Abstract
Antibody-dependent enhancement (ADE) is implicated in severe, usually secondary, dengue virus (DV) infections. Preexisting heterotypic antibodies, via their Fc-gamma receptor (FcγR) interactions, may increase disease severity through enhanced target cell infection. Greater numbers of infected target cells may contribute to higher viremia and excess cytokine levels often observed in severe disease. Monocytes, macrophages, and immature and mature dendritic cells (DC) are considered major cellular targets of DV. Apheresis of multiple donors allowed isolation of autologous primary myeloid target cell types for head-to-head comparison of infection rates, viral output, and cytokine production under direct infection (without antibody) or ADE conditions (with antibody). All studied cell types except immature DC supported ADE. All cells undergoing ADE secreted proinflammatory cytokines (interleukin-6 [IL-6] and tumor necrosis factor alpha [TNF-α]) at enhancement titers, but distinct cell-type-specific patterns were observed for other relevant proteins (alpha/beta interferon [IFN-α/β] and IL-10). Macrophages produced type I interferons (IFN-α/β) that were modulated by ADE. Mature DC mainly secreted IFN-β. Interestingly, only monocytes secreted IL-10, and only upon antibody-enhanced infection. While ADE infection rates were remarkably consistent in monocytes (10 to 15%) across donors, IL-10 protein levels varied according to previously described regulatory single nucleotide polymorphisms (SNPs) in the IL-10 promoter region. The homozygous GCC haplotype was associated with high-level IL-10 secretion, while the ACC and ATA haplotypes produced intermediate and low levels of IL-10, respectively. Our data suggest that ADE effects are cell type specific, are influenced by host genetics, and, depending on relative infection rates, may further contribute to the complexity of DV pathogenesis.
Dengue is the most common arboviral infection worldwide and is a major public health threat in tropical and subtropical regions (37). Clinical dengue virus (DV) infection ranges from asymptomatic or mild illness to life-threatening diseases, including dengue hemorrhagic fever and dengue shock syndrome (DHF/DSS) (19). One proposed pathogenic mechanism contributing to disease severity is antibody (Ab)-dependent enhancement (ADE) (6, 15, 17). ADE was initially defined in the laboratory as subneutralizing concentrations of antibody that enhance virus infection of target cells. Dengue antibodies likely bring the virus-antibody complex into close proximity with the cell surface Fc receptors (FcRs) that, in turn, facilitate viral entry. Various myeloid cell types, including monocytes (22), macrophages (MACs) (34), dendritic cells (DC) (30, 55, 58), mast cells (2), and hepatocytes (20, 52), support direct infection of DV. ADE effects were extensively explored in vitro in monocytes and macrophages with baseline infection ranges of ∼1% and antibody-enhanced infections of 3 to 10% (16, 22, 24, 30). We previously reported that both stages of dendritic cells, immature and mature DC, support the highest levels of direct DV infection (20 to 50% infection without antibody) (1, 30, 39). Furthermore, in the presence of subneutralizing concentrations of dengue antibodies, enhancement was observed only in mature dendritic cells, an effect largely mediated by Fc-gamma receptor IIa (FcγRIIa) (1).
In this study, we systematically and contemporaneously explore ADE in the following autologous myeloid cells: monocytes, macrophages, immature DC (iDC), and mature DC (mDC). We report both qualitative and quantitative differences in ADE effects in each cell type, including infection rates, viral output, and cellular immune responses. Since immunomodulatory cytokines likely influence disease severity (4), we investigated the cytokine patterns produced from these cells as they undergo ADE. High levels of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) were released from all cell types under ADE conditions, but distinct patterns of type I interferons (IFNs) and IL-10 were observed for each cell type. Of all cells studied here, we observed IL-10 production only in monocytes undergoing ADE. IL-10 levels were maximal at peak enhancement titers (PENT). We noted similar patterns of IL-10 secretion between donors but observed large variations in the amounts of released protein. We observed an ADE-associated IL-10 secretion pattern but noted some variability in the magnitudes of protein levels detected between donors. Using restriction fragment length polymorphism (RFLP) and sequencing techniques, we identified an association between known IL-10 promoter polymorphisms and the levels of IL-10 production in these in vitro ADE studies. Our data suggest that antibody-dependent DV infection and replication trigger distinct responses in different human primary target cells that are genetically regulated and potentially linked to clinical disease outcome.
MATERIALS AND METHODS
Virus.
The Burma DV-2 isolate S16803 was used for all experiments. The preparation and titers of virus stock were described previously (55). Briefly, the dengue virus 2 strain S16803 was grown in an African green monkey Vero cell line (American Type Culture Collection), and cell-free supernatants with titers of 106 to 107 PFU/ml were used as virus stocks.
Primary human myeloid cells.
An Institutional Review Board-approved clinical protocol was used for apheresis of normal healthy donors after provision of informed consent, thereby providing large numbers of peripheral blood mononuclear cells (PBMC) from multiple (n > 20) donors. Apheresis products were diluted with phosphate-buffered saline (PBS) and layered over Ficoll-Hypaque to isolate the PBMC. The mononuclear cells were washed extensively with large volumes of PBS in order to minimize platelet contamination. PBMC isolated from leukapheresis of healthy donors were cryopreserved, allowing repeat experiments.
Monocyte isolation.
Primary human monocytes were prepared using a Dynal monocyte negative isolation kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 107 PBMC were incubated with blocking reagent and antibody mixture for 10 min at 2 to 8°C. Depletion Dynal beads were incubated with the PBMC for 15 min at 2 to 8°C. The labeled cells were removed with a magnet (Dynal MPC), leaving purified monocytes (90 to 95% CD14+ as determined by flow cytometry).
Monocyte-derived macrophages.
PBMC were cultured as described previously (31), with some modifications. PBMC (2 × 106) were adhered to 24-well tissue culture plates for 60 min. After several washes, adherent cells were cultured in 2 ml of complete medium (CM; RPMI medium supplemented with 10% heat-inactivated fetal calf serum (Gemini Bio-Products, Sancramento, CA), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Quality Biological, Gaithersburg, MD) containing 50 ng/ml recombinant human macrophage colony stimulation factor (rhM-CSF; Peprotech, Inc., Rocky Hill, NJ) to allow differentiation into macrophages. On day 5, rhM-CSF (1 ml at 50 ng/ml) was added to the culture. The macrophages were harvested for experiments on day 6. The phenotypes of all macrophages were confirmed by flow cytometry before use. CD163 (hemoglobin-haptoglobin complex scavenger receptor) was used as a specific marker for macrophages (58).
Monocyte-derived dendritic cells.
Dendritic cells were cultured from monocytes as described previously (1). PBMC were adhered to tissue culture plates for 60 min, and after several RPMI medium washes, adherent cells were placed into 10 ml of complete medium (CM) with 2 × 104 U/ml recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; Peprotech, Rocky Hill, NJ) and 2 × 104 U/ml recombinant human interleukin-4 (rhIL-4) (R&D Systems, Minneapolis, MN) for 7 days at 37°C with 5% CO2. On day 6, 50 μl of MCM mimic (15 μg/ml IL-6 [Peprotech], 500 ng/ml IL-1β, 500 μg/ml tumor necrosis factor alpha [TNF-α] [Sigma, St. Louis, MO], 100 μg/ml protaglandin E2 [Cayman Chemical, Ann Arbor, MI]) was added to mature the cells. The phenotypes of all DC were confirmed by flow cytometry before use. Note that DC lack CD3, CD19 or CD20, and CD14 but express high levels of CD11c, HLA-DR, and DC-SIGN. Mature cells additionally express CD25, CD83, and CD86 but much lower levels of DC-SIGN, as previously reported (55).
Flow cytometry.
A FACSCalibur instrument (BD Bioscience, San Jose, CA) was used to monitor cell surface staining with a panel of phycoerythrin-conjugated monoclonal antibody (MAb) to HLA-DR, CD80, CD86, CD3, CD14, CD20, CD25, CD1a (BD Biosciences, San Jose, CA), CD83 (Beckman Coulter, Fullerton, CA), fluorescein isothiocyanate (FITC)-conjugated CD163 (Acris Antibodies GmbH, Herford, Germany), and FITC-conjugated Factor XIIIa (Enzyme Research Laboratories, Inc., South Bend, IN). FcγR surface staining was performed using phycoerythrin (PE)-conjugated CD16 (BD Biosciences, San Jose, CA), Alexa 467-conjugated CD32a (clone IV.3; ATCC, Manassas, VA), FITC-conjugated CD32b (clone 2B6, kindly provided by Macrogenics, Rockville, MD), and FITC-conjugated CD64 (BD Biosciences, San Jose, CA). For detection of intracellular de novo DV protein production, cells were permeabilized with Cytofix/Cytoperm (BD Biosciences, San Jose, CA) and stained with 2H2 (anti-DV prM MAb, kindly provided by Robert Putnak, WRAIR, Silver Spring, MD) at 48 h postinfection. For intracellular staining of adherent cells (macrophages), the cells were treated with 500 μl Accutase (Innovative Cell Technology, Inc., San Diego, CA) for 15 min at 37°C. These cells were washed twice with PBS before being stained for cell surface markers or intracellular staining. The cell phenotypes were confirmed by flow cytometry before use in the ADE assay.
