFP could be developed into a system to facilitate the safe, rapid, and sensitive detection of virus-specific neutralizing antibodies and antiviral drugs against human pathogenic arenaviruses without the requirement of facilities of high biosafety levels because the production of infectious progeny of a GP-deficient, GFP-expressing recombinant pathogenic arenavirus would be restricted to GP-expressing cell lines. To explore this idea, we used GP-pseudotyped rLCMVΔGP/GFP to develop a GFP-based microneutralization assay for the characterization of neutralizing antibodies against LCMV and LASV. This GFP-based microneutralization assay and a conventional FRNT assay had similar sensitivities. However, the GFP-based microneutralization assay offered the advantage of rapid and quantitative determinations by using a fluorescent plate reader. In contrast, the FRNT assay required 3 to 4 days of incubation for plaque development and subsequent staining of foci with an appropriate antibody. In addition, the conventional FRNT assay is based on the use of live virus capable of propagation via multiple rounds of infection, which would require the use of BSL-4 in the case of LASV or other highly pathogenic human arenavirus. The GFP-based microneutralization assay we have developed would also be advantageous in cases where only limited amounts of the biological sample being tested are available. This approach to screen for neutralizing antibodies against pathogenic arenaviruses could be expanded to libraries of antibodies and peptides, as recently described for influenza virus (20, 22, 23, 30). Likewise, using ribavirin as a proof of concept, we showed that GP-pseudotyped rLCMVΔGP/GFP could be used very efficiently to assess the activities of candidate antiviral drugs. An advantage of our approach would be that it is amenable to being optimized for high-throughput screening (HTS) to identify antiviral drugs targeting different steps of the arenavirus life cycle without the requirement of using an infectious virus able to undergo multiple rounds of replication that would raise human risk exposure. Recently, a system based on the use of a HA-deficient reporter-expressing influenza virus was used for the screening of small interfering RNAs affecting influenza viral replication (13). Our results support the feasibility of using a similar approach to uncover cellular proteins required for the completion of the arenavirus life cycle.
Old World arenaviruses (LCMV and LASV) use α-dystroglycan (α-DG) as a receptor for cell entry (14), whereas pathogenic New World arenaviruses have been reported to use the human transferrin receptor 1 (TfR1) as receptor for cell entry (21). Our results have shown the possibility of pseudotyping rLCMVΔGP/GFP with the GP of LASV. The possibility, still to be determined, that rLCMVΔGP/GFP could be efficiently pseudotyped with GPs from pathogenic New World arenaviruses would expand the utility of our system. Likewise, such a finding would support the feasibility of rescuing replication-competent rLCMV expressing GPs from New World arenaviruses that could be studied under BSL-2 conditions, as previously described for rLCMV expressing LASV GP (25). This would facilitate the identification of lead compounds and antibodies with specific or general antiarenavirus activities.
Compared to traditional pseudotyped retrovirus- or lentivirus-based systems, our approach offers the advantage that GP-pseudotyped rLCMVΔGP/GFP would recreate all the steps of the arenavirus life cycle in GP-expressing cells, making this system suitable for the screening of inhibitors targeting steps other than virus cell entry. Our single-cycle-replicating rLCMV may also represent a valuable platform for the development of safer arenavirus vaccine candidates (9). Development of Old World and New World GP-pseudotyped rLCMVs limited to single-cycle replication would overcome concerns about potential out-of-control replication of vaccine strains that might lead to disease. In addition, appropriate immunomodulatory genes aimed at augmenting protective host immune responses could be engineered in place of the viral GP. A GP-pseudotyped virus would be expected to induce a complete repertoire of both humoral and cellular host immune responses against different viral components that may improve host protection while limiting risk to the host since virus propagation would be very limited.
Acknowledgments
We thank members of the L.M.-S. laboratory for their discussions and Snezhana Dimitrova for technical support. We acknowledge Randal J. Schoepp (U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD) for kindly providing the LASV GP1-specific L52-74-7A MAb. We also thank Karen Bentley (Electron Microscopy Core Facility) and Danielle C. Alcena (Department of Microbiology and Immunology) at The University of Rochester, Rochester, NY, for their expert technical assistance.