ADE assay.
The ADE assay was performed as described previously (1), using polyvalent dengue virus-immune serum obtained from the Pediatric Dengue Vaccine Initiative (PDVI) in all experiments. The dengue virus-immune serum was tested in a plaque neutralization assay and found to neutralize all four DV serotypes (DV-2 50% plaque reduction neutralization test [PRNT50] = 10,199; PDVI, personal communication). Briefly, DV-immune serum was serially diluted from 1:10 to 1:163,840 in a volume of 50 μl. Virus, at a multiplicity of infection (MOI) of 1, unless otherwise noted, was placed into the antibody dilution tubes for 60 min at 37°C with 5% CO2 to allow immune complex (IC) formation. The contents of each tube were then added to 0.5 × 106 cells and incubated for 2 h. The cells were washed with CM to remove excess DV-immune complexes. The cells were resuspended in CM and incubated for an additional 48 h. For adherent cells (macrophages), the ICs were overlaid on a macrophage monolayer and incubated for 2 h. The ICs were pipetted off and discarded from the monolayer. The attached monolayer was then carefully rinsed with CM to remove residual ICs. Finally, 500 μl of CM was added to the monolayer and incubated for an additional 48 h (unless otherwise noted).
Viral RNA quantitation.
Quantitative real-time PCR (qRT-PCR) was performed using primers, probes, RNA standards, and conditions described previously (1). Briefly, viral RNA was extracted from culture supernatant using a QIAamp viral RNA kit (Qiagen, Velencia, CA). Amplification was performed using an ABI prism 7500 detection instrument (Applied Biosystems, Foster City, CA). The reverse transcription-PCR thermal cycles were performed as follows: 50°C for 30 min and 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. RNA copy numbers were calculated from a standard curve generated by an in vitro-transcribed RNA standard.
Plaque assays.
The Vero cell plaque assay was used to determine infectivity and viral titer as previously described (1, 10). Six 10-fold serial dilutions (10−1 to 10−6) from each supernatant sample were incubated in quadruplicate wells of a 6-well plate containing confluent Vero cells. Virus was absorbed for 1 h, and the Vero monolayer was overlaid with complete minimal essential medium (Cellgro, Manassas, VA) containing low-melting-temperature agarose (Invitrogen, Carlsbad, CA) to restrict dissemination of progeny virions. The cells were incubated for 5 days and overlaid with the vital stain neutral red (Sigma, St. Louis, MO). Plaques were counted by visual inspection at 24 h to calculate the number of PFU of DV/ml of supernatant.
Quantitation of type I interferons (IFN-α and IFN-β).
Cellular production of IFN-α and IFN-β were measured using commercial enzyme-linked immunosorbent assay (ELISA) kits (PBL InterferonSource, Piscataway, NJ) according to the manufacturer's specifications. Briefly, duplicate samples were diluted 1:2 in dilution buffer, and duplicate 6-and 7-point standards for rhIFN-α (156 to 5,000 pg/ml) and rhIFN-β (25 to 2,000 pg/ml), respectively, were added to the assay plate and incubated at room temperature for 1 h. The plates were washed, and then IFN-α- or IFN-β-specific antibody was added for 1 h. After 3 washes, the horseradish peroxidase (HRP)-conjugated secondary antibody was added and incubated for 1 h. The plates were washed, and then 100 μl of tetramethylbenzidine (TMB) substrate was added for 15 min. Next, 100 μl of stop solution was added, and the plates were analyzed using a microplate reader (Molecular Devices, Sunnyvale, CA) at 450 nm within 5 min of stopping the reaction.
Quantitation of cytokines using a multiplex assay.
Cytokines produced by the myeloid cells during ADE assays were quantified from supernatants using the Q-Plex human cytokine infrared (IR) array (Quansys Biosciences, Logan, UT) according to the manufacturer's protocol. Samples were run in triplicate and diluted 1:2 in sample dilution buffer, and duplicate 8-point standards curves were added to the plate and incubated at room temperature for 1 h. Following 3 washes, the plate was incubated with detection reagent for 1 h. After additional washes, IRDye800CW streptavidin was added to the plate for 15 min. The plate was washed again, followed by a rinse with deionized water (diH2O). Images were taken with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE) and analyzed using Quansys Q-view Plus software (Quansys Biosciences).
Determination of IL-10 promoter polymorphisms.
Three biallelic IL-10 promoter polymorphisms, at positions −1082 (G/A), −819 (C/T), and −592 (C/A), were detected by a PCR-restriction fragment length polymorphism (PCR-RFLP) method described previously (56). Amplifications of the fragment spanning position −1082 were performed by PCR using forward primer 5′-CTC GCT GCA ACC CAA CTG GC-3′ and reverse primer 5′-TCT TAC CTA TCC CTA CTT CC-3′. The resulting 139-bp PCR products were digested with MnlI (New England Biolabs, Ipswich, MA), yielding 33-bp and 106-bp fragments only when G was present. A 209-bp fragment spanning position −819 was amplified using forward primer 5′-TCA TTC TAT GTG CTG GAG ATG-3′ and reverse primer 5′-TGG GGG AAG TGG GTA AGA GT-3′. Digestions using MaeIII (Roche diagnostic GmbH, Penzberg, Germany) yielded 125 bp and 84 bp only when C was present. Amplifications of the 412-bp fragment spanning position −592 were performed using forward primer 5′-CCT AGG TCA CAG TGA CGT GG-3′ and reverse primer 5′-GGT GAG CAC TAC CTG ACT AGC-3′. The presence of an RsaI (New England Biolabs, Ipswich, MA) yielded 176 bp and 236 bp when A was present. The PCRs were performed with 50-μl reaction volumes that contained 100 ng genomic DNA. The reaction mixtures were subjected to a first cycle at 94°C for 5 min, followed by 40 cycles of 30 s at 94°C and 45 s at 58°C for position −1082 (A/G), 59°C for position −819 (T/C), and 62°C for position −592 (A/C) and a final elongation at 72°C for 1 min.
Stimulation of monocytes with LPS for measurement of IL-10 levels.
Monocytes (0.5 × 106) were stimulated with Ultrapure lipopolysaccharide (LPS) from Escherichia coli K-12 (InvivoGen, San Diego, CA) at concentrations of 1, 10, and 100 ng/ml. Supernatants were harvested 24 h after stimulation and subjected to cytokine analysis.
Relative quantitation of IL-10 mRNA.
In a subset of samples, IL-10 mRNA was measured as described previously (27). qRT-PCRs were carried out with a 25-μl final volume, using the following reaction mixture: 400 nM forward and reverse IL-10 primers (IL-10_forward, 5′-TCA AGG CGC ATG GAA CTC-3′; IL-10_reverse, 5′-CGG CCT TGC TCT TGT TTT C-3′) and 75 nM 6-carboxyfluorescein [FAM]-labeled IL-10 probe (FAM-CGG CGC TGT CAT CGA TTT CTT CC-Black Hole Quencher [BHQ]) (Sigma, St. Louis, MO). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal gene control in the reaction. The thermal cycle conditions were 49°C for 45 min and 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 2 min.
UV-irradiated DV preparation.
The DV stock was placed into a petri dish and exposed to short-wavelength UV light (540 nm) for 20 min (1). The distance between the petri dish and the UV source was 5 in. Lack of infectivity (<1% of cells) of UV-irradiated DV was confirmed for a highly susceptible Raji-DC-SIGN cell line (1).
Blocking of monocyte FcγRs.
Purified monocytes were pretreated with 10 μg/ml anti-human FcγRIIa blocking antibody (clone IV.3; ATCC, Manassas, VA), anti-FcγRI, anti-FcγRIII (R&D Systems, Minneapolis, MN), anti-FcγRIIb (clone 2B6; kindly provided by Macrogenics, Rockville, MD), or IgG1 control for 1 h at 37°C. Treated cells were washed with complete medium before use in the ADE assays.
Blocking of IFN-α/β in macrophages.
Macrophages cultured on 24-well plates were pretreated with complete medium containing 104 units/ml of sheep (polyclonal) anti-human IFN-α and sheep (polyclonal) anti-IFN-β (Invitrogen, Carlsbad, CA) for 1 h at 37°C. The treated cells were exposed to the dengue virus-immune complex for 1 h at 37°C and then washed and resuspended with CM containing 104 units/ml of blocking antibodies for an additional 48 h at 37°C.
Statistical analysis.
All data were analyzed using Prism software (Prism 5; GraphPad, San Diego, CA) using Student paired t tests. P values of <0.05 were considered significant.
RESULTS
Isolation and differentiation of primary human myeloid lineage cells.
We studied primary human myeloid cells (monocytes, macrophages, and dendritic cells from autologous donors), which were later used as targets for ADE studies. Macrophages and dendritic cells were differentiated from freshly isolated monocytes (95% purity) using macrophage colony-stimulating factor (M-CSF) and IL-4-granulocyte-macrophage colony-stimulating factor (GM-CSF), respectively. Expected morphological differences were noted between monocytes, macrophages, and DC upon culture (data not shown). Additionally, we used a panel of cell surface markers (CD11c expressed on myeloid cells, HLA-DR, CD80, CD86, CD3, CD14, CD20, CD25, CD1a, CD83, CD163, and factor XIIIa) to distinguish cell types. Figure 1 A shows the characteristic phenotypes observed in monocytes, macrophages, iDC, and mDC. High levels of CD14 were observed on monocytes and macrophages but not on iDC or mDC. We used CD163 (hemoglobin-haptoglobin complex scavenger receptor) as a marker to differentiate macrophages from the other myeloid cells, as it is reported to be the only marker uniformly coexpressed by Factor XIIIa+ cells but not by CD11c+ cells (59). Dendritic cells and macrophages express dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)/CD209 to various degrees (iDC > mDC > MACs). Unique Fc-gamma receptor (FcγR) expression patterns were observed in these primary myeloid cells (Table 1). Monocytes express high levels of FcγRIIa (CD32a) and modest levels of FcγRI (CD64) and FcγRIIb (CD32b). Dendritic cells mainly express FcγRIIa and FcγRIIb, which are regulated upon maturation. Interestingly, only macrophages expressed all 4 FcγRs at modest-to-high levels: FcγRI, FcγRIIa and IIb, and FcγRIII.
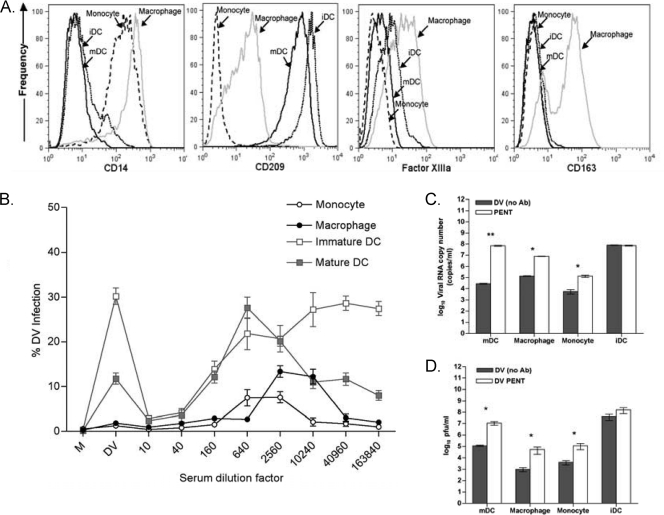
FIG. 1.
Primary human myeloid cells undergoing ADE show higher cellular infection rates, viral output, and infectivity. (A) Phenotype of primary human myeloid cells distinguishing monocytes, macrophages, and dendritic cells (immature and mature). (B) ADE assays: medium only (M), dengue virus/no sera (DV), or serial 4-fold dilutions of DV immune serum was premixed with DV (MOI, 1) and added to autologous iDC, mDC, monocytes, and macrophages. Cells were analyzed by flow cytometry 48 h later for de novo synthesis of DV-prM antigen as defined by intracellular 2H2 expression. The data are expressed as percents infection along the y axis compared to the level for the no-immune-serum control. Data represent the means of results from 5 donors. (C) Viral output: Culture supernatants from panel B were collected for qRT-PCR. The data are expressed on a log scale as viral RNA copy numbers, comparing numbers of copies produced under direct infection (no Ab; “DV”) and numbers of copies produced at the peak enhancement titer (PENT). All data points shown are means ± SD of results from one representative donor among five donors from experiments performed in triplicate. The results from each group were compared using Student's t test. **, P < 0.01; *, P < 0.05. (D) Plaque assays. Culture supernatants from panel B were added to a Vero cell monolayer, and plaque formation was counted 6 days after infection. The data from a representative donor among five donors are shown and expressed as log10 numbers of infectious PFU per ml. All data points shown are means ± SEM for experiments performed in quadruplicate. The results from each group were compared using Student's t test. *, P < 0.05.
TABLE 1.
FcγR expression on primary human monocytes, macrophages, and dendritic cellsa
| Target cell | Expression level of: |
|||
|---|---|---|---|---|
| FcγRI | FcγRIIa | FcγRIIb | FcγRIII | |
| Monocyte | ++ | +++ | ++ | +/− |
| Macrophage | ++ | +++ | ++ | +++ |
| Immature DC | +/− | +++ | ++ | +/− |
| Mature DC | +/− | +++ | + | +/− |
Quantitative expression of FcγR with specific monoclonal antibodies is denoted as follows: +/−, <20% expression; +, 20 to 30% expression; ++, 31 to 60% expression; +++, 61 to 95% cells showing expression.
Primary human myeloid cells support ADE in DV infections.
Well-characterized autologous primary monocytes, macrophages, iDC, and mDC were subjected to the ADE assay using the same dengue virus-immune serum and virus stock. iDC characteristically show high direct-infection rates (30 to 50% of cells) but do not support ADE, while mDC demonstrate both modest direct infection (10% of cells) and a 2.5-fold increase in infection (25% of cells) at peak enhancement titers (Fig. 1B), consistent with our previous reports (1, 55). As expected, we found low levels of direct infection of both monocytes and macrophages (≤0.8 to 2% of cells). However, both cell types undergo ADE, with ∼5- to 10-fold enhancement of cellular infection (5 to 15% of cells) at peak enhancement titers (Fig. 1B, lower curves). Using quantitative real-time RT-PCR (qRT-PCR) to analyze supernatants, we next determined viral production rates from each cell type. As shown in Fig. 1C, we observed a ∼3-log increase in viral copy number from direct infection to peak ADE in mDC, a ∼2 log increase in macrophages, and a 1.5-log increase in monocytes (all P values were <0.05). Note that there was no difference in viral copy number output from iDC (Fig. 1C) with or without antibody, consistent with their inability to support ADE. These data further indicated that mDC released the greatest number of virions on a per-cell basis when a comparison was made between direct infections (10% infected mDC released 104.5 copies/ml) and ADE infections (25% infected mDC released 107.5 copies/ml) relative to other target cells (Fig. 1C). To determine if the released virions in the supernatants retained infectivity, we performed traditional plaque assays. Confluent monolayers of Vero cells were exposed to supernatants collected from each condition (with or without dengue virus-immune sera) at the peak enhancement titer in the ADE assays (Fig. 1D). Macrophages, monocytes, and mDC showed approximately 1.5- to 2-log increases in viral infectivity under ADE conditions, whereas no differences in supernatant infectivity were observed in iDC (Fig. 1D). There was concordance between the results for qRT-PCR and the biological Vero-based plaque assay, as shown in Fig. 1C and D.
ADE in myeloid cells results in IL-6 and TNF-α production.
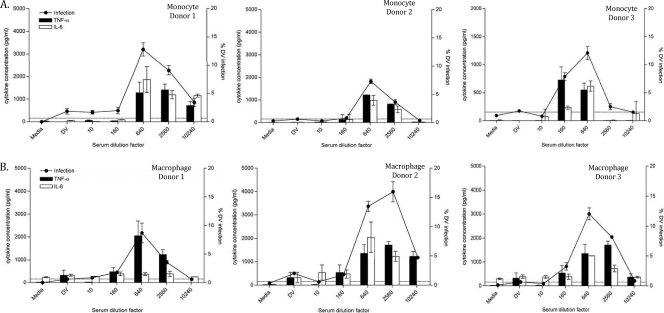
We previously reported high levels IL-6 and TNF-α production in mDC under ADE conditions (1). In this study, similar trends for IL-6 and TNF-α proteins were generally observed, as shown in Fig. 2 A and B, representing monocytes and macrophages, respectively. The levels of these proinflammatory cytokines often coincided with infection rates observed in the ADE curves from three representative donors (Fig. 2).
FIG. 2.
Enhanced proinflammatory cytokine production in primary human myeloid cells undergoing ADE. Supernatants from ADE assays with 5 donors using 2 different cell types: monocytes (A) and macrophages (B) were harvested at 48 h postinfection and assayed for TNF-α and IL-6 protein. On the x axis, the condition marked DV is virus alone (no sera) and the conditions to the right indicate the dilution factor of the DV sera. The lower detection limit for the TNF-α and IL-6 assay was 218.75 pg/ml. Data are shown as the means ± SD from 3 experiments representative of 5 different donors.
Type I interferons are modulated by ADE of dengue virus.