W.W.S.I.R. is a Rochester Vaccine Fellowship recipient (2009). Research in the laboratory of L.M.-S. was partially funded by the NIAID grant RO1 AI077719. Research at the laboratory of J.C.D.L.T. was supported by grants RO1 AI047140, RO1 AIO77719, and RO1 AI079665 from NIH/NIAID.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Barton, L. L. 1996. Lymphocytic choriomeningitis virus: a neglected central nervous system pathogen. Clin. Infect. Dis. 22:197. [DOI] [PubMed] [Google Scholar]
- 2.Battegay, M., et al. 1991. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods 33:191-198. [DOI] [PubMed] [Google Scholar]
- 3.Blechschmidt, M., and R. Thomssen. 1976. Electron-microscopic identification of infectious particles of lymphocytic choriomeningitis. Med. Microbiol. Immunol. 162:193-199. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, M. J., J.-C. de la Torre, and C. J. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1791-1827. In P. David Knipe, Peter Howley, Diane Griffin, Robert Lamb, Malcolm Martin, Bernard Roizman, and Stephen Straus (ed.), Fields virology, 5th ed., vol. II. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Charrel, R. N., and X. de Lamballerie. 2003. Arenaviruses other than Lassa virus. Antiviral Res. 57:89-100. [DOI] [PubMed] [Google Scholar]
- 6.Cosset, F. L., et al. 2009. Characterization of Lassa virus cell entry and neutralization with Lassa virus pseudoparticles. J. Virol. 83:3228-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emonet, S. F., L. Garidou, D. B. McGavern, and J. C. de la Torre. 2009. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc. Natl. Acad. Sci. U. S. A. 106:3473-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, S. A., et al. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 354:2235-2249. [DOI] [PubMed] [Google Scholar]
- 9.Flatz, L., et al. 2010. Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat. Med. 16:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunther, S., and O. Lenz. 2004. Lassa virus. Crit. Rev. Clin. Lab. Sci. 41:339-390. [DOI] [PubMed] [Google Scholar]
- 11.Halfmann, P., et al. 2008. Generation of biologically contained Ebola viruses. Proc. Natl. Acad. Sci. U. S. A. 105:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illick, M. M., et al. 2008. Uncoupling GP1 and GP2 expression in the Lassa virus glycoprotein complex: implications for GP1 ectodomain shedding. Virol. J. 5:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konig, R., et al. 2010. Human host factors required for influenza virus replication. Nature 463:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz, S., P. Borrow, and M. B. Oldstone. 2002. Receptor structure, binding, and cell entry of arenaviruses. Curr. Top. Microbiol. Immunol. 262:111-137. [DOI] [PubMed] [Google Scholar]
- 15.Lee, A. M., et al. 2008. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J. Biol. Chem. 283:18734-18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Sobrido, L., et al. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller, G., M. Bruns, L. Martinez Peralta, and F. Lehmann-Grube. 1983. Lymphocytic choriomeningitis virus. IV. Electron microscopic investigation of the virion. Arch. Virol. 75:229-242. [DOI] [PubMed] [Google Scholar]
- 19.Palacios, G., et al. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358:991-998. [DOI] [PubMed] [Google Scholar]
- 20.Pitaksajjakul, P., P. Lekcharoensuk, N. Upragarin, C. F. Barbas III, M. S. Ibrahim, K. Ikuta, and P. Ramasoota. 2010. Fab MAbs specific to HA of influenza virus with H5N1 neutralizing activity selected from immunized chicken phage library. Biochem. Biophys. Res. Commun. 395:496-501. [DOI] [PubMed] [Google Scholar]
- 21.Radoshitzky, S. R., et al. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajik, M., et al. 2009. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol. J. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajik, M., A. R. Omar, A. Ideris, S. S. Hassan, and K. Yusoff. 2009. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int. J. Biol. Sci. 5:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez, M., J. B. McCormick, and M. C. Weissenbacher. 1986. Antiviral effect of ribavirin on Junin virus replication in vitro. Rev. Argent Microbiol. 18:69-74. [PubMed] [Google Scholar]
- 25.Rojek, J. M., A. B. Sanchez, N. T. Nguyen, J. C. de la Torre, and S. Kunz. 2008. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J. Virol. 82:7677-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Jarabo, C. M., C. Ly, E. Domingo, and J. C. de la Torre. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37-47. [DOI] [PubMed] [Google Scholar]
- 27.Ruo, S. L., et al. 1991. Antigenic relatedness between arenaviruses defined at the epitope level by monoclonal antibodies. J. Gen. Virol. 72(Pt. 3):549-555. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, A. B., and J. C. de la Torre. 2006. Rescue of the prototypic arenavirus LCMV entirely from plasmid. Virology 350:370-380. [DOI] [PubMed] [Google Scholar]
- 29.Snell, N. 1988. Ribavirin therapy for lassa fever. Practitioner 232:432. [PubMed] [Google Scholar]
- 30.Sun, L., et al. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber, E. L., and M. J. Buchmeier. 1988. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology 164:30-38. [DOI] [PubMed] [Google Scholar]
- 32.Whitby, L. R., A. M. Lee, S. Kunz, M. B. Oldstone, and D. L. Boger. 2009. Characterization of lassa virus cell entry inhibitors: determination of the active enantiomer by asymmetric synthesis. Bioorg. Med. Chem. Lett. 19:3771-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright, K. E., M. S. Salvato, and M. J. Buchmeier. 1989. Neutralizing epitopes of lymphocytic choriomeningitis virus are conformational and require both glycosylation and disulfide bonds for expression. Virology 171:417-426. [DOI] [PubMed] [Google Scholar]