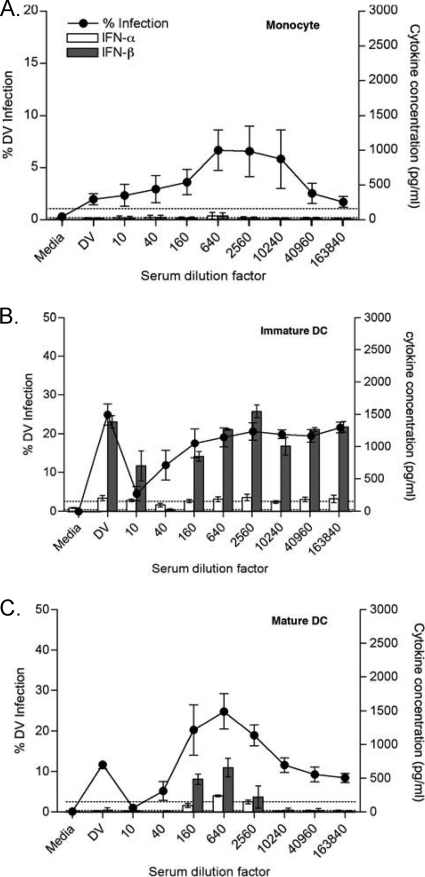
We measured type I interferons in supernatants from all cells in the ADE assay. No IFN-α/βs were observed in assays using primary monocytes as targets (Fig. 3 A), either with direct infection or under ADE conditions. Although both types of DC produced IFN-β, the IFN-β levels were considerably higher in immature DC and generally trended with infection levels. Interestingly, IFN-β levels were often observed only under ADE conditions in mDC (Fig. 3B and C). Note that IFN-α was not appreciably detected in DC experiments under any dengue virus infection conditions (Fig. 3B and C), as previously reported (18).
FIG. 3.
Type I interferon (IFN-α/β) levels in primary DC and monocytes in ADE experiments. ADE assays were performed for each cell type, and supernatants were tested for type I interferons (IFN-α/β) using ELISA kits. Open bars indicate IFN-α levels, and filled bars indicate IFN-β levels from monocytes (A), iDC (B), and mDC (C). Data are shown as the means ± SD from 5 different donors. Samples were run in duplicates. The lower detection limits of IFN-α and IFN-β were 156 pg/ml and 25 pg/ml, respectively.
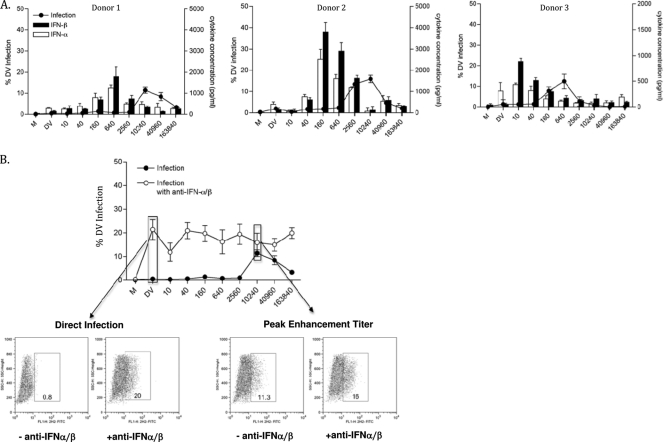
Interestingly, we noted an inverse relationship between the IFN-α/β levels and peak enhanced viral infection in macrophages with serum dilution (Fig. 4 A), suggesting modulation of type I interferons with ADE in these cells. Next, we conducted blocking studies using specific anti-IFN-α/β antibodies, as shown for a representative, donor 1, in Fig. 4B and in tabulated data from all 3 donors in Table 2. In Fig. 4B, note the significant increases in DV infection obtained with interferon-blocking Abs under non-ADE “direct-infection” conditions (e.g., 0.6 versus 21.5% for DV; P < 0.001). However, at peak enhancement titers, the infection levels are not statistically different in the presence of IFN-α/β blocking antibodies (1:10,240; 11.5 versus 16%; P = 0.44). Although, both types of DC produced IFN-β, the IFN-β levels were considerably higher in immature DC tracking directly with infection levels. Interestingly, detectable levels of IFN-β were generally observed only under ADE conditions in mDC (Fig. 3B and C). Note that IFN-α was not detected in DC experiments under any dengue virus infection conditions (Fig. 3B and C), as previously reported (18).
FIG. 4.
Type I interferon (IFN-α/β) effects are modulated by ADE in macrophages. ADE assays were performed on macrophages, and supernatants were tested for type I interferons (IFN-α/β) using ELISA kits. Open bars indicate IFN-α levels, and filled bars indicate IFN-β levels from 3 different donors' macrophages (A). (B) The results for a representative experiment, using donor 1, are shown for the blocking studies using specific anti-IFN-α/β antibodies in macrophages. Flow cytometry results obtained under selected conditions with or without anti-interferon Abs (arrows) are shown below the ADE curve.
TABLE 2.
DV infection levels obtained with and without IFN-α/β-blocking antibodiesa
| Condition or serum dilution | DV infection rate (%) |
|||||
|---|---|---|---|---|---|---|
| Donor 1 |
Donor 2 |
Donor 3 |
||||
| − anti-IFN-α/β (PENT = 1:640) | + anti-IFN-α/β | − anti-IFN-α/β (PENT = 1:10,240) | + anti-IFN-α/β | − anti-IFN-α/β (PENT = 1:10,240) | + anti-IFN-α/β | |
| Medium | 0.13 ± 0.09 | 0.38 ± 0.29 | 0.34 ± 0.11 | 0.23 ± 0.20 | 0.14 ± 0.10 | 0.24 ± 0.21 |
| DV*** | 0.56 ± 0.23 | 21.47 ± 3.52 | 2.0 ± 0.24 | 21.13 ± 3.33 | 1.27 ± 0.30 | 13.73 ± 0.82 |
| 1:10 | 0.38 ± 0.31 | 11.89 ± 3.22 | 0.55 ± 0.19 | 9.6 ± 2.41 | 1.28 ± 0.12 | 5.86 ± 0.65 |
| 1:40 | 0.59 ± 0.07 | 20.96 ± 2.77 | 1.83 ± 0.33 | 17.19 ± 0.86 | 1.4 ± 0.63 | 8.86 ± 1.16 |
| 1:160 | 1.48 ± 0.56 | 19.77 ± 2.74 | 1.84 ± 0.12 | 15.01 ± 2.75 | 7.9 ± 1.19 | 8.68 ± 2.82 |
| 1:640 | 0.83 ± 0.26 | 16.27 ± 4.10 | 2.46 ± 0.76 | 21.07 ± 3.44 | 12.48 ± 0.77† | 10.83 ± 1.87† |
| 1:2,560 | 1.04 ± 0.63 | 19.46 ± 3.45 | 13.47 ± 1.23 | 16.87 ± 1.33 | 2.69 ± 0.12 | 12.26 ± 3.3 |
| 1:10,240 | 11.47 ± 2.0† | 15.97 ± 3.22† | 15.97 ± 2.45† | 17.23 ± 3.0† | 0.76 ± 0.10 | 12.93 ± 1.73 |
| 1:40,960 | 8.47 ± 2.71 | 15 ± 2.16 | 4.72 ± 0.21 | 13.03 ± 2.13 | 0.13 ± 0.10 | 10.17 ± 3.0 |
| 1:163,840 | 3.35 ± 1.37 | 19.93 ± 1.88 | 2.17 ± 0.54 | 13.83 ± 2.78 | 0.31 ± 0.13 | 12.47 ± 0.52 |
Tabulated data from all conditions in the blocking experiments for the 3 representative donors in Fig. 4A. Bolded values highlight the blocking effects under direct DV infection and at peak enhancement. The results from each group were compared using Student's t test. ***, P < 0.001; †, not significant (P > 0.05).
Input virus titration for comparison of direct and ADE infection rates.
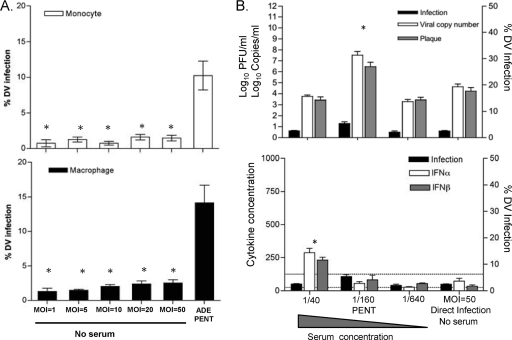
We tried to match direct- and ADE infection levels in monocytes and macrophages by varying input virus (MOI) to optimize comparisons of interferon production and viral output in supernatants. As shown in Fig. 5 A, we titrated DV from an MOI of 1 to an MOI of 50 to “push” direct-infection levels but could not match the ADE infection at PENT (MOI, 1) in either monocytes (top) or macrophages (bottom). Next, we used a two-pronged approach and titrated down the MOI for the ADE infections (0.1 to 0.001) and used the maximum MOI of 50 for direct infections (Fig. 5B and Table 3). Thus, we needed 500 to 50,000 times more input virus for direct infections to approach ADE infections (Fig. 5B, black bars). In Fig. 5B, we show infection data under four different conditions: over a range of serum dilutions (1/40, 1/160, and 1/640) and for the direct infection without sera. The direct-infection rates (MOI, 50; no sera) ranged from 2.5 to 3.8%, whereas in the presence of serum (MOI, 0.01 to 0.001), non-PENT infections ranged from 1.7 to 4.5%. For these two conditions, with reasonably matched infection rates, we found similar output virus levels in Vero plaque assays and qRT-PCR, as shown in the top panel of Fig. 5B. Type I interferon levels were consistently and significantly higher at lower dilutions of serum: 1/40 for donor 1, 1/160 for donor 2, and 1/40 for donor 3 (see lower panel of Fig. 5B and bolded values in Table 3 under the IFN-α/β columns). Notably, a relatively small (4-fold) dilution of serum resulted in a maximal doubling of infection rate at PENT, with a mean of 6.5% (range, 4 to 9%), with a disproportionately large (2- to 3-log) increase in virus output by PCR and Vero plaque assays.
FIG. 5.
Input virus titrations maximize direct infections for comparison to ADE. (A) Monocytes (upper) and macrophages (lower) were exposed to increasingly high MOIs (1 to 50) without serum, and infection rates were compared to those observed at PENT with an MOI of 1. (B) Direct infection using an MOI of 50 and downward titrations of MOI (1 to 0.001) for ADE in macrophages were compared. Four-fold dilutions of serum concentrations (1/40, 1/160, and 1/640) were compared to the level for direct infection at an MOI of 50 (no serum). Viral outputs were measured using qRT-PCR (copy numbers) and in plaque assays (titer) to confirm infectivity (upper). The lower panel shows the IFN-α (white bar) and IFN-β (black bar) detected under these conditions. The lower detection limits of IFN-α and IFN-β were 156 pg/ml and 25 pg/ml, respectively. The results from each group were compared using Student's t test. *, P < 0.05.
TABLE 3.
Input virus titration for comparison of viral output and interferon production in direct versus ADE infectionsa
| Donor | Serum dilution or condition | % infection rate | No. of viral RNA copies/ml | No. of PFU/ml for plaque assay | IFN-α concn (pg/ml) | IFN-β concn (pg/ml) |
|---|---|---|---|---|---|---|
| 1 | 1:40 (MOI = 0.01) | 2.4 ± 0.3 | 5.3 × 103 ± 2.4 × 103 | 2.5 × 103 ± 2.4 × 103 | 288.0 ± 32.5 | 232.1 ± 22.5 |
| 1:160 (PENT) (MOI = 0.01) | 5.4 ± 1.1 | 3.2 × 107 ± 4.2 × 107 | 5.5 × 106 ± 1.2 × 106 | 58.3 ± 11.9 | 84.8 ± 32.5 | |
| 1:640 (MOI = 0.01) | 2.2 ± 0.6 | 6.9 × 103 ± 1.9 × 102 | 2.7 × 103 ± 1.9 × 103 | 27.0 ± 5.2 | 55.5 ± 15.2 | |
| Direct infection (MOI = 50) | 2.5 ± 0.4 | 4.3 × 104 ± 1.6 × 104 | 1.7 × 104 ± 1.5 × 104 | 73.7 ± 22.3 | 35.57 ± 10.1 | |
| 2 | 1:160 (MOI = 0.001) | 1.7 ± 0.2 | 3.3 × 102 ± 1.1 × 102 | 5.5 × 103 ± 1.1 × 103 | 399.0 ± 66.1 | 329 ± 44.6 |
| 1:640 PENT (MOI = 0.001) | 4.9 ± 0.5 | 5.9 × 107 ± 2.4 × 107 | 4.3 × 106 ± 2.2 × 106 | 142.9 ± 14.69 | 104 ± 23.9 | |
| 1:2,560 (MOI = 0.001) | 2.4 ± 0.5 | 2.3 × 103 ± 1.1 × 103 | 4.6 × 103 ± 2.4 × 103 | 98.7 ± 40.3 | 152 ± 32.4 | |
| Direct infection (MOI = 50) | 2.7 ± 0.1 | 6.7 × 104 ± 2.3 × 104 | 7.1 × 104 ± 1.1 × 104 | 78.9 ± 13.7 | 146 ± 34.7 | |
| 3 | 1:40 (MOI = 0.01) | 3.4 ± 0.5 | 5.7 × 102 ± 1.1 × 102 | 8.1 × 103 ± 2.4 × 103 | 406.3 ± 61.9 | 379.0 ± 54.6 |
| 1:160 (PENT) (MOI = 0.01) | 9.2 ± 0.8 | 8.9 × 107 ± 4.1 × 107 | 7.8 × 106 ± 1.1 × 106 | 138.3 ± 34.7 | 82.0 ± 6.08 | |
| 1:640 (MOI = 0.01) | 4.5 ± 1.0 | 6.8 × 103 ± 2.3 × 103 | 3.6 × 103 ± 1.6 × 103 | 101.7 ± 36.9 | 138.0 ± 9.6 | |
| Direct infection (MOI = 50) | 3.8 ± 0.3 | 4.3 × 104 ± 1.2 × 104 | 3.4 × 104 ± 2.1 × 104 | 78.5 ± 43.1 | 112.7 ± 24.5 |
Data are shown for all conditions for 3 representative donors, including infection rates, viral output, infectivity, and IFN-α and IFN-β production. High copy numbers and high cytokine levels are shown in bold. Data show the means ± SD from 3 independent experiments. The lower detection limits of IFN-α and IFN-β were 156 pg/ml and 25 pg/ml, respectively. The results from each group were compared using Student's t test. *, P < 0.05.
Monocytes produce IL-10 under ADE conditions.
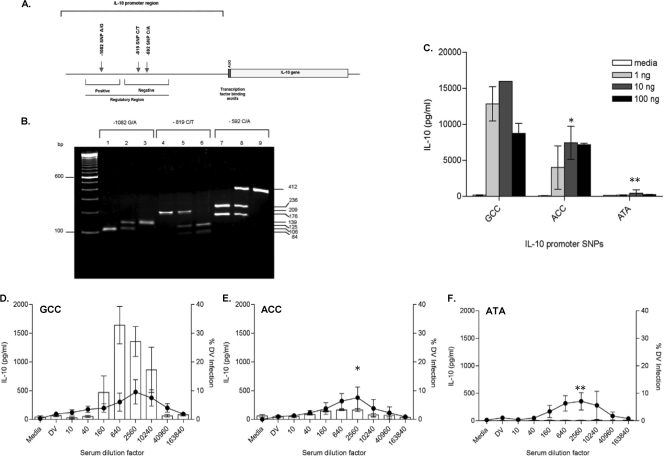
We observed IL-10 production only in monocytes and only under ADE conditions. Although macrophages and mDC supported ADE, neither of these cell types produced IL-10 under any conditions. Interestingly, the magnitudes of IL-10 levels, though reproducibly detected, differed between monocyte donors, raising the possibility that this effect was influenced by genetic polymorphisms. IL-10 promoter polymorphisms at positions −1082, −819, and −592 (Fig. 6 A) were proposed as common haplotypes known to affect IL-10 production capacity in individuals (39, 43, 54). We screened 22 donors for the presence of GCC (frequency, 3/22), ACC (frequency, 3/22), or ATA (frequency, 3/22) haplotypes using PCR-RFLP (Fig. 6B). We confirmed IL-10 production differences among these classic haplotypes by stimulating monocytes from homozygous GCC, ACC, and ATA donors with LPS and measured IL-10 protein in culture supernatants (Fig. 6C) and corresponding mRNA levels (data not shown). An LPS dose-response curve showing high-level (GCC), intermediate (ACC), and low-level (ATA) IL-10 producers, as reported previously (56), was observed with the use of monocytes in this study (Fig. 6C).
FIG. 6.
IL-10 promoter polymorphisms regulate IL-10 production. (A) Schematic diagram of human IL-10 gene showing the exon, AUG start, untranslated regions and SNPs at positions −1082, −819, and −592, modified from the method of Shin et al. (45). (B) PCR-RFLP analysis performed with primers and restriction enzyme specific for positions −1082 (lanes 1 to 3), −819 (lanes 4 to 6), and −592 (lanes 7 to 9). At position −1082, a 139-bp PCR fragment was amplified and digested with MnlI. The −1082*G allele with a MnlI site demonstrated a 106-bp fragment. At position −819, a 209-bp PCR fragment was amplified and digested with MaeIII. The −819*C allele demonstrated two fragments, 125 and 84 bp, whereas −819*T showed no splitting. At position −592, a 412-bp PCR fragment was amplified and digested with RsaI. The −592*A allele with a RsaI site demonstrated two fragments, 236 and 176 bp, whereas the −592*C allele without RsaI site produced cut pattern. (C) monocytes isolated from homozygous GCC, ACC and ATA were stimulated with 1,10 and 100 ng/ml LPS. Culture supernatants were assayed for the presence of IL-10 and data are shown as the means ± SEM for 3 donors from each haplotype. (D to F) Monocytes isolated from homozygous GCC (D), ACC (E), and ATA (F) donors were subjected to the ADE assay and culture supernatants were analyzed for IL-10 protein. The results are shown as means ± SEM for at least 3 donors from each haplotype. The cytokine assay was performed in triplicate. The lower detection limit of the IL-10 assay was 125 pg/ml. *, P < 0.05; **, P < 0.01.
IL-10 promoter polymorphisms influence the IL-10 production in monocytes undergoing ADE.
To test the influence of IL-10 promoter polymorphisms on ADE of DV infection, monocytes were isolated from homozygous GCC (n = 3), ACC (n = 3), and ATA (n = 3) donors and subjected to the ADE assay using the same dengue virus-immune serum and virus stock. Culture supernatants were tested for the presence of IL-10. Remarkably, all donors showed similar degrees of viral infection at the peak enhancement titer (5 to 15%) but demonstrated statistically different IL-10 levels (Fig. 6D to F). The GCC haplotype individuals produced the highest levels of IL-10 that coincided with ADE infection levels (Fig. 6D). Individuals with homozygous ACC showed intermediate IL-10 levels while low to undetectable levels of IL-10 protein were observed in homozygous ATA donors undergoing ADE (Fig. 6E and F, respectively).
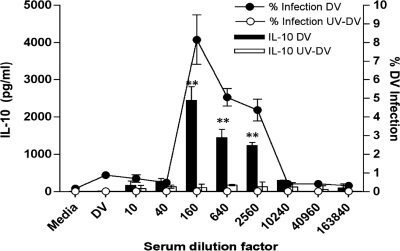
ADE-associated IL-10 production in monocytes requires viral replication.
We wondered if the IL-10 production under ADE conditions was simply triggered by the interaction of immune complexes with the Fc receptors on the monocytes' cell surfaces. To test this, we isolated monocytes from high-level IL-10 producers (GCC) and subjected these cells to the ADE assay using intact DV in parallel with UV-irradiated DV. Stimulation of monocytes with immune sera alone did not induce cytokines (data not shown). Figure 7 shows the resultant ADE patterns. IL-10 production was observed only at enhancement titers with intact DV. Using UV-irradiated DV abrogated ADE and IL-10 production (P < 0.01).
FIG. 7.
IL-10 production requires viral replication and ADE. Monocytes were isolated from homozygous GCC donors (high-level IL-10 producers) and assayed for ADE. The supernatants were tested for IL-10 production using either live DV-2 stock virus (black bars) or UV irradiated DV-2 (white bars). Results from one experiment representative of 3 independent experiments performed in triplicate are shown. All data points are means ± SD. **, P < 0.01.
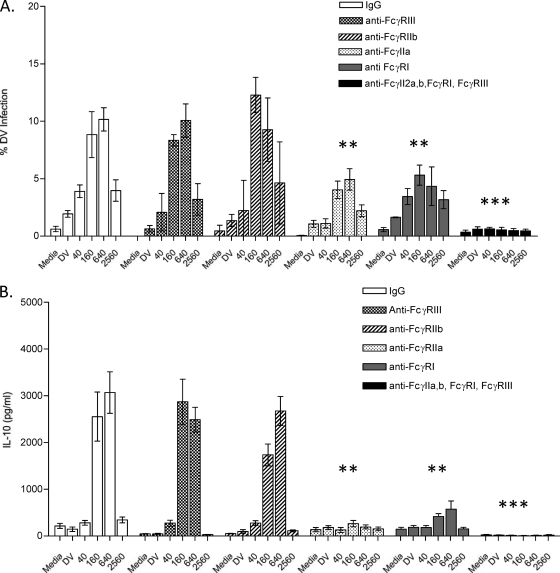
FcγRIIa and FcγRI mediate ADE and IL-10 production in primary monocytes.
FcγRs reportedly mediate ADE in monocytes/macrophage cell lines (26, 28, 36). Here, we tested GCC haplotype donors (n = 3) in this experiment in order to determine the role of these receptors in ADE and IL-10 production in primary human monocytes. Purified monocytes were treated with MAbs individually against FcγRI, FcγRIIa, FcγRIIb, or FcγRIII or against all four receptors or with control Ab (IgG1 antibody). IgG1-treated monocytes and anti-FcγRIII-treated monocytes exhibited typical ADE while cells pretreated with anti-FcγRI or anti-FcγRIIa showed significantly inhibited ADE (Fig. 8 A). Combining all specific anti-FcR antibodies abrogated ADE altogether (Fig. 8A). Furthermore, using anti-FcγRI and anti-FcγRIIa together was sufficient to abrogate both infection and IL-10 production (not shown). We next tested the supernatants collected from these FcR-blocking studies to assess their role in IL-10 production (Fig. 8B). As expected, IL-10 production was observed in monocytes pretreated with IgG1 control and in anti-FcγRIII-treated monocytes. However, IL-10 levels were significantly reduced in monocytes pretreated either with anti-FcγRIIa or anti-FcγRI antibodies. No IL-10 was observed when the monocytes were pretreated with all 4 (anti-FcγRI, -FcγRIIa, -FcγRIIb, and -FcγRIII) (Fig. 8B) or with both anti-FcγRI and anti-FcγRIIa MAbs (not shown).
FIG. 8.
Blocking FcγRI and FcγIIa abrogate ADE-associated IL-10 production in primary human monocytes. Shown are ADE infection patterns (A) and IL-10 production levels (B) under ADE conditions in monocytes treated with control IgG and specific anti-FcγRI, -IIa, -IIb, and -III MAbs. The experiments were performed in triplicate, and the bars represent means ± SEM for 3 donors. The results from each group were compared using Student's t test. **, P < 0.01; ***, P < 0.001.
DISCUSSION
To our knowledge, this is the first detailed characterization of dengue virus ADE (DV/ADE) in a primary cell culture system in which all myeloid human DV target cells were derived from single donors. We and others have described the high degree of permissiveness of primary human dendritic cells to DV infection in the absence of enhancing antibodies (1, 30, 39, 55, 58). Here, we observed an unexpected differential spectrum of ADE responses between primary human monocytes, macrophages, and DC. Macrophages and DC derived from monocytes obtained from the same “autologous” donors were identified morphologically and phenotypically (Fig. 1). These cells expressed different arrays of FcγR (Table 1), which may influence responses to DV/ADE. FcγRIIa was expressed dominantly in all cell types (Table 1); this receptor has frequently been associated with DV/ADE (1, 26, 28). All autologous myeloid cell types other than iDC supported ADE. Our prior publication focused on ADE in DC and showed that the refractoriness of immature DC to ADE resulted from a high level of DC-SIGN and an equivalent (1:1) FcγRIIa/FcγRIIb ratio. FcγRIIb contains an inhibitory motif that may impede ADE. The DC maturation process increases the expression of FcγRIIa and may lower DC-SIGN expression level, permitting ADE (1).
We initiated studies of the mechanism of dengue virus ADE in primary human myeloid cells by noting that in the presence of enhancing concentrations of polyclonal human dengue antibodies and at relatively low multiplicities of DV-2 infection (biologically plausible doses), infection with monocytes, macrophages, and mDC produced disproportionately greater output of viral RNA and virions, as assayed using Vero cells, than infection with DV-2 only. ADE in mDC resulted in a 3-log-higher viral output (copy number) than direct infection and a 2-log increase in virion production per cell but only a 2.5-fold increase in the number of cells infected. Macrophages and monocytes showed similar 2-log increases in both viral RNA and virions (plaque assay) (Fig. 1C and D) associated with 2- to 10-fold increases in cellular-infection rates. We specifically addressed the possibility that the observed increased production of dengue virus simply resulted from the initiation of multiple replicating dengue viruses via attachment and internalization of infectious immune complexes by FcR-bearing cells. This was approached by infecting primary human monocytes or macrophages using a range of increasing input or multiplicity of infection (MOI) of D2-V up to a maximum of 50. In the absence of antibodies, high DV-2 MOIs did not appreciably change cellular-infection rate in comparison with that achieved by ADE (Fig. 5A). Further, viral output per cell measured by RNA production or plaque count always achieved higher outputs at peak enhancing antibody titer at a low MOI than did infection at an MOI of 50 (Fig. 5B).
In our study, type I IFNs were not detected in monocyte supernatants in cells infected via DV/ADE (Fig. 3A) by direct assay, nor were biologically active type I IFNs demonstrated by anti-interferon blocking studies. Macrophages, however, showed a different pattern of type I IFN production (Fig. 4). IFN-α/β levels were high in macrophage supernatants in the presence of DV-immune complexes formed at low (neutralizing) dilutions of antibodies, but IFN levels fell at peak enhanced infection (ADE) antibody dilutions (Fig. 4A). Significant increases in macrophage DV-2 infection rates at low serum dilutions (0.8 versus 20%; P < 0.001) (Fig. 4B, left lower panel) were observed with the blocking studies (Table 2), whereas blocking IFN under peak ADE conditions resulted in no significant changes in infection levels (11.5 versus 16%; P = 0.44) (Fig. 4B). Our observations suggest that type I IFNs suppress infection by DV-2 in the absence of antibody in macrophages (e.g., “anti-viral state”), but at peak DV/ADE, the IFN production and effects were downmodulated. In DC, as illustrated in Fig. 3B and C, IFN-β is released in the presence of high levels of DV-2 infection. Others have previously reported that DV antagonizes IFN-α production via the downregulation of Tyk2-STAT signaling (8, 21, 18, 46). In summary, each myeloid cell type exhibited different type I interferon profiles after exposure to DV-immune complexes. Differences in interferon production at the cellular level could be an important determinant of viremia, subsequent immune responses, and potentially disease severity, depending on the predominant infected cell type(s) and its context and location (48, 50, 51).
FcR ligation can modulate patterns of cytokine production, most notably IL-12 and IL-10, with complete abrogation of IL-12 transcription after FcγR ligation (13, 53) and rapid and dramatic increase in IL-10 transcription (12, 54). In our studies, we did not detect IL-12 under any conditions. IL-10 protein production was restricted to monocytes and mainly detected in cells during ADE (Fig. 6). We consider detection of IL-10 protein as important because many cell types contain IL-10 transcripts but do not secrete protein (42). However, confirmatory IL-10 mRNA detection experiments were conducted with GCC donors after stimulation with LPS or under ADE conditions, both of which supported our findings (not shown). In general, the maximum IL-10 levels correlated with peak enhancement of infection (Fig. 6D and E and 7). Our data indicate that in monocytes, ADE-induced IL-10 production required viral replication (Fig. 7) and was not mediated solely by Fc receptor ligation or by potential LPS contamination in the viral preparation (Fig. 7), as the UV-irradiated samples did not induce IL-10. Several studies link elevated plasma/serum IL-10 levels to severe dengue disease (5, 11, 14, 40), while other studies show that circulating monocytes are highly infected late during clinical DV infections or after vaccination with recombinant dengue virus vaccines (9). Two reports have documented higher serum IL-10 levels in patients with severe disease than in those with mild secondary infections (33, 43). Several viruses that target monocytes/macrophages induce host IL-10 and/or produce their own IL-10 homologues to dampen the host immune response and thereby delay their elimination from the host (44). Elevated levels of IL-10 are associated with susceptibility to a variety of infectious diseases (47). Recent in vitro studies of infection by Ross River virus and DV-immune complexes showed that entry via the FcγR pathway suppressed the expression of LPS-induced antiviral genes and enhanced IL-10 production (3, 25, 29). We observed in monocytes infected with enhancing immune complexes that IL-10 production had little effect on DV/ADE infection of cells. It is possible that the well-described IL-10 immune modulation is mediated through bystander effects (7), as IL-10 inhibits DC maturation, antigen presentation, and T cell proliferation and generally suppresses immune responses (38, 42, 49). These data raise the question of whether IL-10 secretion to DV/ADE occurs in vivo.
In heterogeneous human populations, the capacity for IL-10 production differs between individuals and correlates with the genetic composition of the IL-10 locus. Approximately 75% of the variation in IL-10 secretion capacity derives from genetic factors, and these genetic differences, in turn, contribute to disease susceptibility (23, 35, 57). While we observed qualitatively consistent IL-10 secretion patterns in all donors, quantitatively the protein levels differed. Variations in human IL-10 secretion are influenced by genetic promoter polymorphisms, with the most intensively studied single nucleotide polymorphisms (SNPs) at positions −1082, −819, and −592 5′ of the transcriptional start site. The −819 polymorphism is in linkage disequilibrium with the −592 polymorphism; −819C and −592C are inherited together, and the same is true for −819T and −592A. Together, these SNPs combine to form the “classic” haplotypes GCC, ACC, and ATA; the haplotype GTA is indeed extremely rare (56).
Here, we report the first association between IL-10 promoter polymorphisms and the level of IL-10 production in an in vitro primary monocyte DV/ADE infection model. Using dengue virus, we proposed that the IL-10 promoter haplotype (GCC, ACC, or ATA) was linked to the IL-10 protein levels detected in culture supernatants from individual donors' monocytes undergoing ADE, recalling that IL-10 was not detected in the absence of serum. The GCC donors showed the highest levels, while ACC donors showed intermediate levels and ATA donors produced very low levels of IL-10 (Fig. 6). It seems possible that ADE dengue virus infection of monocytes is a potential source of IL-10 in vivo. Of interest, similar DV infection rates were observed in monocytes (10 to 15%) across all donor haplotypes, despite dramatically different levels of IL-10 production.
Since Fc receptors mediate ADE in all FcR-bearing cells (1, 26, 28) and FcγRIIa mediates ADE in DC, we explored which receptors were involved in ADE and IL-10 production in monocytes. As monocytes express both FcγRI and FcγII, we focused our blocking experiments (Fig. 8A and B) using specific anti-FcγRIIa and anti-FcγRI and used anti-FcγRIII as a “negative” control to illustrate the critical function of these receptors on ADE in primary monocytes. Interestingly, for anti-FcγRIIa and anti-FcγRI, blocking of either these receptors alone markedly reduced ADE infection rates (>50%) and essentially abrogated all monocyte IL-10 production under ADE conditions (Fig. 8B). The potent combination of anti-FcγRI, -IIa, and -III antibodies eliminated all infection and IL-10 production. It would appear that FcγRIII does not play a role in ADE in monocytes, as shown in Fig. 8; perhaps this is correlated with the dim FcγRIII expression on the cells' surfaces (Table 1). Some small decreases in direct-infection rates were observed with the use of IgG and or FcγR antibodies that could have nonspecifically sterically hindered infection.
We identified myeloid cell-type-specific ADE responses using an autologous system that allowed direct head-to-head comparisons at the cellular level. Study of different myeloid cell types identified important differences in response to dengue virus infection in the absence or presence of enhancing dengue antibodies. These differences are not observed in human myeloid cell lines, likely due to their inherent clonal and transformed nature. It seems reasonable to verify observations in primary cells when attempting to reach broad mechanistic conclusions from ADE studies with cell lines. In vivo, DV/ADE infection of these different primary cell types may lead to different effects in terms of viremia, cytokine patterns, and disease severity, especially under enhancement conditions. Primary human myeloid cells offer diverse genetic backgrounds and avoid transformation or immortalization effects and thereby may be especially relevant for clinical associations and hypotheses generation. As an example, our model provides a link to a clinical finding in a recent study suggesting a role for IL-10 production and IL-10 promoter polymorphisms in a dengue cohort in Cuba. Differences in the frequencies of the C allele in the position −592 locus observed between DHF patients and other Cubans experiencing a silent second dengue virus infection suggest a relationship between IL-10 production and severity of infection (43). There are wide geographic variations reported for genotype frequencies of IL-10 promoter polymorphisms, and Asian populations, in particular, have relatively low frequencies of the GCC phenotype (3% versus 20%) compared to Caucasian populations (32, 41). Follow-up to our observations should entail the systematic exploration of IL-10 promoter polymorphisms in dengue patients with different clinical outcomes in different geographic locations.
Using primary human myeloid cells, it may be possible to identify DV receptors and address ADE mechanisms by dissecting out requisite molecules for entry and postentry effects and to describe immune responses at the cellular level. But to study dengue pathogenesis in humans, we must understand what is happening in infections in situ where myeloid cells are not alone and do not respond to viral infection in isolation. Care must be taken to keep in vitro observations in context.
Acknowledgments
We thank Bonnie M. Slike and Mark de Souza for critical comments on and helpful additions to the manuscript.
This work was supported by the Pediatric Dengue Vaccine Initiative and in part by the cooperative agreement DAMD17-98-2-8007 between the U.S. Army Medical Research and Materiel Command, the Henry M. Jackson Foundation for the Advancement of Military Medicine, and the Military Infectious Disease Research Program.
The views expressed are those of the authors and should not be construed to represent the position of the U.S. Department of Defense.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Boonnak, K., et al. 2008. Role of dendritic cells in antibody-dependent enhancement of dengue virus infection. J. Virol. 82:3939-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. G., et al. 2009. Dramatic caspase-dependent apoptosis in antibody-enhanced dengue virus infection of human mast cells. J. Leukoc. Biol. 85:71-80. [DOI] [PubMed] [Google Scholar]
- 3.Chareonsirisuthigul, T., S. Kalayanarooj, and S. Ubol. 2007. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 88:365-375. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi, U. C., R. Agarwal, E. A. Elbishbishi, and A. S. Mustafa. 2000. Cytokine cascade in dengue hemorrhagic fever: implications for pathogenesis. FEMS Immunol. Med. Microbiol. 28:183-188. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L. C., et al. 2006. Correlation of serum levels of macrophage migration inhibitory factor with disease severity and clinical outcome in dengue patients. Am. J. Trop. Med. Hyg. 74:142-147. [PubMed] [Google Scholar]
- 6.Dejnirattisai, W., et al. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Smedt, T., et al. 1997. Effect of interleukin-10 on dendritic cell maturation and function. Eur. J. Immunol. 27:1229-1235. [DOI] [PubMed] [Google Scholar]
- 8.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 9.Durbin, A. P., et al. 2008. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology 376:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckels, K. H., W. E. Brandt, V. R. Harrison, J. M. McCown, and P. K. Russell. 1976. Isolation of a temperature-sensitive dengue-2 virus under conditions suitable for vaccine development. Infect. Immun. 14:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fink, J., F. Gu, and S. G. Vasudevan. 2006. Role of T cells, cytokines and antibody in dengue fever and dengue haemorrhagic fever. Rev. Med. Virol. 16:263-275. [DOI] [PubMed] [Google Scholar]
- 12.Gerber, J. S., and D. M. Mosser. 2001. Reversing lipopolysaccharide toxicity by ligating the macrophage Fc gamma receptors. J. Immunol. 166:6861-6868. [DOI] [PubMed] [Google Scholar]
- 13.Grazia Cappiello, M., F. S. Sutterwala, G. Trinchieri, D. M. Mosser, and X. Ma. 2001. Suppression of Il-12 transcription in macrophages following Fc gamma receptor ligation. J. Immunol. 166:4498-4506. [DOI] [PubMed] [Google Scholar]
- 14.Green, S., et al. 1999. Elevated plasma interleukin-10 levels in acute dengue correlate with disease severity. J. Med. Virol. 59:329-334. [PubMed] [Google Scholar]
- 15.Halstead, S. B. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60:421-467. [DOI] [PubMed] [Google Scholar]
- 16.Halstead, S. B., et al. 1983. Comparison of P388D1 mouse macrophage cell line and human monocytes for assay of dengue-2 infection-enhancing antibodies. Am. J. Trop. Med. Hyg. 32:157-163. [DOI] [PubMed] [Google Scholar]
- 17.Halstead, S. B., and E. J. O'Rourke. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 146:201-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, L. J., et al. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J. Immunol. 174:8163-8172. [DOI] [PubMed] [Google Scholar]
- 19.Huisman, W., B. E. Martina, G. F. Rimmelzwaan, R. A. Gruters, and A. D. Osterhaus. 2009. Vaccine-induced enhancement of viral infections. Vaccine 27:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jindadamrongwech, S., C. Thepparit, and D. R. Smith. 2004. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch. Virol. 149:915-927. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, A. J., and J. T. Roehrig. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kliks, S. C., A. Nisalak, W. E. Brandt, L. Wahl, and D. S. Burke. 1989. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 40:444-451. [DOI] [PubMed] [Google Scholar]
- 23.Knapp, S., et al. 2003. Interleukin-10 promoter polymorphisms and the outcome of hepatitis C virus infection. Immunogenetics 55:362-369. [DOI] [PubMed] [Google Scholar]
- 24.Kou, Z., et al. 2008. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 80:134-146. [DOI] [PubMed] [Google Scholar]
- 25.Lidbury, B. A., and S. Mahalingam. 2000. Specific ablation of antiviral gene expression in macrophages by antibody-dependent enhancement of Ross River virus infection. J. Virol. 74:8376-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Littaua, R., I. Kurane, and F. A. Ennis. 1990. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J. Immunol. 144:3183-3186. [PubMed] [Google Scholar]
- 27.Lopatin, U., et al. 2001. Increases in circulating and lymphoid tissue interleukin-10 in autoimmune lymphoproliferative syndrome are associated with disease expression. Blood 97:3161-3170. [DOI] [PubMed] [Google Scholar]
- 28.Mady, B. J., D. V. Erbe, I. Kurane, M. W. Fanger, and F. A. Ennis. 1991. Antibody-dependent enhancement of dengue virus infection mediated by bispecific antibodies against cell surface molecules other than Fc gamma receptors. J. Immunol. 147:3139-3144. [PubMed] [Google Scholar]
- 29.Mahalingam, S., and B. A. Lidbury. 2002. Suppression of lipopolysaccharide-induced antiviral transcription factor (STAT-1 and NF-kappa B) complexes by antibody-dependent enhancement of macrophage infection by Ross River virus. Proc. Natl. Acad. Sci. U. S. A. 99:13819-13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marovich, M., et al. 2001. Human dendritic cells as targets of dengue virus infection. J. Invest. Dermatol. Symp. Proc. 6:219-224. [DOI] [PubMed] [Google Scholar]
- 31.Marovich, M. A., et al. 2002. Preparation of clinical-grade recombinant canarypox-human immunodeficiency virus vaccine-loaded human dendritic cells. J. Infect. Dis. 186:1242-1252. [DOI] [PubMed] [Google Scholar]
- 32.Meenagh, A., et al. 2002. Frequency of cytokine polymorphisms in populations from western Europe, Africa, Asia, the Middle East and South America. Hum. Immunol. 63:1055-1061. [DOI] [PubMed] [Google Scholar]
- 33.Mege, J. L., S. Meghari, A. Honstettre, C. Capo, and D. Raoult. 2006. The two faces of interleukin 10 in human infectious diseases. Lancet Infect. Dis. 6:557-569. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. L., et al. 2008. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 4:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyazoe, S., et al. 2002. Influence of interleukin-10 gene promoter polymorphisms on disease progression in patients chronically infected with hepatitis B virus. Am. J. Gastroenterol. 97:2086-2092. [DOI] [PubMed] [Google Scholar]
- 36.Moi, M. L., C. K. Lim, T. Takasaki, and I. Kurane. 2010. Involvement of the Fc gamma receptor IIA cytoplasmic domain in antibody-dependent enhancement of dengue virus infection. J. Gen. Virol. 91:103-111. [DOI] [PubMed] [Google Scholar]
- 37.Morens, D. M., and A. S. Fauci. 2008. Dengue and hemorrhagic fever: a potential threat to public health in the United States. JAMA 299:214-216. [DOI] [PubMed] [Google Scholar]
- 38.Muller, G., et al. 2002. Interleukin-10-treated dendritic cells modulate immune responses of naive and sensitized T cells in vivo. J. Invest. Dermatol. 119:836-841. [DOI] [PubMed] [Google Scholar]
- 39.Navarro-Sanchez, E., et al. 2003. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4:723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, T. H., et al. 2004. Dengue hemorrhagic fever in infants: a study of clinical and cytokine profiles. J. Infect. Dis. 189:221-232. [DOI] [PubMed] [Google Scholar]
- 41.Opdal, S. H. 2004. IL-10 gene polymorphisms in infectious disease and SIDS. FEMS Immunol. Med. Microbiol. 42:48-52. [DOI] [PubMed] [Google Scholar]
- 42.Palmer, D. R., et al. 2005. Differential effects of dengue virus on infected and bystander dendritic cells. J. Virol. 79:2432-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez, A. B., et al. 2004. IL-10 levels in Dengue patients: some findings from the exceptional epidemiological conditions in Cuba. J. Med. Virol. 73:230-234. [DOI] [PubMed] [Google Scholar]
- 44.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86-92. [DOI] [PubMed] [Google Scholar]
- 45.Shin, H. D., et al. 2000. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc. Natl. Acad. Sci. U. S. A. 97:14467-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shresta, S., et al. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, A. J., and S. E. Humphries. 2009. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 20:43-59. [DOI] [PubMed] [Google Scholar]
- 48.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 49.Steinbrink, K., M. Wolfl, H. Jonuleit, J. Knop, and A. H. Enk. 1997. Induction of tolerance by IL-10-treated dendritic cells. J. Immunol. 159:4772-4780. [PubMed] [Google Scholar]
- 50.Stetson, D. B., and R. Medzhitov. 2006. Antiviral defense: interferons and beyond. J. Exp. Med. 203:1837-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stetson, D. B., and R. Medzhitov. 2006. Type I interferons in host defense. Immunity 25:373-381. [DOI] [PubMed] [Google Scholar]
- 52.Suksanpaisan, L., A. Cabrera-Hernandez, and D. R. Smith. 2007. Infection of human primary hepatocytes with dengue virus serotype 2. J. Med. Virol. 79:300-307. [DOI] [PubMed] [Google Scholar]
- 53.Sutterwala, F. S., G. J. Noel, R. Clynes, and D. M. Mosser. 1997. Selective suppression of interleukin-12 induction after macrophage receptor ligation. J. Exp. Med. 185:1977-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sutterwala, F. S., G. J. Noel, P. Salgame, and D. M. Mosser. 1998. Reversal of proinflammatory responses by ligating the macrophage Fcgamma receptor type I. J. Exp. Med. 188:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tassaneetrithep, B., et al. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner, D. M., et al. 1997. An investigation of polymorphism in the interleukin-10 gene promoter. Eur. J. Immunogenet. 24:1-8. [DOI] [PubMed] [Google Scholar]
- 57.Westendorp, R. G., J. A. Langermans, T. W. Huizinga, C. L. Verweij, and A. Sturk. 1997. Genetic influence on cytokine production in meningococcal disease. Lancet 349:1912-1913. [DOI] [PubMed] [Google Scholar]
- 58.Wu, S. J., et al. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]
- 59.Zaba, L. C., J. Fuentes-Duculan, R. M. Steinman, J. G. Krueger, and M. A. Lowes. 2007. Normal human dermis contains distinct populations of CD11c+BDCA-1+ dendritic cells and CD163+FXIIIA+ macrophages. J. Clin. Invest. 117:2517-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]